Abstract
Genetic generalized epilepsy (GGE), formerly known as idiopathic generalized epilepsy, is the most common form of epilepsy and is thought to have predominant genetic etiology. GGE are clinically characterized by absence, myoclonic, or generalized tonic-clonic seizures with electroencephalographic pattern of bilateral, synchronous, and symmetrical spike-and-wave discharges. Despite their strong heritability, the genetic basis of generalized epilepsies remains largely elusive. Nevertheless, recent advances in genetic technology have led to the identification of numerous genes and genomic defects in various types of epilepsies in the past few years. In the present study, we performed whole-exome sequencing in a family with GGE consistent with the diagnosis of eyelid myoclonia with absences. We found a nonsense variant (c.196C>T/p.(Arg66*)) in RORB, which encodes the beta retinoid-related orphan nuclear receptor (RORβ), in four affected family members. In addition, two de novo variants (c.218T>C/p.(Leu73Pro); c.1249_1251delACG/p.(Thr417del)) were identified in sporadic patients by trio-based exome sequencing. We also found two de novo deletions in patients with behavioral and cognitive impairment and epilepsy: a 52-kb microdeletion involving exons 5–10 of RORB and a larger 9q21-microdeletion. Furthermore, we identified a patient with intellectual disability and a balanced translocation where one breakpoint truncates RORB and refined the phenotype of a recently reported patient with RORB deletion. Our data support the role of RORB gene variants/CNVs in neurodevelopmental disorders including epilepsy, and especially in generalized epilepsies with predominant absence seizures.
Introduction
Epilepsy is one of the most frequent neurological disorders, affecting up to 4% of people during their lifetime. In recent years, significant progress has been made in the identification of genetic defects underlying various epileptic syndromes.1 The search for the numerous as-yet unidentified genetic defects in human epilepsy remains an important task.2 Genetic generalized epilepsies (GGEs), formerly known as idiopathic generalized epilepsy, represent the most common form of epilepsies and have a predominantly genetic etiology.3, 4 GGEs are clinically characterized by the occurrence either as unique seizure types or in various combinations of absence, myoclonic, and generalized tonic-clonic seizures (GTCS) with electroencephalographic (EEG) pattern of bilateral, synchronous, and symmetrical spike-and-wave or polyspike-and-wave discharges. Photosensitivity, an abnormal cortical response to intermittent photic stimulation (IPS) that manifests as spikes or spike-and-wave discharges on the EEG, is typically associated with GGE.5 The genetic determinants underlying common epilepsies, for which pedigree data suggest complex inheritance and incomplete penetrance, remain largely unknown. Recent access to powerful technologies and approaches such as array comparative genomic hybridization (aCGH) and next-generation sequencing, allowing individual whole-exome (WES) or whole-genome sequencing, has facilitated the identification of part of the genetic factors underlying these disorders. In recent genome-wide studies, rare recurrent copy number variants (CNVs) that are important risk factors for other neuropsychiatric disorders were present in about 4% of the generalized epilepsies.6, 7, 8 Several single cases with non-recurrent deletions have also been reported in otherwise common epilepsies accounting for up to 5% of patients studied. The genes located within these CNVs deserve further study given that they are potential candidates for susceptibility to GGE.9, 10 In this study, we used a WES approach in a family with several individuals affected with photosensitive GGE. This led to the identification of an inherited nonsense variant in the RORB gene, which encodes the retinoid-related nuclear receptor ROR-beta. Family-based exome sequencing in two sporadic patients with generalized epilepsy and intellectual disability (ID) led to the identification of two de novo RORB sequence variants. Moreover, two CNVs and a de novo balanced reciprocal translocation were detected by interrogation of current databases/projects for potentially pathogenic genomic events encompassing this gene.
Subjects and methods
Patients
We performed exome sequencing in a large non-consanguineous French family (Figure 1a), which was first reported >20 years ago11; through a network of collaborating geneticists and epileptologists, we recruited two additional sporadic patients (patients AG1 and RO1) with trio-based exome sequencing identifying de novo mutations. By consultation of the aCGH database of Lyon and Geneva and the Breakpoint mapping consortium, we identified three unrelated female patients: a 25-year-old woman (Case no. 9A1117, Lyon, France), a 9-year-old girl (Case no. GE0705, Geneva, Switzerland) both carrying de novo microdeletions including RORB; and a 10-year-old girl (DK8393) with a de novo balanced reciprocal translocation seq[GRch37]t(9;19)(q21;q12)dn. We also added detailed clinical data on a previously reported patient with childhood absence epilepsy (EC-CAE300), who was revisited during this study.8 Finally, a follow-up cohort of 100 patients with GGE was screened with Sanger sequencing.
Figure 1.
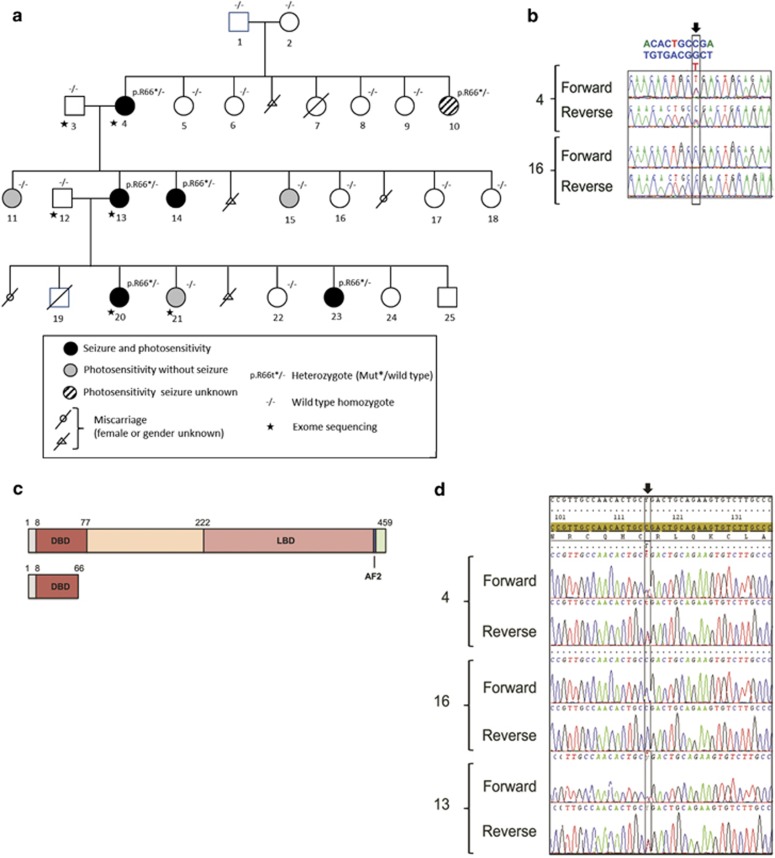
Inherited nonsense RORB variant segregating in a family with GGE with eyelid myoclonia (family 1). (a) Pedigree with GGE. DNA numbers are indicated under each corresponding family member. Empty symbols represent unaffected individuals. ILS, intermittent light stimulation; PPR, photoparoxysmal response. Patients 19–25 have been phenotyped and collected after the initial report on the family.11 Note that the variant was not found in neither individual 1 nor 2, which – together with the genetic exclusion of non-paternity (data not shown) – suggest germline mosaicism. (b) DNA sequence traces (both strands) corresponding to the heterozygous c.196C>T variant (arrow) in exon 3 of the RORB gene (patient no. 4, top) and to a wild-type unaffected individual (family member no. 16, bottom). (c) Predicted mutant p.Arg66* RORβ protein. The different domains of the wild-type RORβ protein (Genbank NP_008845) (top) and the corresponding truncated mutant protein (bottom) are schematically represented. Numbers at the top of each protein indicate amino-acid positions from the N-terminus (left) to the C-terminus (right). DBD, DNA-binding domain; LBD, ligand-binding domain; AF2, activation function domain. (d) cDNA sequence traces showing the presence of the heterozygous c.196C>T variant (arrow) in RORB transcripts extracted from lymphoblastoid cell lines of patients 4 (top) and 13 (bottom) and of control individual no. 16 (middle). No difference was observed upon treatment with puromycin (data not shown).
Experiments were conducted in accordance with the Declaration of Helsinki, and all procedures were carried out with the adequate understanding and written consent of the subjects or their parents or legal guardians in the case of minors or those with ID and according to the appropriate ethical committees (no. CPP 09/40 – no. AC-2008-438/no. DC-2009-1002, France; University of Washington, USA; Austin Health, Melbourne, Australia; H-KF-2006-5901, Denmark; University of Kiel A115/02 and A 116/02, Germany; no. 13/50/508; University of Antwerp, Belgium).
Whole-exome sequencing
Genomic DNA was captured using Agilent in-solution enrichment methodology (SureSelect Human All Exon, Agilent, Santa Clara, CA, USA) with the appropriate biotinylated oligonucleotides probe library (Human All Exon v4+UTR-70 Mb, Agilent). Enriched DNA samples were sequenced on an Illumina HiSEQ 2000 as paired-end 75-bp reads (Illumina, San Diego, CA, USA). Image analysis and base calling was performed using Illumina Real Time Analysis pipeline version 1.12.4.2 with default settings.12, 13 Both sporadic patients with de novo mutations were sequenced as previously described.14
Sanger sequencing and targeted sequencing in additional cohorts of patients
Coding exons and intronic boundaries of RORB (NM_006914.3) were PCR-amplified from genomic DNAs, purified, and sequenced using the Big-dye terminator Cycle Sequencing Kit v1.1 and a 3130 × l genetic analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were analyzed using the Seqscape software (Applied Biosystems) and compared with the RORB GenBank reference sequence (NM_006914.3) (Table 1).
Table 1. Direct RORB sequencing in French and Australian cohorts of patients with various GGE.
| Family or case | No. of patients | Technology applied | Phenotype | Center |
|---|---|---|---|---|
| 18 single cases | 18 | Molecular Inversion Probes | GGE | University of Washington |
| 56 families | 56 | Sanger | GGE | Neurology Unit Strasbourg |
| 26 single cases | 26 | Sanger | GGE | Neurology Unit Strasbourg |
In addition, RORB was sequenced in probands with GGE from 18 single cases from Australia, using Molecular Inversion Probes (MIPs: Integragen DNA Technologies, Coralville, IA, USA).15
Mate-pair sequencing
Mate-pair sequencing on a HiSeq2500 (Illumina) (2 × 100 bp) and detection of discordant breakpoint spanning reads were performed essentially as described.16 For amplification and Sanger sequencing of the breakpoints to sequence level, the forward and reverse primer pairs F1_der9 5′-TGACCACATTGCAAACCATT-3′ R2_der9 5′-TTGTTTGTGGGCAAATTTCA-3′ and R1_der19 5′-AATACACTTTATGTGGATGTGT-3′ F2_der19 5′-CGTGGAAGCCTCCTCAGATA-3′ were used to define the chromosomes 9 and 19 breakpoints.
aCGH and fluorescent in situ hybridization (FISH)
Patients' DNA was extracted with the QIAmpDNA Blood Midikit or the DNeasy Blood and Tissue Kit (Qiagen, Courtaboeuf, France). Oligonucleotide aCGH was performed on DNA extracted from lymphocytes with an 180 000 oligonucleotide microarray (Sure Print G3 Human 4 × 180k CGH Microarray Kit, Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's protocol. Control DNA consisted of three other DNAs according to the loop model except in the case of GE0705, whereas reference DNA was the human control female DNA from Promega (Promega, Madison, WI, USA).17 For patient 9A1117, array-CGH results were completed by FISH using commercial centromeric probe (D9Z1) or subtelomeric probe (9pter), according to the manufacturer's instructions (Cytocell, Cambridge, UK). BAC clone (RP11-404E6 in 9q21.3) was labeled with TRITC by nick translation and hybridized as previously described.18
Online links
All variants from this study are submitted in publicly funded database.
http://databases.lovd.nl/shared/individuals/00056405- ID variant: 0000086604 (family variant).
https://decipher.sanger.ac.uk/ ID 322547 – Patient 9A1117; ID 262761 – Patient GE075; ID 314381 – Patient DK8393.
https://www.ncbi.nlm.nih.gov/clinvar ID223147 – Patient EC-CA300; ID a87-693582 – Patient AG1; ID aC0010 – Patient RO1.
Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/).
ExAC database (http://exac.broadinstitute.org/).
1000 Genomes database (http://browser.1000genomes.org/index.htlm).
Database of Genomic Variants (DGV) (http://dgv.tcag.ca/gb2/gbrowser/dgv2_hg19).
ISCN/HGVS proposal for nomenclature of translocations determined by sequencing: http://www.hgvs.org/mutnomen/comments004.html.
Results
Four patients from family 1 had GGE (patients 4, 13, 14, and 20, Figure 1a). Onset occurred between 3 and 9 years of age. All patients underwent prolonged video-EEG polygraphic recordings, including IPS. They had typical absence seizures and rare GTCS. EEGs showed generalized polyspikes followed by typical 3-Hz spike-and-wave absence seizure with loss of consciousness, ocular elevation, eyelid myoclonia, and neck and shoulder myoclonia of limited amplitude, mostly triggered by IPS at eye closure. For all four patients, the photosensitivity spectrum ranged between 8- and 40-Hz frequencies. Three patients were treated with sodium valproate with good efficacy, and one received a combination of ethosuximide and phenobarbital, with good efficacy. Clinical features, in these four patients, could be consistent with the diagnosis of eyelid myoclonia with absence epilepsy. Individuals 11, 15, and 21 were photosensitive, but contrast with the four aforementioned patients, photoparoxysmal response (PPR) on eye closure was seen for a single frequency of stimulation only, and they never experienced seizures. Neuropsychological assessment showed mild ID in patients 4, 13, and 14 (Supplementary Table S1). aCGH, performed in patient 13, did not show any chromosomal imbalance.
We found 53 variants, located in coding regions or splice site, shared by the three patients who were studied by WES (4, 13, and 20) (Supplementary Table S2). Among these, 10 were not found in databases of control individuals, including ExAC. Of the remaining 43 variants, there was a single-nucleotide alteration, located in exon 3 of the RORB gene leading to a nonsense variant (c.196C>T/p.(Arg66*)) that was confirmed in patient 4 by Sanger sequencing (Figure 1b). This variant was also present in patient 14, who had GGE, and in patient 10, who only had isolated high-amplitude spike during IPS at the time of EEG recording but whose seizure state could not be confirmed. Patient 23, who also carried the variant, denied EEG investigations. Three family members (11, 15, and 21) who had isolated PPR during IPS did not carry the variant. None of the other potentially pathogenic variants found by WES were shared by all four affected individuals. In addition, none of the variants of potential interest identified by WES that were common to the four family members with PPR (4, 13, 20, 21) were found in the remaining family members with PPR (10, 11, 14, 15, and 23) by Sanger sequencing. Further details are in the Supplementary Table S2. The RORB variant led to the N-terminal truncation of the two beta retinoid-related orphan nuclear receptor (RORβ) isoforms (RORβ1, Genbank NP_008845: p.(Arg66*); RORβ2, Genbank Q92753: p.(Arg77*)) (Figure 1c). The variant was also detected in the reversely transcribed RORB mRNA transcripts extracted from lymphoblastoid cell lines of patients 4 and 13, indicating the absence of nonsense-mediated mRNA decay (Figure 1d, Supplementary Figures S1A and B). Constructs for the expression of RORβ fusion wild-type or mutant proteins were then transfected in COS-7 cells. The wild-type protein was detected in the nucleus as expected; in contrast, no mutant protein was detected in the nucleoplasm (Supplementary Figures S1C–H), which, together with the loss of important functional domains of RORB (Figure 1c), was consistent with the loss-of-function effect. The BRCA2 frameshift variant (NM_00005) c.9699_9702delTATG (p.Cys3233Trpfs*15), which was found in patients 4, 13, and 20, can be considered an incidental finding.19
Patients with de novo mutations involving RORB
Patient AG1
This girl was the child of unrelated parents. She had a positive family history for febrile seizures, which were present in patient's brother, father, and paternal uncle.
She had her first seizure at the age of 3 years in the setting of fever. After the initial presentation, she continued to have febrile and afebrile GTCS until the age of 5 years, when seizures were eventually controlled on medication. She was initially treated with valproic acid, which was switched to carbamazepine owing to lack of seizure control.
She had an unremarkable early development until the age of 15 months, when her development plateaued and regressed in social and language domains. She subsequently developed prominent stereotypies, compulsive behaviors, and aggression. She had episodes of rapid behavioral cycling with disruptive behavior and pica and was given the diagnosis of autism spectrum disorder at the age of 2 years. For behavioral issues, she was treated with clonidine and risperidone. On formal neuropsychological testing, she was found to have moderate-to-severe ID.
Diagnostic trio-based exome sequencing was performed and a de novo c.218T>C(p.Leu73Pro) mutation was identified. This mutation was predicted to interfere with DNA binding of the RORB protein (Supplementary Figure S2A).
Patient RO1
This boy was the first child of non-consanguineous Caucasian parents. Seizures started at the age of 4 months with brief bilateral clonic movements or the arms and legs and fixed gaze. At the age of 1 year, the seizure type changed to daily brief tonic seizures, which then transitioned into clonic-atonic seizures, resulting in frequent falls. He also had atypical absences. He was treated with valproic acid, levetiracetam, lamotrigine, topiramate, clobazam, clonazepam, and rufinamide without effect on the seizure burden.
Development was unremarkable until the age of 10 months, when his development slowed. At the age of 4 years, he was found to be at the level of a 2-year-old child (developmental quotient of 50%) and was diagnosed with severe ID.
He was found to have a de novo c.1249_1251delACG (p.Thr417del) in-frame deletion, which was predicted to interfere with proper RORB cofactor binding potentially resulting in a disruption to DNA ligand binding (Supplementary Figure S2C).
Screening of additional cohorts of patients
We did not find any additional DNA variation or CNV involving RORB in 100 probands from French and Australian families with GGE (Table 1).
Patients with de novo microdeletions and balanced translocation involving RORB
Case no. 9A1117
This 25-year-old woman was the first child of healthy unrelated parents. Her younger sister was healthy. A maternal uncle and two of her first cousins had GTCS in adulthood. Neonatal history and motor development were normal. At age 3 years, speech delay and learning difficulties were apparent. She went to a special school. Cognitive evaluation by WISC-III, at age 5.5 years, confirmed mild intellectual impairment. Between the ages of 5.5 and 6 years, she experienced three bilateral clonic seizures during febrile episodes. Interictal EEG recording showed normal background rhythm and bilateral centrotemporal spikes. She was treated with Carbamazepine, Valproate, Ethosuximide, and Vigabatrin. At age 10 years, EEG recording revealed typical 3-Hz spike and wave absence seizures, characterized by complete impairment of consciousness and oral or limb automatisms, and was activated by hyperventilation. She was not photosensitive. Clobazam replaced Vigabatrin because of its potentially aggravating effect. A 24-h ambulatory EEG recording demonstrated persistent absence seizures, lasting 6–10 s and interictal generalized spike or poly spike waves, lasting 0.5–3 s. At age 13 years, three tonic-clonic seizures occurred on awakening, precipitated by sleep deprivation. In the following year, four more convulsive seizures occurred, starting with absences or absence status with eyeball elevation.
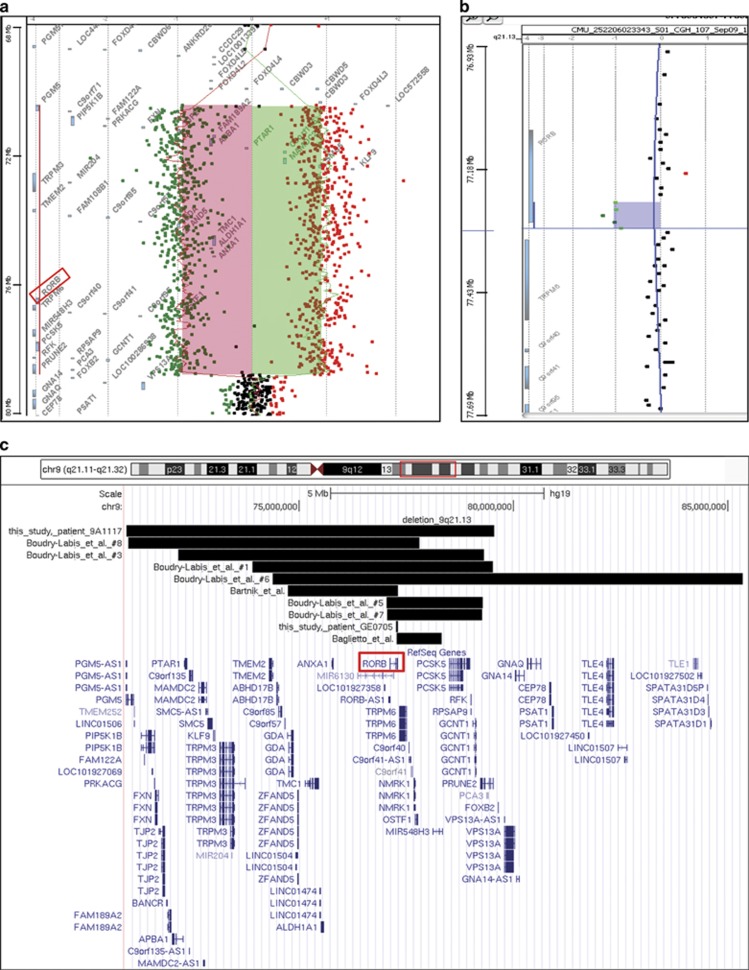
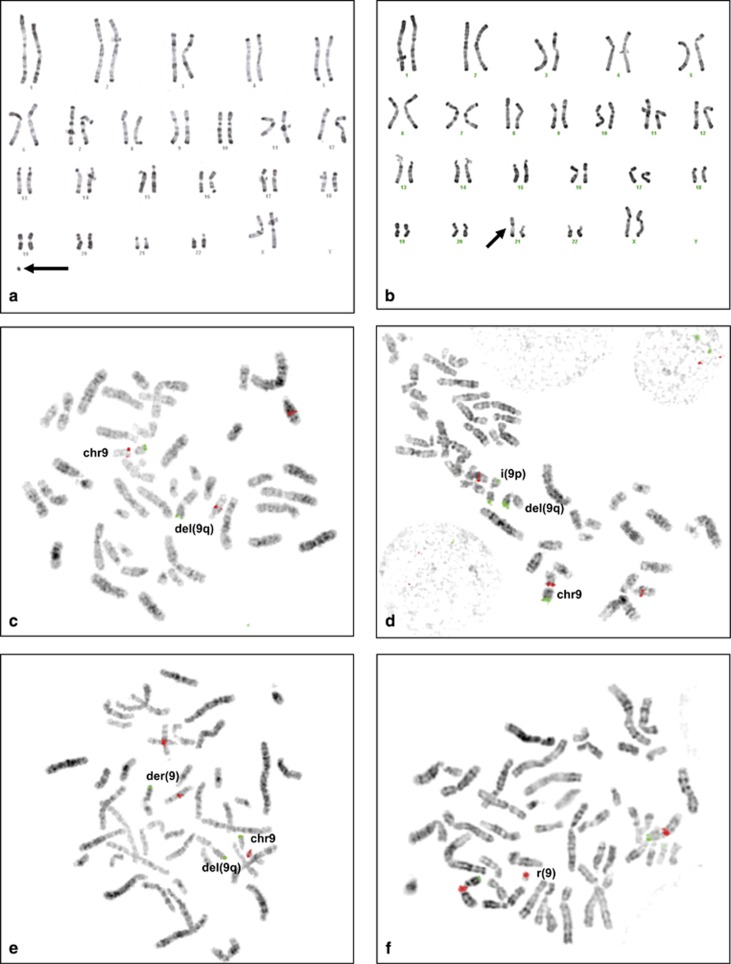
Standard karyotype on 50 mitoses revealed a complex mosaic chromosomes imbalances composed of a small supernumerary ring marker chromosome in 25 mitoses (50%), a derivative chromosome 9 from a t(9;21) translocation in one mitose (2%), and a normal 46,XX population in 24 mitoses (48%): mos 47,XX,+r[20]/46,XX,−21,+der(9)t(9;21)[5]/46,XX[25]. In addition, aCGH analysis revealed a homogeneous 8.5-Mb deletion of the long arm of chromosome 9 (chr9:g.70984481_79549501del), containing 47 genes including RORB and a mosaic gain involving the whole short arm of chromosome 9 (Figure 2a). FISH analysis confirmed the homogeneous 9q13q21.13 deletion with the RP11-404E6 BAC clone. It also showed an isochromosome 9p in 7% of cells, the derivative 9 der(9)t(9;21) in 8% of cells and a small supernumerary ring chromosome 9, mainly composed of heterochromatin in 23% of cells (Figure 3). All these rearrangements were absent in the parents' blood samples.
Figure 2.
The de novo 9q21 microdeletions found in epileptic patients converge on RORB. (a) aCGH profile of the 8.5-Mb microdeletion event (from 70 984 481 to 79 549 501 nt, hg19 coordinates) detected in patient 9A1117. (b) aCGH profile of the 52-kb de novo partial microdeletion event (from 77 261 322 to 77 313 598 nt, hg19 coordinates) detected in patient GE0705. (c) Map of all microdeletions comprising RORB reported to date in epileptic patients. Note the existence of two de novo truncating events on either 5′21 and 3′23 sides of RORB, in addition to the partial microdeletion of exons 5–10 reported here (patient GE0705).
Figure 3.
Conventional (RHG) karyotype and fluorescent in situ hybridization (FISH) showing the complex chromosomal rearrangement in patient 9A1117. (Top) Conventional karyotype. (a) Small supernumerary ring chromosome (arrow) (b) Derivative chromosome 9 from a t(9;21) translocation replacing a normal chromosome 21 (arrow). (Bottom) FISH analysis. (c–e) FISH with RP11-404E6 (9q21.3) probe in TRITC (red) and 9pter probe (Cytocell) in FITC (green) showing the normal chromosome 9 (chr9), the 9q13q21.13 deletion (del9q) lacking the RP11-404E6 signal. Note that RP11-404E6 probe cross-hybridizes on chromosome 14. Additional rearrangements included (d) isochromosome 9p i(9p) showing two 9pter green signals and (e) a derivative 9 chromosome from a t(9;21) translocation der(9) showing a single 9pter green signal. (f) FISH with centromeric chromosome 9 probe in Texas Red and 9pter probe in FITC (green) (Cytocell) showing the supernumerary small ring chromosome 9r(9) with a centromeric red signal but no telomeric signal. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Case no. GE0705
This 10-year-old female patient was the second child of healthy unrelated parents. There was no family history of seizures. She presented with brief absence seizures at the age of 4 years 9 months. EEG recordings showed absence seizures lasting 5–10 s, occasionally with eyelid myoclonia, and triggered by IPS. Ethosuximide was efficacious but poorly tolerated. The treatment was changed to sodium valproate, which was not effective, then to Lamotrigine and Levitiracetam, which had greater efficacy although seizures persisted. At the age of 10 years, she presented with her first nocturnal GTCS. A nocturnal EEG was performed that displayed significant worsening with multiple generalized 3-Hz discharges, most often without clinical manifestation, occurring in the wake and sleep state. She is currently on Lamotrigine and Levetiracetam and a ketogenic diet has just been started. She displayed global developmental delay with early learning difficulties and predominant speech impairment. At the age of 9 years, her IQ was <50.
Array-CGH analysis showed a homogeneous 52-kb microdeletion of exons 5–10 of RORB (chr9:g.77261322_77313598del) (Figure 2b). This heterozygous deletion was confirmed by qPCR (Supplementary Figure S4).
Case no. DK8393
This 10 year-old female was the first child of healthy non-consanguineous parents. There was no family history of seizures. The patient was seen for neuropediatric evaluation at 8 months of age because of delayed motor development and poor eye contact. Examination showed hypermobility and general hypotonia.
At age 3.5 years, the patient was tested by Bayley III and Vineland Adaptive Behavior Scales and found to have moderate ID and infantile autism. She has attended care and school for children with special needs since the age of 4 years.
She currently has severely delayed language development using single words, sounds, and pictures for communication.
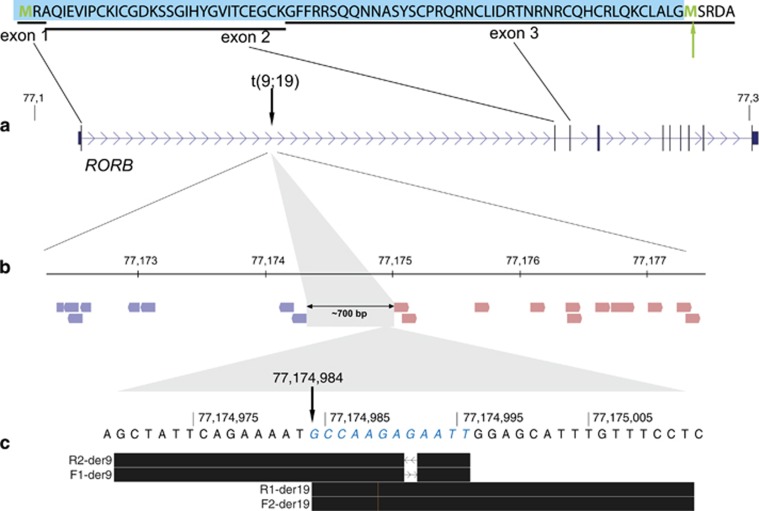
aCGH analysis was normal. Conventional cytogenetic analysis revealed a de novo balanced reciprocal translocation, t(9;19)(q21;q12)dn. The breakpoints were mapped by mate-pair sequencing: out of the 34 071 005 read pairs passing the alignment score, 19 discordant mate-pair reads defined the breakpoints to chr9:g.77174312_77175018 (hg19), within the first intron of RORB (Figures 4a and b), and chr19: g.31139347_31139735, truncating transcript ENST00000592773, distal to the annotated part of ZNF536. Sanger sequencing refined the breakpoints to chr19: g.31139486_31139488, with a single nucleotide (T) deletion (nt 31139487), and to chr9: g.77174984, with two small duplications of chr9:77174985–77174991 and chr9:77174993–77174996 (Figure 4c). The final karyotype according to the proposed ISCN/HGVS nomenclature is: seq[GRCh37] t(9;19)(q21;q12)dn g.[chr9:pter_cen_77174984::chr19:31139488_qter] g.[chr19:pter_cen_31139486::chr9:77174985_77174991dup::chr9:77174993_77174996dup_qter].
Figure 4.
Mapping of seq[GRch37]t(9;19)(q21;q12)dn breakpoint in case DK8393. (a) Mate-pair mapping place the chromosome 9 breakpoint (vertical arrow) within the first intron of RORB, between two oppositely oriented clusters of 19 mate-pair reads (b). (c) Sanger sequencing with forward (F) and reverse primer (R) pairs reveal two small duplications (blue bases in italics) associated with the chromosome 9 breakpoint at nt 77174984 (arrow). The black bars illustrate the BLAT results of the forward and reverse sequences in the UCSC Genome Browser. The amino acids (AA) encoded by the first three exons are shown on the top. If the gene could be transcribed despite the loss of the highly conserved and complex promoter (unlikely, see Supplementary Figure S3), the use of the next start codon (green arrow) distal to the translocation breakpoint (in exon 3) will result in the deletion of the first 74 AA of RORB (blue box). A full color version of this figure is available at the European Journal of Human Genetics journal online.
Case EC-CAE300
This patient is the child of non-consanguineous Caucasian parents. GTCS started at the age of 2–3 years. He was initially started on valproic acid, which controlled the seizures and could be weaned in the interim. At the age of 13 years, nocturnal GTCS recurred and valproate was started again. Valproic acid monotherapy eventually led to good seizure control at the time of the last visit at the age of 28 years. In addition to the GTCS, he had absence seizures during childhood. He has some behavioral problems with aggressive features and is known to ‘act out'. Affymetrix SNP 6.0 array showed a microdeletion removing exon 1 (Figure 2c) of RORB gene (chr9:g.76601085_77182821del), which was validated with an in-house developed technique for Multiplex Amplicon Quantification. The variant was not present in the patient's mother. Paternal DNA was not available for testing.
Structural modelling of the de novo RORB mutations
Both the Leu73Pro (p.L73P) and Thr417del (p.T417del) variants were found to potentially affect the DNA and ligand-binding domains of RORβ (Supplementary Figures S2A and B). The Leu73Pro variant introduces a radically different backbone atom arrangement resulting in severe steric constraints on the DNA-binding domain (Supplementary Figure S2B). The Thr417del alteration occurs within a ligand-binding amphipathic helix near the coactivator-binding site (Supplementary Figure S2B). Thr417del participates in stabilization of the tertiary structure by interacting with Glu312 (E312: Supplementary Figure S2C) on an adjacent helix. Loss of a residue within the helix would cause a shift in the hydrophobic/hydrophilic register, likely resulting in it becoming more destabilized.
Discussion
In this study, we report both familial and de novo variation in the RORB gene, which encodes RORβ. Our study implicates RORB as a novel genetic etiology for neurodevelopmental disorders with a strong predilection for generalized epilepsies with absence seizures. We present the first family with an inherited nonsense variant (c.196C>T/p.(Arg66*)) in the RORB gene in the four family members with photosensitive GGE and in two additional family members: patient 10 who had an as-yet uncharacterized episode of absence-like discharge on EEG but whose seizure state could not be confirmed and patient 23 with probable GGE (narrated by his sister) but who denied EEG investigations. Neither this nor any other truncating RORB variant has been reported to date in databases of control individuals, that is, the EVS, the ExAC database, and the 1000genomes database. One control individual (NSV 517655) among 2026 healthy individuals is reported in DGV with a RORB microdeletion,20 but detailed phenotype of this individual is not available. Taken together, our data strongly suggest that RORB variation leading to haploinsufficiency is virtually absent in control populations and that mutations or deletions involving RORB are associated with a range of neurodevelopmental phenotypes.
The identification of the truncating RORB mutation in the initial family led us to interrogate various exome and CNV databases for additional patients with de novo mutations and microdeletion involving RORB. We identified two additional patients with microdeletions involving RORB through an in-house CNV database at the University of Lyon (9A1117) and Geneva Hospital (GE0705). We obtained detailed phenotypic information on a previously reported patient (EC-CAE300).8
The common phenotypic features of both patients identified through the Lyon and Geneva database were absences and ID. Patient 9A117 has a very complex cytogenetic profile including, in addition to the homogeneous deletion involving RORB, abnormal chromosomes in mosaic state. A participation of these chromosomal imbalances to the phenotype cannot be excluded. However, their role may have been limited since the patient has a clinical presentation very similar to the other patients, with no additional birth defect. This is probably due to low mosaic rates.
We also identified a patient (DK8393) with a seq[GRch37]t(9;19)(q21;q12)dn balanced translocation with a breakpoint disrupting the RORB gene in intron 1, removing the first coding exon (Figure 4), and the highly conserved RORB promoter (Supplementary Figure S3). In contrast to most other patients with pathogenic variations in RORB, the patient did not have seizures but presented with autism and severe disruption of her sleep cycle. We believe that this patient demonstrates the variability of the RORB phenotypes that can be observed in patients.
Furthermore, we identified two patients with de novo RORB mutations affecting the DNA ligand-binding domain, which were predicted to abolish RORB function. In addition to the epilepsy phenotypes that included fever-associated seizures and myoclonic seizures in infancy, both patients shared a common developmental profile with early normal development followed by slowing and plateauing leading to severe ID in late infancy. This feature was not observed in patients with deletions or truncating mutations in RORB and may indicate a more severe effect of these mutations, affecting the DNA-binding region, than haploinsufficiency alone. This observation may be validated if further patients with de novo RORB mutations are identified.
We and others21, 22, 23 have identified de novo 9q21 microdeletions including RORB in patients with epilepsy of variable semiology. Of the 16 patients with 9q21 deletions reported in the literature, all patients have mild-to-moderate ID and speech delay, and a subset of patients had behavioral abnormality (9/16) and epilepsy (14/16). Our data strongly implicate that RORB is one of the potentially causative genes in this region.
We were struck by the high frequency of generalized epilepsies and particularly absence seizures in patients with RORB microdeletions. Data from the present study could sustain this hypothesis, showing that RORB is a strong candidate involved in absence epilepsy spectrum. In humans, genetic association of RORB with bipolar disorder and cognitive functioning has been suggested.24, 25 Mild-to-moderate neurocognitive and behavioral abnormalities mentioned in all patients with RORB dysfunction support that RORB may play a key role in neurocognitive processes.
RORB encodes the RORβ, a ligand-dependent transcription factor. RORβ has important neurodevelopmental functions. It participates in the development of the retina and in the control of circadian rhythms.26 RORβ is involved in neurogenesis and is mainly expressed in murine cerebral cortex27 and is also involved in the guidance of thalamocortical axons, which is of relevant interest given the role of the thalamocortical circuitry in the generation of absence seizures.28, 29
It should be noted that homozygous Rorb knockout mice exhibit retinal degeneration, modified circadian activity, less anxiety-related behavior, and ataxia but unprovoked seizure was not reported so far.30, 31 Such a possible divergence has already been observed for many other genes involved in neurodevelopmental disorders and is representative of the complexity of the human CNS, insufficiently modeled in rodents.
In conclusion, we identify RORB as a new candidate gene for neurodevelopmental disorders, including generalized epilepsy and absence seizures. Nonsense and missense mutations and CNVs of various sizes involving the RORB gene may give rise to RORB haploinsuffiency, resulting in a common phenotypic signature that involves ID and generalized epilepsy, frequently including absences. Our findings suggest that dysfunction of RORβ and related pathways may result in neuronal hyperexcitability, highlighting a novel pathophysiological mechanism for seizure susceptibility.
Acknowledgments
We thank all the patients and families who participated in the study. We thank Professor Jamel Chelly for helpful discussion and comments and Raphaelle Lamy for expert technical assistance. Assistance from the Biological Resource Centre, Hospices Civils, Lyon, France and from the Banque de Génome, Hôpital de Brabois, Vandoeuvre-les-Nancy, France was greatly appreciated. We also thank Tine Deconinck and Tania Djémié for development and analysis of the MAQ assay. We also thank Dr Goedele Malfroid for helpful information for the patient EC-CAE300. This work was supported by a generous grant from UCB-Pharma France, by ANR (Agence Nationale de la Recherche) grant ‘EPILAND' (ANR-2010-BLAN-1405 01) with EuroBiomed label, by the European Union Seventh Framework Programme FP7/2007–2013 under the project DESIRE (grant agreement no. 602531), by INSERM, by ‘Young Hospital Researcher' 2007 grant from Hospices Civils de Lyon, by the Lundbeck Foundation (2013–14290), the UCPH Programme for Interdisciplinary Research (Global Genes, Local Concerns), and The Danish Council for Independent Research – Medical Sciences (4183-00482B).
Author contributions
GR coordinated the collection of samples and of clinical information, coordinated the study design, participated in the overall strategy, and wrote the manuscript. GL coordinated the collection of samples and of clinical information, analyzed genetic data, participated in the study design and overall strategy, and in drafting of the manuscript. AL performed and analyzed genetic data and participated in the study design. MS performed and analyzed genetic data, participated in the functional analyzes, and participated in the study design. MMM performed and analyzed mate-pair data. RSM and NT participated in the study design and final drafting of the manuscript. NB supervised and performed the functional analyzes and participated in the study design. JF, IB, JB, JS, M-PV-H, CS-B, SW, HS, and IS performed phenotyping analysis and clinical data collection. FS-B, PE, CM, GC, and HM participated in the genetic analyses. SC and MA participated in the study design. DS participated in the genetic analyses and in the study design and in drafting of the manuscript. EH participated in the collection of samples and performed phenotyping analysis and clinical data collection. PS coordinated the study design, decided on the overall strategy, directed the follow-up of experiments, supervised data analysis, and wrote the manuscript. IH contributed to and supervised the exome analysis, data collection, and phenotyping. MP contributed to data analysis and phenotyping. DC contributed to phenotyping and sample collection. RH performed the structural modelling and analysis. All authors contributed to the final version of the manuscript.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Dr N Tommerup receives funding from the University of Copenhagen (UCPH), Lundbeck Foundation, and The Danish Council for Independent Research – Medical Sciences. Dr KL Helbig, Dr S Tang, Dr DN Shinde, Dr R Huether, and Dr HM Lu are employed by and receives a salary from Ambry Genetics. Exome sequencing is among its commercially available tests. Dr S Coulbaut and Dr M Abbas are UCB Pharma employees. Dr Scheffer reports grants from NHMRC and NIH during the conduct of the study: Annals of Neurology, Epileptic Disorders, and Neurology; personal fees from UCB, Athena Diagnostics, Transgenomics, and GlaxoSmithKline, outside the submitted work; in addition, Dr Scheffer has a patent A Diagnostic Method for Epilepsy with royalties paid. Hannah Stamberger is PhD fellow of the Fund for Scientific Research Flanders. Dr Heather Mefford receives funding from the National Institutes of Health. Dr Ingo Helbig is supported by intramural funds of the University of Kiel, by a grant from the German Research Foundation (HE5415/3-1) within the EuroEPINOMICS framework of the European Science Foundation and grants of the German Research Foundation (DFG, HE5415/5-1, HE 5415/6-1), German Ministry for Education and Research (01DH12033, MAR 10/012), and German chapter of the International League against Epilepsy (DGfE). He is also supported by the Children's Hospital of Philadelphia (CHOP) with the Genomics Research Initiative Network (GRIN). Dr Szepetowski reports grants from French National Research Agency (ANR), UCB-Pharma France, and European Union FP7 during the conduct of the study. All the other authors declare no conflict of interest.
Supplementary Material
References
- Nabbout R, Scheffer IE: Genetics of idiopathic epilepsies. Handb Clin Neurol 2013; 111: 567–578. [DOI] [PubMed] [Google Scholar]
- Helbig I, Lowenstein DH: Genetics of the epilepsies: where are we and where are we going? Curr Opin Neurol 2013; 26: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, Heron SE, Mulley JC: A polygenic heterogeneity model for common epilepsies with complex genetics. Genes Brain Behav 2007; 6: 593–597. [DOI] [PubMed] [Google Scholar]
- Scheffer IE, Mefford HC: Epilepsy: beyond the single nucleotide variant in epilepsy genetics. Nat Rev Neurol 2014; 10: 490–491. [DOI] [PubMed] [Google Scholar]
- Rubboli G, Gardella E, Capovilla G: Idiopathic generalized epilepsy (IGE) syndromes in development: IGE with absences of early childhood, IGE with phantom absences, and perioral myoclonia with absences. Epilepsia 2009; 50 (Suppl 5): 24–28. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Mulley JC: Genetically complex epilepsies, copy number variants and syndrome constellations. Genome Med 2010; 2: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Depondt C, Cavalleri GL et al: Exome sequecing followed by large-scale genotyping fails to identify single rare variants of large effect in idiopathic generalized epilepsy. Am J Hum Genet 2012; 91: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D, Ruppert AK, Trucks H et al: Burden analysis of rare microdeletions suggests a strong impact of neurodevelopmental genes in genetic generalised epilepsies. PLoS Genet 2015; 11: e1005226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Yendle SC, Hsu C et al: Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol 2011; 70: 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesca G, Rudolf G, Bruneau N et al: GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet 2013; 45: 1061–1066. [DOI] [PubMed] [Google Scholar]
- Grinspan A, Hirsch E, Malafosse A et al: Epilepsie-absences photosensible familiale: un nouveau syndrome ? Epilepsies 1992; 4: 245–250. [Google Scholar]
- Gnirke A, Melnikov A, Maguire J et al: Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 2009; 27: 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimassi S, Labalme A, Ville D et al: Whole-exome sequencing improves the diagnosis yield in sporadic infantile spasm syndrome. Clin Genet 2015; 89: 198–204. [DOI] [PubMed] [Google Scholar]
- Farwell KD, Shahmirzadi L, El-Khechen D et al: Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med 2015; 17: 578–586. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W et al: Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012; 338: 1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca AC, Bonaldi A, Fonseca SA et al: The segregation of different submicroscopic imbalances underlying the clinical variability associated with a familial karyotypically balanced translocation. Mol Cytogenet 2015; 8: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B et al: Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet 2006; 43: 625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluth-Bolard C, Till M, Labalme A et al: TWIST microdeletion identified by array CGH in a patient presenting Saethre-Chotzen phenotype and a complex rearrangement involving chromosomes 2 and 7. Eur J Med Genet 2008; 51: 156–164. [DOI] [PubMed] [Google Scholar]
- Taylor JC, Martin HC, Lise S et al: Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nat Genet 2015; 47: 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaihk TH, Gai X, Perin JC et al: High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res 2009; 19: 1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry-Labis E, Demeer B, Le Caignec C et al: A novel microdeletion syndrome at 9q21.13 characterised by mental retardation, speech delay, epilepsy and characteristic facial features. Eur J Med Genet 2013; 56: 163–170. [DOI] [PubMed] [Google Scholar]
- Bartnik M, Szczepanik E, Derwinska K et al: Application of array comparative genomic hybridization in 102 patients with epilepsy and additional neurodevelopmental disorders. Am J Med Genet B Neuropsychiatr Genet 2012; 159B: 760–771. [DOI] [PubMed] [Google Scholar]
- Baglietto MG, Caridi G, Gimelli G et al: RORB gene and 9q21.13 microdeletion: report on a patient with epilepsy and mild intellectual disability. Eur J Med Genet 2014; 57: 44–46. [DOI] [PubMed] [Google Scholar]
- McGrath CL, Glatt SJ, Sklar P et al: Evidence for gene association of RORB with bipolar disorder. BMC Psychiatry 2009; 9: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersland KM, Christoforou A, Stansberg C et al: Gene-based analysis of regionally enriched cortical genes in GWAS data sets of cognitive traits and psychiatric disorders. PLoS One 2012; 7: e31687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM: Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal 2009; 7: e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kim SY, Fu Y et al: An isoform of retinoid-related orphan receptor beta directs differentiation of retinal amacrine and horizontal interneurons. Nat Commun 2013; 4: 1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabaudon D, Shnider SJ, Tischfield DJ et al: RORbeta induces barrel-like neuronal clusters in the developing neocortex. Cereb Cortex 2012; 22: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadleir LG, Farrell K, Smith S et al: Electroclinical features of absence seizures in childhood absence epilepsy. Neurology 2006; 67: 413–418. [DOI] [PubMed] [Google Scholar]
- André E, Gawlas K, Steinmayr M et al: A novel isoform of the orphan receptor RORβ is specifically expressed in pineal gland and retina. Gene 1998; 216: 277–283. [DOI] [PubMed] [Google Scholar]
- Masana MI, Sumaya IC, Becker-André M et al: Behavioral characterization and modulation of corcadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORβ knockout. Am J Physiol Regul Integr Com Physiol 2007; 292: 2357–2367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.