Abstract
One of the principal mechanisms by which cells differentiate and respond to changes in external signals or conditions is by changing the activity levels of transcription factors (TFs). This changes the transcription rates of target genes via the cell’s TF network, which ultimately contributes to reconfiguring cellular state. Since microarrays provided our first window into global cellular state, computational biologists have eagerly attacked the problem of mapping TF networks, a key part of the cell’s control circuitry. In retrospect, however, steady-state mRNA abundance levels were a poor substitute for TF activity levels and gene transcription rates. Likewise, mapping TF binding through chromatin immunoprecipitation proved less predictive of functional regulation and less amenable to systematic elucidation of complete networks than originally hoped. This review explains these roadblocks and the current, unprecedented blossoming of new experimental techniques built on second generation sequencing, which hold out the promise of rapid progress in TF network mapping.
Keywords: Transcriptional regulatory networks, Regulatory systems biology, Computational methods, Transcription factor activity, Gene expression profiling, Nascent RNA sequencing
NETWORK MAPPING AS ROBUST AND SCALABLE AS SEQUENCING
The development of genome sequencing technologies is the paradigm for the broader group of technologies related to genomics and systems biology. Researchers first set their sights on sequencing a viral genome, then a bacterium, yeast, invertebrate models, and human. Despite much talk of the “post-genomic” era, the publication of the human genome now appears to be a taking-off point in the demand for genome sequencing, starting with other yeasts, invertebrates, and mammals for comparative genomics. This was followed by the sequencing many individuals to sample population diversity. Now that genome sequencing is a reliable, low-cost procedure that can be easily out-sourced, a logical next step is to focus on improving technologies for the elucidation of gene regulation.
In this review, we focus on emerging technologies for mapping transcription factor (TF) networks. A cell’s TF network is the collection of all interactions between its sequence-specific DNA-binding proteins (TFs) and the target genes they bind and regulate – i.e. their direct, functional targets. A full transcriptional regulatory network also includes signaling pathways that activate and inactivate TFs as well as regulatory processes that act on RNA, but for purposes of clarity and focus we define TF networks to include only proteins that act in complex with DNA (Box 2). Because some of the genes regulated by TFs also encode TFs, these interactions form a complex network containing numerous feedback and feed forward loops. A TF network map is a graphical representation, or model, of a cell’s TF network. Such a map consists of nodes, which represent genes and the proteins they encode, and directed edges, which connect TFs to their direct, functional targets. Edges are labeled as either activating (increasing TF activity increases target gene transcription) or repressing (increasing TF activity decreases target gene transcription).
Box 2. Semantics and evaluation of correctness.
Network maps consist of nodes, representing genes and their protein products, and directed edges, linking nodes that represent TFs to nodes that represent their direct targets. Such an edge is correct if, and only if, the following three conditions hold:
Binding The TF interacts physically with the target gene by forming a complex with its promoter or an enhancer that interacts with it.
Regulation The TF functionally regulates the target, meaning that changes in the activity of the TF can change the transcription rate of the target.
Direct causation The physical interaction of the TF with the target plays a causal role in its functional regulation of the target. This implies that eliminating the physical interaction with the target gene would change the transcriptional rate of the target. An interaction can pass the binding and the regulation criteria while failing the direct causation criterion if it regulates the target indirectly, via a pathway that does not depend on the physical interaction.
A more detailed network can be created by adding nodes between each TF and each of its targets to represent the genomic sequences the TF must bind in order to regulate the target [79].
Various types of high-throughput data can be used to check these correctness criteria (see Table 1 for available data sets).
Binding locations ChIP-chip and ChIP-seq with antibodies against TFs (alternatively, transposon calling cards [65, 66]) can be used to determine where in a genome each TF binds. Note that biochemical binding events are not necessarily functional or sequence-specific [70, 71, 82].
Binding potential Models of TF binding specificity obtained from in vitro experiments complement in vivo location methods like ChIP-seq and can provide additional information about whether a physical interaction is sequence-specific.
Functional regulation Transcript abundance data on cells in which a single TF has been perturbed can be used to determine whether the TF functionally regulates each target gene. Note that functional regulation does not imply binding.
Functional binding. If a TF regulates a target by binding to a particular site or sites, TF perturbation should affect the target in wild-type cells, but not in cells where the site(s) have been removed. Such experiments have never been done on a genome-wide scale. A more feasible, if somewhat less definitive experiment is to synthesize pairs of promoters/enhancers, one of which matches a WT genomic sequence and the other of which has a predicted TF binding site disabled. Thousands of pairs can be synthesized in parallel. If these sequences are fused to a minimal promoter driving a reporter gene and the two members of a pair express the reporter at different levels, that supports the hypothesis that the disabled TF binding site is functional (see [83] for a review of related methods).
This review focuses on systematic procedures (“algorithms”) for mapping TF networks, which comprise both data generation and data analysis. We are currently in the midst of an explosion of experimental methods, each of which generates a new type of data. These new data types demand new computational approaches that can effectively analyze and integrate them for network mapping. If the field succeeds in developing TF network mapping algorithms that are as robust and scalable as genome sequencing, we can expect demand for network maps to follow the same trajectory as demand for genome sequences.
APPLICATIONS OF NETWORK MAPS
TF network maps encode basic knowledge about the biochemical functions of molecules, much like metabolic pathway maps. As such, they are a key component of the encyclopedia of molecular cell biology that enables research and development. This knowledge will doubtless have many applications that we cannot foresee, but a few applications have already begun to emerge.
Transcriptome engineering
The problem of transcriptome engineering is this: Given the expression profile of cells of a particular type growing in a particular context, and given a desired expression profile, find a set of TF perturbations (deletions, knockdowns, or over expressions) that will result in the cells having the desired expression profile. Recently, transcriptome engineering has been used to change the expression profile of yeast cells growing in xylose toward that of cells growing in glucose, with the aim of making them produce large quantities of ethanol, like cells growing in glucose do (Michael et. al, unpublished data). Transcriptome engineering has also been applied to regenerative medicine, where the goal is to convert mammalian cells of one type into cells of another type [1–3]. In all these cases, the algorithms used for selecting TF perturbations rely on TF network maps. To date, transcriptome engineering algorithms have not made use of quantitative predictions about the expression levels of genes after a combination of TF perturbations. There have been a few attempts to make such quantitative predictions [4, 5], but this is very much an open research problem.
Quantitative models of TF activity and gene expression
In a quantitative model, the expression level of each target gene is modeled by a function of the expression or activity levels of the TFs that regulate it. In this context, activity refers to the collective effectiveness of all molecules of a TF in activating or repressing its targets. Changes in activity may be caused by changes in the abundance of the TF protein, its localization, its association with other proteins, or its post translational modifications. Typical genome scale models are trained to predict the steady-state mRNA levels of target genes from the expression or activity levels of TFs in the same mRNA sample. This is not the same as predicting target gene levels in a new sample in which the expression of the TF has been artificially perturbed. These models can predict the effects of a TF perturbation on the direct targets of the TF, but not the indirect effects that result from changes in the expression of the direct targets.
Some of these methods also infer changes in the activity level of each TF by analyzing changes in the levels of its target genes. For example, if a TF functions primarily as a repressor and the average expression level of its target genes rises after some treatment, it can be inferred that the activity level of the TF has decreased as a result of the treatment [6]. In principle, such analyses of TF activity can provide powerful insights into the underlying causes of expression differences between samples (Box 1). In practice, such analyses are surprisingly difficult to carry out because many TFs have a very small number of high confidence targets and the targets that are known often give conflicting signals. In addition, multiple TFs often regulate identical or nearly identical sets of target genes, which makes it impossible to determine which TF is responsible for changes in the expression of the target genes [7, 8]. Nonetheless, understanding why a treatment changes the expression profile of a culture in terms of the treatment’s effects on TF activity remains a fundamental problem in computational genomics.
Box 1. Universal maps, condition-specific maps, and TF activity.
In a condition-specific map, an edge from a TF to a target indicates that the TF is actively binding and affecting transcription of the target in the given condition. In a universal map, an edge indicates that the TF would bind and regulate the target gene under some conditions. Network mapping methods that combine gene expression data from many growth conditions (correlation/regression methods) implicitly attempt to construct universal maps. Using other approaches and data sources, it is possible to build condition-specific maps.
It is also possible to view condition-specific maps as special cases that can be derived from a quantitative universal map by specifying the activity levels of the TFs (Fig. I). TF activity levels are not directly observable, but several methods can infer condition-specific TF activity levels from condition-specific expression data (reviewed in [9]). Some methods focus on linking TF binding motifs to the genes that are regulated through them. Each motif is linked to each gene in whose promoter it appears and the link is quantified in terms of the strength of the match. Using this universal quantitative map, parameters representing the “activity level” of each binding motif in a given condition are fit to condition-specific expression data. Motifs are considered “active” to the extent that TFs are exerting regulatory influence by binding to them. The activity of each TF is inferred by the activities of the motifs it binds [11, 75, 76]. A limitation is that each motif may be bound by multiple TFs and each TF may bind multiple motifs. Other methods start with a universal, qualitative map and attempt to simultaneously learn both TF activity levels and the quantitative “influence” strength of each TF on each target [8, 77]. Still others start with only expression data and attempt to learn both the TF activities and the strength with which each TF influences each of its targets [4, 78], but it is not clear whether this is scalable to networks with many TFs. Activity of entire promoters and enhancers has also been inferred from adjacent transcription and linked to TFs through binding specificity models [79].
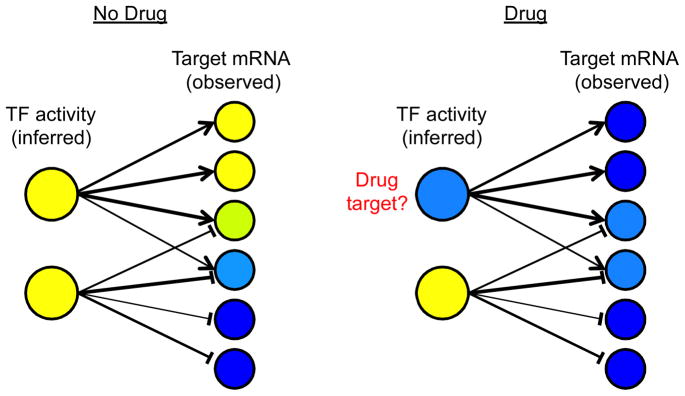
TF activity inference provides significant insights into unobserved biological processes, including drug target identification (Fig. I), the cell cycle [77], changes that occur during dynamic processes such as immune cell differentiation [80, 81], and differences between cell types. Comparison of TF activities to TF mRNA levels can also generate hypotheses about post-transcriptional regulation and upstream signaling networks.
Fig. I.
TF activity inference from universal, quantitative influence maps and condition-specific expression profiles. Large circles: TFs. Small circles: target genes. Line thickness: strength of regulatory influence. Arrow head: Activation. T head: Repression. Blue: Low observed expression level (targets) or inferred activity level (TFs). Yellow: High observed expression level (targets) or inferred activity level (TFs).
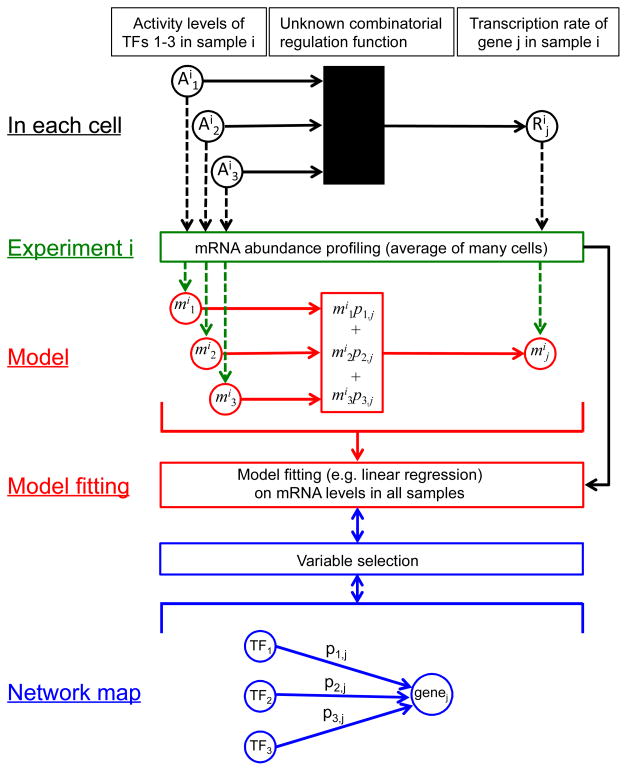
Quantitative models contain parameters that are initially unknown and must be fit to data (Fig. 1). If model fitting is undertaken with no prior knowledge then every TF must be considered a possible regulator of every gene, resulting in millions of unknown parameters. Obtaining good estimates under these conditions without over-fitting the model to noise in the data is extremely challenging. However, if a qualitative TF network map is available, only a handful of TFs must be considered as potential regulators of each target, reducing the number of unknown parameters by two orders of magnitude (Fig. 1; [7–10]). Many methods rely on network maps inferred from binding specificity models of each TF [11, 12] or from genome-wide TF location data (described below) [13–15], but maps derived from any source can be used.
Fig. 1.
Schematic relationship between the true TF network in each cell (black), the experiment typically used to assay the state of the cell (green), a typical linear regression model (red) in which the mRNA abundance of gene j in experiment i (mij) is modeled as a weighted sum of the mRNA abundances of the TFs that regulate it (mi1, mi2, mi3), and a qualitative network map (blue). mRNA abundances serve as proxies for both TF activities and target gene transcription rates. When the goal is to learn the network map, the regression considers all TFs as possible regulators of each gene. After model fitting, the TFs whose mRNA abundances are most predictive of the target gene’s expression level are selected as likely regulators in the qualitative map. When the goal is quantitative modeling and a qualitative map is known in advance, variable selection can feed into model fitting by limiting the potential regulators of each target.
MAPPING ALGORITHMS: DATA GENERATION AND DATA ANALYSIS
Computational methods are often tested by using pre-existing genomic data sets without consideration for the difficulty of generating comparable data for other growth conditions, cell types, or species. These studies contribute to the understanding of specific cell types in specific growth conditions. On the other hand, a generally applicable algorithm for mapping TF networks in any cell type or growth environment must consider the cost, scalability, and reliability of methods for generating the required data. For that reason, the following sections are organized around data types, the experiments required to generate the data, and the computational methods that use the data to map TF networks.
Gene expression profiles
If a TF network is viewed as a control circuit, the transcription rate of each gene is its output. By transcription rate, I mean the number of transcripts per minute that enter productive elongation. Transcription is an intricate process with many steps and the way in which bound TFs affect each step is not fully understood. Given current knowledge, however, transcription rate defined in this way is an appropriate abstraction for TF network mapping. Transcript abundance has traditionally been used as a proxy for the network’s output because it can be measured easily and reproducibly by using spotted arrays, oligonucleotide arrays, or RNA-seq. All these methods can produce good data, but the larger dynamic range and rapidly falling costs of RNA-seq make it increasingly attractive. Newer technologies enable direct measurement of transcription rates (specifically, the density of actively transcribing RNA polymerase molecules in the gene body), but are currently more labor-intensive [16–18]. Measuring transcription rates is important because that is what TFs regulate (Fig. 1); RNA abundances are also affected by RNA degradation rates, which can be specifically regulated by other mechanisms.
The two fundamental approaches to TF network mapping from expression data can be described as co-expression analysis and differential expression analysis. Both approaches require genome-wide mRNA measurements under a range of conditions that differ in the activities of one or more TFs. Changes in TF activity can be experimentally induced by changing growth conditions (e.g. nutrients, temperature, toxins, or drugs) or by single-TF perturbations (e.g. gene deletion, over expression, or RNAi knockdown). When growth conditions are changed, gene expression is most often measured after enough time has elapsed that mRNA levels are thought to have returned to steady state. In principle, measuring gene expression over a series of short intervals during which they are still equilibrating can be useful for learning dynamic models (reviewed in [19]) and for disentangling cause and effect, but this is very difficult to do accurately on a genomic scale. The disadvantage of time series is that measurements at nearby time points tend to be very similar, leading to less information gained per measurement. When growth conditions are varied the effects on the activity of each TF are unknown. Single-TF perturbations are particularly useful because all changes in expression are caused by the perturbation, either directly or indirectly [20, 21]. Perturbation by gene deletion has the additional advantage that the activity of the deleted TF is known to be exactly zero.
Co-expression approaches include pairwise statistical association, regression, and machine learning. Some co-expression methods attempt to detect pairwise statistical associations between the mRNA abundance of each TF (a proxy for TF activity) and the mRNA abundance of each gene (a proxy for its transcription rate). Intuitively, either positive or negative correlation between the mRNA levels of a TF and another gene might be evidence that the TF regulates the gene. Various measures of statistical association have been used, including correlation, mutual information, and variants like context likelihood relatedness [22–25]. All of these approaches share one great weakness – their logic is that the activity of a TF should be correlated with the transcription rates of its target genes, but they rely on the mRNA levels of TFs as proxies for their activity levels. It is well known that a TF’s mRNA level is not a reliable proxy for its activity. For example, it has been estimated that only about 40% of the variation in protein abundance can be explained by mRNA abundance [26]. Beyond their protein levels, the activities of many TFs are regulated by binding to other proteins (e.g. yeast Gal80p binds TF Gal4p and prevents it from interacting with the transcriptional machinery [27]), covalent modification (e.g. yeast Snf1p activates TF Cat8p and inactivates TF Mig1p by phosphorylation), relocalization in response to small ligands (e.g. nuclear hormone receptors), and other mechanisms too numerous to list.
The same basic approach can also be pursued in the framework of linear regression [28–30] (Fig. 1). The expression level of each gene is a dependent variable to be explained as a weighted sum of the expression levels of the TFs, which are the independent predictor variables. The weights, which can be interpreted as abstract representations of the strength with which each TF regulates the target, are initially unknown parameters that are estimated by choosing values that minimize the difference between predicted and observed expression levels. Since the structure of the network is not known in advance, sparse or regularized regression methods such as least angle regression are used to reduce the number of TFs predicted to regulate each target. Although the models and fitting procedures are similar to those used in quantitative modeling, the goal here is not to make accurate predictions of expression levels but to determine which TFs are the best predictors for each target gene (Fig. 1). These TFs are taken to be the direct regulators of the target. Various measures of predictor quality have been used, including the linear regression coefficient – the amount of predicted change in the target per unit change in the TF [28], the regression coefficient weighted by the fraction variance in the target gene that the expression level of the TF can explain [10], and the frequency with which the TF is selected to explain the target in regressions using randomly sampled subsets of the training data [30]. Co-expression analysis has also been pursued in the framework of non-parametric machine-learning algorithms (e.g. [31]).
Differential expression (DE) analysis compares the expression profiles of cells in which a single TF’s activity has been perturbed to the expression profiles of wild type cells grown in the same conditions. Intuitively, a gene whose expression level changes when a TF is perturbed is a candidate direct target of the TF. Because all comparisons are between expression profiles of cells in the same growth conditions, this approach does not require the assumption that TF transcript abundance is a good proxy for activity across growth conditions. Since gene expression data are noisy, DE analysis uses replicate assays to calculate the strength of evidence for a true change in the mean expression level of each gene [32]. Traditionally, these calculations have been used in statistical hypothesis testing to determine which genes show significant evidence of DE and which do not. However, most of the genes showing statistically significant evidence of DE when a TF is deleted do not show evidence of being bound by the TF (reviewed in [33]), so predicting that all DE genes are direct targets would be highly inaccurate. A more recent approach is to use the strength of evidence for DE as a measure of confidence that the TF directly regulates the target. Empirically, the genes that show the strongest evidence of differential expression are highly enriched for direct targets, relative to those with weaker (but still significant) evidence for differential expression [21]. Thus, the effect of deleting a TF on the expression levels of other genes seems to dissipate quickly in the network (also see [34]).
Using the evaluation criteria described in Boxes 2 and 3, co-expression methods have produced reasonable results on both simulated data and real data from bacteria [20] and archaea [28], but they are much less accurate on data from S. cerevisiae [20, 21], one of the simplest eukaryotic organisms. Combining the outputs of multiple co-expression-based algorithms, sometimes called a “wisdom of crowds” approach, improves accuracy and reduces variance on bacterial data, but it is not clear that the same is true for eukaryotes [20]. Methods that score TF-target edges by the strength of evidence that the target is differentially expressed when the TF is deleted do much better on yeast [21] and fruit fly (Kang et al., unpublished data). This may be because co-expression methods, but not DE methods, rely on the mRNA levels of TFs as proxies for their activity levels and the mRNA levels of target genes as proxies for their transcription rates (Fig. 1). In the future, it may be possible to replace these proxies by protein mass spectrometry for TFs [35, 36] and direct transcription rate measurements for targets. Another significant factor hindering the co-expression approach may be the use of mRNA levels from populations of cells rather than individual cells. Averaging artifacts can mask the relationship between mRNA abundance of a TF and that of its targets in individual cells. The recent development of low cost, highly parallel, single-cell RNA-seq (drop-seq, [37]) may eliminate averaging artifacts, but it is not clear that it can be made compatible with protocols for direct measurement of transcription rates.
Box 3. Comparative evaluation of network mapping algorithms.
Progress depends on the ability to determine when a new computational algorithm or a change to an existing algorithm improves accuracy. For several years, community evaluations of algorithms using only gene expression data were held in association with an annual meeting called Dialog on Reverse Engineering and Assessment and Methods (DREAM) [20]. Some evaluations used simulated expression data generated from known network maps (in silico networks [84, 85]), which were used as “gold standards”. Typically, mapping software outputs a list of all possible (TF, target) edges, each associated with a confidence score (Fig. II). Binary networks can be generated by including all edges with scores above some threshold. (Allowing the user to set the inclusion threshold makes sense, as some applications require a small number of high confidence edges while others benefit from a larger number of predicted edges, even at the cost of lower average accuracy.) Binary networks corresponding to many different thresholds were evaluated against the gold standard, producing two numbers for each threshold: precision (the fraction of all included edges that are present in the gold standard) and recall (the fraction of all gold standard edges that are included). For each threshold, a point was plotted on a precision-recall scatterplot, producing a precision recall curve (PRC; Fig. II). The area under the precision recall curve (AUPRC) was used a single figure of merit. DREAM also used data from E. coli and, in the end, from S. cerevisiae, together with “bronze standard” networks assembled from literature, TF binding specificity, and TF location data.
While DREAM was a great motivator and community builder, in retrospect the in silico data were crucially flawed because the activities of TFs were always modeled as being proportional to their mRNA levels. A correlation between activity and mRNA was also seen in E. coli data, but not in the data from yeast, a eukaryote. On yeast data, all algorithms performed poorly and the AUPRC was too small to effectively distinguish algorithms [20]. Furthermore, AUPRC is heavily influenced by the accuracy of very low confidence edges, whereas most users are interested in the highest confidence edges. Finally, absolute precision and recall cannot be accurately estimated because the bronze standard contains false positive edges and is missing many true edges. Nonetheless, the fraction of edges supported by a particular form of evidence, such as ChIP, is a reliable indicator of relative accuracy. We have found it informative to plot the fraction of edges supported (y-axis) versus the number of edges included (x-axis). This plot is useful for estimating the experimental confirmation rate when testing a given number of predicted edges. When plotted this way, the accuracy of different algorithms on yeast expression data can be clearly distinguished [21]. Furthermore, different forms of evidence, such as ChIP and TF binding potential, can be compared to one another in the same way.
Fig II.
Illustration of mapping algorithm output with scores, various inclusion thresholds (colored lines), and corresponding points in a precision-recall plot.
Currently, the most accurate approach seems to be combining sparse regression with the strength of DE evidence while giving greater weight to the DE evidence [21] (see [38] for a similar idea). The regression makes it possible to infer targets for TFs for which DE data are not available, albeit less accurately, and to make use of expression profiles from WT strains subjected to environmental perturbations. For yeast, this combined method seems to identify direct targets with promoter sequences that have binding sites for TFs better than existing ChIP data [21].
For the moment, the application of DE methods is limited by the availability of expression profiles in which a single TF’s activity has been perturbed. There are several large expression data sets in which all non-essential yeast TFs have been deleted, but for animal cells the largest data sets include perturbations of fewer than 10% of TFs (Table 1). Recently, Cas9/CRISPR technology has made it much easier to disable genes in animal genomes [39]. However, the costs and challenges of constructing more than 1,000 individual TF deletion lines still pose a barrier.
Table 1.
Major expression and TF location data sets for eukaryotic organisms. Only TFs with DNA binding domains are counted.
| Data type | TFs | Technology | Cell type | Treatment | URL or accession | Notes |
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae (yeast) | ||||||
|
| ||||||
| Expression | 171 | Spotted array | BY4742 | TF del. | http://deleteome.holstegelab.nl | [86]a |
| Expression | 171 | Spotted array | BY4741 | TF del. | GSE4654 | [34]b |
| Expression | 55 | Spotted array | BY4741 | TF OE, del. | GSE5499 | [87]c |
| Location | 208 | Various | Various | N/A | http://www.yeastract.com | [88]d |
|
| ||||||
| Mus musculus (mouse) | ||||||
|
| ||||||
| Expression | 88 | Agilent array | ESC lines | TF induction | http://esbank.nia.nih.gov | [89]e |
| Expression | 55 | Agilent array | ESC lines | TF KD | http://esbank.nia.nih.gov | [90]f |
| Expression | 42 | Various | ESC lines | LOF/GOF | http://www.maayanlab.net/ESCAPE | [3]g |
| Location | 53 | ChIP-seq | CH12 & MEL | https://www.encodeproject.org | [91]h | |
| Location | 47 | ChIP-X | ESC lines | Various | http://www.maayanlab.net/ESCAPE | [3]i |
|
| ||||||
| Home sapiens (human) | ||||||
|
| ||||||
| Expression | 130 | CodeLink array | HUVEC | TF KD | GSE27869 | [92]j |
| Expression | 42 | Illumina bead chip | THP1 | TF KD | http://fantom.gsc.riken.jp/4 | [81]k |
| Location | 182 | ChIP-X | Various | N/A | https://www.encodeproject.org | [93]l |
| Location | 38 | ChIP-seq | ESC-derived | N/A | GSE61475 | [94]m |
|
| ||||||
| Drosophila melanogaster (fruit fly) | ||||||
|
| ||||||
| Expression | 23 | Affy. array | S2 | TF KD | E-MTAB-453 | [95]n |
| Location | 61 | ChIP-seq | Various | N/A | http://www.modencode.org | [73]o |
|
| ||||||
| Caenorhabditis elegans (round worm) | ||||||
|
| ||||||
| Location | 91 | ChIP-X | whole | N/A | http://www.modencode.org | [96]p |
|
| ||||||
| Cryptococcus neoformans (pathogenic fungus) | ||||||
|
| ||||||
| Expression | 41 | RNA-seq | H99 | N/A | GSE60398 | [97]q |
1,484 mutants including non-TFs profiled in duplicate. SC medium, 2% Glc. 72 TF deletions changed > 3 transcripts.
263 mutants including non-TFs profiled in duplicate. YP medium, 2% Glc.
55 TF overexpression strains profiled of which 46 deemed responsive. Deletion strains for 51 of the same TFs profiled of which 10 deemed responsive. SC medium, 2% Gal.
Yeastract DB. Diverse binding assays; majority are ChIP-chip against Myc-tag in Z1256 from ref.[98]
137 inducible genes including non-TFs. Expression profiling 48 hr after induction by removal of doxycycline.
72 hr after shRNA depletion of 100 targets (including non-TFs); for 89 mRNA < 50% WT.
Aggregated from various sources; loss or gain of TF function; mostly mouse and a few human.
Mouse ENCODE. Most in lymphoma-derived cell lines (CH12 & MEL) and a few in each of a large number of other cells and tissues.
Binary TF-target interactions aggregated from 30 papers (no raw data). Mostly mouse and a few human.
Human umbilical vein epithelial cells; SiRNAs against 400 targets; for 70% mRNA < 40% WT.
Fantom 4 project. SiRNA knockdown of 52 targets including non-TFs.
ENCODE. Many TFs are ChIPped in multiple cell/tissue types. Mostly native epitopes but some eGFP tags. New data is still being produced and released on the web site.
ChIP of all TFs in 4 ESC-derived endoderm, mesoderm, ectoderm, and mesendoderm. Also includes ChIP of histone marks and RNA-seq.
Non-TF cell-cycle regulators were also studied. Affymetrix arrays for all samples and RNA-seq for a subset.
Most TFs studied in embryos at multiple stages and a few in adults.
All TFs ChIPped in whole animals at 3 stages.
90 min after a shift to capsule inducing conditions. The authors plan to add data on all TFs.
DNA-binding specificities of TFs and location of functional binding sites
If the DNA binding specificity of a TF is known it can be used to identify sites in the genome where the TF has the potential to directly bind DNA. If such a site lies within a promoter or enhancer – collectively known as transcriptional regulatory elements (TREs), and if the gene or genes regulated by that TRE can be identified, then the TF can be linked to the target in a network map. Methods for each of these steps are considered below. Methods for TRE identification and target identification are reviewed in more detail in [40].
DNA binding specificity can be determined in vitro, predicted by homology, or predicted from the amino sequence of the DNA binding domain (DBD) by using machine learning methods. In vitro methods have become relatively robust and scalable, although they are significantly more labor intensive than expression profiling. The bacterial one-hybrid assay [41] may be the simplest to scale up because it takes place in E. coli and does not require protein handling, but it comes with the caveat of potential interactions with native E. coli molecules. Protein binding microarrays [42] and HT-SELEX [43] do require purified TF protein but have been successfully scaled up by groups specializing in these techniques [44, 45]. TFs with sufficiently similar DNA-binding domains bind similar DNA sequences, and this has been used to infer specificities for roughly one-third of all known eukaryotic TFs by using existing experimental data on fewer than 3,000 TFs [44]. For any given species, the fraction of TFs whose motifs can be inferred depends on how similar the species is to a well-studied model organism, but this fraction can be expected to grow as more diverse sets of TFs are studied in vitro. An up-to-date collection of both inferred and experimentally determined motifs can be obtained from the Cis-BP database [44] or others reviewed in [46]. One large collection of motifs for human TFs was recently generated by selecting known and novel motifs that are enriched in TF binding locations defined by ENCODE ChIP-seq data (Table 1) [47]. Recently, machine-learning methods have been developed for predicting TF specificities from the amino acid sequences of TFs in the C2H2 zinc finger family [48] [49] and the homeodomain family [50–52]. Ultimately, a single software package may be developed for predicting specificities of families comprising a large fraction of eukaryotic TFs. At present, however, a majority of TFs are not associated with high confidence sequence specificity models.
The next step in using TF specificities to build network maps is to identify the TREs in a genome and scan their sequences for potential to bind each TF. In yeast and other highly compact eukaryotic genomes the primary TREs are thought to be promoters that extend just a few hundred bases upstream of the transcription start sites TSS. In mammalian genomes, which are roughly 100 times larger, promoter regions are thought to be larger and TFs also regulate genes by binding in distal enhancer regions. The locations of potential enhancers can be narrowed down by using several types of high-throughput data. Enhancers that are active in a given condition are generally in open chromatin, which can be identified by DNase-seq [53, 54]. A great deal of DNase-seq data is now available through the ENCODE and modENCODE projects, but carrying out DNase-seq experiments reliably and at scale requires specialized expertise. Newer transposon-based methods such as ATAC-seq [55] and THS-seq [56] are reportedly simpler and more robust. However, many of these open regions are not thought to have TRE function. An exciting new development is the discovery that active enhancers are often sites of bidirectional transcription of unstable RNAs, termed eRNAs. From the transcriptional perspective, the primary difference between enhancers and promoters is the stability and directionality of the transcripts originating from them [18]. eRNAs can be affinity-captured and sequenced by GRO-seq [18, 57] or PRO-seq [16], which simultaneously measure the rate of stable RNA transcription from promoters. CAGE-seq, in which 5′-capped RNAs are affinity-captured and sequenced, also detects transcription from enhancers [58]. Another method, GRO-cap, combines GRO-seq with 5′ cap selection to pinpoint the transcription start sites for both enhancers and promoters [18]. However, since GRO-cap is a more complex procedure, computational methods have been developed for identifying TREs from the simpler PRO-seq protocol [59], the latest versions of which do not require isolation of intact nuclei (supplement to [60] and Charles Danko, p.c.). Thus, it appears that a large fraction of TREs can be detected by RNA-sequencing using affinity-purification protocols which, although more involved than ordinary RNA-seq, are still robust and scalable.
Once active TREs have been located for a given cell type and growth condition, each one must be linked to both the TFs that bind to it and the genes they regulate by binding to it. Even when the DNA-binding specificity of a TF in vitro is known, predicting which TRE sites it will bind to can be challenging due to competition among TFs that bind the same sites and cooperativity among TFs each of which binds only weakly on its own. A more direct way to link a TF to the enhancers through which it acts is to perturb its expression (e.g. via gene deletion or RNAi) and carry out genome wide assays for changes in TRE activity. A good candidate for this approach is to measure changes in the expression of eRNAs from TREs upon TF perturbation, which can potentially be done in the same experiment in which changes in target gene expression are assayed.
The final piece needed for building a TF-TRE-target gene network is to identify the gene or genes regulated by each TRE. In many analyses, TREs are assigned to the nearest transcription start site (TSS) of a stable RNA, but this is probably too simple [61]. Another approach is to assign them based on correlation between their DNA accessibility and that of nearby TSSs across many cell lines [62]. In a similar approach, enhancers have been assigned to all TSSs whose transcript levels correlate with the eRNA transcript level of the enhancer, within a fixed distance. This is reported to result in assignment of 40% of enhancers to the nearest TSS and 64% to a TSS within 500 Kb [58]. The same study [58] reported that assigning enhancers to TSSs based on correlation of DNA accessibility resulted in assignments of which only 4.3% were supported by physical contacts between the enhancer and the TSS in a particular ChIA-PET experiment {Li, 2012 #4661}. Assignment based on correlation of eRNA and stable RNA expression resulted in assignments that of which 20.6% were supported by the ChIA-PET data. These correlation approaches are feasible for the human genome because of the very large number of cell and tissue types that have been subjected to DNAse-seq and CAGE-seq, but it may not be easily reproducible for less studied organisms. Rather than using a 500 Kb window, it would be possible to use interaction domains defined by contacts between different parts of the genome ([63], reviewed in [40]). Another approach that enables assignment of some enhancers is the presence in the enhancer of a genomic variant that is associated with variation in the expression of a target gene (cis eQTL). However, the identification of such eQTLs requires paired genotype and expression data from a large number of individuals with diverse genomes [64], so it is not easily scalable to new organisms. The development of truly robust, scalable, accurate experiments for assigning enhancers to their regulatory targets is an important area for future research.
In vivo binding locations
Complementary to computation of sequence-specific TF binding potential in TREs is measurement of in vivo TF binding locations in TREs. Whereas binding potential is condition-independent, location assays take place in a particular cell type and growth condition (Box 1). In either case, binding does not imply regulatory function, but it does contribute valuable evidence of regulatory potential. However, methods for measuring TF binding locations in vivo are less reliable and scalable than methods that measure TF specificity in vitro.
There are several methods of determining where in the genome of a cell population a TF is bound, including Calling Cards [65, 66] and DAM-ID [67] (reviewed in [68]). By far the most widely used method, though, is chromatin immunoprecipitation followed by microarray hybridization (ChIP-chip) or sequencing (ChIP-seq). This involves crossing-linking physically interacting proteins and DNA with formaldehyde, which fixes and kills the cells, followed by DNA fragmentation, affinity purification of a particular protein together with the DNA it is bound to, and identification of the DNA fragments (by hybridization or sequencing). The primary determinant of the quality of ChIP-seq data is affinity and specificity of the antibody [69]. ChIP-seq often requires significant optimization and may not work or may not work well for certain TFs. Besides yeast, the only organism in which > 100 TFs have been studied by ChIP is human (Table 1), but this required a monumental effort by the ENCODE consortium and still leaves roughly 90% of human TFs unstudied. This effort cannot be easily replicated for other species at this time. Furthermore, many of the binding events identified by ChIP are non-specific [70, 71] and many of those that are specific are non-functional [72, 73].
Although ChIP-chip and ChIP-seq are not robust, easily scalable methods, the data they produce is valuable for network mapping, especially in conjunction with other data sources as described in the next section.
Mapping by integration of multiple data types
We have described methods of linking TFs to the target genes they regulate based on gene expression profiles, TF binding specificities, and in vivo TF location data. It is tempting to think that integrating all available data types will produce the best possible map for a given species. That may be true, but it is very difficult to demonstrate empirically when no significant data type is held out for validation (Box 2). Moreover, the success of these integrative methods depends very much on the mix of data that happen to be available for a given species. Many such methods rely heavily on TF ChIP-seq data, which cannot be easily obtained for a new species, or even for most TFs in well-studied metazoan species. Thus, even when one can show that an integrative mapping method is accurate for one species, it is not clear how well it will generalize to others. The best approach may be to construct maps by integrating only data from the most robust and scalable methods, such as gene expression and TF binding specificity, while validating methods in well-studied organisms by holding out the data that are harder to get.
Concluding remarks
For nearly 20 years, the rising tide of data from high-throughput genomics has inspired many computational methods for mapping and modeling TF networks. There has been no shortage of great ideas. In retrospect, although the data has been copious, it has not been the right data. mRNA abundance is simply not a good proxy for either TF activity or transcription rate in eukaryotes [74]. This is an exciting moment because rapid advances in experimental methods are providing many new or improved data types on a genomic scale (Outstanding Questions box). These include methods for measuring transcription rates (rather than transcript abundances) and measuring gene expression in single cells (rather than population averages), both of which take the data a step closer to the biochemical processes we are trying to model. Improved methods for measuring TF binding specificity have recently been scaled up and new methods for assessing DNA accessibility have been developed. Nascent RNA sequencing is enabling precise identification of enhancers and promoters and helping link enhancers to their target genes. It remains to be seen which of these methods will prove most scalable and robust. Figuring out how best to deploy these methods and the data they produce in a way that makes network mapping as routine as genome sequencing will provide a challenge for years to come.
OUTSTANDING QUESTIONS BOX.
How can we replace mRNA levels of TFs with measurements that more closely approximate total TF activity? Quantitative mass spectrometry using selected reaction monitoring assays can monitor protein levels of hundreds of TFs in a single sample. In principle, it can also quantify each posttranslational modification of each TF. How robust and scalable will these techniques prove?
What are the best mathematical methods for accurately estimating total TF activity from gene expression profiles? Will TF activity inference enable accurate predictions about post-transcriptional regulation that can be tested by mass spectrometry or other methods? To what extent will this enable us to infer the activity levels of specific TF activity modifiers, such as kinases and phosphatases?
How can we best incorporate data from multiplexed single-cell RNA-seq (drop-seq) into network mapping algorithms? What are the fundamental differences between single-cell expression data and population average data in terms of their utility for network mapping?
How robust, scalable, and precise will transposon-based methods of measuring DNA accessibility be? Will the reduced accessibility of DNA bound to TFs (TF “footprints”) radically reduce the genomic space within which we must search for TF binding potential?
How robust and scalable will methods for nascent RNA sequencing be? How sensitively and specifically will they identify active enhancers and link them to the TSSs they regulate?
TRENDS BOX.
TF network maps are now being used for transcriptome engineering applications including directed differentiation of stem cells.
TF network maps are increasingly viewed as dynamic entities in which differences between conditions can be understood in terms of inferred, quantitative TF activity levels.
The specificities of many TFs are now being assayed in vitro and the specificities of many others inferred from in vitro assays of homologous TFs.
Nascent RNA sequence (GRO-seq or PRO-seq) enables simultaneous, direct measurement of mRNA transcription rates from promoters and eRNA transcription rates from enhancers.
Correlations between the activity states of enhancers and nearby promoters, as measured by DNA accessibility or transcription rates, are now being used to link enhancers to the genes they regulate.
Acknowledgments
Thanks to Yiming Kang for help in preparation of Table 1 and to Barak Cohen for a useful discussion about enhancers. I am grateful to Gary Stormo for useful discussions of methods for determining TF binding specificity. M.B. was supported in part by grant GM100452 from the National Institute of General Medical Sciences of the NIH.
GLOSSARY
- Transcription rate
The number of transcripts per minute that enter productive elongation. Transcription rate can be estimated from the density of actively transcribing RNA polymerase complexes on the gene, assuming that productive elongation occurs at an approximately constant rate. The number of transcripts completed per minute depends on the transcription rate and the fraction of productively elongating polymerase complexes that produce complete transcripts.
- Dynamic range
The difference between the lowest and highest measurements that can be returned by a given assay. Microarray spots tend to have minimum and maximum fluorescence levels; for RNA-seq, the dynamic range is determined by the sequencing depth.
- Co-expression analysis
Methods in which a TF is predicted to regulate a target based on statistical association between their expression levels (e.g. correlation) across many expression profiles in which TF activity levels differ.
- Differential expression (DE) analysis
Methods in which a TF is predicted to regulate targets based on changes in the expression level of the target when the activity of the TF is specifically perturbed (e.g. via single gene deletion).
- Pairwise statistical association
Measures of statistical association that compare only two variables, such as the expression levels of a TF and a potential target gene, without considering alternative explanations such as the expression level of another TF.
- Regression
The process of fitting a quantitative model to data by finding parameter values that minimize the difference between predicted and observed values in a training data set.
- Machine learning
A large body of techniques for finding structure in data. Regression can be considered a form of machine learning, but the term machine learning is more typically used to describe non-parametric methods in which the number of unknowns used to explain the data is not limited in advance of the learning procedure.
- Sparse regression or regularized regression
Methods of fitting unknown parameters to data in which parameters are chosen to minimize both the predictive error and the complexity of the model. Complexity is typically measured by the sum of absolute values of the parameters (L1 regression, including least angle regression) or the sum of the squares of the parameters (L2 regression).
- Transcriptional regulatory element (TRE)
A segment of genomic DNA within which TFs bind and thereby regulate gene expression. TREs include regions traditionally referred to as promoters (TREs near the transcription start site of the regulated gene) and enhancers (TREs that regulate genes at a distance).
- DNA binding domain (DBD)
The portion of a protein that confers sequence-specific DNA binding capacity. These well-defined domains fall into large families and can be identified from the full sequence of a protein by using hidden Markov models with software like HMMR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- 1.Cahan P, et al. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rackham OJ, et al. A predictive computational framework for direct reprogramming between human cell types. Nat Genet. 2016 doi: 10.1038/ng.3487. [DOI] [PubMed] [Google Scholar]
- 3.Xu H, et al. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database (Oxford) 2013:bat045. doi: 10.1093/database/bat045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter GW, et al. Prediction of phenotype and gene expression for combinations of mutations. Mol Syst Biol. 2007;3:96. doi: 10.1038/msb4100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter GW, et al. Predicting the effects of copy-number variation in double and triple mutant combinations. Pac Symp Biocomput. 2012:19–30. [PMC free article] [PubMed] [Google Scholar]
- 6.Boorsma A, et al. Inferring condition-specific modulation of transcription factor activity in yeast through regulon-based analysis of genomewide expression. PLoS One. 2008;3:e3112. doi: 10.1371/journal.pone.0003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao JC, et al. Network component analysis: reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci U S A. 2003;100:15522–15527. doi: 10.1073/pnas.2136632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran LM, et al. gNCA: a framework for determining transcription factor activity based on transcriptome: identifiability and numerical implementation. Metabolic engineering. 2005;7:128–141. doi: 10.1016/j.ymben.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Bussemaker HJ, et al. Predictive modeling of genome-wide mRNA expression: from modules to molecules. Annu Rev Biophys Biomol Struct. 2007;36:329–347. doi: 10.1146/annurev.biophys.36.040306.132725. [DOI] [PubMed] [Google Scholar]
- 10.Greenfield A, et al. DREAM4: Combining genetic and dynamic information to identify biological networks and dynamical models. PLoS One. 2010;5:e13397. doi: 10.1371/journal.pone.0013397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balwierz PJ, et al. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24:869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segal E, et al. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 13.Ouyang Z, et al. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci U S A. 2009;106:21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boulesteix AL, Strimmer K. Predicting transcription factor activities from combined analysis of microarray and ChIP data: a partial least squares approach. Theor Biol Med Model. 2005;2:23. doi: 10.1186/1742-4682-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, et al. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics. 2004;5:31. doi: 10.1186/1471-2105-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwak H, et al. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen MA, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46:1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Novere N. Quantitative and logic modelling of molecular and gene networks. Nat Rev Genet. 2015;16:146–158. doi: 10.1038/nrg3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marbach D, et al. Wisdom of crowds for robust gene network inference. Nature Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haynes BC, et al. Mapping Functional Transcription Factor Networks from Gene Expression Data. Genome Res. 2013 doi: 10.1101/gr.150904.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soranzo N, et al. Comparing association network algorithms for reverse engineering of large-scale gene regulatory networks: synthetic versus real data. Bioinformatics. 2007;23:1640–1647. doi: 10.1093/bioinformatics/btm163. [DOI] [PubMed] [Google Scholar]
- 24.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuffner R, et al. Inferring gene regulatory networks by ANOVA. Bioinformatics. 2012;28:1376–1382. doi: 10.1093/bioinformatics/bts143. [DOI] [PubMed] [Google Scholar]
- 26.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traven A, et al. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonneau R, et al. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006;7:R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madar A, et al. The Inferelator 2.0: a scalable framework for reconstruction of dynamic regulatory network models. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:5448–5451. doi: 10.1109/IEMBS.2009.5334018. [DOI] [PubMed] [Google Scholar]
- 30.Haury AC, et al. TIGRESS: Trustful Inference of Gene REgulation using Stability Selection. BMC Syst Biol. 2012;6:145. doi: 10.1186/1752-0509-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh-Thu VA, et al. Inferring regulatory networks from expression data using tree-based methods. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes TR, de Boer CG. Mapping yeast transcriptional networks. Genetics. 2013;195:9–36. doi: 10.1534/genetics.113.153262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z, et al. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 35.Mirzaei H, et al. Systematic measurement of transcription factor-DNA interactions by targeted mass spectrometry identifies candidate gene regulatory proteins. Proc Natl Acad Sci U S A. 2013;110:3645–3650. doi: 10.1073/pnas.1216918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simicevic J, et al. A mammalian transcription factor-specific peptide repository for targeted proteomics. Proteomics. 2015;15:752–756. doi: 10.1002/pmic.201400355. [DOI] [PubMed] [Google Scholar]
- 37.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yip KY, et al. Improved reconstruction of in silico gene regulatory networks by integrating knockout and perturbation data. PLoS One. 2010;5:e8121. doi: 10.1371/journal.pone.0008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sternberg SH, Doudna JA. Expanding the Biologist’s Toolkit with CRISPR-Cas9. Mol Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 40.Shlyueva D, et al. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 41.Christensen RG, et al. A modified bacterial one-hybrid system yields improved quantitative models of transcription factor specificity. Nucleic Acids Res. 2011;39:e83. doi: 10.1093/nar/gkr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y, et al. Inferring binding energies from selected binding sites. PLoS Comput Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Stormo GD. DNA Motif Databases and Their Uses. Curr Protoc Bioinformatics. 2015;51:2, 15, 11–16. doi: 10.1002/0471250953.bi0215s51. [DOI] [PubMed] [Google Scholar]
- 47.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta A, et al. An improved predictive recognition model for Cys(2)-His(2) zinc finger proteins. Nucleic Acids Res. 2014;42:4800–4812. doi: 10.1093/nar/gku132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Persikov AV, Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42:97–108. doi: 10.1093/nar/gkt890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alleyne TM, et al. Predicting the binding preference of transcription factors to individual DNA k-mers. Bioinformatics. 2009;25:1012–1018. doi: 10.1093/bioinformatics/btn645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christensen RG, et al. Recognition models to predict DNA-binding specificities of homeodomain proteins. Bioinformatics. 2012;28:i84–89. doi: 10.1093/bioinformatics/bts202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelossof R, et al. Affinity regression predicts the recognition code of nucleic acid-binding proteins. Nat Biotechnol. 2015;33:1242–1249. doi: 10.1038/nbt.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nature Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buenrostro JD, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods. 2013 doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sos BC, et al. Characterization of chromatin accessibility with a transposome hypersensitive sites sequencing (THS-seq) assay. Genome Biol. 2016;17:20. doi: 10.1186/s13059-016-0882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Danko CG, et al. Identification of active transcriptional regulatory elements from GRO-seq data. Nat Methods. 2015;12:433–438. doi: 10.1038/nmeth.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mahat DB, et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanyal A, et al. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, et al. “Calling cards” for DNA-binding proteins in mammalian cells. Genetics. 2012;190:941–949. doi: 10.1534/genetics.111.137315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayhew D, Mitra RD. Transposon Calling Cards. Cold Spring Harb Protoc. 2016 doi: 10.1101/pdb.top077776. pdb top077776. [DOI] [PubMed] [Google Scholar]
- 67.van Steensel B, et al. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 68.Aughey GN, Southall TD. Dam it’s good! DamID profiling of protein-DNA interactions. Wiley Interdiscip Rev Dev Biol. 2016;5:25–37. doi: 10.1002/wdev.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jain D, et al. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res. 2015;43:6959–6968. doi: 10.1093/nar/gkv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teytelman L, et al. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci U S A. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lenstra TL, Holstege FC. The discrepancy between chromatin factor location and effect. Nucleus. 2012;3:213–219. doi: 10.4161/nucl.19513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roy S, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang IX, et al. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 2014;6:906–915. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conlon EM, et al. Integrating regulatory motif discovery and genome-wide expression analysis. Proc Natl Acad Sci U S A. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Das D, et al. Adaptively inferring human transcriptional subnetworks. Mol Syst Biol. 2006;2:2006. doi: 10.1038/msb4100067. 0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang YL, et al. Inferring yeast cell cycle regulators and interactions using transcription factor activities. BMC Genomics. 2005;6:90. doi: 10.1186/1471-2164-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carter GW, et al. Use of pleiotropy to model genetic interactions in a population. PLoS Genet. 2012;8:e1003010. doi: 10.1371/journal.pgen.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marbach D, et al. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods. 2016;13:366–370. doi: 10.1038/nmeth.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yosef N, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Consortium, F., et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park D, et al. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One. 2013;8:e83506. doi: 10.1371/journal.pone.0083506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White MA. Understanding how cis-regulatory function is encoded in DNA sequence using massively parallel reporter assays and designed sequences. Genomics. 2015;106:165–170. doi: 10.1016/j.ygeno.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haynes BC, Brent MR. Benchmarking regulatory network reconstruction with GRENDEL. Bioinformatics. 2009;25:801–807. doi: 10.1093/bioinformatics/btp068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marbach D, et al. Generating realistic in silico gene networks for performance assessment of reverse engineering methods. J Comput Biol. 2009;16:229–239. doi: 10.1089/cmb.2008.09TT. [DOI] [PubMed] [Google Scholar]
- 86.Kemmeren P, et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell. 2014;157:740–752. doi: 10.1016/j.cell.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 87.Chua G, et al. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci U S A. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teixeira MC, et al. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:D161–166. doi: 10.1093/nar/gkt1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Correa-Cerro LS, et al. Generation of mouse ES cell lines engineered for the forced induction of transcription factors. Sci Rep. 2011;1:167. doi: 10.1038/srep00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nishiyama A, et al. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci Rep. 2013;3:1390. doi: 10.1038/srep01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng Y, et al. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hurley D, et al. Gene network inference and visualization tools for biologists: application to new human transcriptome datasets. Nucleic Acids Res. 2012;40:2377–2398. doi: 10.1093/nar/gkr902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsankov AM, et al. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bonke M, et al. Transcriptional networks controlling the cell cycle. G3 (Bethesda) 2013;3:75–90. doi: 10.1534/g3.112.004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maier EJ, et al. Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome Res. 2015;25:690–700. doi: 10.1101/gr.184101.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]