Abstract
In the vast majority of pediatric patients with dilated cardiomyopathy, the specific etiology is unknown. Studies on families with dilated cardiomyopathy have exemplified the role of genetic factors in cardiomyopathy etiology. In this study, we applied whole-exome sequencing to members of a non-consanguineous family affected by a previously unreported congenital dilated cardiomyopathy syndrome necessitating early-onset heart transplant. Exome analysis identified compound heterozygous variants in the FLNC gene. Histological analysis of the cardiac muscle demonstrated marked sarcomeric and myofibrillar abnormalities, and immunohistochemical staining demonstrated the presence of Filamin C aggregates in cardiac myocytes. We conclude that biallelic variants in FLNC can cause congenital dilated cardiomyopathy. As the associated clinical features of affected patients are mild, and can be easily overlooked, testing for FLNC should be considered in children presenting with dilated cardiomyopathy.
Introduction
Dilated cardiomyopathies (DCMs) are a group of heterogeneous disorders characterized by enlargement of one or both ventricles of the heart, accompanied by left ventricular systolic dysfunction.1, 2 The hereditary DCM can be classified into two forms: isolated or nonsyndromic and syndromic DCMs. Multiple genes have been identified for both syndromic and isolated familial cardiomyopathies. These genes encode a variety of cardiomyocyte proteins, including nuclear envelope, cytoskeletal, sarcomeric, calcium channel regulators, transcription factors and others. Multiple DCM syndromes have been characterized, several of them only in a single family. In some of them, skeletal myopathy is a prominent clinical feature such as in Dystrophin-related Duchenne/Becker muscular dystrophy, LMNA-associated Emery–Dreifuss muscular dystrophies 2+3, and Limb Girdle Muscular Dystrophy 1B, MYH7-associated Laing distal myopathy and mitochondrial cardiomyopathy. Other DCM syndromes can be accompanied by various other clinical manifestations.1, 2 Although numerous genes have been identified for adult-onset DCM, in the vast majority of pediatric patients with DCM, the specific etiology is unknown. In this study, we used whole-exome sequencing to investigate the genetic etiology in a family affected with a previously unreported congenital dilated cardiomyopathy.
Subjects and methods
Subjects
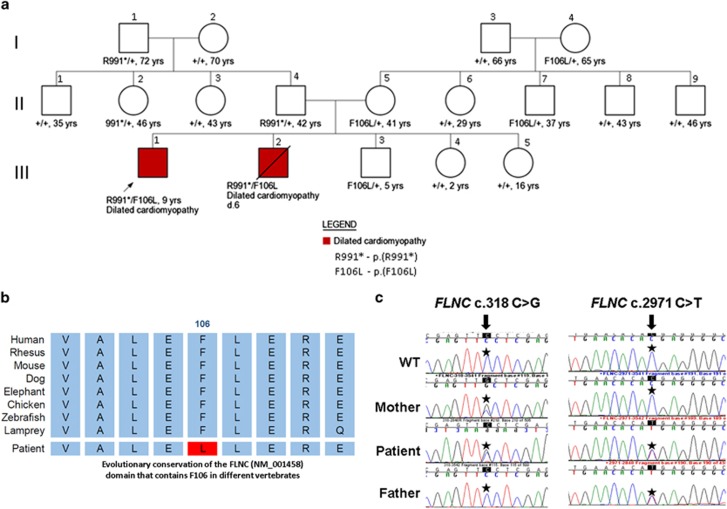
Local approval for this study was provided by the Rabin Medical Center Institutional Review Board, and all participants signed an informed consent. The nuclear family is of Ashkenazi Jewish ancestry, and consists of parents and five children. The parents are non-consanguineous and healthy. The proband is 9 years old. He was born following a pregnancy during which a cystic hygroma was found on sonographic examination at 15 weeks of pregnancy. Fetal echocardiography and karyotype were normal. He was born at term. Physical examination at birth showed mild hypotonia, short webbed neck, low posterior hairline, posteriorly rotated low-set ears and submucous cleft palate. Cognitive and motor developments were normal. As he had a sibling dying at age 6 years because of dilated cardiomyopathy, he underwent echocardiograms every 6 months, which revealed slowly progressing systolic dysfunction. At age 2 years, as advanced dilated cardiomyopathy was demonstrated, he underwent heart transplantation. At age 9 years, physical examination showed additional features including thin and translucent skin, low-set ears, hypotelorism (ICD <3%), microcephaly (HC <2%), micrognathia and normal cognition. He did not show any signs of distal or proximal myopathy. The proband's parents reported a deceased male sibling with the same medical condition. This sibling was born at term following an uneventful pregnancy. At birth, congenital hypotonia and a short webbed neck had been noticed. He was asymptomatic until age 2 years when a chest x-ray, performed because of suspected pneumonia, demonstrated an enlarged cardiac silhouette. A follow-up echocardiogram demonstrated mild systolic dysfunction and he was followed up with repeated echocardiograms until age of 6 years, then, because of rapidly progressing systolic dysfunction, he was placed on the heart transplant waitlist. He had normal coronary angiography, and during the course of pretransplantation workup, he developed arrhythmias and passed away from a cerebrovascular event. Postmortem studies of the myocardium demonstrated fibrosis with no evidence for accumulation of glycogen, iron or amyloid. Electron microscopy studies of the myocardium showed marked reduction in the number of myofibrils and an increased number of swollen, abnormal mitochondria. Skeletal muscle showed no histological abnormalities and normal activity of the mitochondrial electron transport complexes. The parents are in their early 40s, and three additional siblings, age 2, 5 and 16 years, are healthy and have normal echocardiograms (Figure 1a).
Figure 1.
(a) Pedigree of family showing the age of individuals and segregation of FLNC mutant alleles. Wild-type FLNC allele is indicated by plus sign. Filled symbols indicate affected individuals. Diagonal lines across symbols indicate deceased individuals. (b) Evolutionary conservation of the FLNC domain that contains F106L missense variant (Red) in different vertebrates (Vertebrate Multiz Alignment and Conservation, UCSC genome browser). (c) Electropherograms showing the FLNC mutations in the patient and his parents. The full colour version of this figure is available at European Journal of Human Genetics online.
Whole-exome sequencing
Genomic DNA extraction, exome enrichment, sequencing and analysis have been completed as described before.3 Variants were filtered to generate a final list of rare functional variants only (missense, nonsense, splice site variants and indels). Variants with minor allele frequency >0.01 in the Exome Variant Server (release ESP6500) or that have allele count >150 in the ExAC database of European (NFE) samples were removed. Validation and segregation analyses were carried out as described.3 Variants were scored relative to the reference sequences deposited in the National Center for Biotechnology Information (FLNC: NM_001458.4). Variant data was submitted to ClinVar (SUB1474516; http://www.ncbi.nlm.nih.gov/clinvar/).
Directed mutagenesis of full-length Filamin C
A full-length cDNA clone for FLNC (NM_001458.4) with an N-terminal Myc-DDK-Tag was obtained from OriGene (Rockville, MD, USA). Site-directed mutagenesis was performed to generate the clones with the p.F106L, p.R991* variants, respectively, using a QuikChange II XL Site-Directed Mutagenesis Kit according to the manufacturer's recommendations (Stratagene, Santa Clara, CA, USA) and specific oligonucleotides (for c.318C>G,p.(F106L), forward – 5′-GCCCTCGAGTTgCTCGAGCGCGA-3′ and reverse – 5′-TCGCGCTCGAGcAACTCGAGGGC-3′ for c.2971C>T,p.(R991*), forward – 5′-CTGTGAACACAtGAGGGGCTGGC-3′ and reverse – 5′-GCCAGCCCCTCaTGTGTTCACAG-3′ for F106L-sec, 5′-ATGAGCACCTCAAGTGCG-3′ and for R991*-sec, 5′-GGGTGACCATCGTAGGTG-3′). Introduction of the correct variant and absence of additional variants was verified by Sanger sequencing.
Cell culture and immunofluorescence
Rat cardiac myoblast H9C2 cells (ATCC, Manassas, VA, USA; CRL-1446) were grown in DMEM supplemented with 10% new born calf serum and l-glutamine. To express wild-type and mutant FLNC, cells were transfected using Amaxa SF cell line 4D-Nucleofector X Kit by Lonza (Allendale, NJ, USA) following the manufacturer's instructions. For immunofluorescence analysis, cells were fixed in 4% paraformaldehyde solution, rinsed in phosphate-buffered saline (PBS) and permeabilized with 0.5% Triton X-100. Cells were incubated with primary antibodies diluted in 10% goat serum in 1 × PBS for 1–3 h at room temperature. After washing with PBS-T, slides were incubated with 1:500 Alexa 488-conjugated secondary antibody (Life Technologies, Waltham, MA, USA) for 1 h at 25 °C. After washing, nuclei were counterstained with DAPI (Roche, Mannheim, Germany), and slides were mounted in VectaShield mounting medium (Vector Laboratories, Burlingame, CA, USA). To determine the actin stress fiber distribution cells were stained with Phalloidin-TRITC. Micrographs were obtained with an Axioplan-2 Zeiss fluorescent microscope (Zeiss, Oberkochen, Germany), and images were captured with a charge-coupled device camera (Photometrics SenSys, Tucson, AZ, USA). All the images at the precise planes in the z-stack were used to generate a maximum intensity projection.
Western blot analysis and sequential detergent extraction of proteins
Cells were nucleofected with 2 μg of each plasmid and 48 h after transfection in the presence of protein was analyzed. For protein extraction, cultured cells were washed two times with 1 × PBS and lysed in RIPA buffer (100 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10 mM EDTA, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS) supplemented with complete protease inhibitor cocktail without EDTA and PhosStop EASYpack (Roche). Cell lysates were centrifuged at 13 000 g for 5 min, and the supernatant was conserved. Protein concentration was evaluated with the bicinchoninic acid technique (Pierce BCA Protein Assay Kit, Waltham, MA, USA). Proteins were run in 8% SDS-PAGE gels, transferred to nitrocellulose membranes, blocked with 5% non-fat dry milk in TBS-T buffer (20 mM Tris (pH 7.4), 150 mM NaCl, 0.05% Tween-20) and incubated overnight at 4 °C with the following primary antibodies: anti-DDK tag antibody (Cell Signaling, Beverly, MA, USA) and anti-actin (Sigma, St Louis, MO, USA). Primary antibodies were detected with horseradish peroxidase-conjugated species-specific secondary antibodies (Jackson Immunoresearch (West Grove, PA, USA) or Thermo Scientific (Waltham, MA, USA)) with Luminata Forte Western HRP Substrate (Millipore, Billerica, MA, USA) using a LAS-300 FUJIFILM (Tokyo, Japan).
Immunohistochemistry
Tissues were fixed in 4% paraformaldehyde in PBS and stored in 70% ethanol. Fixed tissues were embedded in paraffin by standard procedures. Blocks were sectioned (3 μm) and histochemical techniques including H&E and Masson trichrome were performed following the manufacturer's instructions and as described previously.4 Immunohistochemistry was performed on paraffin 3-μm-thick sections, previously heated at 60 °C, then cleared and dehydrated in xylene and graded alcohols. Antigen retrieval was performed by heating in with a Tris borate EDTA buffer (pH 8.4). Endogenous peroxidase activity was quenched by incubation in TBS-T containing 3% hydrogen peroxide. Each incubation step was carried out at room temperature and followed by three sequential washes in TBS-T. To block nonspecific binding, tissues were incubated for 20 min in TBS-T containing 10% bovine serum albumin. Samples were incubated with a primary rabbit polyclonal antibody against filamin C (Novus Biologicals, Littleton, CO, USA) at 1:25 dilution, followed by incubations with biotinylated OMNImap secondary antibody for 15 min, peroxidase-labeled streptavidin and diaminobenzidine substrate. Control immunohistochemical staining was performed following the same procedure but omitting the primary antibody.
Results
Comparative genomic hybridization study demonstrated no copy number variations. Next, whole-exome sequencing was carried out on the proband, his 16-year-old healthy sister and their parents. Sequencing was performed using the Illumina HiSeq 2500 machinery (San Diego, CA, USA) to generate paired-end reads of 150 bp with an average coverage of 95 ×. Exome results were analyzed for all modes of inheritance, and revealed compound heterozygous of missense and stop-gain variants in the FLNC gene: g.[128471009C>G][128484099C>T]. The first missense variant, c.[318C>G], p.(F106L), according to the ExAC database (http://exac.broadinstitute.org), was found four times in 32 261 Europeans and was inherited from the mother. The phenylalanine in that position is highly conserved in evolution (Figure 1b) (GERP++=4.5, phyloP100way=3.2,5, 6 and was predicted to be damaging by all different prediction algorithms examined (SIFT, Polyphen2, LRT, VariantTaster, VariantAssessor, FATHMM). The second variant, c.[2971C>T], p.(R991*), is a stop-gain/nonsense variant that has not been reported before in any database, and was transmitted from the father. The variants in FLNC were the only ones that were found to segregate as expected in the family. Analysis of the deceased sibling DNA sample revealed the same compound heterozygosity for the FLNC variants (Figures 1a and c).
As heterozygous variants in FLNC have been associated with myofibrillar and distal myopathies,7, 8, 9, 10, 11 a neurological evaluation was performed in all family members who were found to be variant carriers (seven individuals; aged 5–72 years). Neurological examinations and echocardiograms were normal in all subjects.
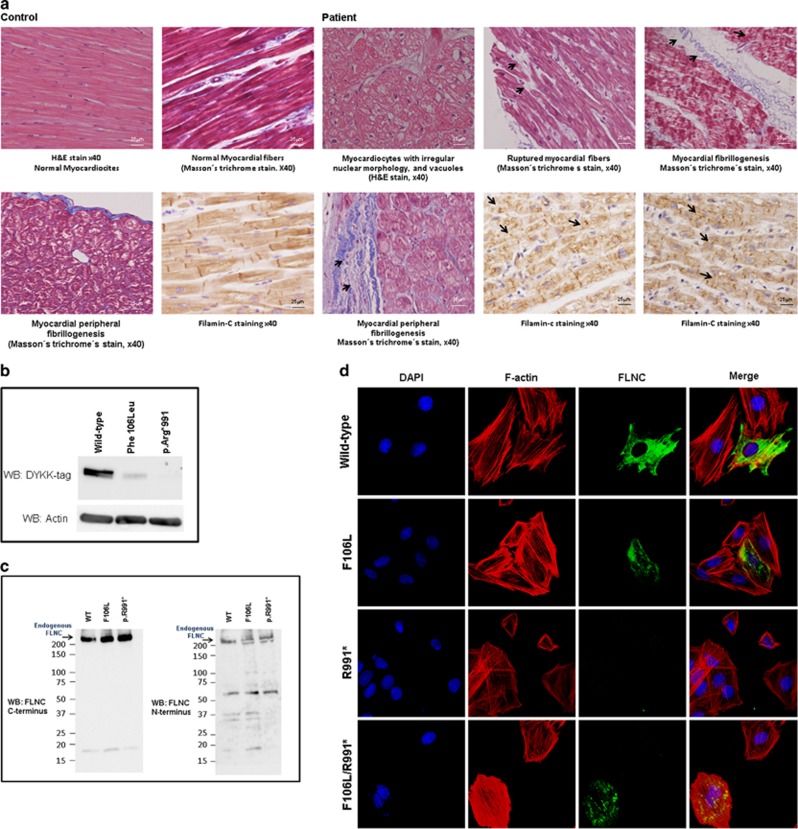
Histological analysis of the probands' cardiac muscle revealed marked sarcomeric abnormalities, including myofibrillar disarray with a reduction of myofibrils, cardiomyocytes with irregular nuclear morphology and vacuoles, ruptured myocardial fibers, sarcomeric aggregates and pericellular and peripheral fibrosis not observed in control hearts (Figure 2a). Immunohistochemical staining confirmed the presence of Filamin C aggregates in cardiac myocytes. To evaluate whether the FLNC variants found in this family could impair the function of this sarcomeric protein, Filamin C full-length cDNA clones containing either the p.(F106L) or the p.(R991*) were generated and nucleofected into H9C2 rat cardiac myoblasts. Western blot analysis, using a flagged antibody, showed high expression levels of Filamin C in the wild type, with a reduction in the expression levels in the p.(F106L) mutant (suggesting that this variant might impair protein stability) and a complete absence of protein expression in the p.(R991*) form (Figure 2b). Western blot analysis, using antibodies directed to the C and N terminus of Filamin C, confirmed that no protein was produced using both variants (Figure 2c). Subcellular distribution analysis of these mutants (assessed by immunofluorescence and confocal microscopy) showed that wild-type Filamin C expression had a normal cytoplasmic distribution, mainly associated with actin fibers. However, the p.(F106L) mutant showed formation of protein aggregates that were not detected in the truncated form p.(R991*). Expression of both variants showed a distribution pattern somewhat similar to the p.(F106L)-expressing cells (Figure 2d). These results are consistent with the biochemical experiments and suggest that p.(F106L) variant impairs the function of Filamin C protein.
Figure 2.
(a) Histological alterations in cardiac tissue of transplanted patient. Hematoxylin and eosin (H&E) staining of heart explants from the transplanted case shows sarcomere disarray, with reduction of myofibrils, cardiomyocytes with irregular nuclear morphology (magnification × 40). Masson's trichrome stain revealed a development of collagen fibrillogenesis in peripheral and perinuclear areas and the rupture of myocardial fibers not observed in the heart of a control subject. Immunohistochemical analysis of the cardiac muscle show an accumulation of Filamin C aggregates (magnification × 40). (b) Expression of wild-type (WT) and mutant Filamin C proteins in transfected cells. Western blot analysis, using a flagged antibody, show high expression levels of Filamin C in the WT, with a reduction in the expression levels in the p.(F106L) mutant and a complete absence of protein expression in the p.(R991*) form. (c) Western blot analysis, using antibodies directed to the C and N terminus of Filamin C showing that no protein was produced using both variants. (d) Subcellular distribution of ectopically expressed Filamin C. Expression of WT and mutants in rat cardiac myoblast was analyzed by immunofluorescence and confocal microscopy. Filamin C expression was detected using an anti-DDK antibody (green), actin fibers were stained with phalloidin (red) and nuclei were counterstained with DAPI (4',6-diamidino-2-phenylindole) (magnification × 100). Expression of mutant FLNC clones resulted in the formation of perinuclear protein aggregates that were not observed in WT FLNC. The full colour version of this figure is available at European Journal of Human Genetics online.
Discussion
We identified biallelic FLNC variants in a family presenting with a previously unreported pediatric dilated cardiomyopathy syndrome. The maternally inherited variant is located in a conserved site within the N-terminal actin-binding domain of Filamin C and all prediction algorithms showed high functional effect (deleterious, damaging or disease causing). As individuals who carry this variant are asymptomatic, haplosufficiency of the functional allele or only mild functional effect of the variant is likely. Based on studies of similar variants leading to distal myopathy,11 the paternally transmitted variant is expected to lead to nonsense-mediated decay of the mRNA, resulting in the absence or a reduction of protein amounts. Although we could not detect protein expression in cells expressing this mutant, variant carriers are asymptomatic suggesting again haplosufficiency of the functional allele or only mild functional effect of the variant. Only recently, a direct connection between dominant variants in FLNC to isolated hypertrophic and restrictive cardiomyopathies has been proposed.4, 12 Interestingly, none of the reported patients had symptoms or clinical findings of myopathy. It was shown previously that disruption of the desmin gene in mice work in a dominant-negative mechanism to compromise myofibril alignment and leads to the appearance of aggregates that are characteristic of desmin-related cardiomyopathy. In addition, expression of a mutant alphaB-crystallin gene in cells resulted in a phenotype that was similar to that observed for the desmin-related cardiomyopathies.13, 14
The mechanisms underlying our observations are not completely clear. Apparently, the mutations described here cause a devastating phenotype only when coexpressed and leading to deposition of Filamin C aggregates ultimately causing disintegration of myofibrils as seen in the explanted heart studies. Haploinsufficiency has been suggested as the most probable reason for the disease phenotype in a number of patients with FLNC-induced myopathy.15 Yet, this is clearly not the case here as the paternal family, expressing only the normal allele, is healthy. In the database of genomic variant, more than 12 cases with full or partial deletion of the FLNC gene have been reported, suggesting that happloinsufficiency may not be a common mechanism in FLNC related disorders. Thus, it appears that the lack of expression of mutant protein precludes the development of major protein aggregates that are a hallmark of myofibrillar myopathty. However, in patients who are heterozygotes for missense mutations in FLNC, there is a later onset disease, perhaps because in addition to the abnormally functioning mutant allele there is also normal protein being produced. It is possible that these missense mutants make the cells susceptible to develop protein aggregation as they interact, through a slowly progressive process, with the normal protein product. This interaction may lead to significant protein aggregation over the years that result in muscular dysfunction and the ensuing phenotype, suggesting a dominant-negative effect. This may explain why our patients have a more severe phenotype because they are essentially compound heterozygous for a deleterious mutation (ie, this is a dominant, but not true dominant mutation). However, if this is a straight dominant-negative effect, which requires interaction of the mutant protein with a normal protein, the phenotype of our patients cannot be fully explained. The lack of phenotype in the mother's family could be because the missense mutant is a mild allele that does not lead to disease by itself. One can speculate that because it is a mild allele, the children were viable, whereas a more severe mutation in the hemizygous state might not be viable. Several Filamin C animal models have been reported.16, 17 Mice homozygous for an FLNC-truncated allele die at birth owing to breathing problems caused by severe abnormalities of their skeletal muscles, whereas the heart had normal appearance. Heterozygous mice were normal and fertile and no abnormalities were reported. One should bear in mind that mouse models are frequently not recapitulating recessive/dominance inheritance in humans. A mutation in one of two FLNC genes of medaka fish17 causes disorganization of skeletal muscle fibers and abnormal heart development associated with a rupture of the myocardial layer. Fish heterozygous for the mutation developed normally. This phenotype was also observed using morpholino-based antisense RNA experiments.
In conclusion, we suggest that Filamin C testing should be considered in children presenting with dilated cardiomyopathy. Timely molecular diagnosis is important for the prediction, treatment and prevention of rapidly progressing heart failure and life-threatening arrhythmias.
Acknowledgments
We thank Rabin Medical Center and Adler cathedra for pediatric cardiology for the support. This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Footnotes
The authors declare no conflict of interest. DMB and CB are compensated and serve as the chief medical officer and the laboratory director of Gene by Gene, respectively.
References
- Towbin JA: Inherited cardiomyopathies. Circ J 2014; 78: 2347–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellefave L, McNally EM: The genetics of dilated cardiomyopathy. Curr Opin Cardiol 2010; 25: 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinstein E, Orvin K, Tayeb-Fligelman E et al: Variants in TAX1BP3 cause dilated cardiomyopathy with septo-optic dysplasia. Hum Mutat 2015; 36: 439–442. [DOI] [PubMed] [Google Scholar]
- Valdés-Mas R, Gutiérrez-Fernández A, Gómez J et al: Variants in filamin C cause a new form of familial hypertrophic cardiomyopathy. Nat Commun 2014; 5: 5326. [DOI] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S: Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 2010; 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS et al: Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005; 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst DO, Goldfarb LG, Kley RA et al: Filamin C-related myopathies: pathology and mechanisms. Acta Neuropathol 2013; 125: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorgerd M, van der Ven PFM, Bruchertseifer V et al: A variant in the dimerization domain of filamin c causes a novel type of autosomal-dominant myofibrillar myopathy. Am J Hum Genet 2005; 7: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff RM, Tay V, Hackman P et al: Variants in the N-terminal actin-binding domain of filamin C cause a distal myopathy. Am J Hum Genet 2011; 88: 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Reardon K, Roberts L et al: A new dominant distal myopathy affecting posterior leg and anterior upper limb muscles. Neurology 2005; 64: 1245–1254. [DOI] [PubMed] [Google Scholar]
- Shatunov A, Olive' M, Odgerel Z et al: In-frame deletion in the seventh immunoglobulin-like repeat of filamin C in a family with myofibrillar myopathy. Eur J Hum Genet 2009; 17: 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodehl A, Ferrier RA, Hamilton SJ et al: Variants in FLNC are associated with familial restrictive cardiomyopathy. Hum Mutat 2016; 37: 269–279. [DOI] [PubMed] [Google Scholar]
- Wang X, Osinska H, Klevitsky R et al: Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res 2001; 89: 84–91. [DOI] [PubMed] [Google Scholar]
- Wang X, Osinska H, Dorn GW II et al: Mouse model of desmin-related cardiomyopathy. Circulation 2001; 103: 2402–2407. [DOI] [PubMed] [Google Scholar]
- Guergueltcheva V, Peeters K, Baets J et al: Distal myopathy with upper limb predominance caused by filamin C haploinsufficiency. Neurology 2011; 77: 2105–2114. [DOI] [PubMed] [Google Scholar]
- Dalkilic I, Schienda J, Thompson TG, Kunkel LM: Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol Cell Biol 2006; 26: 6522–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Mitsuhashi H, Isogai S et al: Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev Biol 2012; 361: 79–89. [DOI] [PubMed] [Google Scholar]