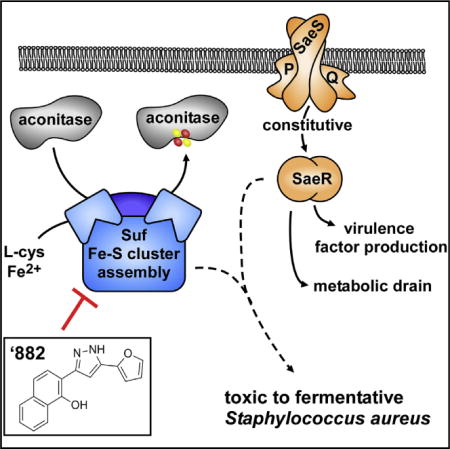
SUMMARY
The rising problem of antimicrobial resistance in Staphylococcus aureus necessitates the discovery of novel therapeutic targets for small-molecule intervention. A major obstacle of drug discovery is identifying the target of molecules selected from high-throughput phenotypic assays. Here, we show that the toxicity of a small molecule termed ’882 is dependent on the constitutive activity of the S. aureus virulence regulator SaeRS, uncovering a link between virulence factor production and energy generation. A series of genetic, physiological, and biochemical analyses reveal that ’882 inhibits iron-sulfur (Fe-S) cluster assembly most likely through inhibition of the Suf complex, which synthesizes Fe-S clusters. In support of this, ’882 supplementation results in decreased activity of the Fe-S cluster-dependent enzyme aconitase. Further information regarding the effects of ’882 has deepened our understanding of virulence regulation and demonstrates the potential for small-molecule modulation of Fe-S cluster assembly in S. aureus and other pathogens.
Graphical abstract
INTRODUCTION
The Gram-positive pathogen Staphylococcus aureus is a leading cause of a wide range of devastating infections, including skin and soft tissue infections, osteomyelitis, infective endocarditis, and bacteremia (Tong et al., 2015). To infect, S. aureus employs a multitude of toxins, exoenzymes, and immune modulators, and its virulence regulators have long been appreciated as vital to pathogenesis. S. aureus virulence regulator SaeRS is a key global regulator of toxin and exoenzyme production (Flack et al., 2014). SaeS is a two-component system (TCS) histidine kinase that responds to molecular components of neutrophils and activates the response regulator SaeR, which upregulates transcription of the Sae regulon (Sun et al., 2010; Palazzolo-Ballance et al., 2008). SaeRS is encoded in the saePQRS operon along with SaePQ, a membrane complex that aids the return of SaeRS to basal levels of activity (Jeong et al., 2012). Complementary to its arsenal of virulence factors, S. aureus has also evolved antibiotic resistance. Methicillin-resistant S. aureus accounts for the majority of clinical isolates (Tong et al., 2015). The dearth of antibiotics in the pharmaceutical development pipeline has led to renewed efforts to discover small molecules that probe the physiology of pathogens for the development of novel antimicrobials. In this regard, central metabolic pathways have become an attractive target in bacterial pathogens (Murima et al., 2014).
S. aureus is a facultative anaerobe, relying on both respiration and fermentation to cause disease (Hammer et al., 2013; Vitko et al., 2015). In aerobic environments, respiration uses oxygen as the terminal electron acceptor and provides higher rates of ATP generation. However, S. aureus experiences many conditions in the host that make fermentation vital. Fermentation is used in hypoxic or anaerobic niches devoid of alternative terminal electron acceptors, including bone and tissue abscesses (Hammer et al., 2013; Wilde et al., 2015). Host-produced nitric oxide directly inhibits the respiratory chain and the tricarboxylic acid (TCA) cycle is inhibited in iron-deplete niches because of the iron dependency of many TCA cycle enzymes (Vitko et al., 2015; Friedman et al., 2006). In addition, respiration-deficient menaquinone or heme auxotrophs, called small colony variants (SCVs), are common isolates and etiological agents of persistent infections, an enormous clinical problem (Proctor et al., 2006). Respiration-deficient SCVs rely on fermentation for growth and are intrinsically resistant to antibiotics that rely on membrane potential to enter the cell. Therefore, targeting of processes essential for fermentation is an exciting therapeutic option for the elimination of these cell populations.
Iron is crucial to the infectious life cycle of S. aureus and a large portion of internalized iron is incorporated into proteins as Fe-S cluster cofactors. Fe-S cluster cofactors are required for a variety of cellular processes and the synthesis of Fe-S clusters is essential for S. aureus viability (Mashruwala et al., 2015). Fe-S cluster-dependent enzymes play key roles in central carbon metabolism (Kennedy et al., 1983), branched chain amino acid synthesis (Flint et al., 1993), antibiotic resistance (Yan et al., 2010), and signal transduction (Sun et al., 2012). S. aureus factors required for Fe-S cluster assembly and maturation of apo-protein targets include SufS, SufBCD, SufU, SufT, SufA, and Nfu. SufS provides sulfur from cysteine to SufU or SufBCD, which synthesize [Fe2-S2] or [Fe4-S4] clusters (Selbach et al., 2014). These Fe-S clusters are inserted into apo-proteins with or without the aid of the Fe-S cluster carriers Nfu or SufA, facilitating the maturation of holo-proteins (Mashruwala et al., 2015; Rosario-Cruz et al., 2015). SufT is an auxiliary factor involved in the maturation of apo-proteins that has an increased role during conditions that impose a high demand for Fe-S clusters (Mashruwala et al., 2016b). The genes encoding for Nfu and SufT display synergism for multiple phenotypes. Analyses of a Δnfu ΔsufT strain have revealed the pleiotropic effects of defective Fe-S assembly upon central metabolism, iron homeostasis, oxidative stress, vancomycin resistance, biofilm formation, and virulence (Mashruwala et al., 2015, 2016b).
Previously, we identified small-molecule VU0038882 (’882), which activates endogenous heme biosynthesis in S. aureus while toxic to S. aureus grown anaerobically (Mike et al., 2013). ’882 exhibits a half maximal inhibitory concentration (IC50) of ~162 μM to aerobic S. aureus and an IC50 of ~5 μM to anaerobic S. aureus (Mike et al., 2013). Further experimentation revealed that ’882 is toxic to S. aureus relying solely on fermentation for energy generation, as ’882 is bacteriostatic to mutants that are respiration deficient, regardless of oxygen availability. Through extensive structure-activity relationship studies, we showed that the toxicity of ’882 can be uncoupled from the capacity to activate heme synthesis, suggesting that ’882 affects two distinct targets in S. aureus (Dutter et al., 2016).
In this work, we sought to identify the mechanism of ’882 toxicity in order to probe the physiology of S. aureus and uncover novel therapeutic targets. Genetic and proteomic approaches uncovered the SaeRS TCS as essential for ’882 toxicity and the Suf Fe-S cluster biogenesis machinery as a likely target of ’882. Here, we identify a unique link between virulence regulation and metabolic fitness in S. aureus. In addition, this work employs a breadth of approaches to understand the effects of a small molecule and emphasizes the importance of Fe-S cluster metabolism in staphylococcal physiology. This study establishes ’882 as a first-in-class manipulator of Fe-S cluster assembly, which may guide the development of new antimicrobials that target this essential pathway.
RESULTS
Constitutive Sae TCS Signaling Is Required for ’882 Toxicity
To identify the cellular target of ’882 toxicity in anaerobic S. aureus, we selected for spontaneously resistant mutants of S. aureus strain Newman (NM) growing fermentatively (anaerobically in the absence of alternative terminal electron acceptors) on medium containing 20 or 40 μM ’882. Seven independently isolated mutants grew under these conditions at a rate of ~7 × 10−7, and were stably ’882 resistant after multiple passages on medium alone. To evaluate the growth of these mutants relative to NM, strains were back-diluted from stationary-phase cultures in medium alone into medium containing 40 μM ’882 and the optical density (OD) was measured after 18 hr of anaerobic growth (Figure 1A). These conditions allowed a demarcation between ’882 sensitivity and resistance based on the growth of NM (Figure S1). To identify the mutations that allowed growth in the presence of ’882, we sequenced the genomes of the seven isolates, and each was found to have a mutation in the saePQRS locus and no other nonsynonymous mutations were found in the genome (Figure 1B). Each mutation is predicted to disrupt the function of the Sae system by altering the protein-coding sequence or disrupting transcription from both promoters (P1, P3), thereby changing expression. Indeed, the ’882-resistant isolates demonstrate phenotypes consistent with inactivation of the Sae system, as evidenced by reduced hemolysis, diminished exoprotein secretion, and undetectable SaeQ expression (Figure S2). These data suggest that Sae activity is required for sensitivity to ’882, and mutations that reduce Sae signaling are sufficient to abrogate ’882 toxicity.
Figure 1. S. aureus Constitutive Sae Function Is Required for ’882 Toxicity.
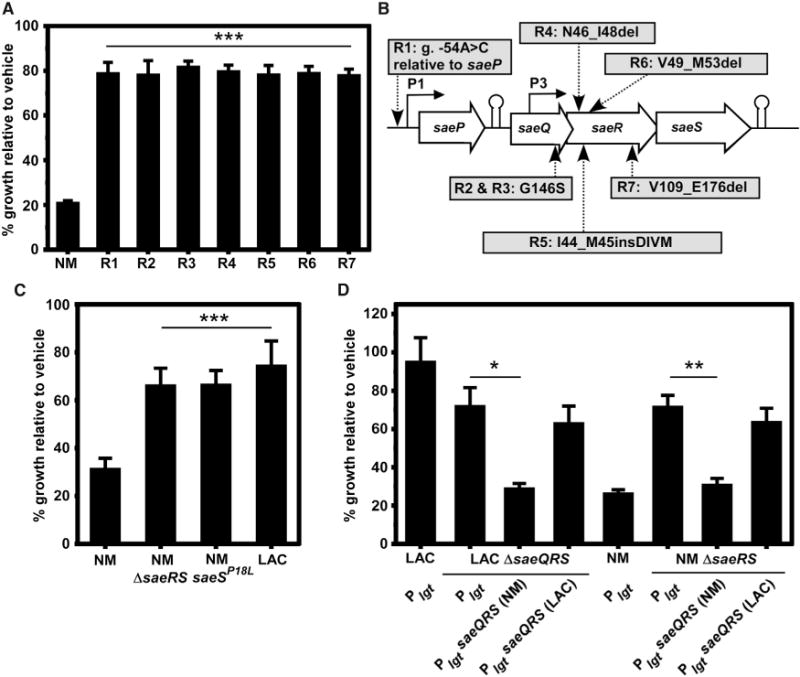
Spontaneously resistant mutants of strain Newman (NM) were isolated in the presence of 20 or 40 μM ’882 in anaerobic conditions.
(A) After four passages on plain medium, the spontaneous mutants still grew robustly by fermentation in medium containing 40 μM ’882 relative to NM, indicating stable resistance to ’882.
(B) Whole-genome sequencing of the isolates in (A) revealed mutations in the saePQRS locus. R1 has a nucleotide substitution at position −54 of the genome sequence (g.) relative to the start of saeP. Mutations resulting in changes to amino acid sequence are shown, where del signifies a deletion of noted amino acids and ins signifies insertion.
(C) Strains that do not encode a constitutive SaeS: ΔsaeRS, saeSP18L, and the USA300 LAC (LAC) clinical isolate are resistant to ’882.
(D) trans-Expression of constitutively active Sae (NM) under control of the lgt promoter in plasmid pOS1 is sufficient to induce ’882 enhanced susceptibility in LAC, and the non-constitutively active saeQRS (LAC) locus provides ’882 resistance to NM.
For (A), (C), and (D), error bars represent SEM from data combined from at least three independent experiments with n > 2 for each. *p < 0.05, **p < 0.01, and ***p < 0.001, calculated by one-way ANOVA with Sidak correction for multiple comparisons. See also Figures S1 and S2.
Importantly, SaeS of S. aureus NM is unique due to an amino acid substitution of leucine to proline at residue 18, located in the first transmembrane helix of SaeS (Adhikari and Novick, 2008). The Leu18 residue is encoded by nearly every other sequenced S. aureus genome. The L18P mutation renders NM SaeS constitutively active and resistant to SaeQ regulation, resulting in high levels of exoprotein transcription, translation, and secretion. Therefore, we hypothesized that constitutive Sae activity may be required for sensitivity to ’882. Consistent with this, strains of S. aureus that do not express constitutively active SaeS: ΔsaeRS (NM) and saeSL18P allele repaired to saeSP18L (NM), as well as the clinical isolate USA300 LAC (LAC), are more resistant to ’882 than NM (Figure 1C). The NM sae locus (carrying saeSL18P) expressed in trans in a LAC ΔsaeQRS strain increased sensitivity to ’882 while the LAC sae locus (carrying saeSL18) expressed in trans did not make NM ΔsaeRS sensitive to ’882, confirming that constitutive Sae activity is sufficient for sensitivity to ’882 (Figure 1D). These data demonstrate that increased Sae activity is required for ’882 toxicity and decreased Sae signaling is sufficient for resistance to ’882.
Inactivation of Genes Implicated in Sae Signaling Provides Increased Resistance to ’882
To identify additional factors required for ’882 toxicity, we again selected for spontaneously resistant mutants that grew by fermentation on medium containing ’882. However, due to the strong selection for mutations in sae, we chose to create NM with a plasmid encoding an additional copy of the native sae locus (psae) to prevent the identification of additional Sae mutants, and increase the likelihood of identifying genes other than sae that could provide ’882 resistance. We isolated eight spontaneously resistant mutants in strain NM carrying psae at a rate of ~1.3 × 10−5, sequenced their genomes, and found mutations in three separate genes. Six of the isolates have changes in the protein-coding sequence of the fatty acid kinase FakA, a seventh isolate encodes a change in an additional fatty acid kinase FakB1, and the eighth has a stop codon incorporated in the coding sequence of the chaperone ClpX (Figure 2A). No other nonsynonymous mutations were identified in these isolates. To determine whether mutations in these genes were sufficient to increase ’882 resistance in an NM background, we tested the growth of NM and strains inactivated for these genes in the presence of ’882 (Figure 2B). These strains all displayed increased resistance to ’882 relative to NM, which indicates that FakA, FakB1, and ClpX are required for ’882 toxicity. Interestingly, previous findings established that the Fak system impacts Sae function in strain USA300 (Parsons et al., 2014). Also, SaeR was identified as a substrate of the ClpXP complex in NM, suggesting SaeR might be affected posttranslationally (Feng et al., 2013). Therefore, we hypothesized that these resistant mutants were identified because they alter Sae activity, and are not themselves the target of ’882.
Figure 2. Genes required for Toxicity Are Implicated in Sae Signaling, which Is a Metabolic Drain to NM.
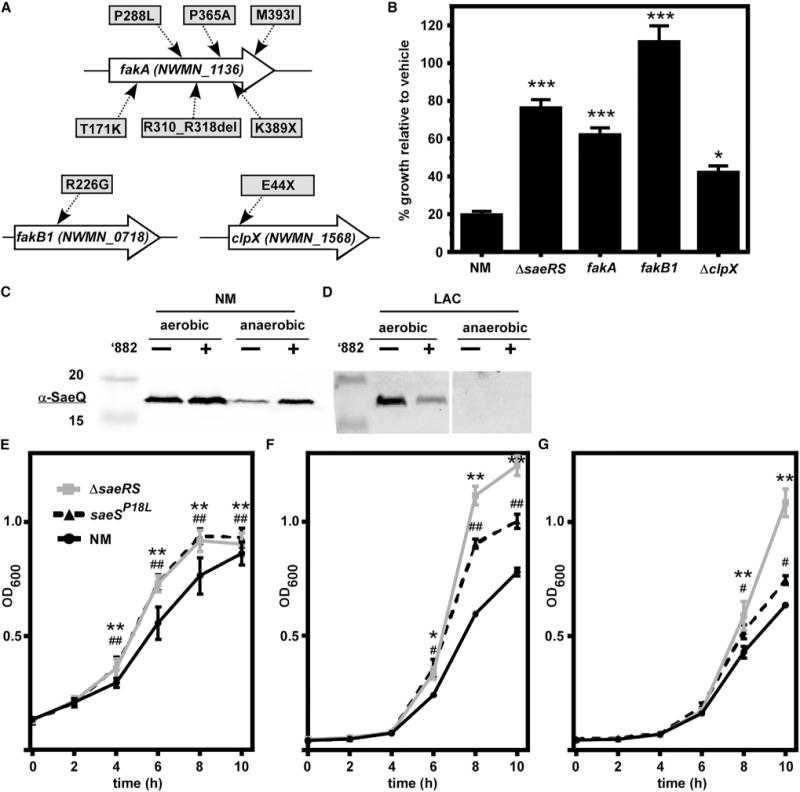
(A) Genomes of eight spontaneously ’882-resistant mutants in the NM pOS1PlgtsaeQRS strain were sequenced. Nonsynonymous mutations were found in fakA, fakB1, and clpX. Changes to the protein sequence are shown in uppercase, as well as a deletion (del).
(B) Strains inactivated for saeRS, fakA, fakB1, and clpX grow better than NM anaerobically in the presence of 40 μM ’882. *p < 0.05, ***p < 0.001, calculated by one-way ANOVA with Sidak correction for multiple comparisons.
(C and D) Immunoblot with α-SaeQ at ~17 kDa from strains NM (C) and LAC (D) during aerobic respiration and anaerobic fermentation in the presence or absence of 40 μM (aerobic) or 4 μM (anaerobic) ’882 or DMSO.
(E–G) Growth of NM, ΔsaeS, and saeSP18L without ’882 anaerobically in TSB (E), and aerobically in carbon-limited medium containing glucose (F) or glycerol (G) as the sole carbon source.
Error bars represent SEM from data combined from at least three independent experiments with n > 3 for each. For (E)–(G), *,#p < 0.05, **,##p < 0.001, calculated by two-way ANOVA with Sidak correction for multiple comparisons, comparing *ΔsaeRS or #saeSP18L with NM at each time point.
Constitutive Sae Is Deleterious to Growth under Energy-Limiting Conditions
The strong selective pressure against Sae indicated that constitutive Sae activity may prevent anaerobic S. aureus from overcoming the toxic effects of ’882. Under growth conditions devoid of a terminal electron acceptor, S. aureus relies on fermentation, which is less efficient at energy generation than respiration. Based on the large SaeR regulon, including the expression and secretion of many exoproteins, we hypothesized that the constitutive Sae activity in NM would be deleterious during fermentative growth. First, SaeQ abundance under aerobic and anaerobic conditions was measured as a proxy for transcription from promoter 1 of two different transcripts: saePQRS (transcript T1) and saeQRS (transcript T2) (Jeong et al., 2011). Indeed, LAC did not express SaeQ during fermentation, indicating that transcription from Sae promoter 1 is inactive regardless of the presence or absence of ’882 (Figures 2C and 2D). This suggested that the increased Sae activity during fermentation may impact growth in NM. As predicted, ΔsaeRS and saeSP18L grew better than NM undergoing fermentation in rich medium (Figure 2E). In addition, ΔsaeRS and saeSP18L displayed enhanced growth in carbon-limited medium using glucose or glycerol as the primary carbon source aerobically (Figures 2F and 2G, respectively). These data confirm that constitutive SaeS activity is deleterious to growth during energy-limiting conditions and are consistent with the hypothesis that the constitutively active SaeS contributes to the toxic effects of ’882.
’882 Disrupts Coenzyme A Pathways in S. aureus
We next hypothesized that Sae may also contribute to ’882 toxicity by transcriptionally activating or repressing expression of the gene encoding the target of ’882. To test this hypothesis, transcriptional differences between NM and ΔsaeRS in the presence of ’882 were identified using microarray (Table S1). In addition to changes in expected virulence factor and exoprotein genes, the transcription of many genes involved in protein synthesis, energy production, and amino acid metabolism were altered. To test whether any of these genes encode the target of ’882, strains lacking each of the non-essential or non-virulence factor genes were tested for resistance to ’882. The inactivation of only two genes, rimJ and NWMN_2021, provided increased resistance to ’882 (Figure S3). RimJ is a ribosomal protein N-acetyltransferase that is predicted to use acetyl-coenzyme A (acetyl-CoA) as its substrate (Tanaka et al., 1989). We hypothesized that the presence of RimJ increases ’882 toxicity due to increased cellular consumption of acetyl-CoA. To further test this hypothesis, we inactivated four other putative acetyl-CoA-consuming enzymes. These proteins were significantly increased in abundance in NM grown in the presence of ’882 relative to DMSO as identified by proteomics (Table S2). Genetic inactivation of two of these four genes also provided resistance to ’882, suggesting that acetyl-CoA consumption by non-essential proteins contributes to the anaerobic toxicity of ’882 (Figure 3A). Together, these data indicate that ’882 disrupts acetyl-CoA or CoA homeostasis. As the effect of CoA limitation by ’882 would affect the growth of NM undergoing fermentation as well as anaerobic respiration, we tested whether increasing CoA abundance would rescue ’882 toxicity. Indeed, the addition of the CoA precursor pantothenate rescued growth of NM anaerobically in rich medium containing ’882 (Figure 3B) as well as in carbon-limited medium containing the non-fermentable carbon source glycerol and terminal electron acceptor nitrate (Figure 3C). In sum, these data suggest that ’882 alters CoA homeostasis, which can be rescued by reducing acetyl-CoA consumption through genetic manipulation or increasing CoA precursor availability.
Figure 3. ’882 Alters CoA Pathways.
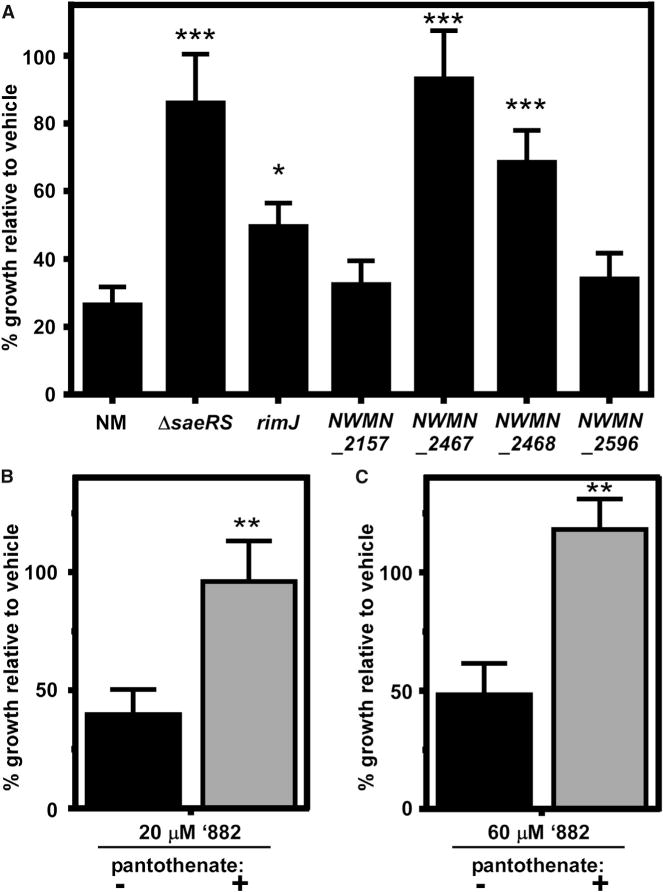
(A) Inactivation of certain acetyltransferases provides resistance to ’882.
(B and C) Anaerobic growth of NM can be rescued by addition of the CoA precursor pantothenate in rich medium containing ’882 (B) or carbon-limited medium containing glycerol (C) as the primary carbon source, ’882, and nitrate as the terminal electron acceptor.
Error bars represent SEM from data combined from at least three independent experiments with n > 3 for each. For (A), *p < 0.05, ***p < 0.001, calculated by one-way ANOVA with Sidak correction for multiple comparisons. For (B) and (C), **p < 0.01, calculated by Student’s t test comparing the absence or presence of pantothenate. See also Figure S3.
’882 Interacts with the Fe-S Cluster Biogenesis Machinery
While these genetic approaches illuminated the role of Sae in ’882 toxicity and fermentative growth, as well as the effects of ’882 on CoA homeostasis, they did not identify a target of ’882 toxicity. Therefore, in an attempt to identify a cellular target of ’882, we performed a pull-down with cellular lysate and biotin-conjugated ’882. One protein band visualized by SDS-PAGE exhibited differential abundance between the ’882-biotin sample and the control. This band was excised and subjected to MudPIT liquid chromatography-tandem mass spectrometry (LC-MS/MS) for peptide identification (Figure 4A). Peptides from three proteins were enriched by the ’882 pull-down, representing putative targets including SufC, NWMN_0632, and PdxS (Figure 4B). We repeated the pull-down experiment and performed proteomics on the entire lanes of the control and ‘882-biotin pull-down in an attempt to identify protein complexes bound by ’882. We found that proteins of the Suf machinery (SufB, SufC, SufD, and SufS) displayed greater enrichment in the ’882 pull-down, while NWMN_0632 and PdxS were not enriched (Figures 4C and 4D).
Figure 4. ’882 Associates with Fe-S Cluster Biogenesis Machinery.
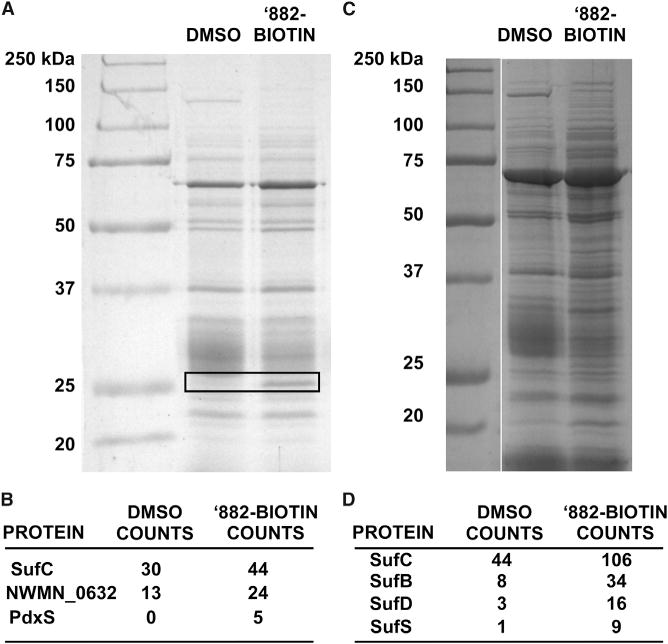
(A) SDS-PAGE of NM proteins collected by streptavidin after pull-down with DMSO or biotinylated ’882. The boxed section of the gel was excised and subjected to proteomics.
(B) Spectral counts of peptides from the boxed area in (A) most enriched in ’882-biotin include SufC; the molecular weight of SufC is ~28 kDa.
(C) SDS-PAGE of NM proteins collected by streptavidin after pull-down with DMSO or biotinylated ’882.
(D) Spectral counts of Suf protein peptides detected by proteomics after whole-lane digest of (C) and mass spectrometry.
To further investigate the interaction between ’882 and SufC, we measured direct binding using biolayer interferometry (BLI). Purified biotinylated SufC bound ’882 with a KD of ~4 μM (Table 1). We then repeated the BLI using purified SufC and biotinylated ’882 and measured binding with a KD of ~2 μM (Table 1). In addition, purified NWMN_0632 did not bind ’882, demonstrating that the SufC-’882 interaction is specific and that the enrichment of NWMN_0632 peptides in the initial ’882 pull-down was likely not the result of direct binding (Table 1). These data are consistent with a direct interaction between ’882 and SufC.
Table 1.
Direct Binding between SufC and ’882
| Biotinylated Ligand | Analyte | KD (μM) | Kon (M−1s−1) [Error] | Koff (s−1) [Error] |
|---|---|---|---|---|
| SufC | ’882 | 4.4 | 3.27 × 104 [5.72 × 103] |
1.45 × 10−1 [1.02 × 10−2] |
| ’882 | SufC | 2.1 | 9.46 × 103 [4.16 × 102] |
1.96 × 10−2 [2.66 × 10−4] |
| NWMN_0632 | ’882 | ND | – | – |
Interactions with low-micromolar KD values between purified SufC and ’882, regardless of which served as the ligand, were observed by biolayer interferometry. Binding was not detected (ND) between ’882 and NWMN_0632.
Assembly of FeS Clusters upon Aconitase Is Disrupted by ’882
SufC is essential in S. aureus and forms a complex with SufB and SufD to assemble Fe-S clusters using sulfur from SufS and SufU (Boyd et al., 2014). Monitoring in vitro Fe-S cluster assembly by the Suf machinery is technically challenging, and can produce non-physiologically relevant Fe-S clusters (Boyd et al., 2014). As an alternative readout, the effects of ’882 supplementation upon the activity of the Fe-S cluster-dependent enzyme aconitase (AcnA) were interrogated. These experiments revealed that ’882 treatment results in decreased activity of AcnA in vivo in strains NM and LAC (Figure 5A). Because the transcription of sufC is decreased during fermentative growth (Mashruwala et al., 2015), we hypothesized that the relative ratio of ’882 to SufC would increase under these conditions and result in greater inhibition of AcnA activity by ’882, relative to aerobic growth. Indeed, as the oxygenation during growth was reduced, the inhibitory effect of ’882 increased (Figure 5B). Likewise, the effect of ’882 on AcnA function was greater during anaerobiosis and a concentration of 25 μM ’882 was sufficient to reduce AcnA activity to 50% (Figure 5C).
Figure 5. ’882 Impairs Aconitase Function.
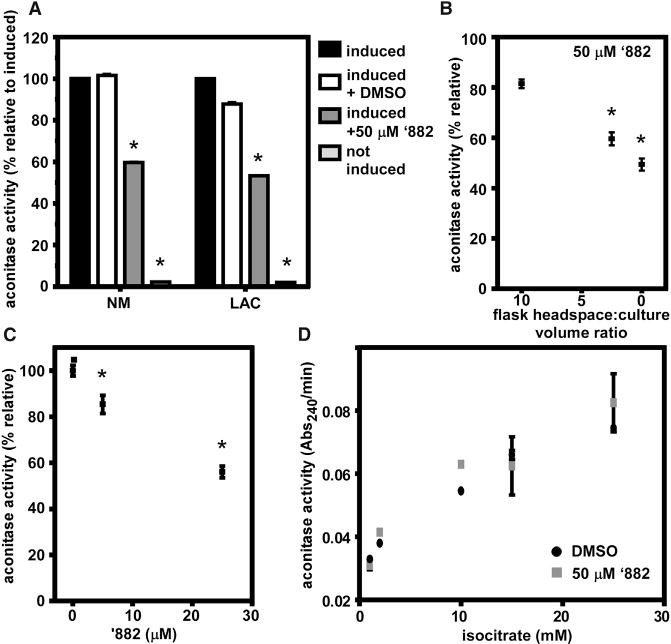
(A) Aconitase activity is reduced in NM and LAC cultured in the presence of ’882 after xylose induction of acnA transcription. *p < 0.001 by Student’s t test with Holm-Sidak correction for multiple comparisons, compared with induced + DMSO.
(B) Aconitase activity in LAC is inhibited by 50 μM ’882 to a greater extent as oxygen availability is reduced by decreasing the volume of gas above growth medium. *p < 0.05 relative to a headspace:culture volume of 10, calculated by one-way ANOVA with Sidak correction for multiple comparisons.
(C) The inhibition of aconitase by ’882 is greater under anaerobic conditions in LAC, where ~50% inhibition is achieved by 25 μM ’882, compared with 50 μM ’882 in (B). *p < 0.05 relative to no ’882 calculated by one-way ANOVA with Sidak correction for multiple comparisons.
(D) ’882 does not inhibit the activity of purified aconitase protein that has been chemically reconstituted with Fe-S cofactor in vitro prior to exposure to ’882.
For (A)–(D), error bars represent SD combined from two independent experiments. For (A)–(D), the acnA::Tn strain carried pacnA, which encodes for acnA under the transcriptional control of a xylose-inducible promoter. See also Figure S4.
Six explanations could underpin the inhibitory effects of ’882 on AcnA activity: (1) decreased transcription of the suf operon, (2) decreased abundance of the Suf machinery or auxiliary factors required for apo-protein maturation, (3) decreased transcription of acnA, (4) decreased abundance of AcnA, (5) direct inhibition of holo-AcnA activity by ’882, or (6) inhibition of assembly of the Fe-S cluster on AcnA. We found that the transcriptional activity of sufC (the first gene in the suf operon) was not decreased upon supplementing media with ’882 (Figure S4A and Table S1) and the abundances of Suf machinery proteins were increased upon treatment with ’882 (Table S2). The abundances of auxiliary factors involved in Fe-S protein maturation such as SufT were also increased upon ’882 supplementation (Table S2). Next, AcnA activity was monitored in an acnA::TN strain containing a plasmid with an acnA_FLAG allele under the transcriptional control of a xylose-inducible promoter (pacnA). Introduction of pacnA allowed for the control of acnA transcription and the determination of AcnA_FLAG abundance (Mashruwala et al., 2015). ’882 supplementation resulted in decreased AcnA activity in the acnA::TN strain carrying pacnA, but did not alter abundance of AcnA_FLAG (Figure S4B). Further, the inhibitory effect of ’882 on AcnA function was not observed if ’882 was added to purified AcnA containing an in vitro reconstituted Fe-S cluster (Figure 5D). Inhibition of protein synthesis prior to supplementation of the medium with ’882 did not result in inhibition of AcnA activity in vivo (Figure S4C). The data presented in Figures 5 and S4 lend support to a model wherein ’882 diminishes AcnA function by disrupting the assembly of the FeS cofactor upon AcnA.
Strains Defective in the Maturation of Fe-S Proteins Are Sensitive to ’882 Intoxication
We hypothesized that if ’882 inhibits Fe-S cluster assembly in vivo, then strains deficient in the maturation of Fe-S proteins would display increased sensitivity to ’882 with respect to AcnA activity, as well as growth. Consistent with our prediction, LAC strains deficient in Fe-S cluster assembly (ΔsufT and ΔsufA) exhibit reduced AcnA activity, which was further decreased by co-culture with ’882 (Figure 6A). Growth upon defined media lacking the amino acids isoleucine and leucine (18AA medium) is reliant upon the Fe-S cluster-dependent enzymes LeuCD and IlvD. Growth of a Δnfu ΔsufT double mutant in 18AA medium was diminished by ’882 (Figure 6B). These data support a model whereby ’882 inhibits the function of Fe-S cluster-dependent processes by targeting the Suf machinery.
Figure 6. Strains Defective in Fe-S Cluster Assembly Are Sensitive to ’882.
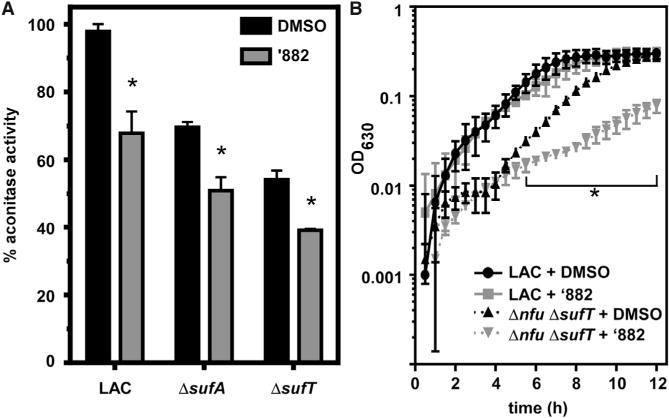
(A) Aconitase activity is reduced in mutants lacking Fe-S cluster assembly factors when cultured in the presence of 50 μM ’882. *p < 0.05 by Student’s t test for each strain comparing DMSO with ’882.
(B) A LAC strain defective in Fe-S protein maturation is sensitive to 10 μM ’882 under microaerobic conditions in defined media lacking Ile and Leu. *p < 0.05 by Student’s t test for each time point comparing Δnfu ΔsufT + ‘882 with Δnfu ΔsufT + DMSO.
For (A) and (B), error bars represent SD combined from two independent experiments.
DISCUSSION
In this study, we sought to characterize the factors responsible for ’882 toxicity. Our attempts using genetic selection offered an insight into the interplay between the SaeRS virulence regulation system and energy generation (Figure 1). The toxicity of ’882 was first observed in S. aureus strain NM, which has a constitutively active SaeS as the result of a mutation resulting in a proline at amino acid 18 (Adhikari and Novick, 2008). The genetic strategies employed here point to a strong selection against SaeS signaling during anaerobic growth. In addition to showing that inactivation of Sae is sufficient to make NM resistant to ’882, our findings (Figures 2A and 2B) that genetic inactivation of fakA, fakB1, and clpX also provide increased resistance to ’882 are consistent with previous work connecting fatty acid kinases (Fak) and the ClpXP protease complex with Sae signaling (Parsons et al., 2014; Feng et al., 2013). FakA (previously called VfrB) and FakB1, along with FakB2, constitute a fatty acid incorporation system required for hemolysis, biofilm formation, and the transcription of SaeR target genes (Bose et al., 2014; Parsons et al., 2014; Sabirova et al., 2015). The direct mechanism of Fak regulation of these virulence determinants is unclear. ClpX is an ATPase that serves as a chaperone as well as forming a complex with the peptidase ClpP (Frees et al., 2007). While the complete interaction between ClpX and Sae signaling has not been elucidated, ClpX likely affects Sae signaling by altering SaeR abundance in NM, as SaeR has been identified as a substrate of ClpXP (Feng et al., 2013). The effects on Sae of ClpX and the Fak system highlight the diverse cellular processes that intersect with Sae virulence regulation.
The inhibition of fermentative growth by ’882 may be observed because the constitutive SaeS activity in NM reduces growth even in the absence of ’882. The finding that LAC and presumably other strains with non-constitutive SaeS alter Sae signaling in anaerobic conditions is consistent with the hypothesis that pathogens have evolved control of metabolically expensive virulence factor production to ensure synthesis occurs only when required (Figures 2C–2G). This is similar to the description of the metabolic cost associated with expression of the Salmonella type III secretion system 1 (Sturm et al., 2011). Cells that express this virulence factor in vitro demonstrate reduced fitness relative to non-expressing cells. It seems that the canonical SaeS of S. aureus has evolved to limit the metabolically expensive synthesis of its regulon during energetically unfavorable conditions.
Our finding that ’882 alters CoA homeostasis (Figure 3) substantiates the effects of Suf disruption on S. aureus undergoing fermentation. Genetic inactivation of rimJ and other non-essential acetyl-CoA-consuming enzymes provided increased resistance to ’882, and based on this we hypothesized that ’882 may reduce available CoA levels during anaerobic growth. The ’882 inhibition of Fe-S cluster assembly are consistent with our model that ’882 disrupts the CoA pool. First, the Fe-S cluster-dependent enzyme dihydroxy-acid dehydratase (IlvD) is required for pantothenate and CoA synthesis. In addition, under anaerobiosis, pyruvate formate lyase (PflB) is the probable source of acetyl-CoA as the transcription of pflA and pflB increase anaerobically while transcription of the pyruvate dehydrogenase complex decreases (Fuchs et al., 2007). PflB is activated by PflA, which is an Fe-S cluster-dependent radical S-adenosyl methionine-dependent activase (Crain and Broderick, 2014). This information is consistent with the model that ’882 disrupts CoA pathways during fermentation by potentially impeding the function of Fe-S cluster-dependent IlvD and PflA enzymes in CoA synthesis and consumption.
The SufBCD proteins were pulled down with biotinylated ’882 and ’882 interacted with purified SufC with high affinity (Figure 4, Table 1), which supports our model that ’882 inhibits Fe-S cluster assembly. The effects of ’882 on Fe-S cluster assembly are widespread. A strain deficient in Fe-S cluster assembly (Δnfu ΔsufT) was more sensitive to ’882 intoxication when cultured in a medium in which the growth is reliant upon the Fe-S cluster-dependent enzymes IlvD and LeuCD (Figure 6) (Flint et al., 1993; Hentze and Argos, 1991). Under these conditions, ’882 inhibits the growth of LAC, suggesting that even in the absence of constitutive Sae activity, the pleiotropic effects of ’882 can be observed, which would be expected if ’882 inhibits Fe-S cluster assembly. The inhibitory effects of ’882 are most pronounced under anaerobic growth, and evidence suggests this is the result of: (1) reduced energy generation and the metabolic burden of constitutive Sae, (2) reduced expression of Fe-S cluster assembly factors, which decreases the effective concentration of ’882 required to inhibit Fe-S cluster assembly, and (3) the dependence of CoA homeostasis on Fe-S cluster requiring enzymes.
The investigations into the effect of ’882 on aconitase (AcnA) support the model that ’882 disrupts the Suf machinery (Figure 5). It was recently demonstrated that AcnA function is reduced in strains with inhibited Fe-S cluster assembly (Mashruwala et al., 2015), and here we show that ’882 decreases AcnA activity. The magnitude of inhibition of AcnA activity by ’882 is amplified as culture dioxygen levels are reduced. These data agree well with the decreased expression of sufC during anaerobic conditions (Mashruwala et al., 2015). Lending further support to the idea that ’882 inhibits the assembly of the Fe-S cluster upon apo-AcnA but does not directly inhibit AcnA, ’882 does not alter the activity of purified AcnA containing a chemically reconstituted Fe-S cluster. Consistent with the in vitro data, we find that in vivo the inhibitory effect of ’882 upon AcnA requires de novo protein synthesis (Campbell et al., 2001). These findings support the conclusion that ’882 decreases AcnA activity through the inhibition of Suf-mediated Fe-S cluster biogenesis.
Proteomic analysis found that cells toxified by ’882 increase the abundance of the core Suf machinery proteins. The transcription of sufC is increased under growth conditions that impose an increased demand for Fe-S cluster biogenesis (Mashruwala et al., 2015, 2016b). SufT is a Fe-S maturation factor selectively used under growth conditions that impose an increased demand for Fe-S cluster biogenesis and the transcription of sufT increases under such conditions (Mashruwala et al., 2015, 2016a, 2016b). The abundance of SufT increases in cells toxified by ’882, further emphasizing that intoxication by ’882 results in an increased need for the Fe-S cluster assembly machinery. Thus, taken with the other findings presented in this study, one explanation is that cells toxified by ’882 increase abundance of the Suf proteins to aid in mediating resistance toward this molecule and bypassing toxicity.
In summary, this work has identified ’882 as an inhibitor of Fe-S cluster assembly and our data suggest that inhibition of Fe-S cluster-dependent enzymes is, in part, responsible for the toxicity of ’882. Fe-S cluster assembly is one of the two distinct cellular targets of ’882 that also include activation of hemebiosynthesis. Our medicinal chemistry efforts have demonstrated the structural components of ’882 required for each of these activities (Dutter et al., 2016). Our previous findings demonstrated the use of ’882 as an inhibitor of S. aureus undergoing fermentation; treatment of mice with a derivative of ’882 reduced the burden of S. aureus in the liver during systemic infection (Mike et al., 2013). Here, we demonstrate a candidate staphylococcal target for this therapeutic effect. The Suf proteins are not conserved in humans, who rely on an E. coli-like Isc system, making Suf an attractive pathogen-specific drug target (Lill, 2009; Rouault, 2012). This work underscores the opportunity to develop small-molecule inhibitors of the Suf machinery as potential novel therapeutics.
SIGNIFICANCE
Antimicrobial resistance in bacterial pathogens is a growing problem, highlighting the need for novel therapeutic targets and inhibitory small molecules. Previous work has demonstrated the toxicity of the small molecule ’882 toward the important human pathogen Staphylococcus aureus. ’882 inhibits the growth of S. aureus that is undergoing fermentation for energy production, a clinically relevant form of metabolism. In this study, we sought to identify the cellular target of ’882 as a means to develop a potential therapeutic strategy. We found that ’882 impairs the assembly of Fe-S clusters in apo-proteins, which is an essential process in S. aureus. In addition, we exhibit a link between ’882 toxicity and the activity of the SaeRS virulence regulator, demonstrating that S. aureus must balance virulence factor production and energy generation in energy-limiting growth conditions. The pleotropic effects of ’882 caused by defective Fe-S cluster assembly demonstrate the potential in small molecules that target the Suf Fe-S cluster machinery. In addition, this work employs a breadth of approaches to understand the effects of a small molecule and emphasizes the importance of Fe-S cluster homeostasis in staphylococcal physiology.
EXPERIMENTAL PROCEDURES
Bacterial Growth Conditions
Strains, plasmids, and primers used are described in Supplemental Experimental Procedures. S. aureus strains Newman (wild-type is referred to throughout as NM), USA300 LAC, and JE2, and their mutants were grown on tryptic soy agar (TSA) or tryptic soy broth (TSB) and at 37°C unless noted otherwise. When appropriate, chloramphenicol or erythromycin were added to a final concentration of 10 μg/mL. ’882 was dissolved in DMSO and added to media at concentrations noted throughout; an equivalent volume of DMSO was added as vehicle control to non-treated cultures. For routine anaerobic growth, a Coy Laboratory Products anaerobic chamber was used.
Generation of Spontaneously Resistant Mutants
Stationary-phase cultures of aerobically grown NM in TSB alone were back-diluted 1:10,000 into TSB and 10 μL was spread onto TSA containing 20 or 40 μM ’882 and moved into an anaerobic jar (Difco). After 24 hr, colonies that appeared were restreaked onto TSA without ’882, allowed to grow for 24 hr, restreaked again on TSA alone, and after 24 hr restreaked on TSA containing ’882 to confirm resistance. For spontaneous resistance in NM pOS1PlgtsaeQRS, mutants were generated in the same manner except media contained chloramphenicol in addition to ’882.
For resistant mutants, genomic DNA was purified using a Wizard Genomic Kit (Promega) and the sae locus from the genome was amplified with primers L202 and L197 and Sanger sequenced (GenHunter) using primers L202, 186, 190, and L191. The plasmid from each resistant strain was purified using a Plasmid Miniprep Kit (Thermo Scientific) and Sanger sequenced (GenHunter) using primers D474, D475, and D476 to check for mutations in the sae locus before whole-genome sequencing. Primer sequences are available in Supplemental Experimental Procedures.
Whole-Genome Sequencing
Genomic DNA were isolated from mutant strains using the Wizard Genomic Kit (Promega) and sequenced along with the parental strain (NM or NM pOS1-PlgtsaeQRS) by PerkinElmer on the MiSeq platform and analyzed for mutations using the Integrated Genomics Viewer available from the Broad Institute. Mutations were confirmed by Sanger sequencing.
Growth Curves
Growth was monitored spectrophotometrically in 96-well plates containing 200 μL volume after stationary-phase cultures were back-diluted into fresh medium. Percent (%) growth relative to vehicle is calculated from the OD600 values for each strain in ’882 compared with DMSO. Additional details are found in Supplemental Experimental Procedures.
Microarray
Strains NM and ΔsaeRS were grown to stationary phase in 5 mL TSB anaerobically, and 100 μL was diluted into 10 mL of fresh, anaerobic TSB. After growth to mid-log phase, ’882 was added to a final concentration of 25 μM. RNA was purified, reverse transcribed, and hybridized to Affymetrix S. aureus GeneChips. Additional details are in the Supplemental Experimental Procedure. The microarray data have been uploaded to the NCBI GEO, accession number GEO: GSE85379.
SaeQ Immunoblot
Sixty micrograms of protein containing membrane and cytoplasmic fractions was subjected to SDS-PAGE, transferred to nitrocellulose membrane, and probed with anti-SaeQ antibodies and goat anti-rabbit secondary antibodies. See Supplemental Experimental Procedures.
’882 and ’882 Biotin Synthesis
’882 was synthesized as described previously (Mike et al., 2013). ’882-biotin was synthesized as described in the Supplemental Experimental Procedures.
’882 Biotin Pull-Down Sample Preparation
Cellular lysate from NM was incubated with biotin ’882 and streptavidin agarose resin was used to affinity purify proteins bound to biotin ’882. The resin was washed, and eluted proteins were subjected to SDS-PAGE for visualization and then to LC-MS/MS for protein identification. See Supplemental Experimental Procedures for more information.
’882 MudPIT Sample Preparation
NM cultures (5 mL) were started from single colonies in TSB containing 20 μM ’882 or DMSO and grown aerobically for 15 hr. The cells were collected by centrifugation. Cell walls were removed by lysostaphin (8 μg/mL final) treatment in TSM and protoplasts were collected by centrifugation. The protoplasts were resuspended in 450 μL PBS + 100 μM (PMSF; Thermo Scientific), and lysed by sonication. The suspension was clarified by ultracentrifugation. The protein was quantified by BCA (Thermo Scientific) and subjected briefly to SDS-PAGE before proteomic analysis.
LC-MS/MS Analysis and Protein Identification
Proteins subjected to SDS-PAGE after ’882 biotin pull-down and ’882 Mud-PIT experiments were excised and subjected to in-gel trypsin digestion and peptide extraction as described previously (Ham et al., 2005). The resulting peptides were analyzed using a Thermo Finnigan LTQ ion trap instrument. Proteins were identified from peptide using S. aureus NM genome and SEQUEST alignment. See Supplemental Experimental Procedures for more details.
’882-SufC Biolayer Interferometry
Binding was monitored using a ForteBio Octet using biotinylated ligands loaded onto streptavidin biosensors. See Supplemental Experimental Procedures for more details.
Fe-S Cluster Reconstitution
All steps were performed under strictly anaerobic conditions inside a Coy Laboratory Products chamber (<1 ppm oxygen). Recombinant purified AcnA (see Supplemental Experimental Procedures) was incubated with reconstitution buffer (50 mM Tris, 150 mM NaCl, and 5 mM DTT [pH 7.5]) for 1 hr. Cluster reconstitution was initiated by the addition of a 5-fold excess of ferrous ammonium sulfate and lithium sulfide as described previously (Mashruwala et al., 2015; Boyd et al., 2009). The reaction mixture was allowed to proceed for 1 hr before excess Fe, S, and DTT were removed by desalting using a PD-10 column (GE Healthcare) that had been pre-equilibrated with reconstitution buffer. Reconstituted protein was concentrated using YM-3 Centriplus Centrifugal Concentrators (Millipore), prior to use in activity assays.
Cell-free Extract and Purified AcnA Enzyme Assays
Aconitase activity from cell lysate and purified AcnA was determined by monitoring the conversion of isocitrate to cis-aconitate spectrophotometrically using a Beckman Coulter DU530 UV visible absorption spectrophotometer (cis-aconitate ε240 nm = 3.6 mM−1 cm−1) (Kennedy et al., 1983). Enzymatic activity was standardized with respect to the total protein concentration and subsequently as indicated in the figure legends. Additional details are available in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
’882 toxicity relies on SaeRS and this is one of two activities of this molecule
Constitutive Sae activity is a metabolic drain on S. aureus
’882 interacts with Suf proteins to inhibit Fe-S cluster assembly
The function of Fe-S cluster-dependent aconitase is inhibited by ’882
Acknowledgments
We thank members of the Skaar laboratory for critical review of this manuscript. We acknowledge Victor Torres (New York University School of Medicine) and Chia Lee (University of Arkansas for Medical Sciences) for the gift of strains and Taeok Bae (Indiana University School of Medicine-Northwest) for the gift of anti-SaeQ antibodies. We thank Hayes McDonald and the Vanderbilt Mass Spectrometry Research Center for mass spectrometry data. We thank Matt Goff and Rob Carnahan for biolayer interferometry data, which were generated by the Vanderbilt Antibody and Protein Resource, supported by the Vanderbilt Institute of Chemical Biology and the Vanderbilt-Ingram Cancer Center (P30 CA68485). Strains NE1321, NE997, NE738, NE72, NE1335, NE598, NE221, and NE1540 were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH: NR-48501. This work was supported by NIH grants R01AI069233 and R01AI073843 (to E.P.S.). J.E.C. and B.F.D were supported by NIH T32GM065086. J.E.C. is supported by NIH F31AI126662. The Boyd Laboratory is supported by Rutgers University, the Charles and Johanna Busch foundation, and USDA MRF project NE–1028. A.M. is supported by the Douglas Eveleigh fellowship from the Microbial Biology Graduate Program and an Excellence Fellowship from Rutgers University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
ACCESSION NUMBERS
The accession number for the microarray data reported in this paper is GEO: GSE85379.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.chembiol.2016.09.012.
AUTHOR CONTRIBUTIONS
Conceptualization, J.E.C., L.A.M., A.A.M., B.F.D., G.A.S., J.M.B., and E.P.S.; Formal Analysis, P.M.D.; Investigation, J.E.C., L.A.M., A.A.M., and B.F.D.; Resources, B.F.D., P.M.D., and G.A.S.; Writing – Original Draft, J.E.C., and E.P.S.; Writing – Review & Editing, J.E.C., L.A.M., A.A.M., B.F.D., P.M.D., G.A.S., J.M.B., and E.P.S.; Supervision, G.A.S., J.M.B., and E.P.S.
References
- Adhikari RP, Novick RP. Regulatory organization of the staphylococcal sae locus. Microbiology. 2008;154:949–959. doi: 10.1099/mic.0.2007/012245-0. [DOI] [PubMed] [Google Scholar]
- Bose JL, Daly SM, Hall PR, Bayles KW. Identification of the Staphylococcus aureus vfrAB operon, a novel virulence factor regulatory locus. Infect Immun. 2014;82:1813–1822. doi: 10.1128/IAI.01655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JM, Sondelski JL, Downs DM. Bacterial ApbC protein has two biochemical activities that are required for in vivo function. J Biol Chem. 2009;284:110–118. doi: 10.1074/jbc.M807003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd ES, Thomas KM, Dai Y, Boyd JM, Outten FW. Interplay between oxygen and Fe-S cluster biogenesis: insights from the Suf pathway. Biochemistry. 2014;53:5834–5847. doi: 10.1021/bi500488r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Crain AV, Broderick JB. Pyruvate formate-lyase and its activation by pyruvate formate-lyase activating enzyme. J Biol Chem. 2014;289:5723–5729. doi: 10.1074/jbc.M113.496877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutter BF, Mike LA, Reid PR, Chong KM, Ramos-Hunter SJ, Skaar EP, Sulikowski GA. Decoupling activation of heme biosynthesis from anaerobic toxicity in a molecule active in Staphylococcus aureus. ACS Chem Biol. 2016;11:1354–1361. doi: 10.1021/acschembio.5b00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, Krieger S, Ohlsen K, Hecker M, Gerth U, et al. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res. 2013;12:547–558. doi: 10.1021/pr300394r. [DOI] [PubMed] [Google Scholar]
- Flack CE, Zurek OW, Meishery DD, Pallister KB, Malone CL, Horswill AR, Voyich JM. Differential regulation of staphylococcal virulence by the sensor kinase SaeS in response to neutrophil-derived stimuli. Proc Natl Acad Sci USA. 2014;111:E2037–E2045. doi: 10.1073/pnas.1322125111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint DH, Emptage MH, Finnegan MG, Fu W, Johnson MK. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J Biol Chem. 1993;268:14732–14742. [PubMed] [Google Scholar]
- Frees D, Savijoki K, Varmanen P, Ingmer H. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol Microbiol. 2007;63:1285–1295. doi: 10.1111/j.1365-2958.2007.05598.x. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2006;2:e87. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol. 2007;189:4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham AJ, Caprioli RM, Gross ML. Proteolytic Digestion Protocols. The Encylopedia of Mass Spectrometry. 2005;2 (Elsevier) [Google Scholar]
- Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Hood MI, Skaar EP. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. MBio. 2013;4 doi: 10.1128/mBio.00241-13. http://dx.doi.org/10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Argos P. Homology between IRE-BP, a regulatory RNA-binding protein, aconitase, and isopropylmalate isomerase. Nucleic Acids Res. 1991;19:1739–1740. doi: 10.1093/nar/19.8.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DW, Cho H, Lee H, Li C, Garza J, Fried M, Bae T. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J Bacteriol. 2011;193:4672–4684. doi: 10.1128/JB.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DW, Cho H, Jones MB, Shatzkes K, Sun F, Ji Q, Liu Q, Peterson SN, He C, Bae T. The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol Microbiol. 2012;86:331–348. doi: 10.1111/j.1365-2958.2012.08198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MC, Emptage MH, Dreyer JL, Beinert H. The role of iron in the activation-inactivation of aconitase. J Biol Chem. 1983;258:11098–11105. [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, Skaar EP, Torres VJ, Nauseef WM, Boyd JM. Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol Microbiol. 2015;95:383–409. doi: 10.1111/mmi.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala A, Roberts CA, Bhatt S, May KL, Carroll RK, Shaw LN, Boyd JM. Staphylococcus aureus SufT: an essential iron-sulfur cluster assembly factor in cells experiencing a high-demand for lipoic acid. Mol Microbiol. 2016a doi: 10.1111/mmi.13539. http://dx.doi.org/10.1111/mmi.13539. [DOI] [PMC free article] [PubMed]
- Mashruwala AA, Bhatt S, Poudel S, Boyd ES, Boyd JM. The DUF59 containing protein SufT is involved in the maturation of iron-sulfur (FeS) proteins during conditions of high FeS cofactor demand in Staphylococcus aureus. PLoS Genet. 2016b;12:e1006233. doi: 10.1371/journal.pgen.1006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mike LA, Dutter BF, Stauff DL, Moore JL, Vitko NP, Aranmolate O, Kehl-Fie TE, Sullivan S, Reid PR, DuBois JL, et al. Activation of heme biosynthesis by a small molecule that is toxic to fermenting Staphylococcus aureus. Proc Natl Acad Sci USA. 2013;110:8206–8211. doi: 10.1073/pnas.1303674110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murima P, McKinney JD, Pethe K. Targeting bacterial central metabolism for drug development. Chem Biol. 2014;21:1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- Palazzolo-Ballance AM, Reniere ML, Braughton KR, Sturdevant DE, Otto M, Kreiswirth BN, Skaar EP, DeLeo FR. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180:500–509. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, Rock CO. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci USA. 2014;111:10532–10537. doi: 10.1073/pnas.1408797111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- Rosario-Cruz Z, Chahal HK, Mike LA, Skaar EP, Boyd JM. Bacillithiol has a role in Fe-S cluster biogenesis in Staphylococcus aureus. Mol Microbiol. 2015;98:218–242. doi: 10.1111/mmi.13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirova JS, Hernalsteens JP, De Backer S, Xavier BB, Moons P, Turlej-Rogacka A, De Greve H, Goossens H, Malhotra-Kumar S. Fatty acid kinase A is an important determinant of biofilm formation in Staphylococcus aureus USA300. BMC Genomics. 2015;16:861. doi: 10.1186/s12864-015-1956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach BP, Chung AH, Scott AD, George SJ, Cramer SP, Dos Santos PC. Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein. Biochemistry. 2014;53:152–160. doi: 10.1021/bi4011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 2011;7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Li C, Jeong D, Sohn C, He C, Bae T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192:2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C. AirSR, a [2Fe–2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J Am Chem Soc. 2012;134:305–314. doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Matsushita Y, Yoshikawa A, Isono K. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol Gen Genet. 1989;217:289–293. doi: 10.1007/BF02464895. [DOI] [PubMed] [Google Scholar]
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitko NP, Spahich NA, Richardson AR. Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. MBio. 2015;6:e00045–15. doi: 10.1128/mBio.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, Hinger SA, Aysanoa EE, Blanchard C, Dunman PM, et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 2015;11:e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


