Abstract
Background
Luminal subtype breast cancer accounts for a predominant number of breast cancers. Considering the heterogeneity of the disease, it is urgent to develop novel biomarkers to improve risk stratification and optimize therapy choices. Long non-coding RNA (lncRNA) represents an emerging and understudied class of transcripts that play a significant role in cancer biology. Growing knowledge of cancer-associated lncRNAs contributes to the development of molecular markers for prognosis evaluation and gene therapy.
Materials and methods
Three pairs of primary luminal subtype breast cancer tissues and adjacent non-cancerous tissues were collected and sequenced. EBseq algorithm was used to identify differentially expressed lncRNAs. RNA sequencing data from The Cancer Genome Atlas (TCGA) database were used to validate the robustness of our RNA-seq results. Kaplan–Meier and Cox regression analyses were utilized to assess the association between the lncRNAs and overall survival of patients in TCGA cohort.
Results
A total of 796 lncRNAs were significantly dysregulated in luminal subtype breast cancer, including 436 upregulated and 360 downregulated lncRNAs. Among them, FAM83H antisense RNA 1 (FAM83H-AS1) was the most upregulated lncRNA, whereas GSN antisense RNA 1 (GSN-AS1) was the most downregulated lncRNA. Moreover, we proved that the high expression level of FAM83H-AS1 indicated unfavorable prognosis not only in luminal subtype breast cancer but also in all subtype breast cancers. To the best of our knowledge, this is the first report indicating that FAM83H-AS1 was involved in luminal subtype breast cancer and was an independent prognostic indicator.
Conclusion
Our study provides a rich resource to the research community for further identifying lncRNAs with diagnostic and therapeutic potentials and exploring biological function of lncRNAs in luminal subtype breast cancer.
Keywords: breast cancer, long non-coding RNA, FAM83H-AS1, prognosis
Background
Breast cancer is the most common cancer worldwide and is the leading cause of cancer death among women.1 Luminal subtype breast cancer, expressing estrogen receptor alpha (ERα) and/or progesterone receptor (PR), represents a predominant part of breast cancers.2 Despite conferring a more favorable prognosis, treatment of luminal subtype breast cancer is still in controversy due to the heterogeneity of the disease and the resistance to endocrine therapy. Similar therapeutic strategies could evoke diverse responses within luminal subtype patients.3,4 Therefore, it is urgent to develop novel biomarkers to improve risk stratification and optimize therapy choice.
Long non-coding RNAs (lncRNAs) are a class of non-coding RNA molecules the transcripts of which are longer than 200 nt.5 Although they are not protein-encoding, lncRNAs play crucial regulatory roles in a diverse range of cellular processes and biological pathways, including cancer initiation, progression and metastasis.6,7 Displaying tissue and cell specific expression, lncRNAs have great potential as biomarkers.8
Shen et al9 have recently reported novel lncRNAs in triple-negative breast cancer. LncRNA expression profile of HER-2-enriched subtype breast cancer has also been analyzed in our previous study.10 However, there is little information on the aberrant expressed lncRNAs in luminal subtype breast cancer. In this study, we aimed to uncover the dysregulated lncRNAs in luminal subtype breast cancer, which might offer potential biomarkers for prognosis evaluation and gene therapy.
Materials and methods
Sample collection and RNA extraction
Three pairs of primary breast cancer tissues and adjacent non-cancerous tissues were collected in the Department of Surgical Oncology at the First Affiliated Hospital of Wenzhou Medical University. Tissue samples were snap-frozen in liquid nitrogen immediately after dissection and then stored at −80°C before RNA extraction. ERα and PR status of the samples was confirmed positive by postoperative immunohistochemistry. Total RNA was extracted from tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. This study was approved by the ethics committee of the First Affiliated Hospital of Wenzhou Medical University. Informed consent was obtained from all individual participants included in the study.
RNA sequencing and identification of differentially expressed lncRNAs
The RNA with integrity number >7.0 was optimum for cDNA library construction. The cDNA libraries were then processed for the Proton Sequencing process according to the commercial protocols. Next, Ion Proton (Life Technologies) was used to perform single-end and polyA-selection sequencing of the three pairs of samples. The MapSplice program (v2.1.6) was used to align clean reads to the human genome (version: GRCH37). We quantified the lncRNA expression as Reads Per Kilobase per Million mapped reads (RPKM) and lncRNAs with sum read counts <10 across all samples were abandoned. EBseq algorithm was used to identify differentially expressed lncRNAs between cancer tissues and adjacent non-cancerous tissues. Differences in RNA expression were regarded as significant if values of the false discovery rate were <0.05 and the fold change was >2. All differentially expressed lncRNAs are listed in Table S1.
The Cancer Genome Atlas (TCGA) data set
The RPKM expression value of lncRNAs in TCGA database was downloaded through The Atlas of Non-coding RNAs in Cancer (TANRIC), which contained 837 breast cancer tissues and 105 non-tumorous tissues.11 Corresponding clinical parameters and follow-up information about these 837 patients were also downloaded from TCGA database.12 According to ERα and PR status, we screened out 626 samples as luminal subtype breast cancer. Clinical characteristics of the 626 luminal subtype patients as well as lncRNAs expression values are listed in Table S2.
Statistical analysis
Mann–Whitney U-test was used to analyze the expression difference in lncRNAs between luminal subtype breast cancer tissues and adjacent non-cancerous tissues in TCGA cohort. Patients were divided into low or high lncRNA expression groups according to the median value. Kaplan–Meier and Cox regression analyses were utilized to assess the association between the lncRNA and overall survival of patients. A P-value <0.05 was considered statistically significant. All statistical analyses were performed using SPSS Version 22.0 (Chicago, IL, USA).
Results
Differentially expressed lncRNAs in luminal subtype breast cancer
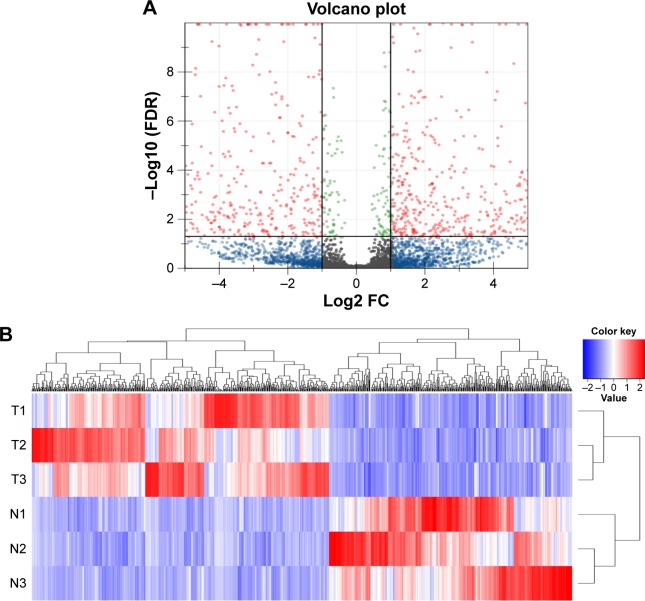
Analysis of global transcriptome expression detected 11,727 lncRNAs (Figure S1). Volcano plot filtering found 796 lncRNAs that were significantly changed in luminal subtype breast cancer tissues compared with their adjacent non-cancerous tissues (Figure 1A). Among them, 436 lncRNAs were upregulated while 360 were downregulated. The hierarchical clustering analysis showed that with the expression of these genes, the samples could be clearly classified into two groups (Figure 1B). The top 20 differentially expressed lncRNAs are listed in Table 1. The results showed that FAM83H antisense RNA 1 (FAM83H-AS1) was the most upregulated lncRNA, whereas GSN antisense RNA 1 (GSN-AS1) was the most downregulated lncRNA.
Figure 1.
Dysregulated lncRNA expression profiles in luminal subtype breast cancer.
Notes: (A) Volcano plot showing variation in lncRNA expression. The negative log of FDR (base 10) is plotted on the Y-axis, and the log of the FC (base 2) is plotted on the X-axis. (B) Heat map of differentially expressed lncRNAs in luminal subtype breast cancer tissues and adjacent non-cancerous tissues. Each column represents a single lncRNA and each row represents one tissue sample. Red indicates high relative expression, and blue indicates low relative expression.
Abbreviations: FDR, false discovery rate; FC, fold change; lncRNA, long non-coding RNA.
Table 1.
Top 20 differential expressed lncRNAs
| Gene symbol | Gene ID | Type of gene | RPKM in tumor | RPKM in normal | Log2 (fold change) | FDR | Status |
|---|---|---|---|---|---|---|---|
| FAM83H-AS1 | 100128338 | lncRNA | 940.932 | 0.430 | 11.097 | 1.12263E-09 | Upregulated |
| ST8SIA6-AS1 | 100128098 | lncRNA | 1,792.825 | 0.900 | 10.960 | 1.3988E-05 | Upregulated |
| HOTAIR | 100124700 | lncRNA | 362.095 | 0.194 | 10.870 | 8.42415E-12 | Upregulated |
| GSN-AS1 | 57000 | lncRNA | 0.264 | 490.184 | −10.859 | 1.43495E-05 | Downregulated |
| LOC100506098 | 100506098 | lncRNA | 63.410 | 0.144 | 8.783 | 0.000104309 | Upregulated |
| LOC101926960 | 101926960 | lncRNA | 0.465 | 204.527 | −8.780 | 1.11022E-16 | Downregulated |
| LINC01405 | 100131138 | lncRNA | 2.445 | 805.872 | −8.365 | 7.76934E-13 | Downregulated |
| LINC01207 | 100505989 | lncRNA | 93.099 | 0.288 | 8.337 | 0.011621155 | Upregulated |
| TRHDE-AS1 | 283392 | lncRNA | 2.962 | 522.324 | −7.462 | 6.85987E-11 | Downregulated |
| ALDH1L1-AS2 | 100862662 | lncRNA | 1.940 | 320.015 | −7.366 | 4.76397E-13 | Downregulated |
| PP14571 | 100130449 | lncRNA | 288.086 | 2.502 | 6.847 | 3.99564E-07 | Upregulated |
| APOC4-APOC2 | 100533990 | lncRNA | 244.790 | 2.214 | 6.788 | 1.49461E-05 | Upregulated |
| MESTIT1 | 317751 | lncRNA | 0.264 | 27.908 | −6.724 | 4.59753E-10 | Downregulated |
| LOC101927793 | 101927793 | lncRNA | 0.570 | 43.984 | −6.271 | 9.22323E-05 | Downregulated |
| LOC101929432 | 101929432 | lncRNA | 0.310 | 23.763 | −6.259 | 0.000459706 | Downregulated |
| LOC101929371 | 101929371 | lncRNA | 0.528 | 39.279 | −6.217 | 0.001171094 | Downregulated |
| LOC101929722 | 101929722 | lncRNA | 1.584 | 116.513 | −6.201 | 7.66724E-07 | Downregulated |
| DRAIC | 145837 | lncRNA | 139.025 | 2.052 | 6.082 | 1.36844E-09 | Upregulated |
| DKFZp451B082 | 401282 | lncRNA | 0.264 | 17.267 | −6.031 | 0.000251753 | Downregulated |
| PGM5-AS1 | 572558 | lncRNA | 1.144 | 66.580 | −5.863 | 1.54321E-14 | Downregulated |
Abbreviations: RPKM, Reads Per Kilobases per Million reads; FDR, false discovery rate.
TCGA cohort validation
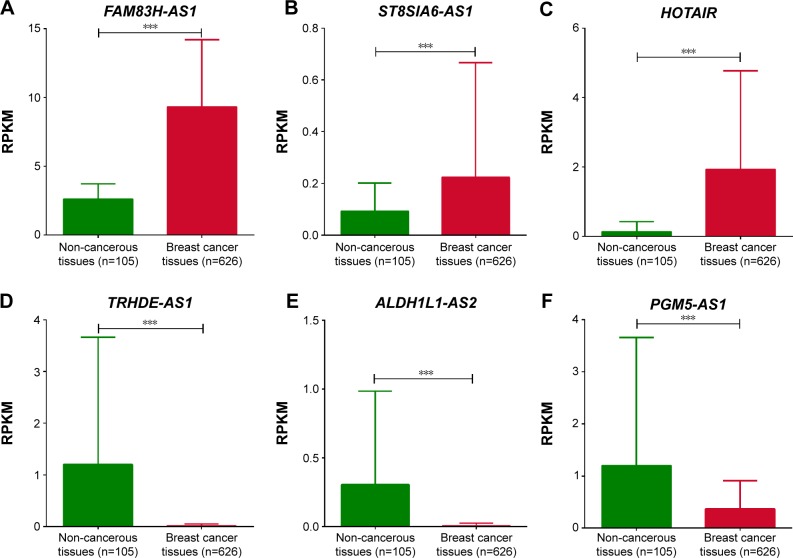
Mining published transcriptome sequencing data was of low cost and feasible for exploring gene expression. To validate the fidelity of our RNA sequencing data, we selected the most differentially expressed lncRNAs and detected these lncRNAs expression in TCGA cohort containing 626 luminal subtype breast cancer samples and 105 non-tumorous samples. We found that six lncRNAs were annotated in the TCGA and the results showed that FAM83H-AS1, ST8SIA6 antisense RNA 1 (ST8SIA6-AS1) and HOX transcript anti-sense RNA (HOTAIR) were upregulated significantly in cancer tissues while TRHDE antisense RNA 1 (TRHDE-AS1), ALDH1L1 antisense RNA 2 (ALDH1L1-AS2) and PGM5 antisense RNA 1 (PGM5-AS1) were downregulated significantly (P<0.001, Figure 2). The consistent variation tendency of each lncRNA showed the robustness of our RNA sequencing.
Figure 2.
LncRNA expression in TCGA cohort.
Notes: The TCGA cohort contained 626 luminal subtype breast cancer tissues and 105 non-cancerous tissues. (A–F) RPKM was represented for expression level of FAM83H-AS1, ST8SIA-AS1, HOTAIR, TRHDE-AS1, ALDH1L1-AS2 and PGM5-AS1 in the TCGA cohort. Data were median ± interquartile ranges of RPKM value. Analysis was done using the Mann–Whitney U-test. ***P<0.001.
Abbreviations: lncRNA, long non-coding RNA; TCGA, The Cancer Genome Atlas; RPKM, Reads Per Kilobase per Million mapped reads.
Identification of novel prognostic marker
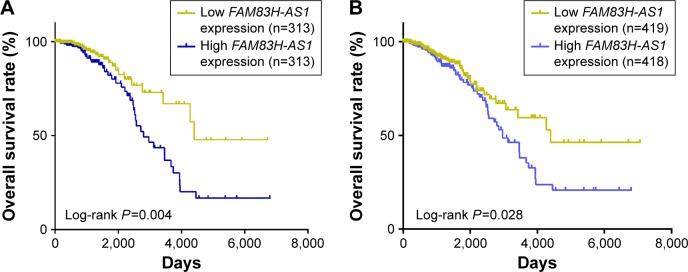
To determine whether lncRNAs could serve as prognostic markers in luminal subtype breast cancer, we analyzed the association between lncRNAs and prognosis in TCGA cohort. We found that high expression level of FAM83H-AS1 was correlated with unfavorable survival. As shown in Figure 3A, the difference between the survival curves of the two groups was statistically significant (P=0.004). The cumulative 10-year overall survival rates were 66.9% and 33.48% in the low expression group and the high expression group, respectively (P=0.012). With respect to the prognostic value of FAM83H-AS1, Cox regression analysis was conducted to adjust for clinical features. It indicated that age (hazards ratio [HR] =1.872, 95% CI =1.028–3.409, P=0.040), high FAM83H-AS1 expression (HR =2.008, 95% CI =1.230–3.544, P=0.006) and lymph node metastasis (HR =1.987, 95% CI =1.145–3.449, P=0.015) were distinctively linked with the prognosis of luminal subtype breast cancer in univariate Cox proportional hazards regression analysis (Table 2). However, high FAM83H-AS1 expression was the only independent prognostic factor after multivariate Cox proportional hazards regression analysis (HR =2.440, 95% CI =1.238–4.807, P=0.010). Furthermore, we found that FAM83H-AS1 was also a prognostic marker for all breast cancer types (P=0.028, Figure 3B). Taken together, these results indicated that FAM83H-AS1 was an independent prognostic indicator and might act as an onco-lncRNA for breast cancer. However, other differential expressed lncRNAs failed to distinguish high-risked luminal subtype breast cancer patients (Figure S2).
Figure 3.
Prognostic value of FAM83H-AS1.
Notes: (A) Kaplan–Meier analysis for the overall survival of 626 luminal subtype breast cancer patients with different expressions of FAM83H-AS1 in TCGA cohort. (B) Kaplan–Meier analysis for the overall survival of 837 breast cancer patients with different expression of FAM83H-AS1 in TCGA cohort.
Abbreviations: FAM83H-AS1, FAM83H antisense RNA 1; TGCA, The Cancer Genome Atlas.
Table 2.
Cox proportional hazards regression analysis
| Variable | Univariate
|
Multivariate
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (<50 years as reference) | 1.872 (1.028–3.409) | 0.040* | ||
| Tumor size (T1 as reference) | 0.975 (0.546–1.743) | 0.933 | ||
| Lymph node metastasis (negative as reference) | 1.987 (1.145–3.449) | 0.015* | ||
| Stage (stage I as reference) | 1.228 (0.620–2.430) | 0.556 | ||
| Estrogen receptor (negative as reference) | 0.352 (0.048–2.578) | 0.304 | ||
| Progestrone receptor (negative as reference) | 0.538 (0.276–1.048) | 0.068 | ||
| HER-2 amplification (unamplification as reference) | 1.726 (0.825–3.763) | 0.144 | ||
| FAM83H-AS1 expression (low expression as reference) | 2.008 (1.230–3.544) | 0.006** | 2.440 (1.238–4.807) | 0.010* |
Note:
P<0.05,
P<0.01.
Abbreviations: CI, confidence interval; HR, hazard ratio.
Discussion
Luminal subtype breast cancer, which accounts for two-thirds of breast cancer, maintains a more differentiated state and confers a more favorable outcome than other subtypes.4,13 The application of adjuvant endocrine therapy has contributed to a remarkable decrease in mortality during recent decades.14 However, breast cancer is highly heterogeneous and it is urgent to distinguish high-risked patients. Cheang et al reported that luminal subtype patients with high expression of HER-2 proteins and the Ki67 index had worse recurrence-free and disease-specific survival.15 It has been declared that among luminal subtype patients who received tamoxifen as their sole adjuvant systemic therapy, the 10-year breast cancer specific survival was 79% for luminal A subtypes and 57% for luminal-HER2 subtypes.15 Therefore, highly sensitive and specific biomarkers would be of great value in the individualized treatment for luminal subtype patients.
Deregulation of lncRNAs has also been shown to contribute to the initiation and progression of several human cancers including breast cancer.16,17 A previous study showed that HOTAIR could promote cancer metastasis by reprogramming chromatin state and was a powerful predictor of eventual metastasis and death.18 In contrast, NF-kappaB interacting long non-coding RNA (NKILA) suppressed breast cancer metastasis by negatively regulating the NF-kB pathway and was associated with poor outcome.19 Besides, FGF14 antisense RNA 2 (FGF14-AS2) has also been proven to be correlated with progression and poorer prognosis in breast cancer in our previous study.20 Hence, increasing knowledge of lncRNAs might help develop novel therapeutic targets and prognostic indicators.
To obtain comprehensive RNA expression profile data, we adopted RNA sequencing in three pairs of luminal subtype breast cancer tissues and their adjacent non-cancerous tissues. Genome-wide analysis revealed a set of lncRNAs with differential expression in cancer tissues compared with non-cancerous tissues. To the best of our knowledge, this is the only study focused on aberrant lncRNAs expression profiling in luminal subtype breast cancer. Furthermore, RNA sequencing data in TCGA database were utilized to validate the reliability of our results. Our study provided a rich resource to the research community for further identifying lncRNAs with diagnostic and therapeutic potentials and exploring biological function of lncRNAs in luminal subtype breast cancer.
Cabanski et al discovered 229 lncRNAs including FAM83H-AS1 with differential expression across multiple cancers and hypothesized that these lncRNAs might have conserved oncogenic and tumor-suppressive functions.21 Consistent with the previous study, we proved that FAM83HAS1 was significantly upregulated in breast cancer and was related to patients’ outcome in a large cohort. It was first reported that FAM83H-AS1 was involved in luminal subtype breast cancer and as an independent prognostic indicator. Further investigation into the functions of FAM83H-AS1 may provide additional target and strategies for treatment.
Conclusion
A total of 796 significantly differentially expressed lncRNAs in luminal subtype breast cancer were identified in the present study. Moreover, FAM83H-AS1 was demonstrated as a novel prognostic marker not only in luminal subtype breast cancer but also in all subtype breast cancers.
Acknowledgments
This study was funded by the Key Project of Science and Technology Innovation Team of Zhejiang Province (2013TD10) and the National Natural Science Foundation of China (no 81372380). We thank dan-dan Sun and her colleagues from Novel Bioinformatics Company for technical support in the bioinformatics analysis process.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Howell A, Cuzick J, Baum M, et al. Results of the ATAC (arimidex, tamoxifen, alone or in combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365(9453):60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 3.Mauri D, Pavlidis N, Polyzos NP, Ioannidis JP. Survival with aromatase inhibitors and inactivators versus standard hormonal therapy in advanced breast cancer: meta-analysis. J Natl Cancer Inst. 2006;98(18):1285–1291. doi: 10.1093/jnci/djj357. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Cong Y, Pan L, et al. Luminal (Her2 negative) prognostic index and survival of breast cancer patients. Cancer Epidemiol. 2014;38(3):286–290. doi: 10.1016/j.canep.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 5.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long non-coding RNA classification. Trends Genet. 2015;31(5):239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim Biophys Acta. 2016;1859(1):16–22. doi: 10.1016/j.bbagrm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Shen X, Xie B, Ma Z, et al. Identification of novel long non-coding RNAs in triple-negative breast cancer. Oncotarget. 2015;6(25):21730. doi: 10.18632/oncotarget.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. Onco Targets Ther. 2016;9:761–772. doi: 10.2147/OTT.S97664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Han L, Roebuck P, et al. TANRIC: an interactive open platform to explore the function of lncRNAs in cancer. Cancer Res. 2015;75(18):3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinstein JN, Collisson EA, Mills GB, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 14.Berry DA, Cronin KA, Plevritis SK, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353(17):1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 15.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Yang F, Liu Y-H, Dong S-Y, et al. A novel long non-coding RNA FGF14-AS2 is correlated with progression and prognosis in breast cancer. Biochem Biophys Res Commun. 2016;470(3):479–483. doi: 10.1016/j.bbrc.2016.01.147. [DOI] [PubMed] [Google Scholar]
- 21.Cabanski CR, White NM, Dang HX, et al. Pan-cancer transcriptome analysis reveals long noncoding RNAs with conserved function. RNA Biol. 2015;12(6):628–642. doi: 10.1080/15476286.2015.1038012. [DOI] [PMC free article] [PubMed] [Google Scholar]