Abstract
Pathology departments routinely process and store formalin-fixed, paraffin-embedded (FFPE) tissue samples for clinical diagnosis. These collections often contain decades’ worth of samples and represent a treasure trove of specimens that can be analyzed for retrospective epidemiological studies, diagnostics, and pathogen discovery. Accurate amplification and sequencing of DNA from these samples is critical for the usability of these FFPE samples. Here we present a collection of protocols that describe extraction of DNA from FFPE tissues, PCR amplification of human papillomavirus DNA, and subsequent genotyping of the infecting virus.
Keywords: HPV, papillomavirus, neoplasia, cervix, cancer, FFPE, formalin, degenerate primers, PCR, sequencing
INTRODUCTION
It has been estimated that over a billion tissue samples are stored in tissue banks, hospitals and labs (Xiao et al., 2015). Most of these specimens have been formalin fixed and embedded in paraffin (FFPE). Importantly, many of these tissue blocks have associated pathological and clinical metadata, and represent a treasure trove of information for further molecular investigation (Halec et al., 2014). These samples can be analyzed readily using immuno-histochemical methods, but formalin fixation crosslinks the nucleic acids, which can also be somewhat degraded, thus complicating molecular analyses.
Infections with papillomaviruses result in pathologies ranging from sub-clinical infection, warts and papillomas, pre-malignant lesions and cancers. Importantly, based on careful epidemiological analyses, only a subset of papillomavirus types are associated with cancer (IARC, 2012), and so it is important to determine the HPV type responsible for the lesion. In addition, many retrospective studies depend on the availability of stored FFPE tissues.
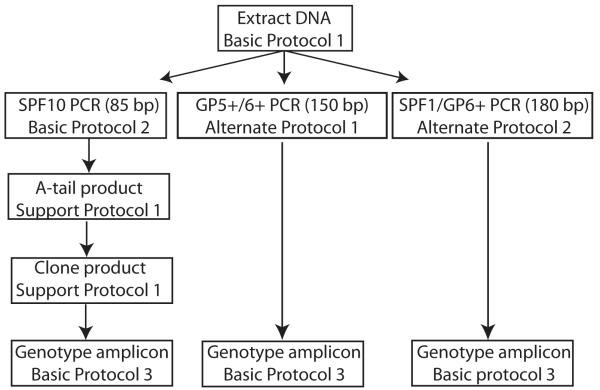
In the present protocol we describe methods used in our labs to isolate viral DNA from FFPE tissue (Basic Protocol 1). In addition, we provide the protocols for three complementary PCR assays that will amplify distinct papillomavirus types and enable their identification (Figure 1). Basic Protocol 2 uses the SPF10 primer set (Quint et al., 2001). This primer set has been shown to be very sensitive, and because it amplifies a short fragment, it is particularly suited for amplification from FFPE. However, due to the size limits, it is unable to distinguish among all known viral types (see Figure 2 and Critical parameters and Troubleshooting section). Furthermore, the small amplicon must be cloned prior to sequencing, significantly complicating the assay (protocols for these steps are included as Supporting protocols). Alternate Protocols 1 and 2 describe the use of alternative primer pairs (GP5+/6+ (GP68) (de Roda Husman et al., 1995) and SPF/GP6+ (Huang et al., 2006) respectively). These alternative primers amplify larger fragments; therefore, their success depends on the quality of the extracted DNA (Greer et al., 1994). Finally, we provide a protocol to determine whether the extracted DNA is suitable for PCR amplification, thereby minimizing the risk for false negative data (Huang et al., 2006).
Figure 1. Flow chart of the different methods in the Protocol.
Following DNA extraction, the Protocol describes three complementary methods aimed at PCR amplifying fragments from the papillomavirus L1 open reading frame. Following PCR, the amplicons are sequenced and typed.
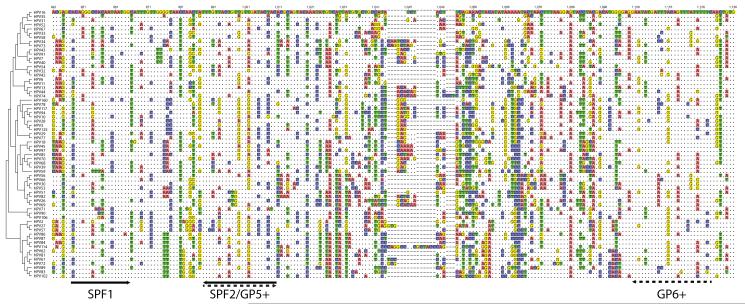
Figure 2. Location of degenerate primers and amplicons within the L1 ORF.
Sequences corresponding to the L1 ORF for all known human Alphapapillomaviruses were downloaded from PaVE (accessed on 7/11/2016)(Van Doorslaer et al., 2013). The sequences were aligned, and a neighbor-joining phylogenetic tree was constructed. The sub-alignment is numbered according to HPV16 (excluding gaps; top) or the alignment (including gaps; bottom). Red lines below the alignment indicate the SPF10 primers, while the green lines show the positions of the GP5+/6+. The alignment highlights the differences between each sequence and HPV34, which was chosen as reference. A dot (“.”) indicates an identical nucleotide, a colored box indicates a single nucleotide polymorphism, while a dash “-” represents an indel.
BASIC PROTOCOL 1 DNA ISOLATION FROM FORMALIN FIXED PARAFFIN EMBEDDED TISSUE
A tissue sample that has been fixed in formalin and embedded in paraffin can be archived for decades. DNA extracted from these tissues can be used for further molecular analyses. Typically, FFPE specimens are subjected to xylene treatment to dissolve the paraffin, next proteinase K digestion removes nucleases and other proteins, and finally the DNA is purified. Excellent reviews, comparing different methods to extract DNA from FFPE tissues were recently published (Alvarez-Aldana et al., 2015; Kocjan et al., 2015). Basic Protocol 1 describes one possible method currently used in our laboratories.
Materials
FFPE tissue block or slides containing tissue sections
Xylene (Sigma, cat. no. 214736)
95% Ethanol (Sigma, cat. no. 493511)
Qiagen DNeasy Blood & Tissue Kit (B&T; Qiagen, cat. no.69504)
DNeasy Mini Spin Columns and Collection Tubes
Tissue Lysis Buffer ATL
Lysis Buffer AL
Wash Buffer AW1 (concentrate; add ethanol before use)
The volume of ethanol (96–100%) to be added is indicated on the bottle. Buffer AW1 is stable for at least 1 year when stored at room temperature.
Wash Buffer AW2 (concentrate; add ethanol before use)
The volume of ethanol (96–100%) to be added is indicated on the bottle. Buffer AW2 is stable for at least 1 year when stored at room temperature.
Elution Buffer AE
Proteinase K
3M Sodium Acetate pH 5.2 (see recipe)
TE buffer (see recipe)
1.5 ml microfuge tube
Razor blades (optional)
Microcentrifuge with rotor for 1.5 ml and 2 ml tubes
Bench top centrifuge capable of 20,000 × g
Thermomixer, shaking water bath, or rocking platform for heating at 56°C
Whenever possible, sample prep should be performed in a room that physically separates the pre- and post-PCR steps. Great care should be taken to minimize cross-contamination between samples.
-
Whenever possible use freshly cut FFPE tissue sections.
Tissue sectioning should be performed using standard practices. Ideally some extra precautions should be taken to minimize cross contamination. The two to three outer sections should be discarded (Kocjan et al., 2011). Prior to use and between different tissue blocks, the microtome blade and working surface should be thoroughly cleaned using an RNA/DNA hydrolysis solution e.g. DNAzap (ThermoFisher, cat. no. AM9890)(Kocjan et al., 2012). HPV negative blocks can be used as negative controls to monitor carry-over during sectioning and downstream processing. The described protocol has been shown to extracts sufficient DNA from a single 4 μm section.
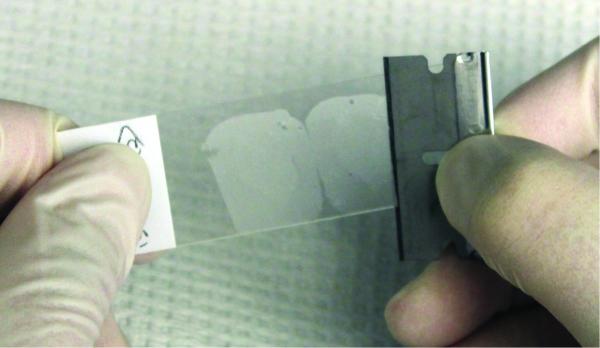
If paraffin blocks are not available for sectioning, a razor blade can be used to remove the tissue-section from a microscope slide (Figure 3). A new razor blade should be used for each sample. Care should be taken to minimize the amount of paraffin.
Collect paraffin sliver containing tissue in a 1.5 ml microfuge tube.
-
Add 1 ml xylene to each 1.5 ml microfuge tube.
Xylene is flammable, and toxic. Perform this and the following steps in a chemical fume hood (Kandyala et al., 2010). Follow local guidelines for disposal of xylene material. Tubes and containers that are resistant to xylene (e.g. polypropylene) should be used for sample extraction and to collect waste.
Vortex for 10 seconds.
Centrifuge in microfuge at top speed for 2 minutes.
Use a pipette tip to remove xylene supernatant without disturbing the pellet.
-
Repeat xylene wash steps (steps 4-7) in order to fully dissolve the tissue.
Typically, two to three wash steps will be sufficient.
Add 1 ml 95% Ethanol.
Vortex for 10 seconds.
Centrifuge in microfuge at top speed for 2 minutes
Use a pipette tip to remove as much ethanol as possible.
Centrifuge in microfuge at top speed for 2 minutes.
Use a pipette tip to remove as much remaining ethanol as possible, then allow pellet to air-dry.
Suspend the dry pellet in 180 μl Qiagen B&T tissue lysis buffer ATL.
Add 20 μl Qiagen B&T proteinase K.
Vortex for 10 seconds.
Incubate for 16 hours at 56°C while shaking at 1000 rpm.
Incubate for one hour at 90°C to reverse cross-linking.
Add 200 μl Qiagen B&T lysis buffer AL.
Vortex for 10 seconds.
Add 200 μl 95% ethanol.
Vortex for 10 seconds.
Use pipette to load entire sample onto a Qiagen B&T column.
Centrifuge for one minute at >6000 × g
Discard flow through.
-
Add 500 μl wash buffer AW1.
Ensure that ethanol has been added to buffer AW1.
Centrifuge for one minute at >6000 × g
Discard flow through.
-
Add 500 μl buffer AW2.
Ensure that ethanol has been added to buffer AW2.
Centrifuge for one minute at >6000 × g
Discard flow through.
Centrifuge for three minutes at 20,000 × g to dry membrane and remove residual AW2 buffer. Discard flow through.
Place the column into a sterile 1.5 ml microfuge tube.
-
Add 200 μl Qiagen B&T elution buffer AE to the column.
Prewarm buffer AE to 56°C
Incubate 5 minutes at room temperature.
Centrifuge for one minute at >6000 × g
-
Repeat steps 34, 35, and 36.
Repeating the elution will increase the total DNA yield, but decrease the concentration.
Precipitate 400 μl eluted DNA by adding 40 μl 3M Sodium Acetate (pH 5.2), mixing and then add 1200 μl 95% ethanol.
Incubate ethanol precipitation for 16 hours at −20°C.
-
Centrifuge in microfuge at top speed for 30 minutes
Equilibrate the samples to room temperature prior to centrifugation.
-
Use a pipet to carefully remove supernatant without disturbing the pellet.
The pellet will likely be very small, so mark the tubes so you know where to expect the pellet.
Add 500 μl 70% ethanol
Centrifuge in microfuge at top speed for 10 minutes
Use a pipet to remove supernatant without disturbing the pellet.
-
Air dry pellet to remove the final traces of ethanol.
Take care not to over dry the pellet.
Resuspend pellet in 20 μl TE buffer.
Incubate DNA at 4°C for 16 hours.
DNA can be stored at −20°C until further use.
Figure 3. Picture illustrating how to remove FFPE slice from slide.
Place a clean razorblade at a 45° angle to the tissue. Carefully scrape the tissue from the slide.
BASIC PROTOCOL 2
Use of Degenerate SPF10 PCR Primers to amplify HPV specific DNA
Type specific papillomaviruses infection is typically diagnosed using molecular tools. In the pre-high-throughput-sequencing era, diagnosis and genotyping have been based on PCR using consensus, and often degenerate primers. Typically, these primers will amplify a broad spectrum of HPV genotypes in a single PCR reaction. Genotyping is based on sequencing, or other sequence specific methods (e.g. dot blots, reverse hybridization line probe assay). Several consensus PCR primer sets have been developed to amplify conserved regions within the viral L1 open reading frame (ORF) (e.g. MY 09/11 primers (Hildesheim et al., 1994), the GP5+/6+ primers (de Roda Husman et al., 1995), and the CPI/II primers (Quint et al., 2001; Tieben et al., 1993)). In this protocol we describe the use of the SPF10 primer set, consisting of 4 forward and 2 reverse primers (Quint et al., 2001). This primer set has the advantage of amplifying a small fragment (65 bp of the L1 region) of the HPV genome, which is useful for DNA extracted from formalin-fixed tissue (Kleter et al., 1999; Kleter et al., 1998). Furthermore, this set is highly sensitive for detection of a broad range of HPV genotypes (Kleter et al., 1999). However, while the small amplicon size increases the odds of amplifying damaged DNA, it complicates genotyping efforts. The SPF10 assay was originally designed in conjunction with a reverse hybridization line probe assay (SPF10-LiPA) (Kleter et al., 1999; Melchers et al., 1999). This assay requires immobilization of type specific probes in parallel lines on nitrocellulose membrane strips, with subsequent hybridization of the amplified DNA to the strips to determine the genotype. While this allows for automation, the commercial kit requires special equipment and only allows for the detection of a limited number of types present on the strips. We provide alternate protocols that involve cloning and sequencing of the SPF10 amplicons. However, another disadvantage of the small amplicon is that the short unique inter-primer region does not allow distinction of all known HPV types (see Figure 2 and Critical parameters and Troubleshooting section).
Materials
SPF10 series of PCR primers (see Table 1; 100 μM stock concentration)
Table 1.
Details of oligonucleotide primers used in this protocol
| sequence | Tm (°C) | reference | |
|---|---|---|---|
| SPF1A | 5′-GCiCAGGGiCACAATAATGG-3′ | 59.0 | (Quint et al., 2001) |
| SPF1B | 5′-GCiCAGGGiCATAACAATGG-3′ | 59.0 | (Quint et al., 2001) |
| SPF1C | 5′-GCiCAGGGiCATAATAATGG-3′ | 56.1 | (Quint et al., 2001) |
| SPF1D | 5′-GCiCAAGGiCATAATAATGG-3′ | 54.0 | (Quint et al., 2001) |
| SPF2A | 5′-GTiGTATCiACAACAGTAACAAA-3′ | 52.7 | (Quint et al., 2001) |
| SPF2B | 5′-GTiGTATCiACTACAGTAACAAA-3′ | 51.8 | (Quint et al., 2001) |
| GP5+ | 5′-TTTGTTACTGTGGTAGATACTAC-3′ | 49.5 | (de Roda Husman et al., 1995) |
| GP6+ | 5′-GAAAAATAAACTGTAAATCATATTC-3′ | 45.3 | (de Roda Husman et al., 1995) |
| GP68 | 5′-TTTCTTACTGTTGTGGATACCAC-3′ | 52.7 | Unpublished |
| GAPDH F | 5′-GGCAGCAGCAAGCATTCCT-3′ | 59.0 | (Huang et al., 2006) |
| GAPDH R | 5′-GCCCAACACCCCCAGTCA-3′ | 60.7 | (Huang et al., 2006) |
“i” represents inosine, will basepair with A, C or T
Do not add 5′ phosphates to your primers for PCR, or the PCR product synthesized will not be suitable for TOPO-TA mediated cloning (Support Protocol 1)
High Fidelity PCR system (we use Roche FastStart High fidelity PCR system; Sigma cat. no. 3553400001)
PCR grade deoxynucleoside triphosphates (dNTPs; Sigma, cat. no. 11969064001)
Nusieve 3:1 agarose (Lonza, cat.no. 50090)
50X TAE running buffer (see recipe)
Agarose gel electrophoresis system
Gel electrophoresis loading buffer (see recipe)
Ethidium bromide (10 mg/ml; Sigma, cat. no. E1510)
Molecular weight markers (e.g. NEB 2-Log DNA ladder; NEB, cat.no. N3200S)
UV light box and camera
200 μl thin-walled PCR tubes (or other appropriate for thermal cycler)
Thermal cycler
-
Dilute each primer to 10 μM working stock.
E.g. Mix 10 μl GP6+ with 90 μl ddH2O
- For each sample, and appropriate controls (Cloned HPV genomes can be used as positive controls and can be obtained from the HPV reference center -www.hpvcenter.se/. Representative HPV types are listed in Table 2) setup the following reaction:
- 29.5 μl ddH2O
- 5 μl 10x PCR buffer 2 (contains 18 mM MgCl2)
- 2 μl each SP1 primer (10 μM stock; 8 μl total)
- 2 μl each SP2 primer (10 μM stock; 4 μl total)
- 1 μl dNTPS (10 mM)
- 0.5 μl FastStart High Fidelity Enzyme Blend (5 U/μl)
- 2 μl template DNA from Basic Protocol 1
Program a thermocycler to perform a preheating step for 10 minutes at 94°C, followed by 40 cycles of 30 seconds at 94°C, 30 seconds at 50°C, and 1 minute at 68°C. Add a final extension step of 10 minutes at 68°C.
-
Prepare a 3% agarose gel in 1X TAE buffer using standard procedures. Ethidium bromide (final concentration 0.5 μg/ml) can be added to the gel mixture at this point.
The 3:1 mixture of high resolution and regular agarose resolves fragments from >10bp to 1000 bp while providing a structurally strong matrix. Ethidium bromide is a known mutagen. Follow local guidelines for disposal of Ethidium bromide containing material.
Mix 5 μl of each PCR product with 1 μl loading buffer and load on the 3:1 Nusieve agarose gel alongside 5 μl of the DNA ladder. Run the gel for 1 hour at 100 V.
-
When adequate migration has occurred, the DNA can be visualized on an ultraviolet transilluminator. An example of a gel is shown in Figure 4.
The expected amplicon is about 65 bp in length (Figure 2). Due to its small size, the amplicon will need to be cloned prior to sequencing (Support Protocol 1). The absence of a specific band cannot be automatically interpreted as a true negative (i.e. no HPV) result. It is essential that it is confirmed that the DNA quality is sufficient to allow for PCR based amplification (Support Protocol 2).
Table 2.
Representative HPV types from each species of alpha HPVs.
| Virus | Species Name | Original accession number |
Reference |
|---|---|---|---|
| HPV32 | Alphapapillomavirus 1 | X74475 | (Delius and Hofmann, 1994) |
| HPV3 | Alphapapillomavirus 2 | X74462 | (Delius and Hofmann, 1994) |
| HPV61 | Alphapapillomavirus 3 | U31793 | Delius, Unpublished |
| HPV2 | Alphapapillomavirus 4 | X55964 | (Hirsch-Behnam et al., 1990) |
| HPV26 | Alphapapillomavirus 5 | X74472 | (Delius and Hofmann, 1994) |
| HPV30 | Alphapapillomavirus 6 | X74474 | (Delius and Hofmann, 1994) |
| HPV18 | Alphapapillomavirus 7 | X05015 | Cole and Danos, 1987 |
| HPV7 | Alphapapillomavirus 8 | X74463 | (Delius and Hofmann, 1994) |
| HPV16 | Alphapapillomavirus 9 | K02718 | (Seedorf et al., 1985) |
| HPV6 | Alphapapillomavirus 10 | X00203 | (Schwarz et al., 1983) |
| HPV34 | Alphapapillomavirus 11 | X74476 | (Delius and Hofmann, 1994) |
| HPV54 | Alphapapillomavirus 13 | U37488 | Delius, Unpublished |
| HPV71 | Alphapapillomavirus 14 | AB040456 | (Matsukura and Sugase, 2001) |
DNA samples can be requested from The HPV Reference Centre http://www.hpvcenter.se/
Updated sequences, and further information, are available from https://pave.niaid.nih.gov/
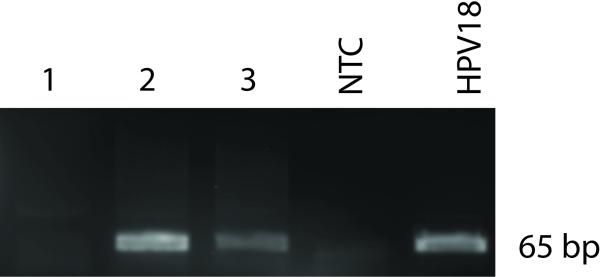
Figure 4. Example of a 3% agarose gel showing SPF10 PCR products.
Five μl PCR product was loaded on a 3% agarose gel and visualized using Ethidium Bromide. Lane 1 contained DNA from an HPV negative sample, lane 2 and 3 were previously typed as HPV16 and HPV18 positive respectively. NTC indicates the no-template negative control (double distilled H2O). 1E-4 ng HPV18 genome cloned in pBR322 (Boshart et al., 1984) was used as a positive control.
BASIC PROTOCOL 3
Genotyping of PCR amplicons
The PCR primers used in this protocol are designed to be conserved enough to amplify a wide array of human papillomaviruses, while generating an amplicon that is diverse enough to allow for genotyping. Support Protocol 1 outlines a method to clone these small amplicons. Basic Protocol 3 describes the steps downstream of the sequencing analysis. Typically, when the amplicons are cloned, sequencing is performed using commonly available primers that flank the cloning site in the vectors (e.g. M13). To directly sequence the amplicon without cloning, the original PCR primers can be used (only possible with longer amplicons; see Alternate Protocols 1 and 2). Several commercial companies offer excellent Sanger based sequencing services. Touchman wrote an exhaustive guide to choosing a company (Touchman, 2009). In addition, many companies will purify DNA from bacterial colonies for a nominal cost.
Materials
Software program capable of displaying sequence chromatograms
We use Geneious (Kearse et al., 2012), however many free alternatives are available for different platforms (Windows, Mac OS, Linux)
Import the chromatogram files into the software. In Geneious this can be accomplished by dragging the chromatogram files (These files typically have an “.abi” extension) into a folder in Geneious.
Visually inspect the chromatogram quality. The occurrence of overlapping peaks may suggest that the original tissue sample was infected with multiple papillomavirus types. In this case, the PCR amplicon should be cloned (Support Protocol 2) and individual clones sequenced.
-
Remove the primer sequence from the chromatogram.
In Geneious this can be done be selecting the sequence, clicking “allow editing” and deleting using the keyboard. Since degenerate primers are used, this step is absolutely essential for accurate genotyping.
-
Export the sequence and save as a FastA file.
The FastA format is a convenient format for representing nucleotide sequences in a form that is recognized by many software programs. A FastA sequence begins with a single-line description (indicated by the “greater-than” sign; “>”), followed by lines of sequence data in single-letter code.
-
Using a Web Browser (preferably Safari, Firefox or Chrome) navigate to the Papillomavirus Episteme (PaVE) BLASTn search page (https://pave.niaid.nih.gov/#search/pv_specific_blast)(Van Doorslaer et al., 2013). This database contains genomic sequences of all papillomaviruses isolated to date. Paste the FastA sequence into the search box, and click submit.
The BLASTn output will show the hits ordered by E-value. The triangle next to each hit can be used to expand the alignment generated by BLAST.
-
The top hit (i.e. lowest E-value) can be used to genotype the virus in your sample.
An E-value of “0”, indicates a perfect match between your sample and the prototype virus stored in the PaVE database. Non-zero values may indicate issues with the sequencing reaction (e.g. bad base calls), double infection, natural sequence variation (e.g. HPV variants), or the presence of a putative novel type.
ALTERNATE PROTOCOL 1
Use of Degenerate GP5+/6+ PCR Primers to amplify HPV specific DNA
Basic Protocol 2 describes the use of the SPF10 set of primers (Quint et al., 2001) to amplify a 65 bp fragment. This short PCR product increases the sensitivity of detection, especially on challenging samples such as DNA extracted from FFPE. However, the short amplicon requires cloning prior to sequencing, dramatically increasing hands-on time. Finally, these short amplicons do not allow discrimination among all unique papillomavirus types. Alternate Protocol 2 describes the use of a different primer set, which was modified from the GP5+/6+ system (de Roda Husman et al., 1995). This amplicon is 150 bp in length, which enables direct sequencing and allows one to distinguish among closely related types.
Materials
GP5+, GP6+, and GP68 PCR primers (see table 1; 100 μM stock concentration)
HotStarTaq Plus DNA Polymerase (Qiagen cat. no. 203601)
Uracyl-DNA Glycosylase, heat labile (Optional; Roche cat no. 11775367001)
PCR grade deoxynucleoside triphosphates (dNTPs; Sigma, cat. no. 11969064001)
PCR nucleotide mixplus (Optional; Roche cat no 11888412001)
Nusieve 3:1 agarose (Lonza, cat.no. 50090)
50X TAE running buffer (see recipe)
Agarose gel electrophoresis system
Gel electrophoresis loading buffer (see recipe)
Ethidium bromide (10 mg/ml; Sigma, cat. no. E1510)
Molecular weight markers (e.g. NEB 2-Log DNA ladder; NEB, cat.no. N3200S)
UV light box and camera
200 μl thin-walled PCR tubes (or other appropriate for thermal cycler)
Thermal cycler
-
Dilute each primer to 10 μM working stock.
E.g. Mix 10 μl GP6+ with 90 μl ddH2O
-
For each sample and appropriate controls (cloned HPV genomes to be used as positive controls can be obtained from the HPV reference center -www.hpvcenter.se/ - representative HPV types are listed in Table 2) setup the following reaction:
- 26.5 μl ddH2O
- 5 μl 10 x CoralLoad PCR buffer (contains 15mM MgCl2)
- 5 μl MgCl2 (25 mM stock)
- 5 μl GP5+ primer (10 μM stock)
- 0.25 μl GP68 primer (10 μM stock)
- 5 μl GP6+ primer (10 μM stock)
- 1 μl dNTPS (10 mM stock)
- 0.25 μl Taq
- 2 μl template DNA from Basic Protocol 1
Note that carry-over contamination can be minimized by the inclusion of 1U uracil-DNA glycosylase (UDG) in the reaction, in which case the PCR nucleotide Mixplus would be used in lieu of the regular dNTPs.
-
Program a thermocycler to perform a touchdown PCR according to the following profile (Evans et al., 2005). A preheating step for 10 minutes at 94°C is followed by 21 cycles each of 1 minute at 94°C, 2 minutes at 50°C, and 1 minute at 72°C. The annealing temperature is reduced by 0.5°C/cycle. Next, 21 cycles are performed of 1 minute at 94°C, 2 minutes at 40°C, and 1 minute at 72°C, with a final extension step of 10 minutes at 72°C.
Touchdown PCR is a method to increase the specificity of PCR reactions. During the first couple cycles, the annealing temperature is gradually reduced from an initial annealing temperature several degrees above the estimated Tm of the primers. Amplification is then continued using the primer specific annealing temperature.
In case UDG was added to the reaction mix (step 2), a 30 minute incubation at 37°C should be included prior to the initial denaturation step.
-
Prepare a 3% agarose gel in 1X TAE buffer using standard procedures. Ethidium bromide (final concentration 0.5 μg/ml) can be added to the gel mixture at this point.
The 3:1 mixture of high resolution and regular agarose resolves fragments from >10bp to 1000 bp while providing a structurally strong matrix. Ethidium bromide is a known mutagen. Follow local guidelines for disposal of Ethidium bromide containing material.
Mix 5 μl of each PCR product with 1 μl loading buffer and load on the 3:1 Nusieve agarose gel alongside 5 μl of the DNA ladder. Run the gel for 1 hour at 100 V.
When adequate migration has occurred, the DNA can be visualized on an ultraviolet transilluminator.
The expected amplicon is about 150 bp in length (figure 2). If electrophoresis identifies a single specific amplicon, the PCR product can be purified and directly sequenced. Otherwise, the 150 bp band should be excised from the gel and purified, followed by sequencing. The absence of a specific band cannot be automatically interpreted as a true negative (i.e. no HPV) result. It is essential that it is confirmed that the DNA quality is sufficient to allow for PCR based amplification (Support Protocol 2).
ALTERNATE PROTOCOL 1
Use of Degenerate SPF1/GP6+ PCR Primers to amplify HPV specific DNA
Basic Protocol 2 describes the use of the SPF10 set of primers (Quint et al., 2001) to amplify a 65 bp fragment. Alternate Protocol 1 uses the GP5+/GP6+ primer set. In Alternate Protocol 2, we describe a PCR approach based on combining four forward SPF10 primers and a reverse GP6+ primer (Huang et al., 2006). Like the GP5+/6+ amplicon (Alternate Protocol 1), this PCR product is long enough to be directly sequenced.
Materials
SPF1 primers (see table 1; 100 μM stock concentration)
GP6+ PCR primers (see table 1; 100 μM stock concentration)
AmpliTaq Gold DNA polymerase (ThermoFisher, cat. no. N8080241)
10 mM PCR grade deoxynucleoside triphosphates (dNTPs; Sigma, cat. no. 11969064001)
25 mM MgCl2
Nusieve 3:1 agarose (Lonza, cat.no. 50090)
50X TAE running buffer (see recipe)
Agarose gel electrophoresis system
Gel electrophoresis loading buffer (see recipe)
Ethidium bromide (10 mg/ml; Sigma, cat. no. E1510)
Molecular weight markers (e.g. NEB 2-Log DNA ladder; NEB, cat.no. N3200S)
UV light box and camera
200 μl thin-walled PCR tubes (or other appropriate for thermal cycler)
Thermal cycler
-
Dilute each primer to 10 μM working stock.
E.g. Mix 10 μl GP6+ with 90 μl ddH2O
- For each sample and appropriate controls (cloned HPV genomes to be used as positive controls can be obtained from the HPV reference center -www.hpvcenter.se/ - representative HPV types are listed in Table 2) setup the following reaction:
- 25.75 μl ddH2O
- 5 μl GeneAmp 10 x PCR buffer II (no MgCl2)
- 6 μl MgCl2 (25 mM stock)
- 2 μl each SP1 primer (10 μM stock; 8 μl total)
- 2 μl GP6+ primer (10 μM stock)
- 1 μl dNTPs (10 mM stock)
- 0.25 μl AmpliTaq Gold
- 2 μl template DNA from Basic Protocol 1
Program a thermocycler to perform a preheating step for 10 minutes at 94°C followed by 40 cycles of one minute at 94°C, one minute at 45°C, and one minute at 72°C. A final extension step of 10 minutes at 72°C.
-
Prepare a 3% agarose gel in 1X TAE buffer using standard procedures. Ethidium bromide (final concentration 0.5 μg/ml) can be added to the gel solution when it has cooled below 60°C.
The 3:1 mixture of high resolution and regular agarose resolves fragments from >10bp to 1000bp while providing a structurally strong matrix. Ethidium bromide is a known mutagen. Follow local guidelines for disposal of Ethidium bromide containing material.
Mix 5 μl of each PCR product with 1 μl loading buffer and load on the 3:1 Nusieve agarose gel alongside 5 μl of the DNA ladder. Run the gel for 1 hour at 100 V.
-
When adequate migration has occurred, the DNA can be visualized on an ultraviolet transilluminator.
The expected amplicon is about 180 bp in length (figure 2). If electrophoresis identifies a single specific amplicon, the PCR product can be purified and directly sequenced. Otherwise, the 180 bp band should be excised from the gel and purified, followed by sequencing. The absence of a specific band cannot be automatically interpreted as a true negative (i.e. HPV negative) result. It is essential that it is confirmed that the DNA quality is sufficient to allow for PCR based amplification (Support Protocol 2).
SUPPORT PROTOCOL 1
Topo Cloning of PCR amplicons
This support protocol will describe two cloning steps. In the first step, a terminal 3’adenine (A) overhang will be added to the purified PCR product. While this is thought to be an optional step when using Taq polymerase (which generates terminal 3’A overhangs), this step is absolutely required when using other high-fidelity polymerases (not Taq). However, even when Taq is used, we find that this additional step dramatically optimizes the downstream ligation step, as the ends of Taq generated fragments are sometimes incomplete. During the tailing step, a non-templated adenine is added to the 3′ end of a blunt, double-stranded DNA molecule, thereby preparing the template for use in TA cloning. TA cloning allows for rapid cloning leveraging the fact that the Taq produced A is stabilized by the complementary T (thymidine) on the T-vector, and the vector has pre-bound topoisomerase that efficiently ligates the insert and vector.
Materials
PCR clean-up kit (e.g. High Pure PCR Product Purification Kit; Roche, cat.no. 11732668001)
Agarose Gel purification kit (e.g. zymoclean Gel DNA recovery kit; Zymo. cat. no. D4007)
10X ThermoPol Buffer (NEB cat. no. B9004)
10 mM dATP
Taq DNA Polymerase (NEB cat. no. M0267)
200 μl thin-walled PCR tubes (or other appropriate for thermal cycler)
TOPO TA cloning kit (ThermoFisher cat. no. K450001)
Topoisomerase I-activated pCR™2.1-TOPO® vector
Salt solution (1.2 M NaCl and 0.06 M MgCl2)
M13 forward and reverse primers
One Shot Chemically Competent E. coli(TOP10; F– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG)
S.O.C. medium (see recipe)
LB plates containing 50 μg/ml Kanamycin or Carbenicillin
Miniprep kit (e.g. Wizard Plus SV Miniprep; Promega, cat. no. A1270)
Thermal cycler
A-Tailing with Taq Polymerase (optional)
-
Purify the PCR fragment from other components of the PCR reaction using a clean-up kit according to the manufacturer’s instructions.
This can be done by using a PCR-column purification protocol, or by purifying a specific band from the agarose gel.
- For each sample setup the following reaction:
- 30 μl purified, PCR-amplified DNA
- 5 μl 10X ThermoPol Buffer
- 1 μl 10 mM dATP
- 13.8 μl ddH2O
- 0.2 μl Taq DNA Polymerase
Incubate the reaction at 72°C for 20 minutes.
-
Continue directly with the ligation step.
To minimize the risk of introducing nucleases, which may degrade the A overhangs on the PCR product, the tailed DNA should be purified using a column based method prior to storing the DNA.
Ligation of A-tailed PCR product to T-tailed vector
- For each sample setup the following reaction
- 4 μl fresh A-tailed product (step 4 above)
- 1 μl salt solution (TOPO TA cloning kit)
- 1 μl TOPO vector (TOPO TA cloning kit)
Mix by gently flicking the tube
Incubate at room temperature for 10 minutes.
-
Place the reaction on ice and continue with bacterial transformation.
At this point, the reaction can be stored at −20°C.
Transformation of Chemically competent cells
-
Add 2 μl of the TOPO Cloning reaction to a vial one shot competent cells and mix gently.
Do not mix by pipetting up and down.
Incubate on ice for 20 minutes.
Heat-shock the cells for 30 seconds at 42°C without shaking.
Immediately transfer the tubes to ice.
Add 450 μl of room temperature S.O.C. medium.
Cap the tube tightly and shake the tube horizontally (200 rpm) at 37°C for 1 hour.
-
At this point incubate LB plates with selective agar at 37°C
Note: the vector contains a marker for selection on Kanamycin and Ampicillin/Carbenicillin containing selecting media. Choice of antibiotic is at your discretion.
Note: the vector allows for blue/white screening, which is a convenient way to select recombinant clones, thereby minimizing sequencing of false positive clones. This technique requires that X-gal and IPTG are added to the LB agar plates (See recipe).
Spread 200 μl from each transformation on a pre-warmed selective plate.
-
Incubate plates at 37°C (overnight incubation is optimal).
An efficient TOPO Cloning reaction results in hundreds of colonies.
For each sample, pick 5 to 10 well isolated colonies, and use it to inoculate 5 ml liquid selective LB medium.
Incubate overnight at 37°C in a shaking incubator.
-
Isolate DNA using manufacturer’s protocol.
DNA is now ready to be sequenced.
SUPPORT PROTOCOL 2
Control Amplification of cellular DNA
The quality of DNA extracted from FFPE specimens is the main limiting step for any of the molecular methods described in this protocol. DNA is usually crosslinked, degraded, and contains mixtures of single- and double-stranded DNA. Therefore, assessing whether the extracted DNA is suited for amplification is critical. This can be done prior to processing any samples or can be performed only on the samples for which HPV specific PCR failed (Huang et al., 2006). This is especially critical in clinical and/or epidemiological studies. Sophisticated methods aimed at estimating fragment length have been developed (e.g. 2100 Bioanalyzer System), however we provide a method to amplify a 137 bp fragment from the GAPDH gene (Huang et al., 2006).
Materials
GAPDH primers (see table 1; 100 μM stock concentration)
AmpliTaq Gold DNA polymerase (ThermoFisher, cat. no. N8080241)
PCR grade deoxynucleoside triphosphates (dNTPs; Sigma, cat. no. 11969064001)
Nusieve 3:1 agarose (Lonza, cat.no. 50091)
200 μl thin-walled PCR tubes (or other appropriate for thermal cycler)
Thermal cycler or heat block capable of maintaining 72°C.
-
Prepare a 200 μl working stock containing 50 μM of each GAPDH primer.
E.g. Mix 100 μl each primer (100 μM stock)
-
For each sample and appropriate controls (a positive control plasmid containing the entire for GAPDH ORF can be obtained from OpenBiosystems, cat. no. LIFESEQ95132246) setup the following reaction:
41.35 μl ddH2O
5 μl GeneAmp 10 x PCR buffer I (15 mM MgCl2)
0.2 μl primer mix
1.2 μl dNTPS (10 mM stock)
2 μl template from Basic Protocol 1
0.25 μl AmpliTaq Gold (5 U/μl)
Program a thermocycler to perform a preheating step for 10 minutes at 94°C followed by 40 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 72°C. A final extension step of 10 minutes at 72°C.
-
Prepare a 3% agarose gel in 1X TAE buffer using standard procedures. Ethidium bromide (final concentration 0.5 μg/ml) can be added to the gel mixture at this point.
The 3:1 mixture of high resolution and regular agarose resolves fragments from >10bp to 1000 bp while providing a structurally strong matrix. Ethidium bromide is a known mutagen. Follow local guidelines for disposal of Ethidium bromide containing material.
Mix 5 μl of each PCR product with 1 μl loading buffer and load on the 3:1 Nusieve agarose gel alongside 5 μl of the DNA ladder. Run the gel for 1 hour at 100 V.
-
When adequate migration has occurred, the DNA can be visualized on an ultraviolet transilluminator.
The expected amplicon is about 137 bp in length (figure 2). The presence of this band indicates that the DNA sample is able to be amplified, providing a higher level of confidence that a sample negative using PV specific primers is not a false negative.
REAGENTS AND SOLUTIONS
100 ml 3M Sodium Acetate (pH 5.2)
Dissolve 40.8 g sodium acetate (m.w. 136.08) in 70 ml of double distilled water.
Adjust the pH to 5.2 using glacial acetic acid.
Add water to bring the total volume of solution to 100 ml.
Filter sterilize (0.2 μm pore), aliquot and store at 4°C.
100 ml solution of TE Buffer
Combine 1 ml of 1 M Tris-HCl (pH 8.0) and 0.2 ml EDTA (0.5 M) in a total volume of 100 ml double distilled water.
1000 ml solution of 50X TAE Buffer
Dissolve 242 g of Tris base in 750 ml double distilled water.
Add 100 ml of 0.5 M EDTA, and 57.1 ml glacial acetic acid.
Adjust the pH 8.3 if required.
Adjust the solution volume to 1000 ml with double distilled water.
Sterilize the solution by autoclaving.
Solution can be stored at room temperature.
10 ml Gel electrophoresis loading buffer
combine
5 ml of 50% glycerol
5 ml of 0.5 M EDTA
0.0276 g bromophenol blue
0.0214 g xylene cyanol FF (Xylene cyanol is an irritant, wear appropriate PPE when preparing this reagent.)
Mix well
Store in 1 ml aliquots at 4°C
100 ml S.O.C. media
Dissolve 2 g Bacto Tryptone and 0.5 g Bacto Yeast Extract in 90 ml double distilled water.
Add the following:
200 μl of 5M NaCl
250 μl of 1M KCl
1 ml of 1M MgCl2
1 ml of 1M MgSO4
2 ml of 1M glucose.
Adjust to 100 ml with distilled H2O.
Sterilize by autoclaving, aliquot and store at 4°C
Agar plates
Dissolve 15 g of Bacto agar (BD; cat. no. 214050) in 1000 ml of LB medium sterilize by autoclaving
Cool to 50°C in a temperature-controlled water bath
Add 1 μl of antiobiotic stock solution (50 mg/ml stock)
Pour into plates
Agar plates for Blue/white screening (optional)
Starting from previously poured plates containing antibiotics:
Add 40 μl of the X-Gal Solution (20 mg/ml stock) (Thermo Scientific, cat. no. R0941).
Add 40 μl of IPTG Solution (100 mM stock) (Thermo Scientific, cat no. R1171)
Spread evenly on the plate with a sterile spatula.
Starting from liquid LB (kept at 50°C). For every ml of media:
Add 1 μl of the X-Gal Solution (20 mg/ml stock) (Thermo Scientific, cat. no. R0941).
Add 1 μl of IPTG Solution (100 mM stock) (Thermo Scientific, cat no. R1171)
Add 1μl of antiobiotic stock solution (50 mg/ml stock)
Mix well.
Pour 25 ml of prepared LB agar into each Petri dish.
Allow agar to solidify at room temperature for about 30 minutes
COMMENTARY
Background Information
Persistent infection with certain, oncogenic papillomaviruses is a necessary prerequisite for the development of cervical cancer (Bodily and Laimins, 2011). To truly understand the clinical importance of individual papillomavirus types, it is important to confidently identify the viral type(s) associated with a specific lesion (Schiffman et al., 2009; Van Doorslaer and Burk, 2010). PCR based approaches have been successfully used to detect and classify HPV types in cervical cytology samples (Schiffman et al., 2011). The majority of these PCR assays target a conserved region within the L1 ORF of evolutionary divergent viruses. This allows for the amplification of a broad set of viral types, while also making it possible to identify the amplified virus by sequence analysis (de Roda Husman et al., 1995; Hildesheim et al., 1994; Quint et al., 2001; Tieben et al., 1993).
FFPE specimens provide a wealth of potential data for molecular studies, however formalin fixation may cause extensive DNA damage (Greer et al., 1991), complicating molecular studies. Nonetheless, several studies have described the ability to genotype viral types from FFPE tissues (Castro et al., 2015; Guerendiain et al., 2016; Kocjan et al., 2016; Lillsunde Larsson et al., 2015). The availability of the world-wide library of FFPE specimens will allow researchers to test the proposed association of specific viruses with specific pathologies.
Critical Parameters
The quality of the extracted DNA is the single most important parameter throughout this protocol. The steps taken prior to fixation have been shown to be critical (Srinivasan et al., 2002). However, most molecular studies will collect tissue blocks, and will not be able to control for their processing. Therefore, the methods used for purification of genomic DNA from FFPE tissues are crucial. The above methods leverage high-quality reagents with easy to use commercial kits, thereby maximizing the chances for reproducible results.
Troubleshooting
Estimation of DNA concentration
The protocol does not require the DNA concentration to be known. However, since FFPE DNA contains a high proportion of single-stranded DNA, optical density (OD) based approaches are not recommended to measure total DNA concentration. DNA concentration should be determined using assays that are specific for double stranded DNA (e.g. Quant-iT picogreen assay; ThermoFisher, cat. no. P7589).
Removal of oligonucleotide primer from sequencing chromatograms
The oligonucleotide primers were designed to match a wide array of papillomavirus types. As a result, primer-template mismatches may introduce PCR generated errors in the amplicons. Therefore, it is essential that the primer sequences are removed prior to Blast based searches.
Multiple infections
Many samples are coinfected with multiple papillomavirus types (Chaturvedi et al.). Direct sequencing of PCR amplicons derived from such multiple infections will return overlapping peaks in the sequencing chromatograms, interfering with genotyping. Figure 5 shows an example of such a mixed chromatogram. In this case, the PCR amplicons will need to be cloned, and DNA from individual colonies sequenced (Supporting Protocol 1).
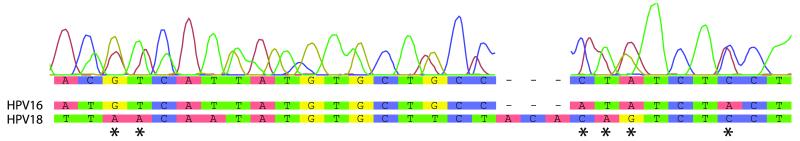
Figure 5. Example of a chromatogram representing a multiple infection.
Multiple overlapping peaks can be seen in the chromatogram indicating a generally poor sequencing reaction. However, upon closer inspection, evidence of both HPV16 and HPV18 DNA can be observed in the traces (indicated by *).
BLASTn based genotyping
In case, the PCR product was sequenced directly, both strands should be blasted independently. If both strands identified the same type, a consensus sequence should be used for final typing. If both strands identify different types, it is recommended that the PCR products are cloned prior to sequencing. The BLASTn based analysis will compare the sample sequence (query) to all HPV prototypes in the PaVE database. The E-value (lower is better) will provide a certain level of confidence that the sequenced amplicon represents a specific papillomavirus type. However, the PaVE database only contains prototype papillomaviruses, not naturally occurring variants. It is therefore possible that the BLASTn results may not return a perfect match between the query (user sequence) and the sequences in PaVE. The BLASTn generated alignments should be manually analyzed prior to calling specific genotypes. If the BLASTn search does not return any hits, the E-value cut-off (default at 1e-10) can be changed, to lower the specificity of the search. If lowering the E-value does not allow type identification, it is possible that the tissue specimen was infected with a novel papillomavirus type.
Tissue contains a putative novel papillomavirus type
If the PaVE BLASTn search does identify a specific known HPV genotype, it is possible that the sample contains a novel virus. To rule out non-specific amplification of non-PV DNA, the sequence should be compared against the NCBI databases using an exhaustive Blast search.
Conserved regions do not enable differentiation of certain viral types
The SPF10 primer set amplifies a short region derived from the L1 ORF. Due to its small size, the amplicon is shared between three different HPV types (HPV68, HPV73, and HPV97). Therefore, samples testing positive for either of these viruses should be typed using another approach (e.g. GP5+/6+ based PCR; Alternate Protocol 1).
Anticipated Results
These protocols are designed to detect and genotype HPVs of the Alphapapillomavirus genus from readily available formalin fixed, paraffin embedded tissues. Most often, the tissue is already sectioned and available on glass slides.
One to three slides are usually sufficient to prepare enough DNA (DNA yield ~ 150 pg/μl (Steinau et al., 2011)) to perform several genotyping reactions as described in this method. The size range of the extracted DNA will be dependent on many factors, such as storage (Srinivasan et al., 2002).
Time Considerations
DNA extraction, including ethanol precipitation and DNA rehydration, is the most labor intensive and takes about three days to complete. If DNA concentration is not a concern, the DNA precipitation step can be omitted, essentially shortening the protocol by day. PCR amplification and sample preparation for sequencing takes another one to two days, depending on whether the PCR amplicons need to be cloned prior to sequencing. The whole protocol, from isolation of DNA to submission of sequencing reactions can be completed in 4 to 5 days. Data analysis takes an additional day. Throughout the protocol, samples can be stored at −20°C, allowing for safe stopping points during the procedure.
ACKNOWLEDGEMENT
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. We are very grateful to Bostjan Kocjan for advice on the protocol and manuscript.
Footnotes
INTERNET RESOURCES
The papillomavirus episteme
The PapillomaVirus Episteme (PaVE) provides highly organized and curated papillomavirus genomics information, and tools, to the scientific community. The PaVE consists of a database and web applications. The PaVE Blast page (https://pave.niaid.nih.gov/#search/pv_specific_blast) can be used to genotype the DNA amplified throughout the protocol
LITERATURE CITED
- Alvarez-Aldana A, Martinez JW, Sepulveda-Arias JC. Comparison of five protocols to extract DNA from paraffin-embedded tissues for the detection of human papillomavirus. Pathol Res Pract. 2015;211:150–155. doi: 10.1016/j.prp.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Bodily J, Laimins LA. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 2011;19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984;3:1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro FA, Koshiol J, Quint W, Wheeler CM, Gillison ML, Vaughan LM, Kleter B, van Doorn LJ, Chaturvedi AK, Hildesheim A, Schiffman M, Wang SS, Zuna RE, Walker JL, Dunn ST, Wentzensen N. Detection of HPV DNA in paraffin-embedded cervical samples: a comparison of four genotyping methods. BMC Infect Dis. 2015;15:544. doi: 10.1186/s12879-015-1281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi AK, Katki HA, Hildesheim A, Rodriguez AC, Quint W, Schiffman M, Van Doorn LJ, Porras C, Wacholder S, Gonzalez P, Sherman ME, Herrero R, Group CVT. Human papillomavirus infection with multiple types: pattern of coinfection and risk of cervical disease. J Infect Dis. 2011;203:910–920. doi: 10.1093/infdis/jiq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76(Pt 4):1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- Delius H, Hofmann B. Primer-directed sequencing of human papillomavirus types. Curr Top Microbiol Immunol. 1994;186:13–31. doi: 10.1007/978-3-642-78487-3_2. [DOI] [PubMed] [Google Scholar]
- Evans MF, Adamson CS, Simmons-Arnold L, Cooper K. Touchdown General Primer (GP5+/GP6+) PCR and optimized sample DNA concentration support the sensitive detection of human papillomavirus. BMC Clin Pathol. 2005;5:10. doi: 10.1186/1472-6890-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CE, Peterson SL, Kiviat NB, Manos MM. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am J Clin Pathol. 1991;95:117–124. doi: 10.1093/ajcp/95.2.117. [DOI] [PubMed] [Google Scholar]
- Greer CE, Wheeler CM, Manos MM. Sample preparation and PCR amplification from paraffin-embedded tissues. PCR Methods Appl. 1994;3:S113–122. doi: 10.1101/gr.3.6.s113. [DOI] [PubMed] [Google Scholar]
- Guerendiain D, Moore C, Wells L, Conn B, Cuschieri K. Formalin fixed paraffin embedded (FFPE) material is amenable to HPV detection by the Xpert((R)) HPV assay. J Clin Virol. 2016;77:55–59. doi: 10.1016/j.jcv.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Halec G, Alemany L, Lloveras B, Schmitt M, Alejo M, Bosch FX, Tous S, Klaustermeier JE, Guimera N, Grabe N, Lahrmann B, Gissmann L, Quint W, Bosch FX, de Sanjose S, Pawlita M, Retrospective International, S., Group, H.P.V.T.T.S. Retrospective International, S., and Group, H.P.V.T.T.S Pathogenic role of the eight probably/possibly carcinogenic HPV types 26, 53, 66, 67, 68, 70, 73 and 82 in cervical cancer. J Pathol. 2014;234:441–451. doi: 10.1002/path.4405. [DOI] [PubMed] [Google Scholar]
- Hildesheim A, Schiffman MH, Gravitt PE, Glass AG, Greer CE, Zhang T, Scott DR, Rush BB, Lawler P, Sherman ME, et al. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- Hirsch-Behnam A, Delius H, de Villiers EM. A comparative sequence analysis of two human papillomavirus (HPV) types 2a and 57. Virus Res. 1990;18:81–97. doi: 10.1016/0168-1702(90)90091-o. [DOI] [PubMed] [Google Scholar]
- Huang SL, Chao A, Hsueh S, Chao FY, Huang CC, Yang JE, Lin CY, Yan CC, Chou HH, Huang KG, Huang HJ, Wu TI, Tseng MJ, Qiu JT, Lin CT, Chang TC, Lai CH. Comparison between the Hybrid Capture II Test and an SPF1/GP6+ PCR-based assay for detection of human papillomavirus DNA in cervical swab samples. J Clin Microbiol. 2006;44:1733–1739. doi: 10.1128/JCM.44.5.1733-1739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 100B: A Review of Human Carcinogens: Biological Agents. IARC Monogr Eval Carcinog Risks Hum. 2012;100:1–441. [PMC free article] [PubMed] [Google Scholar]
- Kandyala R, Raghavendra SP, Rajasekharan ST. Xylene: An overview of its health hazards and preventive measures. J Oral Maxillofac Pathol. 2010;14:1–5. doi: 10.4103/0973-029X.64299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleter B, van Doorn LJ, Schrauwen L, Molijn A, Sastrowijoto S, ter Schegget J, Lindeman J, ter Harmsel B, Burger M, Quint W. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol. 1999;37:2508–2517. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleter B, van Doorn LJ, ter Schegget J, Schrauwen L, van Krimpen K, Burger M, ter Harmsel B, Quint W. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153:1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocjan BJ, Hosnjak L, Poljak M. Commercially available kits for manual and automatic extraction of nucleic acids from formalin-fixed, paraffin-embedded (FFPE) tissues. Acta Dermatovenerol Alp Pannonica Adriat. 2015;24:47–53. doi: 10.15570/actaapa.2015.12. [DOI] [PubMed] [Google Scholar]
- Kocjan BJ, Hosnjak L, Poljak M. Detection of alpha human papillomaviruses in archival formalin-fixed, paraffin-embedded (FFPE) tissue specimens. J Clin Virol. 2016;76(Suppl 1):S88–97. doi: 10.1016/j.jcv.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Kocjan BJ, Maver PJ, Hosnjak L, Zidar N, Odar K, Gale N, Poljak M. Comparative evaluation of the Abbott RealTime High Risk HPV test and INNO-LiPA HPV Genotyping Extra test for detecting and identifying human papillomaviruses in archival tissue specimens of head and neck cancers. Acta Dermatovenerol Alp Pannonica Adriat. 2012;21:73–75. [PubMed] [Google Scholar]
- Kocjan BJ, Seme K, Poljak M. Comparison of the Abbott RealTime High Risk HPV test and INNO-LiPA HPV Genotyping Extra test for the detection of human papillomaviruses in formalin-fixed, paraffin-embedded cervical cancer specimens. J Virol Methods. 2011;175:117–119. doi: 10.1016/j.jviromet.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Lillsunde Larsson G, Carlsson J, Karlsson MG, Helenius G. Evaluation of HPV Genotyping Assays for Archival Clinical Samples. J Mol Diagn. 2015;17:293–301. doi: 10.1016/j.jmoldx.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Matsukura T, Sugase M. Relationships between 80 human papillomavirus genotypes and different grades of cervical intraepithelial neoplasia: association and causality. Virology. 2001;283:139–147. doi: 10.1006/viro.2001.0865. [DOI] [PubMed] [Google Scholar]
- Melchers WJ, Bakkers JM, Wang J, de Wilde PC, Boonstra H, Quint WG, Hanselaar AG. Short fragment polymerase chain reaction reverse hybridization line probe assay to detect and genotype a broad spectrum of human papillomavirus types. Clinical evaluation and follow-up. Am J Pathol. 1999;155:1473–1478. doi: 10.1016/S0002-9440(10)65462-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint WG, Scholte G, van Doorn LJ, Kleter B, Smits PH, Lindeman J. Comparative analysis of human papillomavirus infections in cervical scrapes and biopsy specimens by general SPF(10) PCR and HPV genotyping. J Pathol. 2001;194:51–58. doi: 10.1002/path.855. [DOI] [PubMed] [Google Scholar]
- Schiffman M, Clifford G, Buonaguro FM. Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect Agent Cancer. 2009;4:8. doi: 10.1186/1750-9378-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC, Castle PE. Human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst. 2011;103:368–383. doi: 10.1093/jnci/djq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E, Durst M, Demankowski C, Lattermann O, Zech R, Wolfsperger E, Suhai S, zur Hausen H. DNA sequence and genome organization of genital human papillomavirus type 6b. EMBO J. 1983;2:2341–2348. doi: 10.1002/j.1460-2075.1983.tb01744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf K, Krammer G, Durst M, Suhai S, Rowekamp WG. Human papillomavirus type 16 DNA sequence. Virology. 1985;145:181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinau M, Patel SS, Unger ER. Efficient DNA extraction for HPV genotyping in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2011;13:377–381. doi: 10.1016/j.jmoldx.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieben LM, ter Schegget J, Minnaar RP, Bouwes Bavinck JN, Berkhout RJ, Vermeer BJ, Jebbink MF, Smits HL. Detection of cutaneous and genital HPV types in clinical samples by PCR using consensus primers. J Virol Methods. 1993;42:265–279. doi: 10.1016/0166-0934(93)90038-s. [DOI] [PubMed] [Google Scholar]
- Touchman JW. DNA Sequencing: An Outsourcing Guide. Current Protocols Essential Laboratory Techniques. 2009;2:1–19. [Google Scholar]
- Van Doorslaer K, Burk RD. Evolution of human papillomavirus carcinogenicity. Adv Virus Res. 2010;77:41–62. doi: 10.1016/B978-0-12-385034-8.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, Mohamoud Y, Huyen Y, McBride AA. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 2013;41:D571–578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Sheng ZM, Taubenberger JK. Isolating Viral and Host RNA Sequences from Archival Material and Production of cDNA Libraries for High-Throughput DNA Sequencing. Curr Protoc Microbiol. 2015;37:1E 1–16. doi: 10.1002/9780471729259.mc01e08s37. [DOI] [PMC free article] [PubMed] [Google Scholar]