Abstract
Astrocytes are a morphologically and functionally heterogeneous population of cells that play critical roles in neurodevelopment and in the regulation of central nervous system homeostasis. Studies of human astrocytes have been hampered by the lack of specific molecular markers and by the difficulties associated with purifying and culturing astrocytes from adult human brains. Human neural progenitor cells (NPCs) with self-renewal and multipotent properties represent an appealing model system to gain insight into the developmental genetics and function of human astrocytes, but a comprehensive molecular characterization that confirms the validity of this cellular system is still missing. Here we used an unbiased transcriptomic analysis to characterize in vitro culture of human NPCs and to define the gene expression programs activated during the differentiation of these cells into astrocytes using FBS or the combination of CNTF and BMP4. Our results demonstrate that in vitro cultures of human NPCs isolated during the gliogenic phase of neurodevelopment mainly consist of radial glial cells (RGCs) and glia-restricted progenitor cells. In these cells the combination of CNTF and BMP4 activates the JAK/STAT and SMAD signaling cascades, leading to the inhibition of oligodendrocytes lineage commitment and activation of astrocytes differentiation. On the other hand FBS-derived astrocytes have properties of reactive astrocytes. Our work suggests that in vitro culture of human NPCs represents a valuable cellular system to study human disorders characterized by impairment of astrocytes development and function. Our datasets represent an important resource for researchers studying human astrocytes development and might set the basis for the discovery of novel human-specific astrocyte markers.
Keywords: Human astrocytes, radial glial cells, neural progenitor cells, gliogenesis, astrocyte differentiation, neurogenesis, neurodevelopment, RNAseq, transcriptomic
Graphical Abstract

In this study we isolated and cultured neural progenitor cells (NPCs) from human fetal brain, we differentiated NPCs into astrocyte using different protocols and utilized RNA sequencing to define the characteristics of the differentiated astrocytes. Our datasets is an important resource to study human astrocytes development, identify novel human-specific astrocyte markers and represent valuable tool for future studies of human disorders characterized by impairments in astrocytes.
INTRODUCTION
Astrocytes play a fundamental role in the development of the central nervous system (CNS) as well as in its maintenance in the adult organism. A myriad of functions have been attributed to astrocytes, among them is their ability to regulate the cerebral blood flow, contribute to the maintenance of the extracellular environment, control intercellular communication, and modulate synapse formation and plasticity (Freeman, 2010). During the development of the mammalian CNS, neuroepithelial cells of the neural tube differentiate into radial glial cells (RGCs), which maintain self-renewal and differentiation properties and are responsible for the sequential generation of neurons, astrocytes and oligodendrocytes. In human, at around 12 post conceptual weeks, neuronal production decreases and RGCs begin direct differentiating into astrocytes or start producing glia-restricted intermediate progenitor cells that can differentiate into astrocytes or oligodendrocytes precursor cells (OPCs) (for review see (Rowitch & Kriegstein, 2010)). Studies of astrocytes development have progressed mainly through the use of animal models and in-vitro systems based on non-human cells, however human astrocytes are far more complex and diverse, and their morphology is drastically different from rodent and primate astrocytes (Oberheim et al., 2009; Matyash & Kettenmann, 2010). In order to unravel the extraordinary complexity of the human CNS it is necessary to develop and better characterize new human-specific cellular systems. The use of primary cultures of adult human glial cells is an appealing approach but presents several serious limitations due to the difficulty of purifying and culturing adult human astrocytes and because these primary cultures are often contaminated with other cell types (Newcombe et al., 1988; Becher & Antel, 1996; Gibbons et al., 2007; Jana et al., 2007). Multipotent neural progenitor cells (NPCs) isolated from mammalian fetal CNS, expanded using EGF and FGF containing media have been shown to differentiate into neurons, astrocytes and oligodendrocytes depending on the developmental stage of the fetus and the usage of different culturing protocols (Johe et al., 1996). Even though NPCs have been proven to be useful cellular system to study different aspects of astrocytes function in both physiological and pathological conditions (Liu & Zhang, 2011), the usage of these cells as a model system to study human astrocytes has been hampered by the lack of well defined differentiation protocols and by the poor characterization of the derived cells (Conti & Cattaneo, 2010). The most commonly utilized differentiation procedure to differentiate progenitor cells into astrocytes consists in the use of media containing low percentage of serum (Winkler et al., 1998; Martinez et al., 2012). Nevertheless, exposure to single factors like ciliary neurotrophic factor (CNTF) (Johe et al., 1996; Rajan & McKay, 1998) or bone morphogenic proteins (BMPs) (Gross et al., 1996; Nakashima et al., 2001) have been proven to initiate astrocytic commitment and differentiation of progenitor cells (Haas et al., 2012). Whether these distinctive differentiation paradigms generate astroglial cells with similar or different morphological, functional and molecular properties have not been deeply investigated. Moreover a complete picture of the gene expression programs activated during the differentiation of human progenitor cells into astrocytes is still missing and it would be necessary to better understand human astrocytes development and to lay the groundwork for the use of these cells as a model to study the role of human astrocytes in neurodevelopmental disorders (Molofsky et al., 2012).
MATERIALS AND METHODS
Isolation and culture of human NPCs
NPCs were isolated from five human fetal brains collected from second trimester-aborted fetuses. We received fetal specimens from the Birth Defects Research Laboratory at the University of Washington in Seattle, through a tissue distribution program supported by the National Institutes of Healt (NIH). The Birth Defects Research Laboratory obtained appropriate written informed consent from the parents and the procurement of tissues was monitored by the Institutional Review Board of the University of Washington. All the work was performed with approval by the Human Subject Research Office at the University of Miami. NPCs were cultured as previously described (Magistri et al., 2015). Briefly, fetal brains were mechanically dissociated into single cells and seeded in 75-mm tissue culture flasks in Neurobasal Media supplemented with EGF (20ng/ml), FGF (10ng/ml), B27, Glutamax and Heparin (2μg/ml). After 7–10 days of culture, NSCs cells form neurospheres colonies that can be cultured in suspension for several months. Before any experiments neurospheres underwent at least two passages in order to reduce any possible neuronal contamination. To induce differentiation, neurospheres were disaggregated into single cells using accutase and plated for 1 week in 6-well plates previously coated with poly-L-Ornythine (PLO) for 3 hours and laminin overnight. The differentiation media used consisted of DMEM/F12 supplemented with N2 and Glutamax and containing either 2.5% FBS or CNTF (20ng/ml) and BMP4 (10ng/ml).
RNA extraction and library preparation
RNA was extracted and purified using a combination of TRIzol reagent and QUIAGEN RNeasy columns as previously described (Velmeshev et al., 2013). RNA extraction was performed from each NPCs line at the time cells were seeded for differentiation and after 1 week of culture in the differentiation media (FBS or CNTF/BMP4). 1μg of RNA was used to prepare poly-A selected directional libraries using the NEBNext Ultra Directional RNA Library Prep Kit (New England BioLabs, USA) and libraries were sequenced utilizing the Illumina Hiseq2000 platform at the sequencing core of the University of Miami.
RNAseq data analysis
RNAseq analysis was performed utilizing CANEapp, an application for comprehensive analysis of RNA seq data (Velmeshev et al., 2016). CANEapp is a freely available Java application generated at the University of Miami that allows a completely automated analysis of RNAseq data for the detection of differentially expressed genes (http://psychiatry.med.miami.edu/research/laboratory-of-translational-rna-genomics/CANE-app). In more detail, paired-ends reads generated from sequencing on the Illumina Hiseq2000 were trimmed off the adaptor sequences using a custom script and were aligned to the human genome reference GRCh37 using TopHat 2.0.9. Transcriptome for each sample was assembled using Cufflinks, which also provided transcripts abundance in fragments per kilobase of exon per million fragments mapped (FPKM), and transcripts assemblies were merged using Cuffmerge. Differential gene expression analysis was performed using Cuffdiff and genes having FDR < 0.05 were further filtered based on their expression level (FPKM > 5 in at least one of the group: NPCs or differentiated cells). Genes were annotated according to the ENSEMBL classification and genes that were not annotated as proteins coding genes, lincRNAs or antisense RNAs were filtered out. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE76122.
Gene Ontology (GO) Enrichment Analysis
Differentially expressed genes were used as the input list to perform GO enrichment analysis using PANTHER from the Gene Ontology Consortium. Statistical overrepresentation test analysis was run using default settings and the “GO biological process complete” as reference list.
Gene Set Enrichment Analysis
Over-representation analysis of reactive astrocytes genes was performed using the Gene Set Enrichment Analysis (GSEA) tool (Subramanian et al., 2005). GSEA was applied on the two lists of genes that undergo differential expression during the differentiation of NPCs into astrocytes using FBS and CNTF/BMP4 (Supplementary table 1). Genes were ranked based on their fold changes and the analysis was run in pre-ranked mode. We adopted a classic enrichment statistic, as it is the recommended approach for RNA sequencing data. A list of reactive astrocytes genes were obtained from the study published by Zamanian et al. (Zamanian et al., 2012) and was used to create a reactive-astrocytes gene set that was utilized in the analyses together with the GO biological processes’s gene sets (c5.bp.v5.0) retrieved from the Molecular Signatures Database (MSigDB).
Revere Transcriptase quantitative PCR (RT-qPCR)
1 μg of RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcroption Kit (ThermoFisher Scientific, USA). cDNA was diluted 1 to 5 and used as template for SYBR Green or TaqMan qPCR on the QuantStudio™ 6 Flex Real-Time PCR System. ACTIN (cat# 4310881E), GFAP (Hs00909233_m1 and mm01253033_m1), BLBP (Hs00361426_m1), VIM (Hs00958111_m1) and NESTIN (Hs04187831_g1) TaqMan assays were purchased from Life Technologies. Primers used for SYBR green RT-qPCR were designed spanning exon-exon junction using Primer3 v.0.4.0 (Supplementary table 3). For all RT-qPCR reactions we included three technical replicates. To compare the expression of genes between NPCs and differentiated cells we used Student’s t –test. To compare gene expression changes after spinal cord injury we used GraphPad prism software to perform ANOVA followed by Tukey post-hoc test. A p value of below 0.05 was considered as statistically significant.
Immunostaining
NPCs cells were plated on 8-well glass chamber slides (Millipore, PEZGS0816) coated with poly-L-Ornythine (PLO) for 3 hours and laminin overnight to allow cells to adhere. Cells were fixed with 4% formaldehyde for 10 min, permeabilized with 0.2% triton-X, and incubated for 1 h in blocking buffer containing 20% goat serum to prevent non-specific binding of primary antibodies. Cells were then incubated with primary antibody overnight at 4C, subsequently cells were washed 3 times with PBS and incubated with fluorescently labeled secondary antibodies for 2 h at room temperature. Antibodies used: mouse anti-GFAP (1:5000) (Millipore, MAB360), chicken anti-VIMENTIN (1:1000) (Millipore, AB5733), mouse anti-NESTIN (1:200) (Millipore, MAB5326), rabbit anti-BLBP (1:50) (Santa Cruz Biotech, sc-30088).
Western Blotting
Whole cells were lysed in 2.5% SDS, 250mM Tris-HCl, pH 7.4, at 95°C. Gel electrophoresis and immunoblotting were done as previously described (Yamanaka et al., 2015). Immunoblots were developed using primary antibodies directed to GFAP (Millipore, MAB360) and GAPDH (Santa Cruza Biotech, sc-32233) and HRP-conjugated secondary antibodies.
Spinal Cord Injury
All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Miami and conducted according to specifications of the National Institutes of Health as outlined in the Guide for the Care and Use of Laboratory Animals.
Twelve C57B/L6 female mice were purchased from Jackson Laboratories and the Mutant Mouse Resource and Research Center (MMMRC) at 7- to 9-weeks of age. Eight mice were anesthetized using ketamine/xylazine (100 mg/15 mg/kg i.p.) before receiving mid-thoracic (T8) spinal cord injuries as previously described (Soderblom et al., 2013), while the remaining 4 mice were used as control. Injury (75 kDynes) was performed using the Infinite Horizon impactor device (Precision Systems and Instrumentation). Injured mice received lactated Ringer’s solution, antibiotics (Baytril, 10 mg/kg), and analgesics (buprenorphine, 0.05 mg/kg) subcutaneously for the first week after surgery. All mice were first anaesthetized with isoflurane inhalation, then euthanized by cervical dislocation. Control mice and four injured mice were euthanized one-week post-surgery, while the remaining four injured mice were euthanized two-weeks post-surgery. The spinal cord was collected at the site of injury for RNA extraction and RT-qPCR gene expression analysis.
RESULTS
Evaluation of human fetal neural progenitor cells (NPCs) differentiation into astrocytes
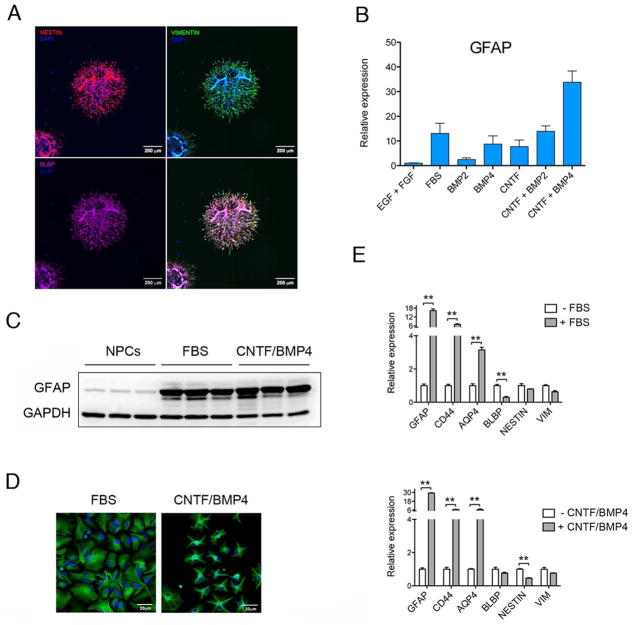
During development, neurons and glia are generated from multipotent neural progenitor cells (NPCs) derived from radial glial cells. Early in development during the so-called neurogenic phase, NPCs mainly produce neurons while later in development, during the gliogenic phase, radial glial cells can differentiate in glial progenitor cells or directly differentiate in glia cells (Kriegstein & Alvarez-Buylla, 2009). In order to study astrocyte differentiation we isolated NPCs from human fetal brains collected from second trimester-aborted fetuses, a time point during neurodevelopment that corresponds with the end of the neurogenic phase and the beginning of gliogenic phase. Immunostaining analysis of isolated NPCs revealed expression of the typical NPCs markers NESTIN, VIMENTIN and BLBP (FABP7) (Figure 1A), thus confirming the undifferentiated state of these cells. In order to differentiate NPCs into astroglial cells we tested several differentiation paradigms and used RT-qPCR to assess the expression of the well-established astrocytic marker glial fibrillary acid protein (GFAP) to compare the efficiency of the different culturing conditions in inducing astroglial differentiation. NPCs cultured in media containing 2.5% FBS had a 10-fold increase in the expression of GFAP, while exposure to the gliogenic cytokine CNTF (Bonni et al., 1997) or the bone morphogenic proteins BMP2 and BMP4 separately induced GFAP expression at a lower degree compared to FBS (Figure 1B). Since CNTF has been shown to act synergistically with BMP2 to induce astrocyte differentiation in rodents (Nakashima et al., 1999b), we decided to test the effects of CNTF/BMPs combinations. CNTF combined with BMP2 induced expression of GFAP at a level comparable with FBS, while the combination of CNTF with BMP4 resulted in even stronger (more than 30-fold) induction of this glial marker (Figure 1B). To validate our results at the protein level, we then used western blot analysis (Figure 1C) and immunohistochemistry (Figure 1D) to attain quantitative and qualitative estimate of GFAP expression in NPCs and cells differentiated with 2.5% FBS and CNTF/BMP4. As shown by western blot, GFAP was expressed, although at a low level, in the NPCs and increased after astroglial differentiation with both paradigms (Figure 1C). GFAP immunostaining revealed differences in the morphology of differentiated cells, where FBS-differentiated cells assumed an elongated fibroblast-like morphology, whereas cells differentiated with CNTF/BMP4 assumed a typical astrocyte-like stellate morphology (Figure 1D). Cells differentiated with CNTF/BMP4 resemble fibrous astrocytes of the white matter, which have a stellate morphology and typically express glial filaments that stain positive for GFAP.
Figure 1. Astroglial differentiation of human Neural Progenitor Cells (NPCs).
A) Immunostaining of human neurospheres stained with anti-NESTIN (red), anti-VIMENTIN (green) and anti-BLBP (magenta) antibodies. Nuclei were stained with DAPI (blue).
B) RT-qPCR analysis of GFAP expression changes in NPCs differentiated for 1 week using different culturing conditions. On the Y axe is depicted the expression of GFAP relative to the housekeeping gene β-ACTIN. GFAP expression level was normalized to NPCs (EGF + FGF).
C) Western blot analysis of GFAP protein expression in NPCs and in cells differentiated for 1 week in the presence of FBS and CNTF/BMP4. The expression of the housekeeping gene GAPDH was used to as loading control.
D) Immunostaining of NPCs differentiated for one 1 week in the presence of FBS and CNTF/BMP4. Cells were stained with anti-GFAP antibody (Green) and with DAPI (blue).
E) RT-qPCR analysis of astrocytes (GFAP, CD44 and AQP4) and progenitor cells (BLBP, NESTIN and VIM) specific-genes in NPCs differentiated for 1 week using FBS (upper panel) and CNTF/BMP5 (lower panel). On the Y axe is depicted the expression of the analyzed gene relative to the housekeeping gene β-ACTIN. Gene expression is normalized to non-differentiated NPCs (-FBS and –CNTF/BMP4).
Error bars are S.E.M.; **p<0.01.
In order to better describe the cells resulting from exposure to FBS and the combination of CNTF and BMP4, we utilized RT-qPCR to measure the expression of a panel of astroglial and NPCs markers. After one week of differentiation in the presence of FBS or CNTF/BMP4 we observed a significant increase in the expression of the astrocytic markers GFAP, AQP4 and CD44 (Figure 1E). In cells differentiated with FBS we noticed a decrease in the expression of the progenitor marker BLBP but not significant changes in NESTIN and VIMENTIN. On the other hand, cells differentiated with CNTF/BMP4 showed a decrease in NESTIN but not in BLBP and VIMENTIN. These data suggest that NPCs isolated from fetal brain differentiate into astroglial cells when exposed to both FBS and CNTF/BMP4, but that the degree of differentiation and the characteristics of these cells might depend on the differentiation paradigm utilized.
RNAseq analysis of NPCs differentiation into astrocytes
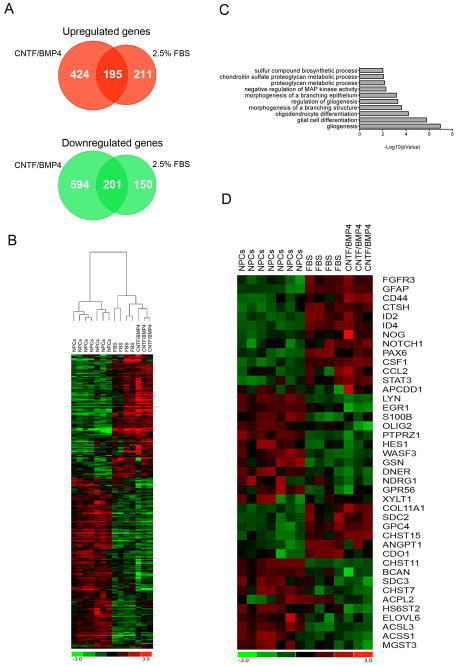
In order to reveal the dynamic changes of gene expression programs activated in NPCs during astrocyte differentiation, we decided to take advantage of next generation sequencing and perform a transcriptomic analysis of NPCs before and after differentiation in the presence of FBS and CNTF/BMP4. From now on we will refer to differentiated cells as FBS-astrocytes and CNTF/BMP4-astrocytes according to the differentiation method utilized. NPCs isolated from 4 different donors (91, 103, 110 and 114 days embryos) were differentiated for 1 week using 2.5% FBS, while 3 NPCs lines (two from 103 and one from 110 days embryo) were differentiated for 1 week in the presence of CNTF/BMP4. RNA was extracted from NPCs before and after differentiation and submitted for sequencing on the Illumina HiSeq 2000 platform. Using our validated bioinformatics analysis pipeline (Magistri et al., 2015) we identified 757 genes to undergo differential expression (406 upregulated and 351 downregulated) in NPCs differentiated with FBS and 1414 genes to be differentially expressed (619 upregulated and 795 downregulated) when NPCs are differentiated in CNTF/BMP4-containing media (Figure 2A and Supplementary table 1). Hierarchical clustering using differentially expressed genes revealed that the two differentiation protocols activate a similar pattern of gene expression (Figure 2B). In support of this observation we identified 396 genes (195 upregulated and 201 downregulated) to be in common between the two lists of differentially expressed genes (Figure 2A), suggesting that these might represent the core set of genes responsible for driving astroglial differentiation process. Among these genes there were the known astrocytic markers GFAP, CD44 and AQP4, which we confirmed by RT-qPCR to be upregulated during the differentiation process (Figure 1E). Beside these markers, and among the most upregulated genes we found the four members of the inhibitor of differentiation (ID) family of helix-loop-helix transcriptional inhibitors, which have been previously implicated in astrocytic cell fate determination and differentiation (Ross et al., 2003; Samanta & Kessler, 2004). Among the most downregulated genes we found the transcription factors OLIG1 and OLIG2, which are crucial for oligodendrocytes lineage commitment (Ligon et al., 2006). These data confirm that NPCs cultured in both FBS and CNTF/BMP4 undergo astrocytic differentiation and that these two different culturing conditions induce the activation of a mutual transcriptional program leading to astrocyte differentiation.
Figure 2. Transcriptomic analysis of differentiated astrocytes.
A) Venn diagram depicting the number of genes that are differentially expressed in NPCs differentiated for one 1 week in the presence of FBS and CNTF/BMP4.
B) Heatmap showing log2(FPKM) values and hierarchical clustering analysis of differentially expressed genes in NPCs, FBS-astrocytes and CNTF/BMPB4-astrocytes.
C) Graphical representation of Gene Ontology (GO) enrichment analysis showing biological processes that are overrepresented in the 398 genes differentially expressed in both FBS- and CNTF/BMP4-astrocytes.
D) Heatmap showing log2(FPKM) values of genes differentially expressed with both culturing conditions (FBS and CNTF/BMP4) and belonging to the most enriched biological processes from GO enrichment analysis (Table1).
Differentiating cells undergo inhibition of oligodendrocytes lineage commitment and activation of astroglial differentiation
To attain a better picture of the common transcriptional program activated by FBS and CNTF/BMP4 we examined the mutual 396 differentially expressed genes using gene ontology (GO) enrichment analysis (http://pantherdb.org/). Gliogenesis and glia cell differentiation were the most significantly enriched biological processes thus confirming that this core set of genes is indeed enriched for genes playing important roles during astrocyte differentiation (Figure 2C, Figure 2D and Table 1). Among the upregulated genes belonging to these processes we found astrocytic markers (GFAP and CD44), astrocytes-associated inhibitors of DNA binding (ID2 and ID4) and several other genes know to play a role in astrocyte maturation. For example, FGFR3 is expressed by neuroepithelial precursor cells in the embryonic brain and spinal cord and is highly enriched in astrocytes compared to other neural cells in the postnatal brain (Cahoy et al., 2008; Young et al., 2010). NOTCH1 is among the upregulated genes, and notch signaling is known to inhibit maturation of neurons and oligodendrocytes from precursor cells while promoting astrocytes differentiation (Givogri et al., 2003; Louvi & Artavanis-Tsakonas, 2006). Among other upregulated genes were APCDD1, which is a negative regulator of Wnt signaling that promotes astrocyte precursor migration (Kang et al., 2012), the chemokine CCL2 that recruit CCR2-expressing monocytes into the CNS and is mainly expressed by astrocytes upon viral infection and TBI (Semple et al., 2010; Zaritsky et al., 2012), and the colony stimulating factor-1 (CSF1) which is expressed by astroglial cells during CNS development and following inflammatory response (Shafit-Zagardo et al., 1993; Alterman & Stanley, 1994). When we looked at the downregulated genes we noticed that the majority of these genes are normally expressed by oligodendrocytes, by they precursor cells (OPCs) or they are genes important for oligodendrocytes differentiation. Among these genes we found the oligodendrocyte lineage transcription factors 1 and 2 (OLIG1 and OLIG2) and the non-receptor tyrosine kinase LYN, which are important regulators of oligodendrocytes differentiation (Colognato et al., 2004; Ligon et al., 2006). We also noticed downregulation of EGR1 and S100β, two genes that are expressed at high level in undifferentiated oligodendrocyte precursor cells (OPCs) (Deloulme et al., 2004; Dugas et al., 2006). Another gene expressed in the OPCs and downregulated in our dataset was PTPRZ1, which codes for the receptor protein tyrosine phosphatase gamma. PTPRZ1 is expressed at the surface of OPCs and plays a dual role in inhibiting their proliferation and promoting their maturation (Lamprianou et al., 2011). The actin-binding protein gelsolin (GSN) is an early marker of oligodendrocytes (Lena et al., 1994; Vouyiouklis & Brophy, 1997) and was also downregulated upon differentiation of the NPCs. Another downregulated gene is the adhesion G protein-coupled receptor GPR56 that is abundantly expressed throughout oligodendrocytes development. Loss of GPR56 causes hypo-myelination in the CNS (Giera et al., 2015). Mutations in NDRG1, another downregulated gene, cause motor and sensory neuropathy characterized by severe peripheral demyelination (Tazir et al., 2013), while in the CNS NDRG1 is predominantly expressed by oligodendrocytes (Okuda et al., 2008). Beside glia cell differentiation we also noticed enrichment for the chondroitin sulfate proteoglycan (CSPG) metabolic process. CSPGs are indeed one of the main components of the extracellular matrix of the central nervous system (CNS) (Rowitch & Kriegstein, 2010) and upon injury CSPGs production is upregulated in astrocytes and results in the formation of a glia scar in the proximity of the lesion (McKeon et al., 1991). Expression of mature astrocyte’s markers ALDH1L1 and SLC1A2 did not significantly increase upon 1 week of differentiation with 2.5% FBS, nor CNTF/BMP4, suggesting that differentiated cells maintain an immature phenotype. Our data demonstrate that in NPCs, 1 week exposure to both FBS and CNTF/BMP4 triggers the concomitant inhibition of oligodendrocyte lineage commitment and activation of astrocyte differentiation but do not generate fully mature astrocytes.
Table 1.
Gene Ontology enrichment analysis of the core set of differentially expressed genes
| GO biological process | UPregulated Genes | DOWNregulated Genes |
|---|---|---|
| gliogenesis | FGFR3 GFAP ID2 NOTCH1 PAX6 CCL2 STAT3 APCDD1 ID4 FGFR3 GFAP ID2 | LYN EGR1 OLIG2 PTPRZ1 S100B HES1 WASF3 GSN DNER NDRG1 GPR56 |
| glial cell differentiation | NOTCH1 PAX6 STAT3 ID4 | LYN EGR1 OLIG2 PTPRZ1 S100B HES1 WASF3 GSN DNER NDRG1 |
| oligodendrocyte differentiation | FGFR3 ID2 NOTCH1 PAX6 ID4 | LYN EGR1 PTPRZ1 OLIG2 WASF3 GSN |
| morphogenesis of a branching structure | CSF1 NOG NOTCH1 SOCS3 CTSH ADAMTS16 CD44 | ETV4 SLC12A2 NPNT SFRP1 ADM CTNND2 CLIC4 SPRY1 NFATC4 SPRY2 |
| regulation of gliogenesis | CSF1 NOG FGFR3 ID2 GFAP NOTCH1 ID4 | LYN PTPRZ1 OLIG2 HES1 |
| morphogenesis of a branching epithelium | CSF1 NOG NOTCH1 SOCS3 CTSH ADAMTS16 CD44 | ETV4 SLC12A2 NPNT SFRP1 ADM CLIC4 SPRY1 NFATC4 SPRY2 |
| negative regulation of MAP kinase activity | LYN HMGCR DUSP4 SPRED2 SFRP1 DUSP6 SPRY4 SPRED1 SPRY4 SPRY1 SPRY2 | |
| proteoglycan metabolic process | COL11A1 XYLT1 SDC2 GPC4 CHST15 | CHST11 BCAN SDC3 CHST7 ACPL2 HS6ST2 |
| chondroitin sulfate proteoglycan metabolic process | XYLT1 SDC2 GPC4 CHST15 | CHST11 BCAN SDC3 CHST7 ACPL2 |
| sulfur compound biosynthetic process | XYLT1 ANGPT1 CHST15 CDO1 | ELOVL6 CHST11 BCAN ACSL3 CTH ACSS1 CHST7 MGST3 ACPL2 HS6ST2 |
Validation of RNAseq results by RT-qPCR
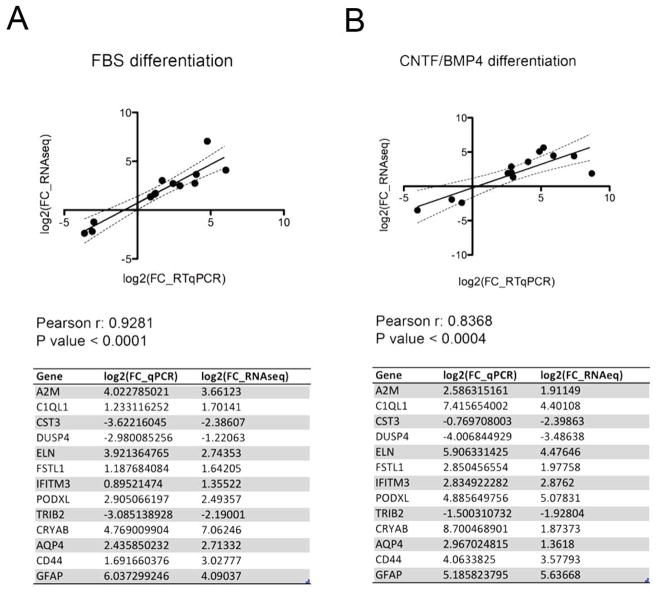
To confirm the accuracy of RNA-seq analysis, we selected a dozen of genes with different expression level and with a wide range of differential expression changes and performed RT-qPCR validation (Figure 3A and 3B). In this list of genes we included known astrocytic markers, as well as genes previously reported to play a role in CNS development and function. Alpha2 microglobulin (A2M) is a secreted protein produced by astrocytes with a neurite-promoting activity (Mori et al., 1990), and its expression increases in the brain of Alzheimer’s disease patients (Kovacs, 2000) and after SCI (Ramer et al., 2005). C1q-like protein 1 (C1QL1) is a synaptic molecules that is expressed by neurons and is involved in activity-dependent synapse plasticity and formation (Eroglu & Barres, 2010). Cystain C (CST3) is required for neural stem cell proliferation (Taupin et al., 2000). Dusp4 (Dual specificity phosphatase 4) promotes neurogenesis (Kim et al., 2015). Elastin (ELN) expression is stimulated by hydrostatic pressure in cultured optic nerve head astrocytes (Hernandez et al., 2000) and in astrocytes of the lamina cribrosa (Pena et al., 2001) Mutations in ELN are reported in patients with intellectual disability or learning problems. IFITM3 is an interferon-induced protein expressed in astrocytes following activation of the innate immune system and responsible for neuronal impairments (Ibi et al., 2013). CRYAB codes for a small heat shock protein, which is also one of the constituents of the crystalline lens. In the central nervous system CRYAB is mainly expressed by oligodendrocytes but its expression was reported to increases in other glial cells after injury (Piao et al., 2005; Klopstein et al., 2012). Podocalyxin (PODXL), a member of sialomucin family of protein, and Follistatin-like 1 (FSTL1) are both upregulated in glioblastoma (Reddy et al., 2008; Wu et al., 2013), while TRIB2 is a pseudokinase protein that has been associated with narcolepsy (Cvetkovic-Lopes et al., 2010). As shown in Figure 3A and 3B differential expression by RT-qPCR highly correlates with RNAseq data in both differentiation experiments, thus validating the accuracy of our analysis.
Figure 3. Quantitative RT-qPCR validation of RNAseq differential expression analysis.
Technical validation of RNAseq differential expression analysis of FBS (A) and CNTF/BMP4 (B) differentiation using RT-qPCR showing high degree of correlation between log2 fold change differences from RNAseq and RT-qPCR data for 13 different genes.
CNTF/BMP4- differs from FBS-astrocytes and represents bona fide astrocytes
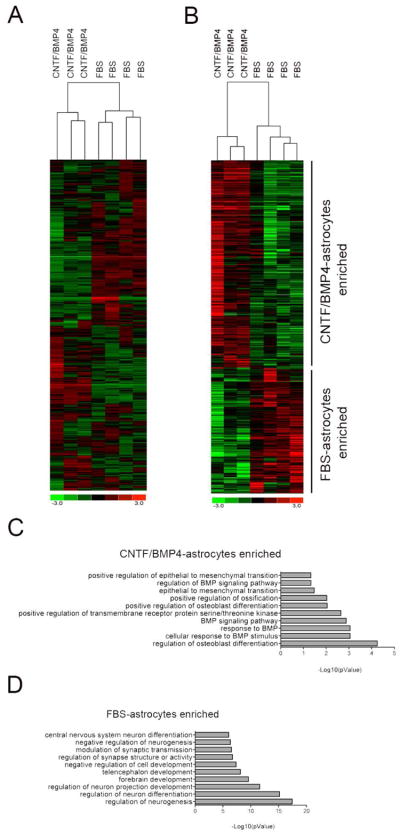
Beside the similarities, CNTF/BMP4-astrocytes differ from FBS-astrocytes as it is shown by unsupervised hierarchical clustering analysis performed on all the genes expressed by the two cell types (Figure 4A). To study these differences we decided to take a closer look at the genes that are upregulated exclusively during differentiation with CNTF/BMP4 or FBS. Gene ontology enrichment analysis of the genes upregulated solely in CNTF/BMP4-astrocytes showed enrichment for “regulation of glial cell differentiation” and “proteoglycan metabolic process” (Supplementary Table 2), suggesting that these cells may represent bona fide astrocytes. On the other hand the genes solely upregulated in FBS-astrocytes were enriched for genes involved in tissue development and extracellular matrix organization, including SPARC and CYR61 that are matricellular proteins known to be secreted by reactive astrocytes (Jones & Bouvier, 2014) (Supplementary Table 2). To better compare CNTF/BMP4-astrocytes and FBS-astrocytes we performed differential gene expression analysis using Cuffdiff. We found 988 protein-coding genes to be differentially expressed, 376 enriched in FBS-astrocytes and 612 enriched in CNTF/BMP4-astrocytes (Figure 5A and Supplementary Table 1). Gene ontology analysis of genes enriched in CNTF/BMP4-astrocytes revealed overrepresentation of genes involved in BMP signaling pathway (Figure 4C and Supplementary Table 2). Among the most upregulated (log2 fc > 1.5) and most abundantly expressed genes (FPKM > 30) we found several genes expressed by astrocytes in normal or pathological conditions, such as CD44 (Hernandez et al., 2000), PODXL (Wu et al., 2013), SPARCL1 (Kucukdereli et al., 2011), ELN (Hernandez et al., 2000), SOCS3 (Okada et al., 2006), CCL2 (Lo et al., 2014) and IGFBP3(Honda et al., 2011). Astrocytes play important roles in synapses formation, maturation and maintenance through the secretion of different proteins including glypicans (GPC4) and hevin (SPARCL1), and these two genes were enriched in CNTF/BMP4-astrocytes (Kucukdereli et al., 2011; Allen et al., 2012). On the other hand gene ontology analysis of FBS-astrocytes enriched genes showed a clear overrepresentation for genes involved in neurogenesis, neuronal differentiation and synapses formation and function (Figure 4D and Supplementary Table 2), pointing out to the presence of neuronal cells among the differentiated cells. Taken together these data demonstrate that exposure to CNTF/BMP4 primarily induces differentiation of NPCs into astrocytes, while low percentage of serum gives rise to a mixed population of neuronal and astroglial cells that express markers of reactive astrocytes (McCarthy et al., 2006).
Figure 4. Comparison of FBS-astrocytes and CNTF/BMP4-astrocytes.
A) Heatmap showing log2(FPKM) values and unsupervised hierarchical clustering analysis of all the genes expressed in FBS- and CNTF/BMP4-astrocytes.
B) Heatmap displaying log2(FPKM) and hierarchical clustering analysis of differentially expressed genes between FBS- and CNTF/BMP4 astrocytes. Graphical representation of Gene Ontology (GO) enrichment analysis showing biological processes that are overrepresented in FBS-astrocytes enriched genes (C) and CNTF/BMP4-astrocytes enriched genes (D).
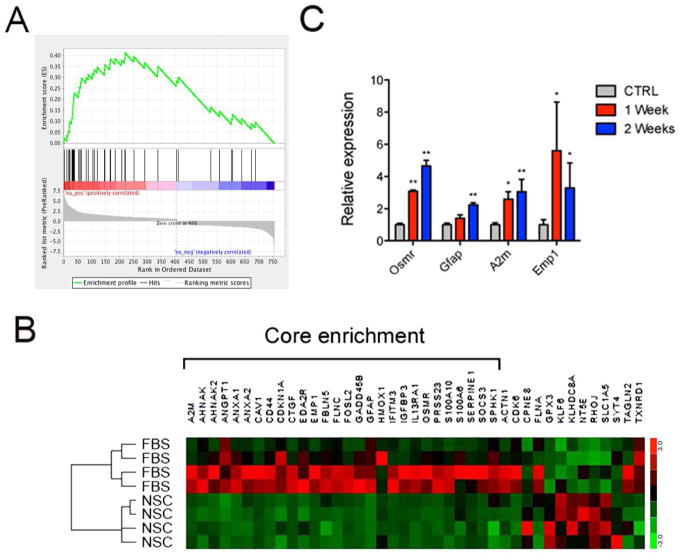
Figure 5. Enrichment of reactive astrocytes genes in FBS-astrocytes.
A) Gene set enrichment analysis (GSEA) performed in pre-ranked mode using differentially expressed genes in NPCs differentiated with FBS. A) Enrichment plot for reactive astrocytes genes signature. Genes related to reactive astrocytes genes signature most strongly associated with the FBS-astrocytes are represented on the far left.
B) Heatmap showing FPKM expression profile of the core enrichment of genes from GSEA.
C) RT-qPCR analysis of FBS-astrocytes-expressed reactive astrocytes genes in the spinal cord of mice control mice (CTRL) or mice after 1 or 2 weeks from injury. RNA was extracted from the site of injury of the spinal cord. On the Y axe is depicted the expression of the analyzed gene relative to the housekeeping gene β-Actin. Gene expression is normalized to CTRL. Error bars are S.E.M.; *p<0.05; **p<0.01.; n=4.
FBS-astrocytes are enriched for markers of reactive gliosis
The role of astrocytes is not only limited to normal development and homeostasis as they play an essential role during diseases and injury of the CNS by adopting a different functional state in the process of astrocyte reactivation. It has been shown that in vitro differentiated or cultured astrocytes undergo morphological and molecular changes that make cultured cells very different from mature in vivo astrocytes (Cahoy et al., 2008). In particular, in vitro cultured rodents astrocytes have been shown to express many reactive astrocyte genes (Zamanian et al., 2012). In our datasets we noticed that CNTF/BMP4- and FBS-astrocytes express immature and reactive astrocytes markers. For example CNTF/BMP4-astrocytes retain the expression of NESTIN and VIM, while FBS-astrocytes express BLBP and VIM (Figure 1E) and express matricellular proteins secreted by reactive astrocytes (Table 3). In order to verify whether there was an enrichment of reactive astrocytes genes among the genes expressed in our cultures of astroglial cells, we decided to compare genes upregulated during differentiation of CNTF/BMP4- and FBS-astrocytes with genes upregulated in reactive gliosis from the study published by Zamanian et al. (Zamanian et al., 2012). Gene set enrichment analysis (GSEA) showed a statistically significant enrichment for reactive astrocytes genes in FBS-astrocytes (FDR < 0.001) (Figure 5A), while there was not statistical significant enrichment in CNTF/BMP4-astrocytes (FDR 0.270). Among the genes forming the core enrichment of our analysis (6B) we selected few to monitor their expression changes during reactive gliosis after spinal cord injury in vivo. In order to induce astrocyte reactivation in vivo, we utilized a spinal cord injury paradigm where it is well established that reactive astrocytes create a scar around the site of injury preventing the spread of further tissue damage and at the same time prevent neuronal regeneration. C57BL6 mice received surgical injury to their spinal cord and mice were sacrificed one or two weeks after injury. Increased GFAP expression is a hallmark of reactive astrocytes and represents a well-studied phenomenon during spinal cord injury (Garrison et al., 1991; Yang & Wang, 2015). Alpha2 microglobulin (A2M) is a secreted protein produced by astrocytes that has a neurite-promoting activity (Mori et al., 1990), and its expression increases in Alzheimer’s disease brain (Kovacs, 2000). EMP1 is indispensable for blood-brain barrier tight junction formation and function and is involved in mechanisms of drug resistance (Bangsow et al., 2008). OSMR encodes for the oncostatin M (OSM) receptor, a member of the type I cytokine receptor family. In astrocytes OSM binds to OSMR to induce IL-6 expression (Van Wagoner et al., 2000). Both OSMR and OSM were reported to be upregulated after spinal cord injury at the site of lesion where they play protective role and promote recovery (Slaets et al., 2014). As shown in Figure 5C, these genes showed significant increase after spinal cord injury, thus confirming their reactive astroglia-specific expression profile. These data show that FBS-astrocytes compared to CNTF/BMP4 astrocytes are enriched for reactive astrocytes markers.
DISCUSSION
Radial glial cells (RGCs) and neural progenitors cells (NPCs) can be isolated from embryonic mammalian brain and cultured in vitro in the presence of mitogens as floating cellular aggregates called neurospheres (Buc-Caron, 1995; Bez et al., 2003). These cells retain the capability to differentiate in different neural lineages, thus they represent an appealing in vitro model system to study development and function of the CNS. In the developing mammalian CNS, asymmetric division of RGCs initially gives rise to neuronal progenitors that differentiate into neurons migrating along the RGCs processes. Later in development RGCs undergo a switch in their developmental program from neuron to glia cells production (Rowitch & Kriegstein, 2010). At the molecular level this switch is underlined by the concomitant repression of pro-neuronal proteins and the activation of pro-glia proteins. Our neurospheres derived from cells isolated during the gliogenic phase of neural development expressed the typical progenitor cells markers (BLBP, NESTIN, VIM and GFAP), confirming the undifferentiated state of these cells and therefore suggesting their capability to undergo in vitro differentiation. Gene expression profiling using RNAseq revealed that neurospheres expressed very low levels of well-established pro-neuronal proteins like the bHLH transcriptional regulators neurogenin 1 (NEUROG1), NEUROG2, NHLH1 and ATOH1 (Bertrand et al., 2002). At the same times these cells expressed high levels of members of the NOTCH signaling pathway (NOTCH1, DLL1 and JAG1) and of the pro-glia transcription factor SOX9, both of which play fundamental role during the switch from neuron to glia in the developing CNS (Stolt et al., 2003; Yoon et al., 2008; Namihira et al., 2009; Kang et al., 2012). Our transcriptomic data showed that neurospheres isolated during the gliogenic phase of CNS development consist mainly of a mixed population of glia-committed progenitor cells and RGCs.
The in vitro usage of embryonic NPCs has been an important model to study neuronal and glial differentiation and to understand the molecular bases underlying CNS development. In vivo studies indicate that gliogenesis initiates when RGCs and progenitor cells are exposed to neurons-secreted cytokines belonging to the interlueukin 6 (IL-6) family (Barnabe-Heider et al., 2005). Among these cytokines, the leukemia inhibitory factor (LIF) (Asano et al., 2009), the ciliary neurotrophic factor (CNTF) (Rajan & McKay, 1998) and cardiotrophin 1 (CT1) (Barnabe-Heider et al., 2005) have been shown to promote astroglia differentiation of NPCs through the activation of the JAK-STAT signaling pathway. Astrogenesis requires also the activation of the SMAD signaling cascade by bone morphogenic proteins (BMPs) that has been shown to converge with the JAK-STAT pathway and results in the activation of astrocyte-specific genes (Nakashima et al., 1999a). BMPs also repress NPCs differentiation into neurons and oligodendrocytes by controlling the expression of bHLH factors (Nakashima et al., 2001). In vitro, the fate choice decision of NPCs can be modulated by the exposure to specific growth factors that mimic the extracellular cues driving differentiation in vivo. For example, members of the IL-6 family of cytokine and BMPs molecules are also effective in driving in vitro differentiation of progenitor cells into astrocytes. In our study, we showed that human neurospheres exposed to the combination of CNTF and BMP4 differentiated into astrocytes through the activation of the JAK/STAT and SMAD signaling pathways. RNAseq revealed that signaling through CNTF receptor culminates with the activation of STAT3, which is a direct transcriptional regulator of GFAP expression, while, signaling through BMPs receptors and SMAD transducers activates the expression of all four members of the ID family of proteins (Hollnagel et al., 1999), which negatively regulate oligodendrocytes transcription factors OLIG1 and OLIG2 (Samanta & Kessler, 2004). Our findings showed that human astrocyte differentiation of glia-committed progenitors requires the concomitant activation of the JAK/STAT and SMAD signaling pathways, resulting in the inhibition of oligodendrocyte lineage commitment and activation of astrocytes differentiation. Our results are in agreement with previous studies (Raff et al., 1983; Rao et al., 1998; Herrera et al., 2001) suggesting that glia-committed progenitor cells may retain the capability to differentiate into astrocyte and oligodendrocyte and that their fate decision is dictated by the stimulation of diverse signaling pathways and the activation of lineage-specific genetic programs.
FBS has been widely used to induce astrocytic differentiation of NPC (McCarthy et al., 2000; Brunet et al., 2004; Haas et al., 2012) but the signal transduction mechanisms underlying the differentiation process and the characteristics of the differentiated cells remained poorly investigated. To better highlight the differences and fully understand the nature of FBS- and CNTF/BMP4-astrocytes we compared their transcriptional profiles attained with RNAseq. In comparison to FBS-differentiated cells, CNTF/BMP4-astrocytes are enriched for genes involved in glia cell differentiation and astrocytes-specific metabolic processes and express high level of gene important for astrocytes function. For examples, CNTF/BMP4-astrocytes express glypicans (GPC4) and hevin (SPARCL1), which are astrocytes-secreted proteins playing a crucial role in the formation, maturation and maintenance of synapses (Kucukdereli et al., 2011; Allen et al., 2012). Differently, FBS-astrocytes when compared to CNTF/BMP4-astrocytes showed enrichment for genes involved in neurogenesis, neuronal differentiation and synapses formation and function. This observation is in line with previous studies showing that NPCs exposed to low percentage of serum differentiate into a mixed population of neuronal and glial cells (McCarthy et al., 2000; McCarthy et al., 2006), and suggests that neurospheres isolated from second trimester embryonic bran might contain a small percentage of progenitors capable of differentiating into neuronal cells in response to FBS but not CNTF/BMP4 stimulation. Our data imply that CNTF/BMP4 stimulation is a more controlled and specific experimental paradigm than FBS to differentiate human NPCs into astrocytes.
In response to CNS injury astrocytes undergo different molecular, morphological and functional changes in a process called reactive gliosis. For example after spinal cord injury, reactive astrocytes create a scar around the site of injury preventing the spread of further tissue damage (Faulkner et al., 2004) but at the same time releasing extracellular matrix components and pro-inflammatory molecules that are detrimental for neuronal regeneration (Silver & Miller, 2004). Reactive astrocytes are characterized by the upregulation of GFAP, increased secretion of CSPGs (Dyck & Karimi-Abdolrezaee, 2015) and the expression of immature markers like NESTIN (Clarke et al., 1994) and VIMENTIN (Liu et al., 2014) but more complex molecular changes occur during reactive gliosis. The study of Zamanian et al. was instrumental to reveal genome-wide gene expression changes occurring during mouse reactive gliosis, providing a transcriptome database of reactive astrocytes (Zamanian et al., 2012). The transcriptional profile of primary rodents astrocytes cultured in vitro have been shown to drastically differ from acutely purified mature astrocytes, with the in vitro culture being more similar to immature astrocytes or astrocytes in the reactive state (Cahoy et al., 2008; Zamanian et al., 2012). In concordance with these studies we noticed enrichment for reactive-astrocytes genes in FBS-astrocytes but not in CNTF/BMP4 astrocytes, thus suggesting that FBS-astrocytes might have a phenotype more similar to reactive astrocytes which might explain the fibroblast-like phenotype of astrocytes differentiated with FBS compared to the more stellate phenotype obtained with CNTF/BMP4. Our findings demonstrated that the combination of CNTF and BMP4 triggered human NPCs to restrict their fate and to differentiate into astrocytes through the activation of signaling pathways that recapitulate in vivo development of astrocytes: the JAK/STAT and the SMAD signaling cascades. Differently, NPCs cultured in the presence of FBS differentiated in astrocytes with a more reactive-like phenotype.
Our datasets and culture system are an important resource to study human astrocytes development and may be of help to identify novel human-specific astrocyte markers. Our study consist of a valuable tool for future research of human disorders characterized by impairment of astrocyte development and function and represents a step forward for the therapeutic application of RGCs-derived astrocytes.
Supplementary Material
Acknowledgments
This work was supported by the US NIH NINDS R01NS081208-01A1 awarded to Mohammad Ali Faghihi.
ABBREVIATIONS
- NPCs
Neural progenitor cells
- RGCs
radial glial cells, CNS, central nervous system
- OPCs
oligodendrocyte progenitor cells
- CNTF
ciliary neurotrophic factor
- BMPs
bone morphogenic proteins
- RNAseq
RNA sequencing
Footnotes
All authors indicate no potential conflicts of interest.
Authors Contribution:
Marco Magistri: conception and design, collection and assembly of the data, data analysis and interpretation, manuscript writing.
Nathalie Khoury: conception and design, collection and assembly of the data, manuscript reviewing and editing.
Emilia Mazza: data analysis and interpretation, manuscript reviewing and editing.
Dmitry Velmeshev: data analysis and interpretation, manuscript reviewing and editing.
Jae K, Lee: provision of study material, manuscript reviewing and editing.
Silvio Bicciato: data analysis and interpretation, manuscript reviewing and editing.
Pantelis Tsoulfas: conception and design, manuscript reviewing and editing.
Mohammad Ali Faghihi: financial support, conception and design, manuscript reviewing and editing, final approve of the manuscript
References
- Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alterman RL, Stanley ER. Colony stimulating factor-1 expression in human glioma. Mol Chem Neuropathol. 1994;21:177–188. doi: 10.1007/BF02815350. [DOI] [PubMed] [Google Scholar]
- Asano H, Aonuma M, Sanosaka T, Kohyama J, Namihira M, Nakashima K. Astrocyte differentiation of neural precursor cells is enhanced by retinoic acid through a change in epigenetic modification. Stem Cells. 2009;27:2744–2752. doi: 10.1002/stem.176. [DOI] [PubMed] [Google Scholar]
- Bangsow T, Baumann E, Bangsow C, Jaeger MH, Pelzer B, Gruhn P, Wolf S, von Melchner H, Stanimirovic DB. The epithelial membrane protein 1 is a novel tight junction protein of the blood-brain barrier. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2008;28:1249–1260. doi: 10.1038/jcbfm.2008.19. [DOI] [PubMed] [Google Scholar]
- Barnabe-Heider F, Wasylnka JA, Fernandes KJ, Porsche C, Sendtner M, Kaplan DR, Miller FD. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 2005;48:253–265. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Becher B, Antel JP. Comparison of phenotypic and functional properties of immediately ex vivo and cultured human adult microglia. Glia. 1996;18:1–10. doi: 10.1002/(SICI)1098-1136(199609)18:1<1::AID-GLIA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bez A, Corsini E, Curti D, Biggiogera M, Colombo A, Nicosia RF, Pagano SF, Parati EA. Neurosphere and neurosphere-forming cells: morphological and ultrastructural characterization. Brain Res. 2003;993:18–29. doi: 10.1016/j.brainres.2003.08.061. [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Brunet JF, Grollimund L, Chatton JY, Lengacher S, Magistretti PJ, Villemure JG, Pellerin L. Early acquisition of typical metabolic features upon differentiation of mouse neural stem cells into astrocytes. Glia. 2004;46:8–17. doi: 10.1002/glia.10348. [DOI] [PubMed] [Google Scholar]
- Buc-Caron MH. Neuroepithelial progenitor cells explanted from human fetal brain proliferate and differentiate in vitro. Neurobiol Dis. 1995;2:37–47. doi: 10.1006/nbdi.1995.0004. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SR, Shetty AK, Bradley JL, Turner DA. Reactive astrocytes express the embryonic intermediate neurofilament nestin. Neuroreport. 1994;5:1885–1888. doi: 10.1097/00001756-199410000-00011. [DOI] [PubMed] [Google Scholar]
- Colognato H, Ramachandrappa S, Olsen IM, ffrench-Constant C. Integrins direct Src family kinases to regulate distinct phases of oligodendrocyte development. J Cell Biol. 2004;167:365–375. doi: 10.1083/jcb.200404076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L, Cattaneo E. Neural stem cell systems: physiological players or in vitro entities? Nat Rev Neurosci. 2010;11:176–187. doi: 10.1038/nrn2761. [DOI] [PubMed] [Google Scholar]
- Cvetkovic-Lopes V, Bayer L, Dorsaz S, Maret S, Pradervand S, Dauvilliers Y, Lecendreux M, Lammers GJ, Donjacour CE, Du Pasquier RA, Pfister C, Petit B, Hor H, Muhlethaler M, Tafti M. Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J Clin Invest. 2010;120:713–719. doi: 10.1172/JCI41366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloulme JC, Raponi E, Gentil BJ, Bertacchi N, Marks A, Labourdette G, Baudier J. Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Mol Cell Neurosci. 2004;27:453–465. doi: 10.1016/j.mcn.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyck SM, Karimi-Abdolrezaee S. Chondroitin sulfate proteoglycans: Key modulators in the developing and pathologic central nervous system. Exp Neurol. 2015;269:169–187. doi: 10.1016/j.expneurol.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Eroglu C, Barres BA. Regulation of synaptic connectivity by glia. Nature. 2010;468:223–231. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner JR, Herrmann JE, Woo MJ, Tansey KE, Doan NB, Sofroniew MV. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci. 2004;24:2143–2155. doi: 10.1523/JNEUROSCI.3547-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR. Specification and morphogenesis of astrocytes. Science. 2010;330:774–778. doi: 10.1126/science.1190928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison CJ, Dougherty PM, Kajander KC, Carlton SM. Staining of glial fibrillary acidic protein (GFAP) in lumbar spinal cord increases following a sciatic nerve constriction injury. Brain Res. 1991;565:1–7. doi: 10.1016/0006-8993(91)91729-k. [DOI] [PubMed] [Google Scholar]
- Gibbons HM, Hughes SM, Van Roon-Mom W, Greenwood JM, Narayan PJ, Teoh HH, Bergin PM, Mee EW, Wood PC, Faull RL, Dragunow M. Cellular composition of human glial cultures from adult biopsy brain tissue. J Neurosci Methods. 2007;166:89–98. doi: 10.1016/j.jneumeth.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Giera S, Deng Y, Luo R, Ackerman SD, Mogha A, Monk KR, Ying Y, Jeong SJ, Makinodan M, Bialas AR, Chang BS, Stevens B, Corfas G, Piao X. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat Commun. 2015;6:6121. doi: 10.1038/ncomms7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givogri MI, Schonmann V, Cole R, De Vellis J, Bongarzone ER. Notch1 and Numb genes are inversely expressed as oligodendrocytes differentiate. Dev Neurosci. 2003;25:50–64. doi: 10.1159/000071468. [DOI] [PubMed] [Google Scholar]
- Gross RE, Mehler MF, Mabie PC, Zang Z, Santschi L, Kessler JA. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron. 1996;17:595–606. doi: 10.1016/s0896-6273(00)80193-2. [DOI] [PubMed] [Google Scholar]
- Haas C, Neuhuber B, Yamagami T, Rao M, Fischer I. Phenotypic analysis of astrocytes derived from glial restricted precursors and their impact on axon regeneration. Exp Neurol. 2012;233:717–732. doi: 10.1016/j.expneurol.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez MR, Pena JD, Selvidge JA, Salvador-Silva M, Yang P. Hydrostatic pressure stimulates synthesis of elastin in cultured optic nerve head astrocytes. Glia. 2000;32:122–136. doi: 10.1002/1098-1136(200011)32:2<122::aid-glia20>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Herrera J, Yang H, Zhang SC, Proschel C, Tresco P, Duncan ID, Luskin M, Mayer-Proschel M. Embryonic-derived glial-restricted precursor cells (GRP cells) can differentiate into astrocytes and oligodendrocytes in vivo. Exp Neurol. 2001;171:11–21. doi: 10.1006/exnr.2001.7729. [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- Honda T, Fujino K, Okuzaki D, Ohtaki N, Matsumoto Y, Horie M, Daito T, Itoh M, Tomonaga K. Upregulation of insulin-like growth factor binding protein 3 in astrocytes of transgenic mice that express Borna disease virus phosphoprotein. J Virol. 2011;85:4567–4571. doi: 10.1128/JVI.01817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibi D, Nagai T, Nakajima A, Mizoguchi H, Kawase T, Tsuboi D, Kano S, Sato Y, Hayakawa M, Lange UC, Adams DJ, Surani MA, Satoh T, Sawa A, Kaibuchi K, Nabeshima T, Yamada K. Astroglial IFITM3 mediates neuronal impairments following neonatal immune challenge in mice. Glia. 2013;61:679–693. doi: 10.1002/glia.22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Jana A, Pal U, Pahan K. A simplified method for isolating highly purified neurons, oligodendrocytes, astrocytes, and microglia from the same human fetal brain tissue. Neurochem Res. 2007;32:2015–2022. doi: 10.1007/s11064-007-9340-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- Jones EV, Bouvier DS. Astrocyte-secreted matricellular proteins in CNS remodelling during development and disease. Neural Plast. 2014;2014:321209. doi: 10.1155/2014/321209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, Deneen B. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Han YM, Oh M, Kim WK, Oh KJ, Lee SC, Bae KH, Han BS. DUSP4 regulates neuronal differentiation and calcium homeostasis by modulating ERK1/2 phosphorylation. Stem Cells Dev. 2015;24:686–700. doi: 10.1089/scd.2014.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopstein A, Santos-Nogueira E, Francos-Quijorna I, Redensek A, David S, Navarro X, Lopez-Vales R. Beneficial effects of alphaB-crystallin in spinal cord contusion injury. J Neurosci. 2012;32:14478–14488. doi: 10.1523/JNEUROSCI.0923-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs DM. alpha2-macroglobulin in late-onset Alzheimer's disease. Exp Gerontol. 2000;35:473–479. doi: 10.1016/s0531-5565(00)00113-3. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, Barres BA, Eroglu C. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci U S A. 2011;108:E440–449. doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprianou S, Chatzopoulou E, Thomas JL, Bouyain S, Harroch S. A complex between contactin-1 and the protein tyrosine phosphatase PTPRZ controls the development of oligodendrocyte precursor cells. Proc Natl Acad Sci U S A. 2011;108:17498–17503. doi: 10.1073/pnas.1108774108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena JY, Legrand C, Faivre-Sarrailh C, Sarlieve LL, Ferraz C, Rabie A. High gelsolin content of developing oligodendrocytes. Int J Dev Neurosci. 1994;12:375–386. doi: 10.1016/0736-5748(94)90021-3. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Fancy SP, Franklin RJ, Rowitch DH. Olig gene function in CNS development and disease. Glia. 2006;54:1–10. doi: 10.1002/glia.20273. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang SC. Specification of neuronal and glial subtypes from human pluripotent stem cells. Cell Mol Life Sci. 2011;68:3995–4008. doi: 10.1007/s00018-011-0770-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li Y, Cui Y, Roberts C, Lu M, Wilhelmsson U, Pekny M, Chopp M. Beneficial effects of gfap/vimentin reactive astrocytes for axonal remodeling and motor behavioral recovery in mice after stroke. Glia. 2014;62:2022–2033. doi: 10.1002/glia.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo U, Selvaraj V, Plane JM, Chechneva OV, Otsu K, Deng W. p38alpha (MAPK14) critically regulates the immunological response and the production of specific cytokines and chemokines in astrocytes. Sci Rep. 2014;4:7405. doi: 10.1038/srep07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Magistri M, Velmeshev D, Makhmutova M, Faghihi MA. Transcriptomics Profiling of Alzheimer's Disease Reveal Neurovascular Defects, Altered Amyloid-beta Homeostasis, and Deregulated Expression of Long Noncoding RNAs. J Alzheimers Dis. 2015;48:647–665. doi: 10.3233/JAD-150398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Chunjing W, Geffin R, McCarthy M. Depressed neurofilament expression associates with apolipoprotein E3/E4 genotype in maturing human fetal neurons exposed to HIV-1. J Neurovirol. 2012;18:323–330. doi: 10.1007/s13365-012-0079-0. [DOI] [PubMed] [Google Scholar]
- Matyash V, Kettenmann H. Heterogeneity in astrocyte morphology and physiology. Brain Res Rev. 2010;63:2–10. doi: 10.1016/j.brainresrev.2009.12.001. [DOI] [PubMed] [Google Scholar]
- McCarthy M, Auger D, Whittemore SR. Human cytomegalovirus causes productive infection and neuronal injury in differentiating fetal human central nervous system neuroepithelial precursor cells. J Hum Virol. 2000;3:215–228. [PubMed] [Google Scholar]
- McCarthy M, Vidaurre I, Geffin R. Maturing neurons are selectively sensitive to human immunodeficiency virus type 1 exposure in differentiating human neuroepithelial progenitor cell cultures. J Neurovirol. 2006;12:333–348. doi: 10.1080/13550280600915347. [DOI] [PubMed] [Google Scholar]
- McKeon RJ, Schreiber RC, Rudge JS, Silver J. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J Neurosci. 1991;11:3398–3411. doi: 10.1523/JNEUROSCI.11-11-03398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Krencik R, Ullian EM, Tsai HH, Deneen B, Richardson WD, Barres BA, Rowitch DH. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Iijima N, Kitabatake K, Kohsaka S. Alpha 2-macroglobulin is an astroglia-derived neurite-promoting factor for cultured neurons from rat central nervous system. Brain Res. 1990;527:55–61. doi: 10.1016/0006-8993(90)91059-p. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takizawa T, Ochiai W, Yanagisawa M, Hisatsune T, Nakafuku M, Miyazono K, Kishimoto T, Kageyama R, Taga T. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc Natl Acad Sci U S A. 2001;98:5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999a;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Taga T. Astrocyte differentiation mediated by LIF in cooperation with BMP2. FEBS Lett. 1999b;457:43–46. doi: 10.1016/s0014-5793(99)00997-7. [DOI] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16:245–255. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Newcombe J, Meeson A, Cuzner ML. Immunocytochemical characterization of primary glial cell cultures from normal adult human brain. Neuropathol Appl Neurobiol. 1988;14:453–465. doi: 10.1111/j.1365-2990.1988.tb01337.x. [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, Ransom BR, Goldman SA, Nedergaard M. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Nakamura M, Katoh H, Miyao T, Shimazaki T, Ishii K, Yamane J, Yoshimura A, Iwamoto Y, Toyama Y, Okano H. Conditional ablation of Stat3 or Socs3 discloses a dual role for reactive astrocytes after spinal cord injury. Nat Med. 2006;12:829–834. doi: 10.1038/nm1425. [DOI] [PubMed] [Google Scholar]
- Okuda T, Kokame K, Miyata T. Differential expression patterns of NDRG family proteins in the central nervous system. J Histochem Cytochem. 2008;56:175–182. doi: 10.1369/jhc.7A7323.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JD, Agapova O, Gabelt BT, Levin LA, Lucarelli MJ, Kaufman PL, Hernandez MR. Increased elastin expression in astrocytes of the lamina cribrosa in response to elevated intraocular pressure. Invest Ophthalmol Vis Sci. 2001;42:2303–2314. [PubMed] [Google Scholar]
- Piao CS, Kim SW, Kim JB, Lee JK. Co-induction of alphaB-crystallin and MAPKAPK-2 in astrocytes in the penumbra after transient focal cerebral ischemia. Exp Brain Res. 2005;163:421–429. doi: 10.1007/s00221-004-2197-2. [DOI] [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- Rajan P, McKay RD. Multiple routes to astrocytic differentiation in the CNS. J Neurosci. 1998;18:3620–3629. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer LM, Ramer MS, Steeves JD. Setting the stage for functional repair of spinal cord injuries: a cast of thousands. Spinal Cord. 2005;43:134–161. doi: 10.1038/sj.sc.3101715. [DOI] [PubMed] [Google Scholar]
- Rao MS, Noble M, Mayer-Proschel M. A tripotential glial precursor cell is present in the developing spinal cord. Proc Natl Acad Sci U S A. 1998;95:3996–4001. doi: 10.1073/pnas.95.7.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SP, Britto R, Vinnakota K, Aparna H, Sreepathi HK, Thota B, Kumari A, Shilpa BM, Vrinda M, Umesh S, Samuel C, Shetty M, Tandon A, Pandey P, Hegde S, Hegde AS, Balasubramaniam A, Chandramouli BA, Santosh V, Kondaiah P, Somasundaram K, Rao MR. Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin Cancer Res. 2008;14:2978–2987. doi: 10.1158/1078-0432.CCR-07-4821. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Semple BD, Frugier T, Morganti-Kossmann MC. CCL2 modulates cytokine production in cultured mouse astrocytes. J Neuroinflammation. 2010;7:67. doi: 10.1186/1742-2094-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafit-Zagardo B, Sharma N, Berman JW, Bornstein MB, Brosnan CF. CSF-1 expression is upregulated in astrocyte cultures by IL-1 and TNF and affects microglial proliferation and morphology in organotypic cultures. Int J Dev Neurosci. 1993;11:189–198. doi: 10.1016/0736-5748(93)90078-r. [DOI] [PubMed] [Google Scholar]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci. 2004;5:146–156. doi: 10.1038/nrn1326. [DOI] [PubMed] [Google Scholar]
- Slaets H, Nelissen S, Janssens K, Vidal PM, Lemmens E, Stinissen P, Hendrix S, Hellings N. Oncostatin M reduces lesion size and promotes functional recovery and neurite outgrowth after spinal cord injury. Mol Neurobiol. 2014;50:1142–1151. doi: 10.1007/s12035-014-8795-5. [DOI] [PubMed] [Google Scholar]
- Soderblom C, Luo X, Blumenthal E, Bray E, Lyapichev K, Ramos J, Krishnan V, Lai-Hsu C, Park KK, Tsoulfas P, Lee JK. Perivascular fibroblasts form the fibrotic scar after contusive spinal cord injury. J Neurosci. 2013;33:13882–13887. doi: 10.1523/JNEUROSCI.2524-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taupin P, Ray J, Fischer WH, Suhr ST, Hakansson K, Grubb A, Gage FH. FGF-2-responsive neural stem cell proliferation requires CCg, a novel autocrine/paracrine cofactor. Neuron. 2000;28:385–397. doi: 10.1016/s0896-6273(00)00119-7. [DOI] [PubMed] [Google Scholar]
- Tazir M, Bellatache M, Nouioua S, Vallat JM. Autosomal recessive Charcot-Marie-Tooth disease: from genes to phenotypes. J Peripher Nerv Syst. 2013;18:113–129. doi: 10.1111/jns5.12026. [DOI] [PubMed] [Google Scholar]
- Van Wagoner NJ, Choi C, Repovic P, Benveniste EN. Oncostatin M regulation of interleukin-6 expression in astrocytes: biphasic regulation involving the mitogen-activated protein kinases ERK1/2 and p38. J Neurochem. 2000;75:563–575. doi: 10.1046/j.1471-4159.2000.0750563.x. [DOI] [PubMed] [Google Scholar]
- Velmeshev D, Lally P, Magistri M, Faghihi MA. CANEapp: a user-friendly application for automated next generation transcriptomic data analysis. BMC Genomics. 2016;17:49. doi: 10.1186/s12864-015-2346-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmeshev D, Magistri M, Faghihi MA. Expression of non-protein-coding antisense RNAs in genomic regions related to autism spectrum disorders. Mol Autism. 2013;4:32. doi: 10.1186/2040-2392-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vouyiouklis DA, Brophy PJ. A novel gelsolin isoform expressed by oligodendrocytes in the central nervous system. J Neurochem. 1997;69:995–1005. doi: 10.1046/j.1471-4159.1997.69030995.x. [DOI] [PubMed] [Google Scholar]
- Winkler C, Fricker RA, Gates MA, Olsson M, Hammang JP, Carpenter MK, Bjorklund A. Incorporation and glial differentiation of mouse EGF-responsive neural progenitor cells after transplantation into the embryonic rat brain. Mol Cell Neurosci. 1998;11:99–116. doi: 10.1006/mcne.1998.0674. [DOI] [PubMed] [Google Scholar]
- Wu H, Yang L, Liao D, Chen Y, Wang W, Fang J. Podocalyxin regulates astrocytoma cell invasion and survival against temozolomide. Exp Ther Med. 2013;5:1025–1029. doi: 10.3892/etm.2013.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Faghihi MA, Magistri M, Alvarez-Garcia O, Lotz M, Wahlestedt C. Antisense RNA controls LRP1 Sense transcript expression through interaction with a chromatin-associated protein, HMGB2. Cell Rep. 2015;11:967–976. doi: 10.1016/j.celrep.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang KK. Glial fibrillary acidic protein: from intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 2015;38:364–374. doi: 10.1016/j.tins.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Young KM, Mitsumori T, Pringle N, Grist M, Kessaris N, Richardson WD. An Fgfr3-iCreER(T2) transgenic mouse line for studies of neural stem cells and astrocytes. Glia. 2010;58:943–953. doi: 10.1002/glia.20976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritsky LA, Gama L, Clements JE. Canonical type I IFN signaling in simian immunodeficiency virus-infected macrophages is disrupted by astrocyte-secreted CCL2. J Immunol. 2012;188:3876–3885. doi: 10.4049/jimmunol.1103024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.