Abstract
Cordon-bleu (COBL) is a multifunctional WASP-Homology 2 (WH2) domain-containing protein implicated in a wide variety of cellular functions ranging from dendritic arborization in neurons to the assembly of microvilli on the surface of transporting epithelial cells. In vitro biochemical studies suggest that COBL is capable of nucleating and severing actin filaments, among other activities. How the multiple activities of COBL observed in vitro contribute to its function in cells remains unclear. Here, we used live imaging to evaluate the impact of COBL expression on the actin cytoskeleton in cultured cells. We found that COBL induces the formation of dynamic linear actin structures throughout the cytosol. We also found that stabilizing these dynamic structures with the parallel actin bundling protein espin slows down their turnover and enables the robust formation of self-supported protrusions on the dorsal cell surface. Super-resolution imaging revealed a global remodeling of the actin cytoskeleton in cells expressing these two factors. Taken together, these results provide insight as to how COBL contributes to the assembly of actin-based structures such as epithelial microvilli.
Keywords: protrusion, epithelia, polarity, brush border, cytoskeleton, WASP homology 2
INTRODUCTION
A major goal in actin cytoskeleton biology is to identify factors responsible for assembling specific subcellular networks and arrays. Investigators have uncovered a vast collection of regulatory factors, which function together to drive the assembly of actin arrays using a range of activities including filament nucleation, elongation, capping, bundling, and severing [Lee and Dominguez, 2010]. One functionally diverse class of regulatory factors are the WASP-Homology 2 (WH2) domain-containing molecules, which include Cordon-bleu (COBL), SPIRE, JMY, the bacterial effector proteins VopF and VopL, TARP, Sca2, Leiomodin, and WASP family nucleation promoting factors [Namgoong et al., 2011]. WH2 domains are 17–20 amino acid sequences characterized by an amphipathic α-helix, an LKKV motif, and a variable C-terminus [Dominguez, 2016]. In isolation, WH2 domains can bind G-actin, but much more complex functions are possible when these domains are linked in tandem.
A prime example is provided by COBL, which is comprised of an N-terminal proline rich region with several PxxP motifs that bind SH3 domain-containing proteins, a central weak coiled coil that may play a role in oligomerization, and three tandem C-terminal WH2 domains. COBL was first identified in a gene trap study in mouse as a novel gene expressed along the midline during embryogenesis [Gasca et al., 1995]. Subsequently, it was shown that COBL functions as an actin nucleator, critical for neurite outgrowth [Ahuja et al., 2007]. In zebrafish, COBL is important for the proper development of the Kupffer’s vesicle [Ravanelli and Klingensmith, 2011], and necessary to form functional ciliated sensory hair cells [Schuler et al., 2013]. More recently, COBL was shown to be involved in the assembly of F-actin bundles that support microvilli on the surface of intestinal and placental epithelial cells [Wayt and Bretscher, 2014; Grega-Larson et al., 2015]. In these contexts, COBL targets near the base of microvillar actin bundles, where the pointed ends of actin filaments reside [Hirokawa et al., 1982]. Intestinal epithelial cells lacking COBL are deficient in brush border assembly, whereas overexpression of this factor drives microvillar growth and expansion of the brush border [Grega-Larson et al., 2015].
Numerous studies indicate that COBL is a multifunctional regulator of actin dynamics. Based on the spacing between its WH2 domains, COBL was initially proposed to stabilize a short pitch trimeric actin nucleus from which spontaneous filament assembly can occur [Ahuja et al., 2007]. However, a subsequent study using purified fragments of COBL showed that only one WH2 domain was necessary for nucleation [Husson et al., 2011]. This study also revealed that COBL constructs with at least two WH2 domains exhibit filament severing activity and sequestered ADP-actin in pyrene-actin polymerization assays. In terms of mechanisms underpinning these activities, a crystal structure of the first two WH2 domains of COBL in complex with two actin monomers suggests that a COBL-actin nucleus initially grows slowly in the pointed-end direction because barbed-end growth is blocked [Chen et al., 2013]. ATP hydrolysis of bound actin monomers is proposed to induce a conformational change that in turn allows for rapid barbed-end elongation [Chen et al., 2013]. Thus, if COBL does nucleate actin filaments in cells, it is expected to enrich near the pointed-ends of the resulting actin filaments. This prediction is consistent with the studies in epithelial cells alluded to above [Wayt and Bretscher, 2014; Grega-Larson et al., 2015].
Whereas COBL activities that impact actin dynamics are localized to its C-terminal WH2 domains, the N-terminal proline rich domain has been implicated in protein/protein interactions and subcellular targeting. COBL polyproline motifs bind the SH3 domain of the F-BAR protein syndapin, which plays a role in targeting COBL to the membrane in neurons and intestinal epithelial cells [Schwintzer et al., 2011; Grega-Larson et al., 2015]. Additionally, COBL contains numerous calmodulin binding sites, calcium and calmodulin regulate COBL actin binding, and the effect of COBL on dendritogenesis is dependent on its interaction with calmodulin [Hou et al., 2015].
How COBL contributes to cytoskeletal remodeling in cells remains unclear, in part due to a lack of live cell imaging data. To determine how COBL impacts the dynamics and architecture of the actin cytoskeleton in live cells, we examined the impact of expressing fluorescent protein-tagged COBL in cultured B16F1 mouse melanoma cells. We found that COBL expression induces the formation of dynamic linear actin structures, which associate with COBL-enriched puncta at their pointed ends. Stabilization of these features with the actin bundling protein, espin, allows for the formation of long-lived, self-supporting plasma membrane protrusions that resemble microvilli. Our results reveal that WH2 domain actin regulatory factors such as COBL hold the potential to drive assembly of F-actin-supported finger-like protrusions. These findings also provide insight on how epithelial cells create and maintain the elaborate cellular morphologies that are required for physiological function.
RESULTS
COBL induces formation of dynamic linear actin structures in melanoma cells
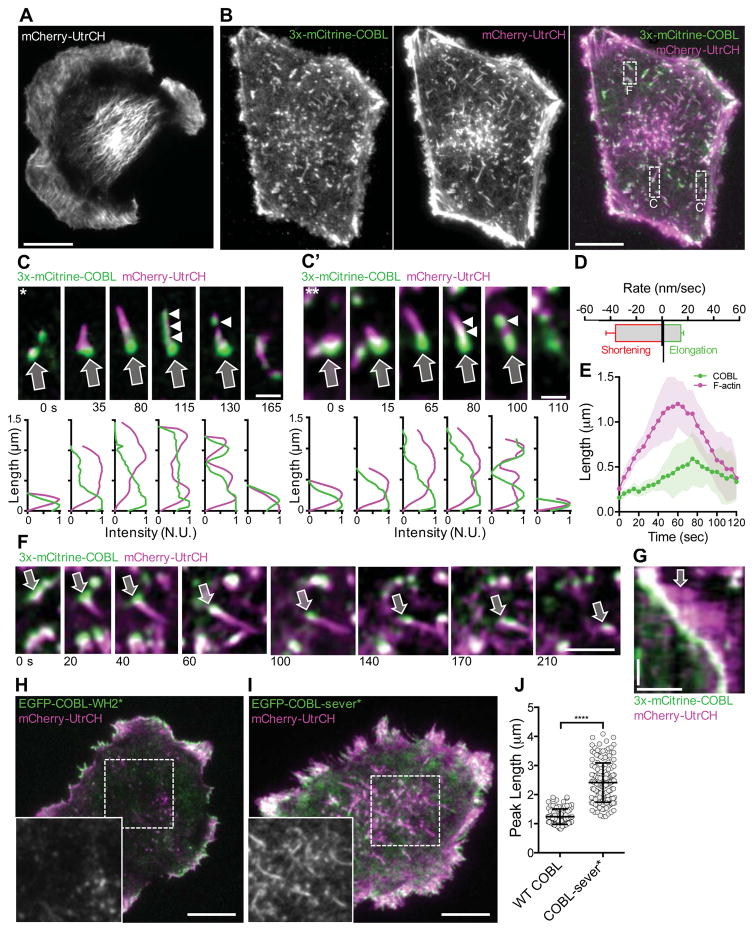
Localization analysis, along with overexpression and KD studies, suggest that COBL is necessary and sufficient to drive microvillar growth on the apical surface of intestinal epithelial cells [Grega-Larson et al., 2015]. If COBL is a key factor in microvillar assembly, we hypothesized that its expression in cells that do not normally make these protrusions would lead to the production of microvillus-like features. To test this hypothesis, we transfected B16F1 mouse melanoma cells with 3x-mCitrine-tagged COBL and mCherry-UtrCH [Burkel et al., 2007], and examined cells using near-TIRF microscopy (Figure 1). The 3x-mCitrine-tag enabled us to visualize low levels of COBL and avoid confounding dominant negative effects that can be associated with strong over-expression. B16F1 cells have been used in numerous previous studies on actin cytoskeleton dynamics [Svitkina et al., 2003; Mejillano et al., 2004; Vignjevic et al., 2006; Schober et al., 2007; Starke et al., 2014] and lack appreciable levels of endogenous COBL (Supplemental Figure 1A). These cells assemble a broad range of actin-rich networks including stress fibers, filopodia, and lamellipodia (Figure 1A, Supplemental Figure 1B). Expression of 3x-mCitrine-COBL had little impact on the number of finger-like protrusions. However, we did note the aberrant formation of F-actin-rich structures throughout the cytoplasm (Figure 1B; Movie S1). Due to their high labeling intensity, we reasoned that these were composed of numerous actin filaments. Control experiments revealed that an EGFP-tagged variant of COBL produced an identical effect (Supplemental Figure 2A). One end of each linear structure was capped by a prominent COBL punctum (Figure 1C, C’). COBL-induced structures were highly dynamic (Figure 1C, C’; Movie S2), growing (14.6 ± 1.5 nm/s) and shrinking (36 ± 5.3 nm/s) over the course of minutes (Figure 1D), with a mean lifetime of 162 ± 20 s. Interestingly, COBL appeared to accumulate along the length of the structure during the time leading up to disassembly (Figure 1C, C’, E). Such oscillatory dynamics are consistent with the unique behavior exhibited by COBL in pyrene-actin assembly assays [Husson et al., 2011].
Figure 1. COBL expression induces the formation of linear cytosolic actin structures in B16F1 cells.
Live near-TIRF imaging of a B16F1 cell expressing mCherry-UtrCH alone (A) or 3x-mCitrine-COBL (green) and mCherry-UtrCH (magenta) (B). Scale bar 5 μm. (C & C’) Montages reveal the dynamics of single linear actin structures that form in the dashed boxes in merge image from A. Arrows highlight the initial COBL puncta, arrowheads highlight COBL signal along the structure at later time points. Line scans are shown below each image. Scale bar 1 μm. (D) Rates of assembly and disassembly measured for intracellular actin structures induced by COBL overexpression. n = 42 structures from 10 cells. (E) Graph depicting the ensemble averages of the length of COBL-induced structure (magenta) and the length of the structure occupied by COBL (green) relative to time for ten individual structures ~110–115 sec in duration. Shaded area represents the standard deviation at each time point (F) Montage of a single linear actin structure from the boxed region in (A) that grows and moves through the cytosol before disassembly. Arrow highlights the COBL-associated end of the structure. Scale bar 2 μm. (G) Kymograph of actin structure in (F). Scale bars are 2 μm (horizontal) and 60 seconds (vertical). (H) Near-TIRF image of a B16F1 cell co-expressing EGFP-COBL-WH2* and mCherry-UtrCH. Zoom in lower left-hand corner is of the mCherry-UtrCH channel alone from boxed region. Scale bar is 5 μm. (I) Near-TIRF image of a B16F1 cell co-expressing EGFP-COBL-sever* and mCherry-UtrCH. Zoom in lower left-hand corner is of the mCherry-UtrCH channel alone from boxed region. Scale bar is 5 μm. (J) Graph of peak linear structure length in COBL expressing cells vs. COBL-sever* expressing cells.
A subset of COBL-induced structures appeared to glide through the cytosol (Figure 1F, G; Movie S3). In these cases, actin grew from a COBL punctum to a steady state length. Fiduciary marks in kymographs revealed that after initial elongation, the COBL-associated end of the structure shortened whereas the opposite end continued growing, producing an effective translocation of the structure through the cytosol. After a period of time, the COBL-associated end began to shorten faster than growth at the opposite end, leading to complete disassembly of the structure. The observed linear F-actin structures did not form in cells expressing a COBL construct with WH2 domain mutations that abolish actin binding [Chen et al., 2013], although this mutant still appeared to co-localize with F-actin in small puncta (EGFP-COBL-WH2*; Figure 1H; Movie S4). Additionally, the peak length of these structures increased by almost two fold when cells were expressing EGFP-COBL-sever*, a COBL mutant predicted to exhibit normal nucleation but defective severing activity (1.24 ± 0.27 vs. 2.41 ± 0.68; Figure 1I, J; Movie S5) [Jiao et al., 2014]. Together, these data suggest that COBL can induce the formation of short-lived linear actin structures, that functional WH2 domains are required for this activity, and that the lengths of these structures are limited by COBL severing activity.
COBL localizes to the pointed ends of dynamic linear actin structures
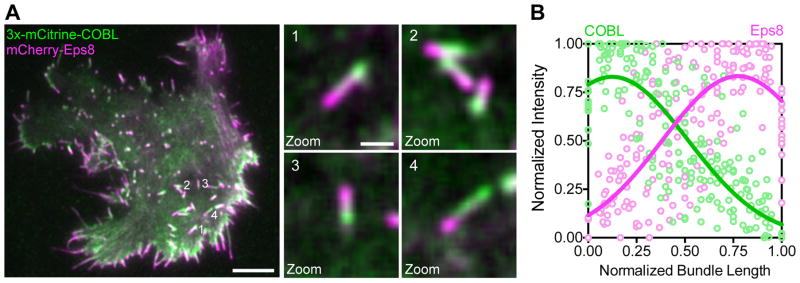
We sought to determine the polarity of COBL-induced linear actin structures. To differentiate between barbed- and pointed-ends, we co-expressed COBL with Eps8, which is known to target to the tips of actin bundles that support microvilli, stereocilia, and filopodia, where barbed ends are located [Croce et al., 2004; Disanza et al., 2006]. As expected for a WH2 domain-containing nucleator [Dominguez, 2009], the COBL-associated end of the structure was identified as the pointed-end based on the fact that Eps8 was enriched on the opposite end (Figure 2A,B; Movie S6). This is also consistent with data from epithelial cells where COBL localizes near the pointed-ends of microvillar actin bundles [Wayt and Bretscher, 2014; Grega-Larson et al., 2015].
Figure 2. COBL localizes to the pointed ends of assembling linear actin structures.
(A) Near-TIRF image of a B16F1 cell co-expressing 3x-mCitrine COBL and mCherry-Eps8. Zoom panels highlight intracellular structures with COBL and Eps8 on opposite sides. Numbers to the right of the structures in the first image correspond to the zoom numbers. Scale bar is 5 μm for left image and 1 μm for zoom images. (B) Line scans (n = 15 scans from 8 cells) along linear intracellular actin structures show the distribution of 3x-mCitrine-COBL (green) and mCherry-Eps8 (magenta) from cells co-expressing 3x-mCitrine-COBL and mCherry-Eps8.
Co-expression of COBL and espin induces robust formation of finger-like protrusions
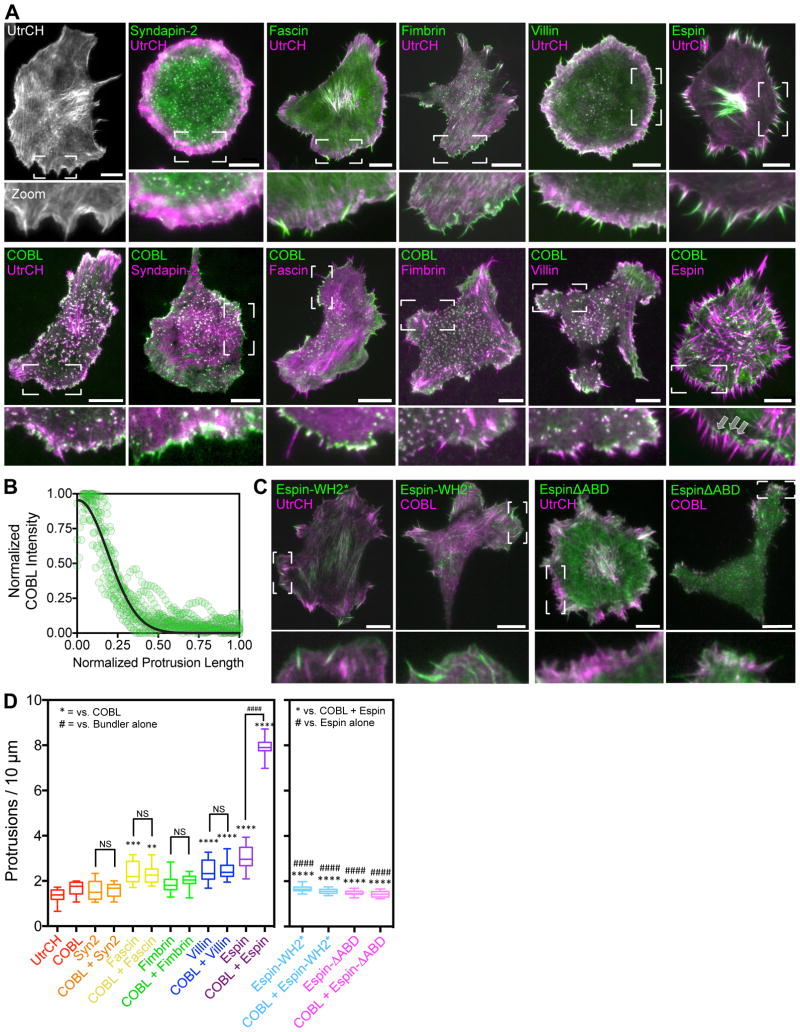
Although these results indicate that full length COBL can promote the assembly of linear actin structures, these features fail to protrude from the surface of B16F1 cells (Figure 1; Figure 3A). One reason could be that COBL/actin complexes are poorly anchored at the plasma membrane. Because syndapin-2 (also known as PACSIN2) has been shown to target COBL to sites of protrusion formation in epithelial cells [Grega-Larson et al., 2015], we reasoned that co-expression of COBL with syndapin-2 should promote anchoring of COBL-induced F-actin structures at the cell membrane. While we did observe prominent co-localization between these probes when co-expressed, we did not find a significant effect on protrusion per se; protrusion number was unchanged in co-expressing cells (Figure 3A,D). Thus, anchoring COBL at the membrane with syndapin-2 is not sufficient to generate finger-like protrusions in B16F1 cells.
Figure 3. Espin stabilizes COBL-induced linear actin structures.
(A) Near-TIRF images of B16F1 cells expressing the constructs indicated, zoomed images highlight the morphology of the cell edge. All scale bars are 10 μm. Arrows in COBL/Espin zoom panel show COBL puncta at the base of peripheral protrusions, where the pointed-ends of actin filaments are expected to reside. (B) Line scans (n = 15 scans from 7 cells) along protrusions of cells co-expressing COBL and espin showing the distribution of 3x-mCitrine-COBL. (C) Neaer-TIRF images of B16F1 cells expressing the constructs indicated, zoomed images highlight the morphology of the cell edge. All scale bars are 10 μm. (D) Number of protrusions per 10 microns perimeter for cells expressing the constructs indicated. ANOVA with Tukey’s multiple comparison’s test was used for comparisons and to determine significance. (#### & **** p<0.0001). For both graphs, the boxes represent the 25th and 75th percentiles around the median, and the whiskers represent the 5th and 95th percentiles. n = 15 cells/condition.
We next explored the possibility that the rapid turnover dynamics and short lifetime of COBL-induced actin structures (162 ± 20 s) prevented their protrusion. To this end, we co-expressed 3x-mCitrine-COBL with factors that stabilize microvillar actin bundles including espin [Bartles et al., 1998], villin [Bretscher and Weber, 1979], and fimbrin [Bretscher and Weber, 1980] (Figure 3A). Interestingly, only co-expression of COBL with espin generated a significant increase in the number of protrusions as compared to each bundling protein tested alone (Figure 3A,D). Protrusions in COBL/espin co-expressing cells were long-lived (874 ± 34 s in espin/COBL co-expressing cells vs. 162 ± 20 s with COBL alone), with many persisting for the full fifteen-minute duration of our time-lapse acquisitions. In COBL/espin co-expressing cells, we observed COBL puncta at the base of protrusions, where the pointed-ends of actin filaments are expected to reside (Figure 3A, COBL/espin; Figure 3B). Similar to villin and fimbrin, the actin bundling protein fascin, which is important for filopodial formation [Vignjevic et al., 2006], failed to increase protrusion number when co-expressed with COBL when compared to fascin alone (Figure 3A,D). Interestingly, COBL did not enrich on fascin-containing filopodia, suggesting that these factors segregate onto distinct populations of F-actin.
To investigate the domains of espin responsible for promoting robust protrusion when co-expressed with COBL, we generated two espin constructs, one with inactivating mutations in the WH2 domain (Espin-WH2*), and a second lacking the C-terminal actin bundling domain (EspinΔABD) (Figure 3C, D). Both of these constructs were unable to promote protrusion at levels comparable to wild-type espin, with protrusion numbers near control levels. Additionally, when these constructs were co-expressed with COBL, they were unable to induce the robust increase in protrusion formation that was observed with wild-type espin. Taken together, these data suggest that both the WH2 domain and C-terminal actin bundling domain of espin are required to synergize with COBL to drive the robust growth of finger-like protrusions.
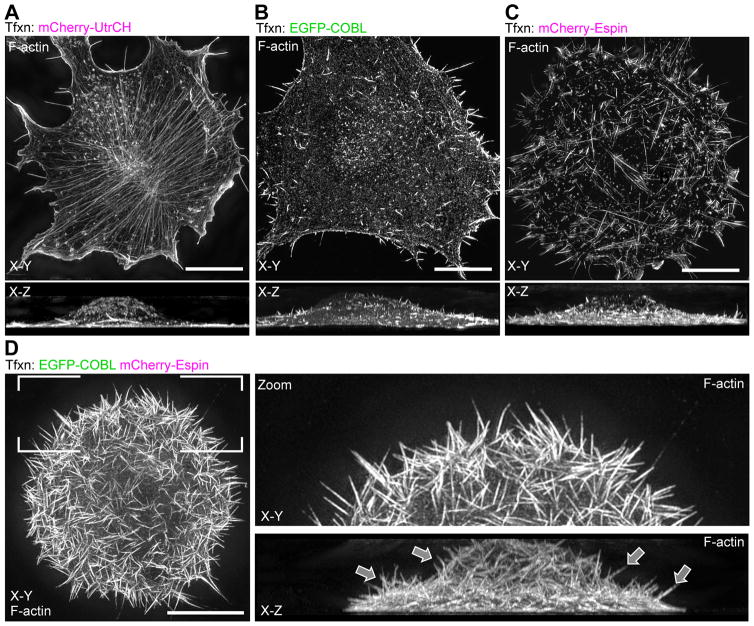
Because the near-TIRF imaging approach used here only provides a limited view of the ventral cell surface, we turned to super-resolution structured-illumination microscopy (SIM) to assess how COBL co-expression with bundling proteins impacts the architecture of the actin cytoskeleton on a whole cell level (Figure 4). Strikingly, SIM imaging of COBL/espin co-expressing cells revealed numerous self-supported protrusions extending from the dorsal surface (Figure 4D), structures that were largely absent from the surface of cells expressing either factor alone (Figure 4B, C). We also noted an apparent lack of ventral stress fibers in COBL/espin co-expressing cells, suggesting a significant re-allocation of actin into the induced surface protrusions. These data suggest that although COBL expression is capable of inducing linear actin based structures in both epithelial [Grega-Larson et al., 2015] and non-epithelial (Figure 1) cell types, COBL activity must be coordinated with bundlers such as espin to allow cells to assemble long-lived surface protrusions (Figure 3 & 4).
Figure 4. Espin and COBL co-expressing cells build microvillus-like protrusions.
SIM X–Y and X–Z projections of phalloidin-stained cells transfected with mCherry-UtrCH alone (A), EGFP-COBL alone (B), or mCherry-espin alone (C) or co-transfected with EGFP-COBL and mCherry-espin (D). All scale bars are 5 μm.
DISCUSSION
In this report, we show that COBL expression in B16F1 mouse melanoma cells drives the formation of dynamic linear actin structures in the cytoplasm (Figure 1). These structures fail to give rise to plasma membrane protrusions, but they do exhibit COBL-enrichment at their pointed-ends and undergo dramatic cycles of elongation and shortening over the course of minutes (Figure 1B–F; Movies S2 & 3). Co-expression of COBL with the actin bundling protein espin was sufficient to drive the formation of long-lived finger-like protrusions on the dorsal surface of the cell, free from any substrate attachment (Figures 3A, COBL/espin; 4D). Thus, by expressing only these two factors, a cell type that does not normally make microvilli is induced to form microvillus-like protrusions on its surface.
The cycles of elongation and shortening observed here for COBL-induced actin structures are consistent with previous in vitro studies, which demonstrated that COBL drives oscillatory kinetics in pyrene-actin assembly assays [Husson et al., 2011]. At a superficial level, the behavior of COBL-induced linear actin structures is reminiscent of the dynamic instability originally described for microtubules [Mitchison and Kirschner, 1984]. In this case, switching between elongation and shortening is controlled by subunit nucleotide hydrolysis and the concentration of free subunits available for incorporation into polymers. While these parameters may also impact the behavior of the COBL-induced structures we describe here, our live cell observations suggest some clear distinctions from classic dynamic instability. Most importantly, fiduciary marks in structure kymographs indicate that elongation is due to assembly at barbed-ends, whereas shortening is a consequence of disassembly at pointed-ends (COBL puncta-associated) or in some cases, severing along the length of the structure. Thus, the observed dynamics are more likely driven by a treadmilling mechanism, rather than classic dynamic instability, where assembly and disassembly take place at the same end of the polymer.
How does COBL contribute to the formation and subsequent dynamics of these linear actin structures? Given that COBL has been implicated as an actin nucleator [Ahuja et al., 2007; Husson et al., 2011; Chen et al., 2013], one straightforward scenario is that COBL puncta nucleate filaments that then elongate from their barbed ends. Here, COBL would remain behind at the pointed ends as filaments elongate, similar to what has been reported for filaments nucleated by APC [Okada et al., 2010; Breitsprecher et al., 2012]. Such localization is consistent with the fact that COBL targets to the pointed ends of microvillar actin bundles on the surface of polarized epithelial cells [Wayt and Bretscher, 2014; Grega-Larson et al., 2015]. Based on their strong labeling intensity, we suggest that COBL-induced structures are composed of numerous filaments. Given that Eps8, which enriches at the tips of the parallel actin bundles that support microvilli and stereocilia [Croce et al., 2004; Disanza et al., 2006], is also enriched at one end, we suspect these structures are parallel actin bundles, but confirmation of this point will require ultrastructural analysis. After elongating for over a minute, COBL appears to accumulate along the structure’s length, which is then followed by rapid shortening. Because expression of a COBL mutant deficient in severing (EGFP-COBL-sever*) [Jiao et al., 2014] leads to an increase in the length of these structures, COBL severing activity is likely involved in the disassembly phase. This proposal would be consistent with previous in vitro studies showing that COBL WH2 domains can bind to and sever ADP-actin [Husson et al., 2011].
Previous studies have shown that numerous cell types use syndapin family F-BAR proteins to target COBL to the plasma membrane [Schwintzer et al., 2011; Schuler et al., 2012; Grega-Larson et al., 2015]. Somewhat unexpectedly, our co-expression experiments revealed that formation of COBL/syndapin-2 complexes at the membrane is not sufficient for persistent protrusion formation. However, we did find that co-expressing COBL with the filament bundling protein espin led to a dramatic increase in peripheral protrusion number relative to cells expressing COBL or espin alone. Additionally, co-expression of these factors allowed B16F1 cells to assemble microvillus-like protrusions on their dorsal surface. In intestinal epithelial cells, which express endogenous espin, expression of COBL alone is sufficient to promote the robust growth of finger-like protrusions (in this case microvilli) [Grega-Larson et al., 2015]. The specificity of espin exerting this effect versus other bundling proteins in the current study is intriguing; espin is unique among actin bundlers in that it contains a WH2 domain in addition to its C-terminal actin-bundling domain [Loomis et al., 2006]. Interestingly, we found that an espin construct with inactivating mutations in the WH2 domain, was unable to increase protrusion number when expressed alone or co-expressed with COBL. Espin has been implicated in the early stages of stereocilia formation [Sekerkova et al., 2006], suggesting that this bundling protein is important for the initial assembly of actin-based protrusions. In line with our data, the actin bundling protein fimbrin has been shown to be important for maintenance but not formation of stereocilia [Taylor et al., 2015].
Whether COBL induces the formation of dynamic linear actin structures independently or in cooperation with other actin regulatory factors remains an open question. Other actin nucleators and elongation factors are known to synergize activities to form actin filaments. Indeed, the tandem WH2 domain nucleator spire cooperates with the formin cappuccino in actin meshwork assembly in developing Drosophila oocytes [Dahlgaard et al., 2007; Quinlan et al., 2007]. Moreover, the nucleation promoting factor Bud6 works with the formin Bni1 in the assembly of actin cables in budding yeast [Graziano et al., 2011]. Finally, actin nucleator APC and the formin mDia1 are known to cooperate in building the actin networks that drive normal directional cell motility [Okada et al., 2010; Breitsprecher et al., 2012]. Future studies are needed to determine if COBL cooperates with formins or other elongation factors when building cellular actin features.
Another outstanding question relates to the role of dimerization/oligomerization in the regulation of COBL activity. COBL contains a weak coiled coil motif, but how this domain contributes to COBL activity remains unclear. Previous studies showed that dimerization is critical for boosting the activity of other WH2 domain nucleators including VopL and spire [Namgoong et al., 2011]. Spire most likely dimerizes through its interaction with the formin cappuccino in Drosophila and formin-2 in mammals [Dahlgaard et al., 2007; Montaville et al., 2014]. Interestingly, while VopL and spire require dimerization for strong nucleation activity, COBL displays nucleation activity without dimerization [Ahuja et al., 2007; Husson et al., 2011]. This may be related to the fundamental mechanism of nucleation in these different cases, i.e. stabilization of long pitch (VopL and spire) vs. short pitch (COBL) actin nuclei [Namgoong et al., 2011]. Future investigations will need to determine if COBL dimerization/oligomerization is required for assembling the linear actin structures described here.
The data presented here provides insight on the mechanisms employed by COBL in the assembly of actin-based structures in cells. In concordance with in vitro kinetic studies of COBL fragments [Ahuja et al., 2007; Husson et al., 2011], our live cell imaging revealed the formation of dynamic linear actin structures, which emanate from COBL puncta. Based on findings presented here as well as results from previous studies [Grega-Larson et al., 2015], we propose that COBL/syndapin-2 complexes cooperate with actin bundling proteins such as espin, to drive the formation of persistent microvillus-like cell surface protrusions. Future studies using in vitro reconstitution approaches will be needed to test this concept directly.
METHODS
Cell culture and transfections
B16F1 cells (ATCC) were cultured in DMEM with high glucose and 2mM L-glutamine supplemented with 10% FBS. All cells were grown at 37°C and 5% CO2. One day before imaging, B16F1 cells were transfected in a T25 flask at 80% confluency using Lipofectamine 2000 as described by the manufacturer (Invitrogen, 11668). On the day of imaging, 35 mm glass bottom dishes (In Vitro Scientific, D35-20-1.5-N) were coated with 25 μg/ml laminin (Sigma, L2020) in PBS. B16F1 cells were plated sparsely on laminin-coated dishes (TIRF) or coverslips (SIM) and allowed to adhere for 2 hours before imaging (TIRF) or fixation (SIM).
Microscopy
Live cell imaging was performed on a Nikon TiE inverted light microscope equipped with 488 and 561 excitation LASERs, a 100x/1.49 NA TIRF objective, and Roper Evolve EM-CCD (Photometrics) and Andor Neo sCMOS detectors. Imaging was performed using near-TIRF illumination, where the incident angle of the LASER was adjusted to increase the depth of penetration of the excitation field, which we estimate to extend up to 400 nm into the cytoplasm. Cells were maintained in a humid environment at 37°C and 5% CO2 using a stage top incubation system (Tokai Hit). Images were acquired every 5 seconds for 15 minutes, and image acquisition was controlled with Nikon Elements software. For SIM, cells were first washed once with warm PBS and then fixed with 4% PFA in PBS for 15 minutes at 37°C. Following fixation, cells were washed three times with PBS and permeabilized with 0.1% Triton X-100/PBS for 15 minutes at room temperature (RT). Cells were then washed 3X with PBS and incubated with fluorescent phalloidin (Life Technologies) diluted 1:200 in PBS and incubated with cells at 37°C for 30 minutes. Cells were then washed 4X with PBS and mounted on slides in ProLong Gold (Life Technologies). SIM images were acquired using an Applied Precision DeltaVision OMX (GE Healthcare) equipped with a 60x Plan-Apochromat N/1.42 NA oil immersion objective (Olympus) located in the Vanderbilt Cell Imaging Shared Resource, and processed using softWorx software (GE Healthcare).
Plasmids and molecular biology
pENTR(tm)221 COBL (Invitrogen, IOH536) corresponding to full length human COBL was purchased and shuttled into pEGFP-C1 and pmCherry-C1 vectors (Clontech) adapted for Gateway cloning using the Gateway conversion kit (Invitrogen) to generate pEGFP- and pmCherry-COBL. PCR was used to generate a full length COBL fragment flanked by 5’ XhoI and 3’ BamHI restriction sites, which was subsequently ligated into linearized p3x-mCitrine-C1 (a kind gift from K. Verhey), producing p3x-mCitrine-COBL. To generate pEGFP-COBL-WH2*, mutations L1113A and L1166A were introduced to pEGFP-COBL using QuickChange site-directed mutagenesis (Agilent). QuickChange site-directed mutagenesis was also used to generate mutations L1153A, L1154A, I1157A and L1166A in the full length EGFP-COBL construct to generate EGFP-COBL-sever*. pmCherry-Eps8 (mouse) was purchased from Addgene (29779, deposited by C. Merrified) along with pmCherry-syndapin-2 (mouse) (27681, deposited by C. Merrifield) [Taylor et al., 2011] as well as EGFP- and mCherry-tagged variants of UtrCH (26737 and 26740, deposited by W. Bement) [Burkel et al., 2007]. Syndapin-2 was excised from pmCherry-syndapin-2 using 5’ EcoRI and 3’ BamHI and ligated into pEGFP-C1 to generate pEGFP-syndapin-2. A full length fimbrin construct was purchased from the mammalian gene collection (MGC, clone 33811). PCR was used to create a full length fimbrin fragment, which was then ligated into linearized pEGFP-C1 to generate pEGFP-C1-Fimbrin. A full length mouse villin-1 was construct was purchased from the MGC (clone 18506). PCR was used to generate a full length fragment which was then cloned into pCR8/GW/TOPO (Invitrogen) using TOPO cloning, and subsequently shuttled into pmCherry-C1 and pEGFP-C1 vectors adapted for Gateway cloning described above. pTYB12-fascin was a kind gift from Dr. Roberto Dominguez, and was used a PCR template for cloning into pCR8/GW/TOPO as described above, and shuttled into pmCherry-C1 adapted for Gateway cloning. The pmCherry-espin construct [Loomis et al., 2003] was a kind gift from Dr. James Bartles, Quickchange site-directed mutagenesis was used to make mutations L302A, L303A and L312A to generate Espin-WH2*. EspinΔABM (1-385 A.A.) was generated using PCR with pmCherry-espin used as the template. tdTomato-F-tractin was a kind gift from Dr. Alissa Weaver. mCherry-zyxin was a kind gift from Dr. Irina Kaverina. All newly generated plasmids were verified by sequencing.
Quantification and Image Analysis
For measurements number of protrusions per 10 microns, ANOVA with the Tukey’s multiple comparisons HSD test was used to determine significance. All graphs were generated and statistical analyses performed using Prism v.6 or v.7 (GraphPad). All image adjustments (contrast enhancement and smooth function) and analyses were performed using ImageJ (NIH). For line scans in Figure 1, intensities were normalized to the first image of each montage. Kymographs were generated by making a maximum intensity projection of the entire time series drawing a line along the resulting linear actin structure of interest and using the ‘Reslice’ function in ImageJ. For the graph in Figure 1E, total structure length and total COBL occupied structure length was measured for each time point that the structure was visible and plotted. To perform line scan analysis of linear actin structures, a line was drawn along the structure axis beginning on the side of COBL signal; the intensity of the COBL and Eps8 signal along that line was then recorded. Intensities from individual line scans were normalized such that the maximum value was equal to 1. The length axis from individual scans was also normalized such that the COBL-enriched side of the linear structure was set equal to 0 and the opposite side 1. Normalized line scans for COBL and eps8 were then plotted together and fit to a single Gaussian using non-linear regression (Prism v.6, GraphPad). Elongation and shortening rates were determined using the slopes of kymographs generated as stated above. The number of protrusions per 10 microns was calculated by first measuring the cell perimeter excluding protrusions and then counting the number of protrusions that the cell contained. The number of protrusions was then divided by the perimeter and multiplied by 10.
Supplementary Material
Acknowledgments
The authors thank all members of the Tyska laboratory, Vanderbilt Microtubule and Motors Club, and Vanderbilt Epithelial Biology Center for advice and support. Acquisition of the OMX super-resolution microscope was supported by 1S10-OD012324 (MJT). This work was also supported by VUMC CBMS training grant (NGL), American Heart Association Predoctoral Fellowship (NGL), American Heart Association Postdoctoral Fellowship (SWC), and National Institutes of Health grants R01-DK075555 and R01-DK095811 (MJT).
References
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131(2):337–50. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartles JR, Zheng L, Li A, Wierda A, Chen B. Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol. 1998;143(1):107–19. doi: 10.1083/jcb.143.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336(6085):1164–8. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Weber K. Villin: the major microfilament-associated protein of the intestinal microvillus. Proc Natl Acad Sci U S A. 1979;76(5):2321–5. doi: 10.1073/pnas.76.5.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Weber K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol. 1980;86(1):335–40. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64(11):822–32. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ni F, Tian X, Kondrashkina E, Wang Q, Ma J. Structural basis of actin filament nucleation by tandem W domains. Cell Rep. 2013;3(6):1910–20. doi: 10.1016/j.celrep.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce A, Cassata G, Disanza A, Gagliani MC, Tacchetti C, Malabarba MG, Carlier MF, Scita G, Baumeister R, Di Fiore PP. A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol. 2004;6(12):1173–9. doi: 10.1038/ncb1198. [DOI] [PubMed] [Google Scholar]
- Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13(4):539–53. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Mantoani S, Hertzog M, Gerboth S, Frittoli E, Steffen A, Berhoerster K, Kreienkamp HJ, Milanesi F, Di Fiore PP, et al. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol. 2006;8(12):1337–47. doi: 10.1038/ncb1502. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44(6):351–66. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. The WH2 Domain and Actin Nucleation: Necessary but Insufficient. Trends Biochem Sci. 2016;41(6):478–90. doi: 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasca S, Hill DP, Klingensmith J, Rossant J. Characterization of a gene trap insertion into a novel gene, cordon-bleu, expressed in axial structures of the gastrulating mouse embryo. Dev Genet. 1995;17(2):141–54. doi: 10.1002/dvg.1020170206. [DOI] [PubMed] [Google Scholar]
- Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22(21):4016–28. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grega-Larson NE, Crawley SW, Erwin AL, Tyska MJ. Cordon bleu promotes the assembly of brush border microvilli. Mol Biol Cell. 2015;26(21):3803–15. doi: 10.1091/mbc.E15-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Tilney LG, Fujiwara K, Heuser JE. Organization of actin, myosin, and intermediate filaments in the brush border of intestinal epithelial cells. J Cell Biol. 1982;94(2):425–43. doi: 10.1083/jcb.94.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Izadi M, Nemitz S, Haag N, Kessels MM, Qualmann B. The Actin Nucleator Cobl Is Controlled by Calcium and Calmodulin. PLoS Biol. 2015;13(9):e1002233. doi: 10.1371/journal.pbio.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson C, Renault L, Didry D, Pantaloni D, Carlier MF. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol Cell. 2011;43(3):464–77. doi: 10.1016/j.molcel.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Walker M, Trinick J, Pernier J, Montaville P, Carlier MF. Mutagenetic and electron microscopy analysis of actin filament severing by Cordon-Bleu, a WH2 domain protein. Cytoskeleton (Hoboken) 2014;71(3):170–83. doi: 10.1002/cm.21161. [DOI] [PubMed] [Google Scholar]
- Lee SH, Dominguez R. Regulation of actin cytoskeleton dynamics in cells. Mol Cells. 2010;29(4):311–25. doi: 10.1007/s10059-010-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis PA, Kelly AE, Zheng L, Changyaleket B, Sekerkova G, Mugnaini E, Ferreira A, Mullins RD, Bartles JR. Targeted wild-type and jerker espins reveal a novel, WH2-domain-dependent way to make actin bundles in cells. J Cell Sci. 2006;119(Pt 8):1655–65. doi: 10.1242/jcs.02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis PA, Zheng L, Sekerkova G, Changyaleket B, Mugnaini E, Bartles JR. Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J Cell Biol. 2003;163(5):1045–55. doi: 10.1083/jcb.200309093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118(3):363–73. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312(5991):237–42. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Montaville P, Jegou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier MF. Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biol. 2014;12(2):e1001795. doi: 10.1371/journal.pbio.1001795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S, Boczkowska M, Glista MJ, Winkelman JD, Rebowski G, Kovar DR, Dominguez R. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct Mol Biol. 2011;18(9):1060–7. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189(7):1087–96. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179(1):117–28. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanelli AM, Klingensmith J. The actin nucleator Cordon-bleu is required for development of motile cilia in zebrafish. Dev Biol. 2011;350(1):101–11. doi: 10.1016/j.ydbio.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober JM, Komarova YA, Chaga OY, Akhmanova A, Borisy GG. Microtubule-targeting-dependent reorganization of filopodia. J Cell Sci. 2007;120(Pt 7):1235–44. doi: 10.1242/jcs.003913. [DOI] [PubMed] [Google Scholar]
- Schuler S, Hauptmann J, Perner B, Kessels MM, Englert C, Qualmann B. Ciliated sensory hair cell formation and function require the F-BAR protein syndapin I and the WH2 domain-based actin nucleator. Cobl J Cell Sci. 2012 doi: 10.1242/jcs.111674. [DOI] [PubMed] [Google Scholar]
- Schuler S, Hauptmann J, Perner B, Kessels MM, Englert C, Qualmann B. Ciliated sensory hair cell formation and function require the F-BAR protein syndapin I and the WH2 domain-based actin nucleator Cobl. J Cell Sci. 2013;126(Pt 1):196–208. doi: 10.1242/jcs.111674. [DOI] [PubMed] [Google Scholar]
- Schwintzer L, Koch N, Ahuja R, Grimm J, Kessels MM, Qualmann B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011;30(15):3147–59. doi: 10.1038/emboj.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekerkova G, Zheng L, Mugnaini E, Bartles JR. Differential expression of espin isoforms during epithelial morphogenesis, stereociliogenesis and postnatal maturation in the developing inner ear. Dev Biol. 2006;291(1):83–95. doi: 10.1016/j.ydbio.2005.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke J, Wehrle-Haller B, Friedl P. Plasticity of the actin cytoskeleton in response to extracellular matrix nanostructure and dimensionality. Biochem Soc Trans. 2014;42(5):1356–66. doi: 10.1042/BST20140139. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160(3):409–21. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9(3):e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R, Bullen A, Johnson SL, Grimm-Gunter EM, Rivero F, Marcotti W, Forge A, Daudet N. Absence of plastin 1 causes abnormal maintenance of hair cell stereocilia and a moderate form of hearing loss in mice. Hum Mol Genet. 2015;24(1):37–49. doi: 10.1093/hmg/ddu417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174(6):863–75. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayt J, Bretscher A. Cordon Bleu serves as a platform at the basal region of microvilli, where it regulates microvillar length through its WH2 domains. Mol Biol Cell. 2014;25(18):2817–27. doi: 10.1091/mbc.E14-06-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.