Abstract
Recently, germline variants of the transcriptional co-regulator gene TCF20 have been implicated in the aetiology of autism spectrum disorders (ASD). However, the knowledge about the associated clinical picture remains fragmentary. In this study, two individuals with de novo TCF20 sequence variants were identified in a cohort of 313 individuals with intellectual disability of unknown aetiology, which was analysed by whole exome sequencing using a child–parent trio design. Both detected variants – one nonsense and one frameshift variant – were truncating. A comprehensive clinical characterisation of the patients yielded mild intellectual disability, postnatal tall stature and macrocephaly, obesity and muscular hypotonia as common clinical signs while ASD was only present in one proband. The present report begins to establish the clinical picture of individuals with de novo nonsense and frameshift variants of TCF20 which includes features such as proportionate overgrowth and muscular hypotonia. Furthermore, intellectual disability/developmental delay seems to be fully penetrant amongst known individuals with de novo nonsense and frameshift variants of TCF20, whereas ASD is shown to be incompletely penetrant. The transcriptional co-regulator gene TCF20 is hereby added to the growing number of genes implicated in the aetiology of both ASD and intellectual disability. Furthermore, such de novo variants of TCF20 may represent a novel differential diagnosis in the overgrowth syndrome spectrum.
Introduction
Recent research by Babbs and colleagues1 has implicated variants of the transcription factor 20 gene (TCF20, MIM *603107) in the aetiology of autism spectrum disorders (ASD). TCF20 is strongly expressed in premigratory neural crest cells2 and the developing mouse brain,3 especially in the hippocampus and cerebellum.4 The gene encodes the transcriptional co-regulator TCF20 (also known as AR1, SPBP). The nuclear factor TCF20 probably acts as a coactivator of various structurally and functionally disparate transcription factors binding to target sequences in promoters or enhancers, such as c-Jun, Ets, Sp1 and Pax6.5 TCF20 is paralogous to RAI1 and also interacts with RAI1,5 mutations and deletions of which underlie Smith–Magenis syndrome (MIM #182290) while duplications encompassing RAI1 cause Potocki–Lupski syndrome (MIM #610883). Functionally essential regions of the transcriptional co-regulator TCF20 include an N-terminal transactivation domain; three nuclear localisation signals; and several C-terminal DNA- and chromatin-binding domains – including a zinc finger domain – as well as three PEST domains.1, 6 Using cytogenetic techniques, Babbs et al identified a pericentric inversion of chromosome 22 in two brothers with ASD, one of whom also presented with intellectual disability (ID). Further breakpoint characterisation demonstrated that this inversion in fact is a more complex balanced intrachromosomal rearrangement involving an inversion and the transposition of a segment to one of the inversion breakpoints. One of the breakpoints was then shown to disrupt TCF20. In addition, the authors identified a de novo frameshift variant in a proband with craniosynostosis, ASD and moderate ID and a de novo missense variant in a proband with ASD and normal intelligence.
Here, we report two independent individuals with ID in whom de novo nonsense and frameshift variants of TCF20 were identified by trio whole exome sequencing (WES). We considerably expand the clinical picture of individuals with de novo variants of TCF20, in particular regarding growth anomalies and the incidence of ID.
Materials and Methods
Subjects
Written informed consent for study participation was obtained from the legal representatives of all participants and written permission for the publication of clinical photographs from the parents of individuals 1 and 2. All investigations were performed in accordance with the Declaration of Helsinki and were approved by the local institutional review board (Ethics Committee of the Medical Faculty of the University of Bonn, approvals 131/08 and 024/12). All 313 individuals selected for the study presented with ID/developmental delay (DD) with or without additional features (eg, craniofacial dysmorphism, organ malformation and so on) that could not be attributed to a clinically recognisable syndrome by experienced clinical geneticists. Chromosomal microarray analyses excluded clinically relevant chromosomal aberrations in all subjects. All data were interpreted using the GRCh37/hg19 genome assembly.
Whole exome sequencing
Genomic DNA was extracted from peripheral blood of affected individuals and their parents using standard methods. Exomes were enriched in solution using the SureSelect XT Human All Exon 50 Mb kit, version 5 (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's instructions in an automated manner using the Bravo Liquid Handling Platform (Agilent Technologies) and 3 μg of input material. Sequencing was performed as 101 bp paired-end reads on HiSeq2500 systems (Illumina, San Diego, CA, USA). Reads were aligned using BWA v 0.6.2. Variant calling was performed using SAMtools (v 0.1.18), PINDEL (v 0.2.4t) and ExomeDepth (v 1.0.0). Variants were then filtered using the SAMtools varFilter script with default parameters except for the maximum read depth (-D) and the minimum P-value for base quality bias (-2), which were set to 9999 and 1e-400, respectively, and custom scripts. A custom script was applied to mark all variants with adjacent bases of low median base quality. Variant annotation was performed using custom scripts and included known transcripts, known variants, type of mutation and – if applicable – amino acid changes. The annotated variants were integrated into an in-house database. To discover putative de novo variants, variants present in the parents of an affected individual, in the 1000 Genomes Project or in more than 4 of 5165 in-house controls, which had a variant quality of <30, or which did not pass the filter scripts were filtered out. Raw read data of the remaining variants were then checked using the Integrative Genomics Viewer. To discover putative homozygous and compound heterozygous variants or X-linked variants that may be disease-causing, we filtered out variants that were already present in frequencies of at least 1% in our 5165 in-house control exomes, the 1000 Genomes Project or in the ExAC database. We also filtered variants with a variant quality less than 30, or a read depth below 7 and variants that did not pass the filter scripts. For the compound heterozygous variants, the frequency filters were applied to both variants and the variants were only filtered out if both compound heterozygous variants had frequencies >1%. Raw read data of the remaining variants were then checked using the Integrative Genomics Viewer. For the remaining variants, the affected genes were checked to see if they were listed as disease-associated in the OMIM database or in an in-house curated list of autosomal recessive and X-linked recessive genes including, but not limited to, the DDG2P gene list7 or the gene list published by Kochinke et al.8 Prediction algorithms such as SIFT, Polyphen2, MutationTaster or Combined Annotation Dependent Depletion (CADD) were used to determine potential pathogenicity of variants.9, 10
Validation by Sanger sequencing
Bidirectional Sanger sequencing of TCF20 variants was performed using the ABI BigDye Terminator v.3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) to verify the variants and their de novo status. For individual 1 and his parents, whole blood genomic DNA was analysed, and for individual 2 and his parents, genomic DNA extracted from whole blood, buccal swaps and saliva. Primer sequences are available upon request.
Results
Clinical reports
Individual 1
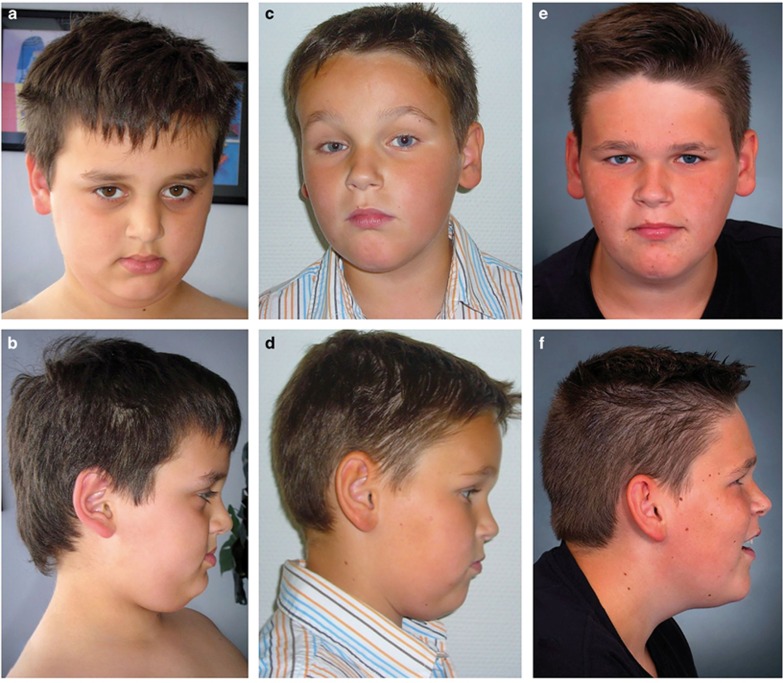
This 14-year-old boy was the third child of healthy non-consanguineous parents with unremarkable family histories. After a normal pregnancy, he was born at gestational week 42 with normal measurements (weight 4050 g (71st percentile, +0.5 SD); length 54 cm (60th percentile, +0.3 SD); OFC 36.5 cm (61st percentile, +0.3 SD)). The patient sat at the age of 9 months, and walked without support at the age of 17 months. He spoke his first words at the age of 2½ years. Subsequent speech development was also considerably delayed and speech comprehension was limited. During his first 2 years of life, he developed obesity. At the age of 4 years, he displayed delayed psychomotor development, muscular hypotonia, and atactic and stereotypic movements. SON-R testing at the age of 5 years revealed mild ID (IQ 54). Investigations including brain MRI, EEG and metabolic screening gave normal results except for slightly increased homocysteine and triglyceride blood plasma concentrations. Hand radiographs showed an accelerated bone age (corresponding to 7 years at the age of 4½ years). No seizures were reported. Physical examination at the age of 7 years revealed an adipose and borderline macrocephalic boy of tall stature (weight 46 kg (>97th percentile, +3 SD, BMI 24.2); length 138 cm (>97th percentile, +2.4 SD); OFC 55 cm (97th percentile, +1.9 SD)). No significant craniofacial dysmorphism was apparent (Figures 1a and b). Minor findings comprised a prominent forehead, downturned corners of the mouth and a prominent lower lip. He also showed mild scoliosis, pseudogynecomastia with inverted nipples, a small penis and early pubic hair. Neuropaediatric evaluation at the age of 13 years revealed persistent hypotonia and no evidence of ASD. He still showed tall stature (182 cm, >97th percentile, +2.1 SD); obesity (106.8 kg, >97th percentile, +3 SD, BMI 32.3); and macrocephaly (59.5 cm, >97th percentile, +3 SD). Parental height was in the normal range (father: 180 cm (46th percentile, −0.1 SD); mother: 160 cm (10th percentile, −1.3 SD)).
Figure 1.
(a–f) Facial phenotypes of the two individuals with de novo nonsense and frameshift variants of TCF20: Facial images of Individual 1 at the age of 7 years (a, b) and Individual 2 at the age of 7 (c, d) and 12 years (e, f). No major facial dysmorphism is apparent.
Individual 2
This 14-year-old boy was the second child of healthy, non-consanguineous parents with unremarkable family histories. After a normal pregnancy, he was born at gestational week 42 with normal measurements (weight 3890 g (58th percentile, +0.2 SD); length 54 cm (60th percentile, +0.3 SD); OFC 36 cm (47th percentile, −0.1 SD). DD became apparent during his first year of life. He was first able to sit at the age of 2 years and to walk without support at the age of 2½ years. He often fell without any reflex of stabilisation, and showed muscular hypotonia. He spoke his first words at the age of 1 year and three-word-sentences at the age of 3 years. At the age of 6 years, he presented with multiple dyslalia, dysgrammatism and language development delay with respect to expressive and receptive speech. At the age of 9 years, he showed mild ID (IQ 62, HAWIK-IV). A brain MRI was unremarkable. He showed behavioural anomalies, with his attitude varying from impulsive and aggressive to very friendly and sociable. Until the age of 5 years, he had sleep disturbances with approximately eight sleep disruptions every night. Neuropaediatric evaluation confirmed the presence of ASD. Epileptic seizures commenced at the age of 10 years. At the age of 12 years, he displayed tall stature, obesity and macrocephaly (height 171.5 cm (>97th percentile, +2.1 SD); weight 80.7 kg (>97th percentile, +2.5 SD, BMI 27.4); OFC 58.5 cm (>97th percentile, +2.8 SD)). His macrocephaly may have been partly familial (maternal OFC: 59 cm, >97th percentile, +2.4 SD, father: 59 cm, 90th percentile, +1.3 SD). No facial dysmorphism was apparent (Figures 1c and f). He displayed inverted nipples, tapering fingers and sandal gaps. Muscular hypotonia and problems with writing and coordinating movements were still apparent. Hand radiographs had not been performed. Parental height was in the normal range (father: 185 cm (74th percentile, +0.6 SD); mother: 172 cm (73rd percentile, +0.6 SD)).
Both in individuals 1 and 2, conventional karyotyping, subtelomeric FISH, chromosomal microarray analysis and fragile X testing gave normal results.
TCF20 sequence variants
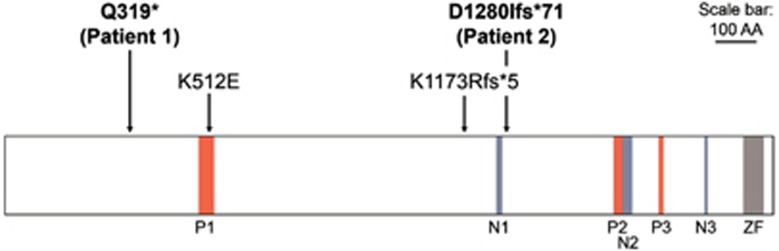
Exome sequencing of 313 child–parent trios identified de novo TCF20 variants in two individuals in whom no other variants that obviously affect function (see below) were detected. Exomes were sequenced to high depth (average: 125 × coverage, median 111 ×), resulting in an at least 20-fold coverage for approximately 97% of the target region. The average coverage in individual 1 was 132 × (median 116 ×) and in his mother and father was 150 × (median 132 ×) and 122 × (median 107 ×), respectively. The average coverage in individual 2 was 118 × (median 103 ×), in his mother 116 × (median 103 ×) and in his father 110 × (median 97 ×). Both de novo TCF20 (NM_005650.3) variants were confirmed in the child and excluded in the parents by Sanger sequencing. Individual 1 carried a nonsense variant (hg19 chr22:g.42610357G>A, c.955C>T, (p.(Gln319*))). The variant was present in 64 out of 141 high quality bases at the position in question (base phred quality⩾30, mapping quality 60). The variant was absent in 174 and 108 bases in the mother's and the father's reads, respectively. Individual 2 carried a frameshift variant leading to a premature stop codon at position 1350 (hg19 chr22:g.42607475_42607475delT, c.3837del, (p.(Asp1280Ilefs*71))) (Table 1, Figure 2). The single base pair deletion was present in 71 out of 134 high quality bases of individual 2 at the position in question (base phred quality⩾30, mapping quality 60 for the bases themselves for wild-type reads and for adjacent bases for deletion reads). The variant was absent in 159 and 143 reads of the mother and the father, respectively. The results of Sanger sequencing of buccal mucosa and saliva DNA of individual 2 were identical to the results of blood DNA sequencing (Supplementary Figure 1), giving no evidence of a variant limited to the haematopoietic cell compartment and pointing to a germline or early postzygotic origin of the variant. No evidence for parental low level mosaicism was found neither in buccal mucosa or saliva DNA of the parents nor in the high-coverage WES data of the parents.
Table 1. Clinical findings in the two reported individuals with de novo TCF20 variants.
| Individual 1 | Individual 2 | Babbs et al., Family 1, Individual II-4 | Babbs et al., Family 1, Individual II-2 | Babbs et al., Family 2 | Babbs et al., Family 6 | |
|---|---|---|---|---|---|---|
| TCF20 variant | nonsense | frameshift | complex balanced intrachromosomal rearrangement disrupting TCF20 (parental germline mosaicism) | missense | frameshift | |
| c.955C>T (p.(Gln319*)) | c.3837del (p.(Asp1280Ilefs*71)) | seq[GRCh37] inv(22)(pter→p11.?2::q13.2→q13.1::q13.1→p11.?2::q13.1→q13.1::q13.2→qter g.[chr22:(8,300,001_12,200,000)ins40,709,620_42,634,698?inv;(8,300,001_12,200,000)_(39,507,139_39,511,083)inv;40,709,620_42,634,698del] | c.1534A>G (p.(Lys512Glu)) | c.3518delA (p.(Lys1173Argfs*5)) | ||
| Age at last examination (years) | 13 | 12 | n.r. | n.r. | n.r. | n.r. |
| Gender | male | male | male | male | male | female |
| Gestational weeks | 42 | 42 | n.r. | n.r. | n.r. | n.r. |
| Birth weight (g) | 4,050 (+0.5 SD) | 3,890 (+0.2 SD) | n.r. | n.r. | n.r. | n.r. |
| Birth length (cm) | 54 (+0.3 SD) | 54 (+0.3 SD) | n.r. | n.r. | n.r. | n.r. |
| Birth OFC (cm) | 36.5 (+0.3 SD) | 36 (−0.1 SD) | n.r. | n.r. | n.r. | n.r. |
| Height (cm) at last examination | 182 (+2.1 SD) | 171.5 (+2.1 SD) | n.r. | n.r. | n.r. | n.r. |
| Weight (kg) at last examination | 106.8 (+3.0 SD) | 80.7 (+2.5 SD) | n.r. | n.r. | n.r. | n.r. |
| BMI (kg/m2) | 32.3 | 27.4 | n.r. | n.r. | n.r. | n.r. |
| OFC (cm) at last examination | 59.5 (+3 SD) | 58.5 (+2.8 SD) | n.r. | n.r. | n.r. | n.r. |
| Tall stature | postnatal | postnatal | n.r. | n.r. | n.r. | n.r. |
| Advanced bone age | + | n.d. | n.r. | n.r. | n.r. | n.r. |
| Macrocephaly | postnatal | postnatal | n.r. | n.r. | n.r. | n.r. |
| ID | mild | mild | borderline intellectual functioning (IQ 79) | mild - moderate | − | moderate |
| Motor delay | + | + | n.r. | n.r. | n.r. | n.r. |
| Walking age (years) | 1.5 | 2.5 | n.r. | n.r. | n.r. | n.r. |
| Speech delay | + | + | n.r. | n.r. | n.r. | n.r. |
| First words (years) | 2.5 | 1 | n.r. | n.r. | n.r. | n.r. |
| Muscular hypotonia | + | + | n.r. | n.r. | n.r. | n.r. |
| Seizures | − | + | n.r. | n.r. | n.r. | n.r. |
| Brain anomalies (MRI) | − | − | n.r. | n.r. | n.r. | n.r. |
| ASD | − | + | + | + | + | + |
| Behavioural anomalies | atactic/stereotypic movements | aggressive, impulsive, attention-demanding | n.r. | n.r. | n.r. | n.r. |
| Organ malformations | − | − | − | − | − | − |
| Major craniofacial dysmorphism | − | − | craniosynostosis causing trigonocephaly, hypotelorism, mild exorbitism | − | n.r. | craniosynostosis |
| Somatic findings | mild scoliosis, gynaecomastia, inverted nipples, early pubic hair growth, small penis | inverted nipples, tapering fingers, sandal gap | n.r. | n.r. | n.r. | n.r. |
Abbreviations: ASD, autism spectrum disorder; BMI, Body mass index;ID, intellectual disability; MRI, cranial magnetic resonance imaging; n.d., not determined; n.r., not reported; OFC, head circumference; SD, standard deviations.
Figure 2.
Graphical view of protein TCF20 with the variants c.955C>T (p.(Gln319*)) and c.3837del (p.(Asp1280Ilefs*71)) identified in individuals 1 and 2: the variants c.1534 A>G (p.(Lys512Glu)) and c.3518delA (p.(Lys1173Argfs*5)) identified by Babbs et al1 are highlighted. The previously annotated PEST domains5 (P1-P3), the nuclear localisation signal domains (N1-N3) and the zinc finger domain (ZF) are shown. Scale bar corresponds to 100 amino acids (AA).
All 12 non-synonymous TCF20 sequence variants discovered in the remaining 311 index cases were inherited and considered as unrelated to disease because of their frequency in control populations and/or because they were predicted to be benign by algorithms (Polyphen2, Sift, CADD).
Variant calling for putative homozygous and compound heterozygous variants or X-linked variants yielded no variants with conclusive evidence for pathogenicity (Supplementary Tables 2 and 3). For individuals 1 and 2, respectively, compound heterozygous variants in 4 and 7 genes passed the autosomal recessive filter criteria. Three variants each passed the X-linked filter criteria. The genes affected by the compound heterozygous variants were either (i) not listed as disease-associated in OMIM or in the comprehensive in-house curated list of autosomal recessive genes or (ii) listed as disease-associated in OMIM either for unrelated phenotypes or for phenotypes clinically excluded in the patients. In addition, seven of the variants in question were rather frequent with minor allele frequencies>0.5%. The genes affected by X-chromosomal variants were not listed as disease-associated in OMIM except for one gene (CCDC22, Supplementary Table 3). However, the variant affecting this gene has been classified as not disease-associated because (i) the clinical picture of individual 2 differs considerably from Ritscher-Schinzel syndrome 2 which is known to be associated with mutations of CCDC22 and (ii) the variant is rather frequent and many ExAC control persons are hemizygous for this variant.
Discussion
Clinical spectrum of individuals with de novo TCF20 variants
The two individuals with de novo TCF20 variants presented here share a phenotype of mild ID, secondary tall stature, postnatal macrocephaly, obesity and muscular hypotonia. Only one of them has ASD and seizures. Neither organ malformations nor major dysmorphism were present.
The inclusion criterion of the first study reporting TCF20 variants was ASD; therefore, all probands with de novo TCF20 variants in that report had presented with this phenotype.1 The reported variants included a de novo missense and a de novo frameshift variants (Figure 2) as well as two disruptions caused by an inversion breakpoint, which had arisen from parental germline mosaicism. Normal intelligence was reported for the individual with the missense variant while the remaining, more deleterious variants were associated with ID or borderline intellectual functioning. Interestingly, two of the probands had craniosynostosis. No further clinical data were provided.
Our identification of de novo nonsense and frameshift TCF20 variants in two clinically well-characterised individuals considerably expands the known clinical picture. In addition to their mild ID, both individuals shared previously undescribed physical findings such as tall stature, macrocephaly, obesity and muscular hypotonia. In contrast to the first study, only one of the present individuals had ASD. Amongst all known six individuals with de novo TCF20 variants, ID/DD and ASD are equally prevalent (five out of six, respectively). All three frameshift and nonsense variants were associated with ID and only two of them with ASD. Given that in the present sample two deleterious variants were identified in 313 ID patients compared with one missense variant in the 342 ASD probands reported previously,1 de novo truncating TCF20 variants may be more frequent in ID/DD than in ASD.
Frameshift and nonsense variants of TCF20: a new differential diagnosis in the overgrowth spectrum
The clinical features presented here, for example, postnatal tall stature and macrocephaly, obesity, ID, muscular hypotonia and in one case ASD, are unspecific when considered individually. As a whole, however, they make de novo TCF20 variants a novel differential diagnosis for several disorders, especially those from the overgrowth syndrome spectrum. Some overgrowth syndromes such as Lujan–Fryns syndrome (MIM #309520)11 typically show a certain clinical overlap with the spectrum presented here such as tall stature, macrocephaly, ID, muscular hypotonia and ASD. However, the disproportionate, marfanoid tall stature as well as the craniofacial dysmorphism may distinguish Lujan–Fryns syndrome from the clinical picture of patients with de novo frameshift and nonsense variants of TCF20. The difference is even more obvious in syndromes without macrocephaly, for example, homocystinuria (MIM #236200).12 Consequently, overgrowth syndromes with proportionate tall stature show a greater overlap with the clinical spectrum presented here. Not only Weaver syndrome (MIM #277590),13 but also Sotos syndrome (MIM #117550), Beckwith–Wiedemann syndrome (MIM #130650), Simpson–Golabi–Behmel syndrome (MIM #312870) and Bannayan–Riley–Ruvalcaba syndrome (MIM #153480) all comprise proportionate tall stature, often with advanced bone age. The latter is interesting because one of the TCF20 individuals also presented with an advanced bone age. Amongst these five syndromes, the clinical similarities with the phenotype of the individuals with de novo TCF20 variants presented here are more apparent for Weaver syndrome than for Sotos syndrome, Beckwith–Wiedemann syndrome, Simpson–Golabi–Behmel syndrome and Bannayan–Riley–Ruvalcaba syndrome owing to the distinguishable facial dysmorphism in Sotos syndrome and the usually prenatal or neonatal onset of overgrowth in the latter three syndromes. In addition, muscular hypotonia and especially ID are uncommon in Beckwith–Wiedemann syndrome.12 In general, the pronounced and often characteristic craniofacial dysmorphism, physical findings and/or organ malformations of all these above-named five syndromes differ from the clinical spectrum presented here, which comprises only dysmorphism and somatic findings that are very minor.
Some less frequent disorders such as Macrocephaly, Macrosomia and Facial Dysmorphism Syndrome (MIM #614192), which is caused by variants of RNF135, also show extensive clinical overlap (postnatal overgrowth, ID, ASD).14 However, typical craniofacial dysmorphism is present and obesity usually is absent in Macrocephaly, Macrosomia and Facial Dysmorphism Syndrome. In contrast, MOMO syndrome (MIM #157980) is an overgrowth syndrome in which postnatal obesity is typical. It often also comprises macrocephaly and at least in one case ASD. However, the delayed bone age, ocular abnormalities and craniofacial dysmorphism may distinguish this syndrome from the clinical spectrum presented here.15, 16 Another rare differential diagnosis is Primrose syndrome (MIM #259050), which comprises macrocephaly, ID, craniofacial dysmorphism, large and calcified auricles, sparse body hair, distal muscle wasting, and specific minor abnormalities.17, 18 However, several typical features of Primrose syndrome such as the large calcified auricles and the distal muscle wasting seem to be absent in our patients.
Phelan–McDermid syndrome (MIM #606232) is an example for another important differential diagnosis, which upon first sight may be overlooked because, for example, the typically more pronounced ID and often absent or severely delayed speech may seem dissimilar to the clinical picture presented here. However, it is known that the syndrome has a wide phenotypic variability concerning, for example, growth and language development19 so that an appreciable subset of Phelan-McDermid individuals has, for example, ID/ASD, tall stature, macrocephaly, only minor dysmorphic features and/or muscular hypotonia.
In summary, the phenotype resulting from TCF20 sequence variants is an important novel differential diagnosis to several known syndromes with overgrowth, macrocephaly and ID/ASD such as Weaver syndrome, Sotos syndrome, Macrocephaly, Macrosomia and Facial Dysmorphism Syndrome, and Phelan–McDermid syndrome.
Mutational spectrum of TCF20 in ID and ASD
A de novo origin of the two TCF20 loss-of-function variants is highly probable because no evidence for parental mosaicism was found in high-coverage WES data or by sequencing saliva and buccal mucosa DNA. The sequencing results of buccal mucosa and saliva DNA of individual 2 were identical to the results of the peripheral blood DNA sequencing, pointing to a germline or early postzygotic origin. Comprehensive chromosomal microarray analyses and WES analyses of the two individuals gave no evidence that other genetic factors noticeably contribute to the phenotype. The broad phenotypic overlap of the two individuals may also be regarded as an argument for a monocausal aetiology. However, a modification of the phenotype or minor contribution of other exogenous or genetic factors such as the compound heterozygous or hemizygous X-linked variants discovered by WES cannot be excluded entirely. Other than the TCF20 loss-of-function mutations, no common potential aetiological factors were identified except for compound heterozygous variants in CMYA5 (cardiomyopathy associated 5), a gene without known associations with ID/DD and many compound heterozygous variants amongst in-house controls. Taken together, no convincing evidence for digenic, oligogenic or multifactorial models of inheritance was found and an autosomal dominant mode of inheritance with high penetrance seems highly probable.
Frameshift or nonsense variants of TCF20 are extremely rare. The variants reported here are neither present in 5165 in-house control exomes nor in the ExAC Browser database with 60 706 exomes (http://biorxiv.org/content/early/2015/10/30/030338). No truncating variants are present in the in-house control exomes. Only one sample in the ExAC database carries a heterozygous nonsense variant in TCF20 that is not disputable (Supplementary Table 1). However, given the number of only one most probably healthy carrier with a single unverified loss-of-function variant, it would be premature to draw final conclusions regarding a potentially incomplete penetrance. The Residual Variation Intolerance score, which quantifies gene intolerance to functional mutations,20 of TCF20 is −2.55 (0.85th percentile), suggesting that TCF20 is significantly more intolerant to deleterious variants than known developmental disorder genes (average −0.56; 19.54th percentile). The CADD scores of the present variants are very high (individual 1: 35, individual 2: 24.5), indicating that these variants are amongst the 0.1% or 1% most deleterious substitutions in the human genome.21 The algorithms MutationTaster and SIFT predict nonsense-mediated mRNA decay for both variants. However, the same applies to the frameshift variant reported by Babbs et al.1 Here, cDNA analysis excluded nonsense-mediated mRNA decay so that a truncated TCF20 protein without the PEST motifs 2 and 3 was expected. PEST domains mediate proteasomal destruction of proteins by interacting with the Cullin-RING ubiquitin E3 ligase complex that polyubiquitinates proteins.22 The authors hypothesised that their patients' ASD-associated variants might stabilise the protein rather than causing a haploinsufficiency. As the nonsense and frameshift variant presented here are upstream from the PEST motifs 2 and 3 (Figure 2), these variants may also give rise to truncated proteins without the PEST motifs 2 and 3, although nonsense-mediated mRNA decay cannot be excluded.
To find out whether microdeletions of TCF20 are associated with an overlapping clinical picture, the DECIPHER database was searched for entries with phenotypic data and deletions of up to 3 Mb (in order to limit possible confounding effects of additional genes), which affected coding sequence of TCF20 transcript NM_005650.3. The search yielded six entries (patients 25944, 251708, 274092, 248554, 262531 and 257430). Four of the microdeletions were de novo and two of unknown origin. ID and/or delayed speech and language development was present in all six cases. In addition, one patient had macrocephaly and another one had, amongst other signs, macrocephaly and tall stature. This clinical overlap may point to a possible aetiological role of TCF20 in these microdeletions. Interestingly, there are also three entries of deletions affecting only either the 5' UTR of transcript NM_005650.3 or the shortest TCF20 Ensembl transcript ENST00000515426 which is incomplete, not supported by either an mRNA or an EST and does not contain for example zinc finger domains (patients 251248, 281451 and 281450). These three deletions have been inherited which points to their benignity and a lesser developmental importance of transcript ENST00000515426.
The comorbidity of ID and ASD is well-documented. ID is reported to be present in approximately 55–70% of individuals with ASD23, 24, 25 and the reported incidences of comorbid ASD in ID patients are approximately 10–40%.24, 26, 27 Mirroring this, the genetic aetiology of ASD and ID shows substantial overlap, and many genes are associated with both disorders. Here, we add TCF20 to the list of these genes, in particular to those ID/ASD genes encoding transcription factors or transcriptional regulators such as ADNP or TCF4.28, 29
Web resources
MutationTaster: www.mutationtaster.org. SIFT: http://sift.jcvi.org. OMIM: http://www.omim.org. Combined Annotation Dependent Depletion (CADD): http://cadd.gs.washington.edu/score. Polyphen2: http://genetics.bwh.harvard.edu/pph2/. Leiden Open Variation Database/LOVD v.3.0: http://databases.lovd.nl/shared/. DECIPHER: https://decipher.sanger.ac.uk/. Ensembl: www.ensembl.org
Acknowledgments
We thank the patients and their families for their participation in this study. This work was supported in part by the German Ministry of Research and Education (grant numbers 01GS08164, 01GS08167, 01GS08163, German Mental Retardation Network) as part of the National Genome Research Network. This study makes use of data generated by the DECIPHER community. A full list of centres who contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust.
Variant information The two variants identified in the present study have been submitted to the Leiden Open Variation Database/LOVD v.3.0 (accession numbers 0000078995 and 0000078996).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Babbs C, Lloyd D, Pagnamenta AT et al: De novo and rare inherited mutations implicate the transcriptional coregulator Tcf20/SPBP in autism spectrum disorder. J Med Genet 2014; 51: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MS, Gammill LS, Bronner-Fraser M: Discovery of transcription factors and other candidate regulators of neural crest development. Dev Dyn 2008; 237: 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P et al: Mouse brain organization revealed through direct genome-scale Tf expression analysis. Science 2004; 306: 2255–2257. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N et al: Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007; 445: 168–176. [DOI] [PubMed] [Google Scholar]
- Rekdal C, Sjottem E, Johansen T: The nuclear factor SPBP contains different functional domains and stimulates the activity of various transcriptional activators. J Biol Chem 2000; 275: 40288–40300. [DOI] [PubMed] [Google Scholar]
- Darvekar S, Johnsen SS, Eriksen AB et al: Identification of two independent nucleosome-binding domains in the transcriptional co-activator SPBP. Biochem J 2012; 442: 65–75. [DOI] [PubMed] [Google Scholar]
- Wright CF, Fitzgerald TW, Jones WD et al: Genetic diagnosis of developmental disorders in the DDD Study: a scalable analysis of genome-wide research data. Lancet 2015; 385: 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochinke K, Zweier C, Nijhof B et al: Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 2016; 98: 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramswig NC, Ockeloen CW, Czeschik JC et al: 'Splitting Versus Lumping': Temple-Baraitser and Zimmermann-Laband Syndromes. Hum Genet 2015; 134: 1089–1097. [DOI] [PubMed] [Google Scholar]
- Hempel M, Cremer K, Ockeloen CW et al: De novo mutations in CHAMP1 cause intellectual disability with severe speech impairment. Am J Hum Genet 2015; 97: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma-Carrillo I, Molina JD, Cuevas-Duran T et al: Psychopathology in the Lujan-Fryns Syndrome: report of two patients and review. Am J Med Genet A 2006; 140: 2807–2811. [DOI] [PubMed] [Google Scholar]
- Visser R, Kant SG, Wit JM et al: Overgrowth syndromes:from classical to new. Pediatr Endocrinol Rev 2009; 6: 375–394. [PubMed] [Google Scholar]
- Cohen MMJr.: Mental deficiency, alterations in performance, and CNS abnormalities in overgrowth syndromes. Am J Med Genet J C Semin Med Genet 2003; 117C: 49–56. [DOI] [PubMed] [Google Scholar]
- Douglas J, Cilliers D, Coleman K et al: Mutations in RNF135, a gene within the Nf1 microdeletion region, cause phenotypic abnormalities including overgrowth. Nat Genet 2007; 39: 963–965. [DOI] [PubMed] [Google Scholar]
- Giunco CT, Moretti-Ferreira D, Silva AE et al: MOMO syndrome associated with autism: a case report. Genet Mol Res 2008; 7: 1223–1225. [DOI] [PubMed] [Google Scholar]
- Sabin MA, Werther GA, Kiess W: Genetics of obesity and overgrowth syndromes. Best Pract Res Clin Endocrinol Metab 2011; 25: 207–220. [DOI] [PubMed] [Google Scholar]
- Carvalho DR, Speck-Martins CE: Additional features of unique Primrose syndrome phenotype. Am J Med Genet A 2011; 155 A: 1379–1383. [DOI] [PubMed] [Google Scholar]
- Primrose DA: A slowly progressive degenerative condition characterized by mental deficiency, wasting of limb musculature and bone abnormalities, including ossification of the pinnae. J Ment Defic Res 1982; 26 (Pt 2): 101–106. [DOI] [PubMed] [Google Scholar]
- Sarasua SM, Boccuto L, Sharp JL et al: Clinical and genomic evaluation of 201 patients with Phelan-McDermid syndrome. Hum Genet 2014; 133: 847–859. [DOI] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL et al: Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013; 9: e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P et al: A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Hong Y, Sarge KD: PEST sequences mediate heat shock factor 2 turnover by interacting with the Cul3 subunit of the Cul3-RING ubiquitin ligase. Cell Stress Chaperones 2010; 15: 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird G, Simonoff E, Pickles A et al: Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: The Special Needs and Autism Project (Snap). Lancet 2006; 368: 210–215. [DOI] [PubMed] [Google Scholar]
- Matson JL, Shoemaker M: Intellectual disability and its relationship to autism spectrum disorders. Res Dev Disabil 2009; 30: 1107–1114. [DOI] [PubMed] [Google Scholar]
- Yeargin-Allsopp M, Rice C, Karapurkar T et al: Prevalence of autism in a US metropolitan area. JAMA 2003; 289: 49–55. [DOI] [PubMed] [Google Scholar]
- de Bildt A, Sytema S, Kraijer D et al: Prevalence of pervasive developmental disorders in children and adolescents with mental retardation. J Child Psychol Psychiatry 2005; 46: 275–286. [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Schwartz CE: Intellectual disability and autism spectrum disorders: causal genes and molecular mechanisms. Neurosci Biobehav Rev 2014; 46 (Pt 2): 161–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsmoortel C, Vulto-van Silfhout AT, Coe BP et al: A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet 2014; 46: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD: Pitt-Hopkins syndrome: intellectual disability due to loss of Tcf4-regulated gene transcription. Exp Mol Med 2013; 45: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.