Summary
Filamentous fungi produce a vast array of secondary metabolites (SMs) and some play a role in agriculture or pharmacology. Sequencing of the rice pathogen Fusarium fujikuroi revealed the presence of far more SM-encoding genes than known products. SM production is energy-consuming and thus tightly regulated, leaving the majority of SM gene clusters silent under laboratory conditions. One important regulatory layer in SM biosynthesis involves histone modifications that render the underlying genes either silent or poised for transcription. Here, we show that the majority of the putative SM gene clusters in F. fujikuroi are located within facultative heterochromatin marked by trimethylated lysine 27 on histone 3 (H3K27me3). Kmt6, the methyltransferase responsible for establishing this histone mark, appears to be essential in this fungus, and knock-down of Kmt6 in the KMT6kd strain shows a drastic phenotype affecting fungal growth and development. Transcription of four so far cryptic and otherwise silent putative SM gene clusters was induced in the KMT6kd strain, in which decreased expression of KMT6 is accompanied by reduced H3K27me3 levels at the respective gene loci and accumulation of novel metabolites. One of the four putative SM gene clusters, named STC5, was analysed in more detail thereby revealing a novel sesquiterpene.
Introduction
Fungi are known to produce a plethora of secondary metabolites (SMs). These low-molecular-weight compounds can be detrimental to humankind due to their toxic and carcinogenic properties, as exemplified by the mycotoxins aflatoxin and fumonisins. Some other SMs have been or are used in medicine, such as the antibiotics penicillin and cephalosporin or the immunosuppressant cyclosporine A (Fleming, 1929; Dreyfuss et al., 1976; Keller et al., 2005). Both properties render research on fungal secondary metabolism of great interest. SMs are built from only a few building blocks and their biosynthesis is generally performed by one or several of the following key enzymes: polyketide synthase (PKS), non-ribosomal peptide synthetase (NRPS), terpene cyclase (TC), dimethylallyl tryptophan synthase (DMATS), or hybrids thereof. Only a few SMs are derived from other pathways, e.g. oxylipins originate from fatty acids. Accessory enzymes may further modify the key enzyme-derived product (Brakhage, 2013). The genes involved in the biosynthesis of a particular SM are commonly located in close proximity to one another in gene clusters, thereby facilitating their co-ordinated expression (Keller and Hohn, 1997). SM biosynthesis is energy-consuming and thus typically only initiated when advantageous for the producer, leaving most of the SM genes silent under standard laboratory conditions. In fact, bioinformatic analyses of sequenced fungal genomes indicate the presence of many more as yet uncharacterised putative SM gene clusters than the gene clusters with known products, therefore revealing fungi as ‘treasure chests’ for novel chemical compounds. Only initiated under defined environmental conditions, SMs are subject to a complex regulatory network involving signalling cascades as well as pathway-specific and/or global regulators. Several successful strategies have been applied to awaken ‘silent’ SM gene clusters, including variations in the culture conditions (Bode et al., 2002), or co-cultivation of the fungus with other organisms (Schroeckh et al., 2009; Nützmann et al., 2011; König et al., 2013). The majority of approaches have been based on genetic manipulation, e.g. over-expression of genes encoding a pathway-specific transcription factor (TF) and/or the key enzyme (Bergmann et al., 2007; Brock et al., 2013; Wiemann et al., 2013; Von Bargen et al., 2015; Rösler et al., 2016) or the manipulation of global regulators (Bok and Keller, 2004; Michielse et al., 2014; Studt et al., 2016). However, the manipulation of the fungal epigenome by deleting or over-expressing histone-modifying genes has evolved as another powerful tool more recently (Bok et al., 2009).
Gene expression in eukaryotes functions within the context of chromatin. Chromatin is composed of structural nuclear proteins such as histones and non-histone proteins that package and condense the DNA, thus leaving most of the genes inaccessible for transcriptional activation. Accessibility of DNA is mainly regulated at the level of reversible post-translational modifications (PTMs) of core histones by acetylation, phosphorylation or methylation, the combination of which define the degree of condensation from a loose (euchromatin) to a dense configuration (heterochromatin). All modifications take place at defined amino acid residues and are highly dynamic, changing within minutes upon a received stimulus, thereby promoting rearrangement of the nucleosomes and simultaneously changing the accessibility of DNA (e.g. Jenuwein and Allis, 2001; Bannister and Kouzarides, 2011; Gacek and Strauss, 2012). SM gene expression was shown to depend on the chromatin landscape, and several successful attempts of epigenome manipulation were demonstrated to induce expression of otherwise silent SM gene clusters (Shwab et al., 2007; Williams et al., 2008; Bok et al., 2009; Reyes-Dominguez et al., 2010; Nützmann et al., 2011; Soukup et al., 2012). Recent studies in Neurospora crassa, Fusarium graminearum, Epichloё festucae and Zymoseptoria tritici revealed the presence of a histone PTM associated with gene silencing previously only known for plants and animals, i.e. trimethylation of lysine 27 on histone H3 (H3K27me3) (Connolly et al., 2013; Jamieson et al., 2013; Chujo and Scott, 2014; Schotanus et al., 2015). H3K27me3 is established by the multimeric Polycomb repressive complex 2 (PRC2) that consists of four core proteins with the histone methyltransferase Enhancer of zeste [E(z)], acting as the catalytic subunit (Qian and Zhou, 2006), and was first identified in Drosophila melanogaster as a negative regulator of homeotic gene expression (Jones and Gelbart, 1990; Müller et al., 2002). Notably, PRC2 is absent in the unicellular fungi Saccharomyces cerevisae and Schizosaccharomyces pombe, and also in several filamentous fungi including Aspergillus fumigatus, Aspergillus nidulans and the smut fungus Ustilago maydis (Connolly et al., 2013; Shaver et al., 2014). Genome-wide chromatin immunoprecipitation combined with high-throughput sequencing (ChIP-seq) in F. graminearum indicated that H3K27me3 is localised in subtelomeric regions or regions that may constitute ancestral subtelomeric regions, and these regions were specifically enriched for putative SM gene clusters. Loss of the E(z) homolog, designated as Kmt6 for lysine methyltransferase 6 in F. graminearum, resulted in upregulation of several yet uncharacterised SM-related genes (Connolly et al., 2013). Similarly, in E. festucae deletion of the E(z) homolog, Ezhb, released repression at SM gene clusters involved in the biosynthesis of ergot alkaloids and lolitrem B (Chujo and Scott, 2014), a tremorgenic mycotoxin responsible for the ryegrass staggers syndrome causing a neurological disorder in farm animals (Saikia et al., 2012). This evidence collectively highlights H3K27me3 as a promising target for the induction of otherwise silent SM gene clusters also in other fungi.
In the current study, we analysed the impact of H3K27me3 in the ascomycete F. fujikuroi, a notorious rice pathogen and causal agent of the bakanae (‘foolish seedling’) disease due to its ability to produce gibberellic acid (GA). Besides GA, F. fujikuroi possesses the genomic capacity to produce 46 additional SMs, of which only a few have been identified and linked to the respective SM gene cluster so far (Wiemann et al., 2009, 2013; Studt et al., 2012, 2016; Niehaus et al., 2013, 2014a, 2014b; Von Bargen et al., 2013, 2015; Rösler et al., 2016). We found H3K27me3 to be predominantly, although not exclusively, located at subtelomeric regions where the majority of the predicted SM gene clusters in F. fujikuroi are localised. In marked contrast to other fungal species, deletion of KMT6 in F. fujikuroi is lethal. Not suprisingly, knock-down of Kmt6 in the KMT6kd strain already had a severe impact on fungal development and resulted in an altered gene expression of about one third of the genome with a significant enrichment of mostly cryptic SM gene clusters. Four yet unknown SM key enyzme-encoding genes were up-regulated in the KMT6kd strain accompanied by reduced H3K27me3 levels at the respective gene loci and accumulation of novel metabolites. One of the SM genes, namely STC5 encoding a sesquiterpene cyclase, was analysed in more detail. Being non-functional in the F. fujikuroi strain under study, we performed an in vitro approach using STC5 from the closely related Fusarium mangiferae, which enabled the identification of the STC5-derived product.
Results
H3K27 methylation is predominantly localised in genomic regions with high abundance of SM gene clusters in F. fujikuroi
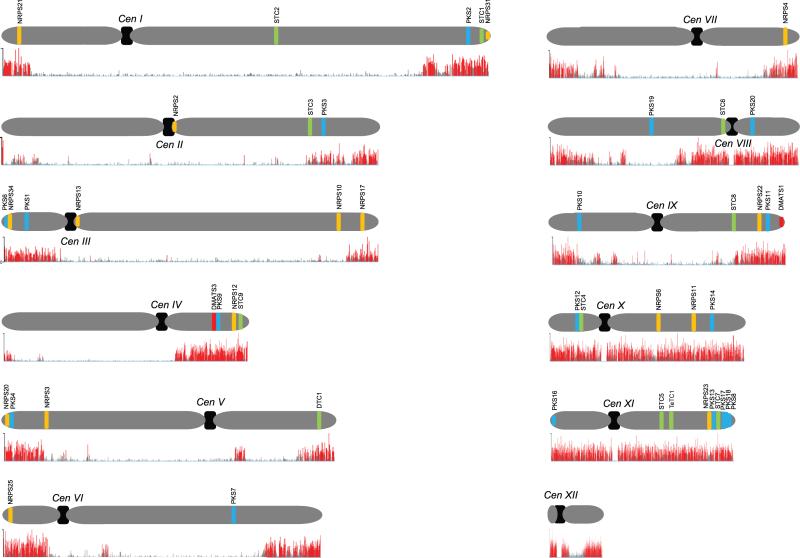
To gain an overview of the genome-wide distribution of H3K27me3 in F. fujikuroi we performed ChIP-seq in liquid synthetic ICI (Imperial Chemical Industries, UK) using an anti-H3K27me3 antibody. H3K27me3 is predominantly localised at, although not restricted to, subtelomeric regions (Fig. 1). Notably, these regions are site of the majority of the putative SM gene clusters identified in this fungus (Wiemann et al., 2013). Only eight out of the 47 identified SM key enzyme-encoding genes are localised in regions with low abundance of H3K27me3 (Fig. 1). Chromosomes ten and eleven that are specifically enriched for putative SM genes show H3K37me3 throughout the entire chromosome arms. Taken together, 39 putative SM gene clusters fall into regions associated with H3K27me3, and several so far cryptic SM gene clusters are enriched for this histone PTM suggesting that genetic manipulation of the involved methyltransferase Kmt6 constitutes a promising target for the induction and subsequent identification of otherwise silent SM gene clusters in F. fujikuroi.
Fig. 1. Genome-wide distribution of H3K27me3 in F. fujikuroi.
The F. fujikuroi wild-type strain was grown in liquid synthetic ICI medium under conditions of nitrogen starvation for 3 days, the mycelium was crosslinked and used for subsequent ChIP as described in the experimental section. ChIP was performed with an anti-H3K27me3 antibody (AM39155) and high-throughput sequencing was carried out on an Illumina GAII genome analyzer by single-end 36- to 50-nt sequencing. Experiments were done in a biological triplicate; red bars indicate enriched reads at the respective genome location. Chromosomes are shown in grey and centromeres are depicted in black; putative SM gene clusters are indicated by bars according to the following colour code: polyketide synthase (PKS), blue; non-ribosomal peptide synthetase (NRPS), orange; (di-/sesqui-/tetra-)terpene cyclase (D/S/TeTC), green; dimethylallyltryptophane synthase (DMATS), red.
Kmt6 and H3K27me3 are essential in F. fujikuroi
The histone methyltransferase Kmt6 (FFUJ_00719) in F. fujikuroi was identified by BLASTp analysis (Altschul et al., 1990) in the F. fujikuroi wild-type strain IMI58289 using the Kmt6 protein sequence from F. graminearum (FGSG_15795) (Connolly et al., 2013). Sequence identity between Kmt6 from F. fujikuroi and F. graminearum is 77.2%. The F. fujikuroi Kmt6 contains the typical domains, i.e. the well conserved SET domain and the cysteine-rich CXC (pre-SET) domain upstream of SET (Fig. S1). The SET (Su(var)3-9; E(z); Trx) domain was initially identified as a shared sequence motif in the D. melanogaster proteins Suppressor of variegation 3-9 (Su(var)3-9), Enhancer of zeste [the Polycomb-group chromatin regulator E(z)] and the homeobox gene regulator Thritorax (Trx) (Jenuwein et al., 1998). A highly conserved stretch at the C-terminus contains an invariable tyrosine residue that is required for SAM-binding and involved in catalysis (Zhang et al., 2002) (Fig. S1).
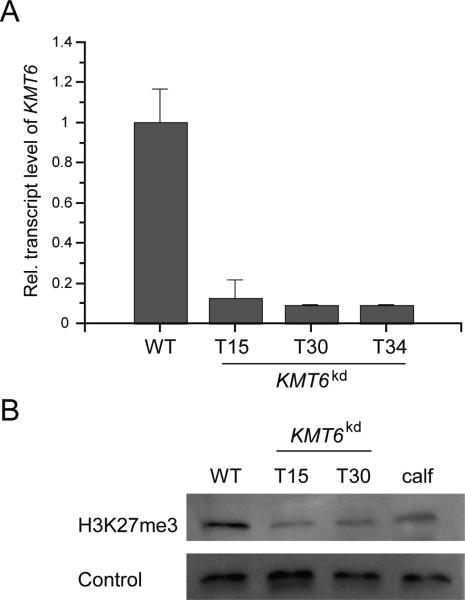
To analyse the function of Kmt6 the gene replacement cassette Δkmt6 was generated and used for targeted deletion of KMT6 in the wild-type background. Several hygromycin-resistant transformants were produced that showed homologous integration events (Fig. S2A). However, attempts to generate homokaryotic mutants by several rounds of single-spore isolation failed. Next, we generated a plasmid for amino acid exchange of lysine 27 for arginine on H3, H3K27R, in order to prevent methylation at this residue (Fig. S2B). However, none of the gained transformants showed the desired amino acid exchange (data not shown) indicating that Kmt6 is most likely essential in F. fujikuroi. Because both the deletion of KMT6 and the H3K27R amino acid exchange did not result in viable mutants, we down-regulated KMT6 expression by using RNA interference (RNAi). The F. fujikuroi wild type was transformed with a plasmid-containing part of the gene as an inverted repeat with a loop region leading to a hairpin in the corresponding mRNA product (Fitzgerald et al., 2004). Gene fragments utilised for KMT6 silencing were under control of the constitutive A. nidulans oliC promoter and contained no conserved domains to exclude off-target effects, such as non-specific silencing of other SET domain-containing proteins (Fig. S2C). Several hygromycin-resistant transformants were generated that contained the knock-down construct (data not shown). Subsequent reverse transcriptase quantitative PCR (RT-qPCR) analysis verified the successful downregulation of the KMT6 wild-type gene copy in nine knock-down mutants (Fig. S2C). Three of these mutants were chosen for further studies, i.e. KMT6kd T15, T30 and T34, with a residual KMT6 transcript level of 12%, 9% and 9%, respectively, relatively to the wild type (Fig. 2A). Subsequent western blot analysis using an anti-H3K27me3 antibody showed reduced H3K27me3 levels in a total protein extract, thereby verifying that Kmt6 is indeed involved in methylation of H3K27 in F. fujikuroi (Fig. 2B).
Fig. 2. Verification of the KMT6 knock-down mutants.
A) Three stable KMT6 knock-down mutants (KMT6kd) were analysed. Reduced KMT6 transcript levels were verified by RT-qPCR. Complementary DNA was retrieved from indicated strains grown for 3 days on solid CM. Experiments were done in technical replicates; mean values and standard deviations are shown. For direct comparison expression of the wild type (WT) was set to 1. B) Western blot analysis of the wild type and two KMT6kd mutants. Total protein extracts were isolated from indicated strains grown on CM and separated by SDS-PAGE. The anti-H3K27me3 antibody (ab6002) was used for blotting. As a loading control the anti-H3, C-terminal antibody (AM39163) was used for detection. Approximately 15 μg of protein extracts or 2 μg of commercial calf thymus histones (calf; Sigma) were loaded. Both expression and western blot analyses were performed in at least two independent biological repeats.
Hyphal growth and asexual development is drastically reduced in the KMT6kd strain
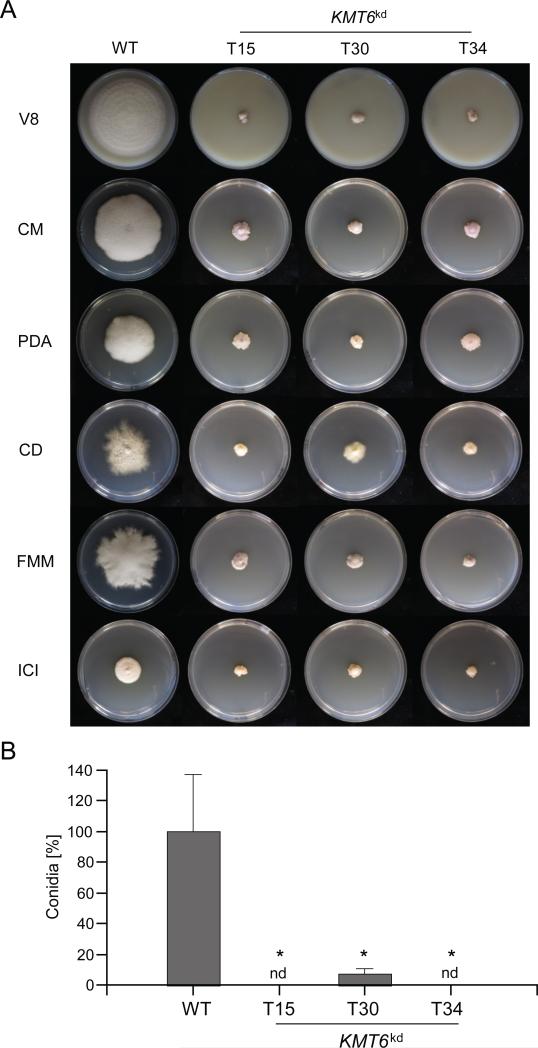
Hyphal growth of the KMT6kd mutants was assessed on different rich media, i.e. V8 (vegetable juice), CM (complete medium) and PDA (potato dextrose agar), and also on minimal media, i.e. CD (Czapek Dox), FMM (Fusarium minimal medium) and synthetic ICI. All fungal strains were grown in triplicate and colony diameters were assessed 7 days post inoculation. On all tested media hyphal growth of KMT6kd mutants was severely reduced (Fig. 3A). To analyse asexual development, the fungal strains were grown on V8 agar and the formation of conidia was quantified after 14 days of growth. In order to compensate for the slow growth of the KMT6kd mutants, six agar plugs were punched out of each plate and vigorously vortexed in water. Conidia formation was significantly reduced in the KMT6kd mutant T30 to about 10% of the wild type, while no conidia were detectable in the KMT6kd mutants T15 and T34 (Fig 3B).
Fig. 3. Hyphal growth and asexual development of KMT6kd mutants.
A) Hyphal growth was assessed on different rich and minimal media: vegetable juice (V8) agar, complete medium (CM), potato dextrose agar (PDA), Czapek dox (CD), Fusarium minimal medium (FMM) and synthetic ICI medium. Colony growth of the wild type (WT) and the indicated KMT6kd mutants was analysed after 7 days of growth at 28 °C in constant darkness. B) Conidia formation of the indicated strains was assessed on V8 agar. Fungal strains were grown for 2 weeks at 20 °C and a 12 h light-dark cycle. Experiments were done in triplicate, mean values and standard deviations are given in the diagram. Conidia formation of the wild type was arbitrarily set as 100%. A Student's t-test (tow-sample assuming equal variances) was performed to determine statistical significance; nd, not detectable; *, p<0.05.
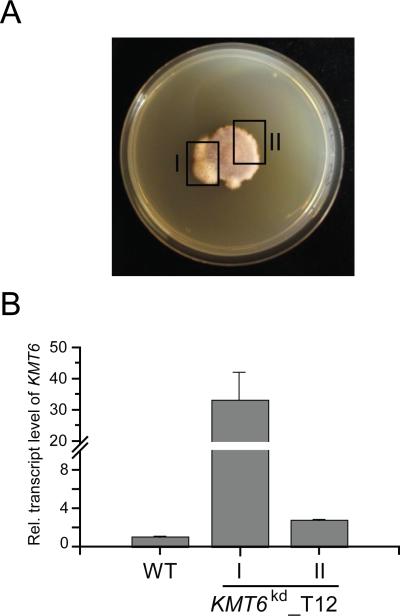
Several KMT6kd mutants showed fast-growing wild type-like sectors after several days on medium without the selecting antibiotic (Fig. 4A). Subsequent KMT6 transcript quantification revealed an about 35-fold increased KMT6 transcript level in these areas of the colony compared to the wild type (Fig. 4B) despite the ability to grow on hygromycin B and presence of the knock-down construct (Fig. S3A), suggesting that suppressor mutation(s) did occur to rescue the wild type-like phenotype. Increased accumulation of KMT6 transcript in the fast-growing sectors of these suppressor mutants led to restoration of the wild-type phenotype with regard to hyphal growth and asexual development (Fig. S3B/C).
Fig. 4. Reversion of the KMT6 phenotype by putative suppressor mutation(s).
A) KMT6kd mutants eventually showed fast growing wild type-like sectors (I). B) Complementary DNA was generated from the wild type-like sectors (I) and the slow growing sectors of KMT6kd (II) as well as the wild type (not shown). Subsequent RT-qPCR revealed induced KMT6 transcript levels in the fast growing wild type-like sectors compared to the wild type. Experiments were performed in duplicate; mean values and standard deviations are given in the diagram.
KMT6kd relieves gene repression of several SM-related genes
The effect of KMT6kd on genome-wide gene expression was studied by microarray analysis. Therefore, the wild type and the stable KMT6kd mutant T15 were grown on solid CM and synthetic ICI media for 3 days to ensure stable growth of the KMT6kd strain. Total RNA was isolated and applied to microarray analysis. From a total set of 14,814 genes, 4,829 and 4,545 showed significantly different transcript concentrations in the mutants on CM and ICI, respectively, using a factor of 2 as cut off. This corresponds to approximately one third of the genome. It is noteworthy that the number of up- and down-regulated genes was similar (Fig. S4). To identify over-represented functional categories we performed FunCat analysis (Ruepp et al., 2004) using the MIPS website interface (http://mips.helmholtz-muenchen.de/funcatDB/). Similar functional groups of genes were found to be enriched on both media, e.g. genes involved in detoxification processes, disease-, virulence- and defence-related genes, as well as genes involved in carbohydrate metabolism. Genes associated with lipid and fatty acid metabolism were found to be significantly enriched and up-regulated in both conditions, whilst the genes involved in general transport were down-regulated in both conditions. A summary of all significantly overrepresented genes is given in Tab. S1. Genes associated with secondary metabolism were among the over-represented groups of de-regulated genes in both environments. This result accords with the fact that the majority of the predicted SM gene clusters are located within HK27me3-enriched regions (Fig. 1), and suggests that H3K27me3 is indeed an important regulatory layer for SM-associated genes in F. fujikuroi. Among the 47 predicted SM gene clusters seven and nine (represented by the key enzyme-encoding gene) are up- or down-regulated, respectively, when grown on solid ICI medium, while nine and six SM key enzyme-encoding genes are up- or down-regulated, respectively, when grown on solid CM (Tab. S2). All up-regulated putative SM key enzyme-encoding genes are associated with H3K27me3-rich regions.
Focusing only on those SM genes which are up-regulated on CM and/or ICI, we identified the following Kmt6-dependent genes: up-regulated on both media are NRPS22 (FFUJ_09296), STC4 (FFUJ_12585), the tetraterpene cyclase-encoding gene TeTC1 (FFUJ_11802), PKS-NRPS1 (FFUJ_02219), STC5 (FFUJ_11739) and STC8 (FFUJ_09423), while DMATS3 (FFUJ_14683) is only up-regulated on ICI and NRPS4 (FFUJ_08113) as well as PKS2 (FFUJ_00118) are only up-regulated when grown on CM (Tab. S2). While for NRPS22, STC4 and TeTC1 the corresponding products, i.e. beauvericin, (+)-koraiol and neurosporaxanthin, respectively, are known (Rodríguez-Ortiz et al., 2009; Brock et al., 2013; Wiemann et al., 2013; E.-M. Niehaus and Tudzynski, in preparation), the products of the remaining six up-regulated SM gene clusters are still cryptic.
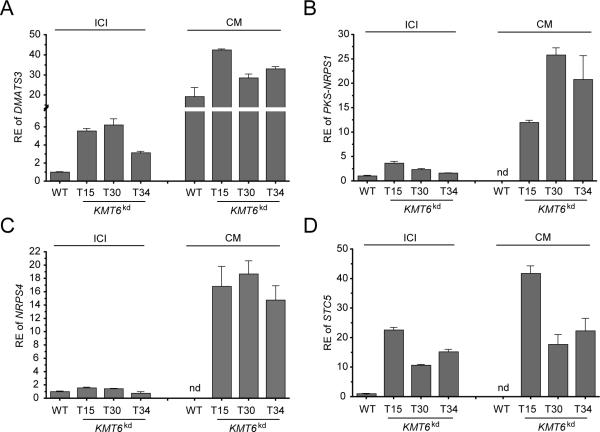
Taken together, we found that 20 out of 47 predicted SM key enzymes were affected by KMT6 knock-down. From these 20 key enzyme-encoding genes 11 (23.4%) were significantly up-regulated either on solid ICI medium, on CM or on both. Six of them (NRPS4, DMATS3, PKS-NRPS1, PKS2, STC5, STC8) constitute so far uncharacterised SM key enzyme-encoding genes. While PKS2 was up-regulated on CM, its expression was down-regulated on ICI medium. Apart from PKS2, adjacent putative SM cluster genes were not simultaneously up-regulated in the KMT6kd mutant (Tab. S2). Furthermore, expression of STC8 was relatively low in the wild type and only slightly up-regulated in the KMT6kd mutant. Therefore, we then focused on the remaining four SM key enzyme-encoding genes. To verify the microarray data we performed RT-qPCR analyses on NRPS4, DMATS3, PKS-NRPS1 and STC5. The wild type and the three KMT6kd mutants, i.e. KMT6kd_T15, KMT6kd_T30 and KMT6kd_T34, were grown on solid CM and ICI medium. Subsequent RT-qPCR analysis verified the obtained microarray data. DMATS3 was (5-fold) up-regulated on ICI, while almost no difference between the wild type and the mutant strains was observed on CM (Fig. 5A). In the case of PKS-NRPS1, expression was slightly (2.5-fold) up-regulated on ICI medium, but about 20-fold elevated on CM (Fig. 5B). Similarly, for NRPS4 gene expression was highly (17-fold) up-regulated in all KMT6kd mutants only on CM, whereas wild-type expression was undetectable (Fig. 5C), in accordance with the results from the microarray analysis. The highest up-regulation in gene expression was observed for STC5. Here, knock-down of KMT6 resulted in 16-fold and 27-fold up-regulation on solid ICI and CM, respectively (Fig. 5D).
Fig. 5. Knock-down of KMT6 induces expression of otherwise silent SM key enzyme-encoding genes.
Expression of the following SM key enzymes was analysed by RT-qPCR: A) DMATS3 (FFUJ_14683), B) NRPS4 (FFUJ_08113), C) PKS-NRPS1 (FFUJ_02219), D) STC5 (FFUJ_11739). RNA was isolated from the wild type (WT) and three independent KMT6kd mutants grown for 3 days on solid CM as well as ICI medium. RT-qPCR experiments were performed in biological and technical replicates giving the same results; only the technical replicate is shown here. Expression of the wild type grown on solid ICI was arbitrarily set as 1 in each experiment. Mean values and standard deviations are shown. Ct values over 32 were calculated as not detectable, nd; RE, relative expression.
Reduced H3K27me3 levels result in an altered SM profile and accumulation of putatively novel compounds
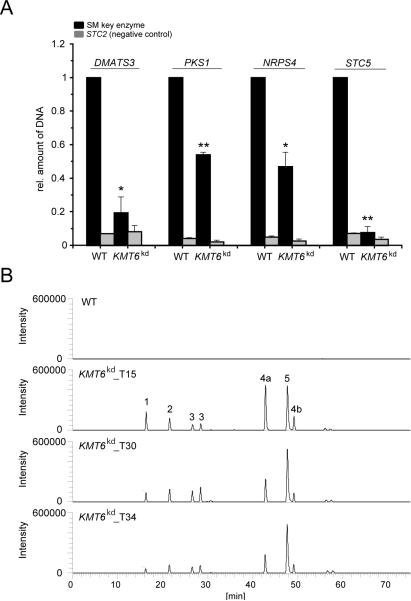
To link the increased transcript concentration of the four SM key enzyme-encoding genes to the KMT6kd mutant and thus H3K27me3, we performed ChIP experiments using an anti-H3K27me3 antibody and subsequently quantified the amount of precipitated DNA by qPCR. Therefore, the wild type and the KMT6kd_T15 mutant were grown on solid CM in accordance to the microarray data. Subsequent ChIP analyses showed that nucleosomes corresponding to all four SM key enzyme-encoding genes, i.e. DMATS3, PKS-NRPS1, NRPS4 and STC5, which are up-regulated in the KMT6kd mutant have significantly less H3K27me3 compared to the wild type (Fig. 6A). A chromosomal region completely devoid of H3K27me3 in the ChIP-seq experiments, i.e. the STC2 locus, was used as a negative control and consequently showed only low H3K27me3 levels in both strains.
Fig. 6. Induced SM gene expression is accompanied by reduced H3K27me3 levels and accumulation of putative novel metabolites.
A) The F. fujikuroi wild-type strain (WT) and KMT6kd mutants were grown for 3 days on solid CM. ChIP experiments were performed using an anti-H3K27me3 antibody (AM39155). The precipitated amount of DNA was quantified at the DMATS3 (FFUJ_14683), PKS-NRPS1 (FFUJ_02219), NRPS4 (FFUJ_08113) and STC5 (FFUJ_11739) gene loci by qPCR. In each case, the amount of DNA in the wild type was arbitrarily set as 1, precipitated DNA at the STC2 (FFUJ_00969) gene locus (grey bars) was used as a negative control and in each case related to the respective SM cluster gene (black bars). Mean values and standard deviations are shown. A Student's t-test (two-sample assuming equal variances) was performed to determine statistical significance. *, p<0.05; **, p<0.01; ***, p<0.001. B) HPLC-HRMS base peak chromatograms of the wild type and KMT6kd mutants. The peak numbers represent the following m/z ratios: 1) m/z 325.2271, 2) m/z 672.4646, 3) m/z 524.3140, 4a) m/z 523.2791, 4b) in source fragment (m/z 523.2791) of beauvericin, as proven with a standard compound, 5) m/z 360.2159.
Changes in the metabolome of the KMT6kd mutants grown on solid CM were analysed by high performance liquid chromatography coupled to high resolution mass spectrometry (HPLC-HRMS) and compared to the wild-type strain. The analyses of extracts from three different mutants, i.e. KMT6kd_T15, KMT6kd_T30 and KMT6kd_T34, revealed the presence of several compounds that did not occur in the wild type (Fig. 6B). The numbered peaks, except 4b which corresponds to beauvericin (Wiemann et al., 2013), do not represent any of the known F. fujikuroi metabolites and are currently under investigation.
Correlated with the production level, the NRPS most likely involved in beauvericin biosynthesis, i.e. NRPS22 (Wiemann et al., 2013; E.-M. Niehaus and B. Tudzynski, in preparation), was highly up-regulated in the microarray data under both conditions (Tab. S2), which was subsequently also verified by RT-qPCR (Fig. S5A). As expected, up-regulation of NRPS22 was accompanied by significantly reduced H3K27me3 levels in all three KMT6kd mutants (Fig. S5B). Taken together, KMT6kd results in induction of otherwise silent SM key enzyme-encoding genes, accompanied by significantly reduced H3K27me3 levels and accumulation of several putatively novel compounds.
The F. mangiferae STC5 homologue synthesises (1R,4R,5S)-guaia-6,10(14)-diene in vitro
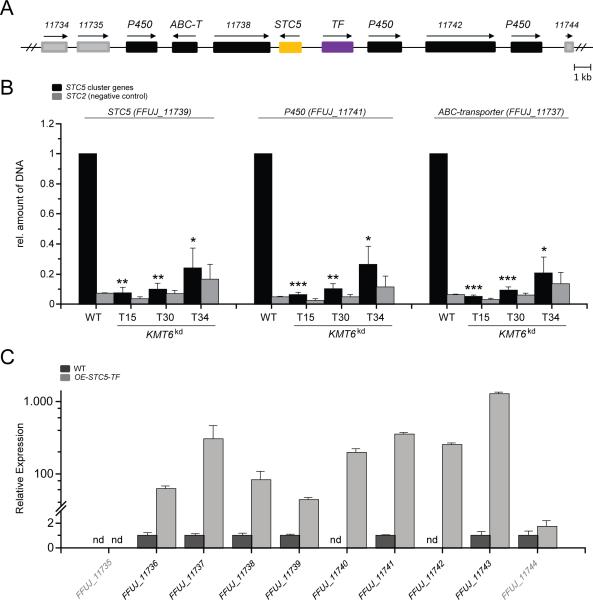
Significant upregulation of STC5 was observed on both tested media, i.e. solid CM and ICI, but was highest when grown on CM (Fig. 5D). Bioinformatic analyses of the genes located in proximity to STC5, combined with synteny analysis with other Fusarium spp., indicated that the putative STC5 cluster consists of at least nine neighbouring genes, i.e. FFUJ_11735-FFUJ_11743. All of these putative cluster genes were co-ordinately up-regulated in the KMT6kd mutant in both growth conditions, while the first gene downstream of this putative cluster, i.e. FFUJ_11744, was not significantly up-regulated under either of the two conditions, and FFUJ_11734, located directly upstream of the cluster, was up-regulated only on CM (Tab. S2). Three of the putative STC5 cluster genes (FFUJ_11736, FFUJ_11741 and FFUJ_11743) encode putative cytochrome P450 monooxygenases (CYPs). FFUJ_11737, FFUJ_11739 and FFUJ_11740 encode a putative ABC-transporter, the key enzyme STC5 and a putative Zn2Cys6 transcription factor (TF), respectively. No putative functions could be drawn from the remaining three genes, i.e. FFUJ_11735, FFUJ_11738 and FFUJ_11742 (Fig. 7A; Tab. S3). The amount of precipitated DNA in the ChIP experiments was also quantified at the putative ABC transporter-encoding gene (FFUJ_11737) and at one of the putative CYP-encoding genes (FFUJ_11741) (Fig. 7B). As expected H3K27me3 levels were also reduced at nucleosomes corresponding to these genes, therefore further emphasising the role of Kmt6 in silencing the STC5 cluster genes. Assumed cluster borders were subsequently verified by constitutive over-expression of STC5 as well as of the putative pathway-specific TF-encoding gene, STC5-TF, in the FfSG139/ΔFFUJ_12585 mutant that fails to produce the terpenes (+)-koraiol and ent-kaurene (Tudzynski et al., 2001; Brock et al., 2013), which are produced in high amounts in the wild type and thus hamper identification of new minor compounds. Transformation yielded two mutants for each approach, i.e. OE-STC5_T1 and OE-STC5_T2 as well as OE-STC5-TF_T1 and OE-STC-TF_T3. Successful over-expression of the respective genes was verified by RT-qPCR (Fig. S6A). Fungal strains were grown on solid CM and expression of the putative STC5 cluster genes was analysed by RT-qPCR. Whilst over-expression of STC5 only resulted in up-regulation of STC5 itself (Fig. S7), over-expression of the TF-encoding gene led to an up-regulation of STC5 and of additional six out of the nine predicted STC5 cluster genes (Fig. 7C, Fig. S7). Thus, FFUJ_11735 does most likely not belong to the STC5 gene cluster. This experimental strategy unravelled the most probable cluster borders, i.e. FFUJ_11736-FFUJ_11743, and verified STC5-TF as a pathway-specific TF positively regulating STC5 cluster genes.
Fig. 7. Identification and analysis of the STC5 gene cluster.
A) Organisation of the putative STC5 gene cluster. The STC5 cluster genes (FFUJ_11736-FFUJ_11743) are depicted as black boxes, genes directly upstream (FFUJ_11734, FFUJ_11735) and downstream (FFUJ_11744) of the cluster are shown in grey. The key enzyme-encoding gene, STC5 (FFUJ_11739), and the cluster-specific transcription factor (TF)-encoding gene, TF (FFUJ_11740), are highlighted in yellow and purple, respectively. Arrows indicate direction of translation. B) ChIP experiments at the STC5 gene cluster. The wild type (WT) and KMT6kd mutants were grown on solid CM. ChIP was performed with an anti-H3K27me3-specific antibody (AM39155) as described in the experimental section. Quantification of precipitated DNA was performed by qPCR at the following gene loci: putative ABC-transporter-encoding gene (FFUJ_11737), STC5 (FFUJ_11739) and a putative CYP-encoding gene (FFUJ_11741). In each case, the amount of precipitated DNA in the wild type was arbitrarily set as 1. Precipitated DNA at the STC2 gene locus was used as a negative control and in each case related to the analysed STC5 cluster gene. Experiments were done in technical and biological replicates. Mean values and standard deviations are given in the diagram. A Student's t-test (two sample assuming equal variances) was performed to determine statistical significance. *, p<0.05; **, p<0.01; ***, p<0.001. C) Determination of the STC5 cluster borders by RT-qPCR using the OE-STC5-TF strain. Complementary DNA was retrieved from the wild type and one of the STC5-TF overexpression mutants (OE-TF), grown for 3 days on solid CM. Expression of all STC5 cluster genes (FFUJ_11735-FFUJ_11743) as well as of the genes directly upstream (FFUJ_11734) and downstream (FFUJ_11744) of the cluster was quantified according to the ΔΔCt method (Pfaffl, 2001). Experiments were done in technical and biological replicates giving the same results; only the technical replicate is shown here. Mean values and standard deviations are given in the diagram. Ct values over 32 were calculated as not detectable, nd. For direct comparison expression levels of the wild type were arbitrarily set as 1.
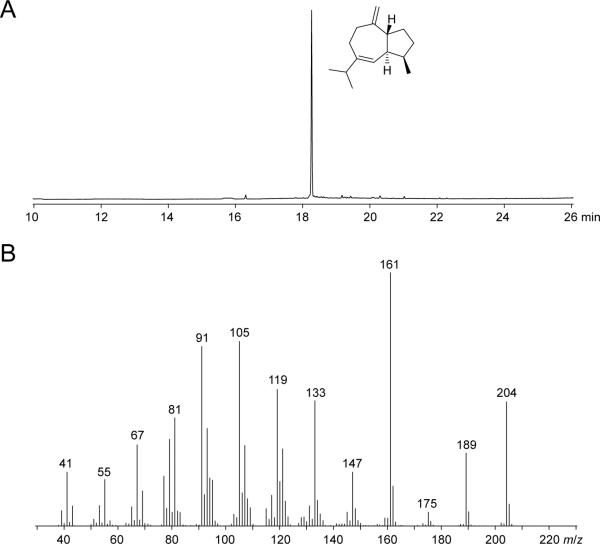
To identify the yet unknown product of this gene cluster, the KMT6kd, the OE-STC5, the OE-STC5-TF mutants and the wild type were grown on solid CM and the emitted volatiles were collected by using a closed-loop stripping apparatus (CLSA), and analysed by gas chromatography-coupled MS (GC-MS). However, no volatile terpene (presumably formed by STC5) could be identified in the headspace extracts of either of the mutant or wild-type strains. A sequence analysis of STC5 revealed a presumably critical amino acid exchange in the highly conserved NSE triad (ND(L,I,V)XSXXX(D,E)) in which the asparagine that is directly involved in binding of the Mg2+ cofactor was exchanged by lysine (KDILSYEKD, Fig. S8). Previous, site-directed mutagenesis experiments on the pentalenene synthase have shown that this exchange is critical for function of the enzyme functioning (Seemann et al., 2002), and suggests that STC5 of the F. fujikuroi strain under study may also be non-functional. This hypothesis was supported by heterologous expression of STC5 in Escherichia coli, because subsequent incubation of the purified enzyme with farnesyl pyrophosphate (FPP) failed to form a terpene. Notably, the critical amino acid exchange (N288K) was not observed in STC5 homologs from other fusaria (Fig. S8A), and is also not present in two different sequenced F. fujikuroi strains, i.e. KSU X-10626 and FGSC 8932 (Chiara et al., 2015) (Fig. S8B). Similarly, only one out of eight recently sequenced F. fujikuroi shows this amino acid exchange (Fig. S8B) (B. Tudzynski and co-workers, unpublished data), suggesting that this mutation is specific to only some F. fujikuroi isolates including the strain used in this study and thus that the STC5 gene cluster is generally functional in F. fujikuroi. To identify the STC5 product, FfSTC5 was exchanged by the putatively intact FmSTC5 gene from the highly related species Fusarium mangiferae driven by the native F. fujikuroi promoter (Fig. S6C). Gene-swapping was performed with simultaneous over-expression of STC5-TF, because STC5 proved to be silent also in F. mangiferae. Due to shortage of resistance markers, we had to first generate a new STC5-TF over-expressing mutant in the SG139 (no ent-kaurene) background. In this strain, SG139/OE-STC5-TF, the F. fujikuroi STC5 gene has been replaced by the F. mangiferae STC5 (Fig. S6C), yielding several FfSG139/FfpromFmSTC5 mutants that showed in loco integration of the STC5 exchange fragment and resulted in over-expression of FmSTC5. This enabled extraction of mRNA and generation of FmSTC5 cDNA from this mutant, which was subsequently cloned into the pYE-Express for heterologous expression in E. coli. The purified enzyme was incubated with FPP, resulting in the production of a sesquiterpene hydrocarbon as indicated by GC-MS analysis ([M]+: m/z = 204, Fig. 8). Structure elucidation via one- and two-dimensional NMR spectroscopic methods in combination with an enantioselective synthesis to establish the absolute configuration of the sesquiterpene hydrocarbon resulted in the structure of (1R,4R,5S)-guaia-6,10(14)-diene. Details of the structure elucidation, enantioselective synthesis and a mechanistic investigation of the terpene cyclase STC5 is published elsewhere (Burkhardt et al., 2016).
Fig. 8. Identification of the STC5 product.
A) Total ion chromatogram of an extract from the enzymatic conversion of FPP by FmSTC5 showing the formation of a single product that was identified as (1R,4R,5S)-guaia-6,10(14)-diene (structure as shown), B) mass spectrum of guaia-6,10(14)-diene.
Discussion
KMT6 is essential in F. fujikuroi
Histones are subject to a variety of PTMs such as acetylation, phosphorylation or methylation and these PTMs confer an important regulatory layer for co-ordinated gene expression in eukaryotes. Not surprisingly, lack of proteins involved in writing, reading or erasing such PTMs have a huge impact on the fungal development (Reyes-Dominguez et al., 2008, 2010, 2012; Connolly et al., 2013; Studt et al., 2013; Cánovas et al., 2014; Chujo and Scott, 2014; Gacek-Matthews et al., 2015; Liu et al., 2015). It is noteworthy, that whilst deletion of the methyltransferase Kmt6-encoding gene was successful in a variety of organisms, including N. crassa (Jamieson et al., 2013), E. festucae (Chujo and Scott, 2014) or the distant relative F. graminearum (Connolly et al., 2013), in this study several attempts to generate homokaryotic mutants in F. fujikuroi failed. Similarly, the exchange of lysine 27 for arginine (H3K27R) did not result in viable transformants, thereby suggesting that Kmt6-mediated gene silencing is crucial for the viability of F. fujikuroi. Besides the catalytic subunit, Kmt6, the PRC2 complex contains three additional core proteins, i.e. the WD40 domain-containing polypeptide Extra Sex Comb (ESC), the C2H2-type zinc finger protein Suppressor of zeste 12 [Su(z)12] and the Nucleosome-remodelling factor Nurf-55. Su(z)12 and Nurf-55 facilitate nucleosome binding, while ESC binds to H3K27me3 thereby stimulating the enzymatic activity of E(z) and therefore is likely to account for the spreading of H3K27me3 on silent chromatin (Hansen et al., 2008; Margueron et al., 2008, 2009; Jamieson et al., 2013; Jiao and Liu, 2015). Homologues for these PRC2 components are present in the genome of F. fujikuroi: FFUJ_12272 (ESC homolog), FFUJ_09784 ([Su(z)12] homolog), FFUJ_03072 (Nurf-55 homolog), and characterisation of these components in the future will shed light on their function in F. fujikuroi.
So far no homokaryotic mutant strains could be generated also for another silencing histone PTM in F. fujikuroi, i.e. the methyltransferase Dim5-encoding gene involved in H3K9 methylation (L. Studt and B. Tudzynski, unpublished data), while lack of the Dim5 homolog was again dispensable for survival in N. crassa, E. festucae and F. graminearum (Connolly et al., 2013; Jamieson et al., 2013; Chujo and Scott, 2014). Recent results in N. crassa showed that the lack of H3K9 methylation resulted in a re-distribution of H3K27me2/3 (Jamieson et al., 2016). Regions of constitutive heterochromatin that normally carry H3K9me3 gained H3K27me2/3 and regions of facultative heterochromatin lost this histone PTM, thereby ensuring maintenance of the constitutive heterochromatic regions. Whether this outcome holds also true for F. fujikuroi still awaits proof. However, it is tempting to speculate that lack and/or re-distribution of H3K27me3 results in de-repression of critical and thus normally silenced gene loci.
Knock-down of KMT6 resulted in differential expression of about one third of the genome, therefore providing an explanation for the pleiotropic effects, such as impaired hyphal growth and the abolished asexual phase of development observed in the mutants. These results emphasise the importance of Kmt6 for normal development. In accordance with this, most of the KMT6kd mutants eventually gained putative suppressor mutation(s) that rescued the wild-type phenotype. Surprisingly, the KMT6 transcript level was much higher in strains with an reversion of the KMT6 phenotype compared to the wild type, and it is tempting to speculate that mutations did occur in a pathway normally limiting KMT6 transcript levels. However, higher KMT6 transcript levels did not lead to a phenotype that is different from the wild type with regard to hyphal growth and asexual development. These results also raise the question as to whether this phenotype is perhaps unique for F. fujikuroi, or if other (more closely related) fungal species also strictly depend on H3K27me3. Notably, H3K9me3 and H3K27me3 appear to be absent in single-celled eukaryotic organisms like S. cerevisiae, while S. pombe only carries H3K9me3 but lacks H3K27me3 (Lachner et al., 2004). Similarly, PRC components are absent in A. nidulans. However, S. pombe and A. nidulans are both still viable upon deletion of the methyltransferase-encoding gene Clr4 and ClrD, respectively (Ivanova et al., 1998; Reyes-Dominguez et al., 2010). In E. festucae both silencing mechanisms, i.e. H3K9me3 and H3K27me3, are dispensable for survival of the fungus (Chujo and Scott, 2014). Therefore, different chromatin-based regulatory mechanisms seem to apply within different fungal clades. Future investigations focusing on additional silencing mechanisms in different fungal species will shed light on these interesting findings.
H3K27me3 is an important layer for the regulation of SM genes in F. fujikuroi
While the association of SM genes with H3K9me3 marks is weak both in F. graminearum (Connolly et al., 2013) and in F. fujikuroi (Wiemann et al., 2013), most of these genes are instead associated with H3K27me3 in both fungi. Only eight out of 47 putative SM gene clusters in F. fujikuroi are located in genomic regions that completely lack H3K27me3, i.e. STC2, NRPS2, NRPS3, NRPS10 and NRPS13, as well as PKS7, PKS10 and PKS19, and accordingly most of these SM genes are not affected by knock-down of KMT6. There are two exceptions, i.e. PKS7 and PKS19. However, both key enzyme-encoding genes are silent in the wild type and downregulated in the KMT6kd mutant, suggesting rather an indirect, if any, regulation by Kmt6. Exceptionally, chromosomes ten and eleven in F. fujikuroi that are specifically enriched for putative SM-related genes show high levels of H3K37me3 almost throughout the whole chromosome arms. Apart from TeTC1 and STC4 involved in the production of neurosporaxanthin and (+)-koraiol (Rodríguez-Ortiz et al., 2009; Brock et al., 2013), respectively, the products of all putative SM gene clusters located on these two chromosomes are cryptic so far and expression is low under laboratory conditions. As expected, SM genes for which gene expression was significantly induced upon KMT6 knock-down were accompanied by reduced H3K27me3 levels at the respective gene loci. Accordingly, several putative novel compounds accumulated in the KMT6kd strains. Therefore, manipulation of the methyltransferase Kmt6 proved to be an effective tool to activate expression of otherwise silent SM gene clusters. More precisely, knock-down of KMT6 activated the expression of four so far cryptic SM key enzyme-encoding genes in F. fujikuroi, i.e. DMATS3, PKS-NRPS1, NRPS4 and STC5.
Heterologous expression of STC5 from F. mangiferae allowed for the identification of a novel fungal metabolite
The highest induction of gene expression upon KMT6 knock-down was observed for STC5 and altogether eight co-regulated cluster genes (FFUJ_11736-FFUJ11743). Along with the key enzyme, this cluster harbours genes encoding for a positively acting pathway-specific transcription factor, a putative transporter and three putative CYPs. Though all relevant cluster genes were up-regulated in the KMT6kd strain, a critical point mutation in the NSE triad rendered STC5 from the F. fujikuroi strain under study non-functional and thus no product was formed. This critical amino acid exchange (N288K) was neither present in STC5 homologues from other fusaria nor observed in most of the sequenced F. fujikuroi strains (Chiara et al., 2015, B. Tudzynski and co-workers, unpublished data), thereby suggesting that the STC5 gene cluster is generally functional in F. fujikuroi. Heterologous expression of the F. mangiferae STC5-homolog in E. coli allowed for the identification of the first pathway-specific intermediate of this so far cryptic gene cluster. FmSTC5 synthesises (1R,4R,5S)-guaia-6,10(14)-diene, a novel sesquiterpene metabolite derived from FPP. Recently, we could show that correction of the F. fujikuroi STC5 via site-directed mutagenesis followed by heterologous expression in E. coli results in the production of the same product (Burkhardt et al., 2016), thereby providing solid evidence that (1R,4R,5S)-guaia-6,10(14)-diene is indeed the first pathway-specific product also in F. fujikuroi. Presence and co-expression of three putative CYP-encoding genes within the gene cluster suggest that (1R,4R,5S)-guaia-6,10(14)-diene is further modified and identification of additional STC5-related metabolites is on-going using generated FfSG139/FfpromFmSTC5 strains. Since the STC5 gene cluster appeared to be silent in the F. fujikuroi wild-type strain, nothing is known about its functions so far. However, presence and conservation of the STC5 gene cluster in several Fusarium spp. suggests that the corresponding metabolite is produced under certain conditions and here might pose an advantage for its producer.
In conclusion, H3K27me3 is a major histone PTM in F. fujikuroi and its absence is fatal for the producing organism. The majority of putative SM-encoding genes fall within regions decorated with H3K27me3 and knock-down of the corresponding histone methyltransferase Kmt6 proved to be an effective tool for their induction. KMT6kd resulted in the activation of four cryptic and otherwise silent SM key enzymes putatively involved in the production of novel and yet to identify SMs and was accompanied by reduced H3K27me3 levels at the respective gene loci, thereby suggesting the direct involvement of Kmt6 in their regulation. Several novel peaks accumulated in the KMT6kd strains and elucidation of their structure is currently in progress. Heterologous expression of a functional homologue of the most strongly up-regulated key enzyme-encoding gene, i.e. STC5, from the related species F. mangiferae enabled identification of the first pathway-specific product (1R,4R,5S)-guaia-6,10(14)-diene.
Experimental Procedures
Fungal strains, media and growth conditions
The wild-type strain F. fujikuroi IMI58289 (Commonwealth Mycological Institute, Kew, United Kingdom) was used as parental strain for deletion and knock-down experiments. Overexpression of STC5 and the respective TF (STC5-TF) were performed in the triple mutant background SG139/ΔΔFFUJ_10353/12585, which no longer produces the most abundant volatile terpenes. To generate this mutant, the key genes for (+)-koraiol and α-acorenol production, STC4 and STC6, respectively (Brock et al., 2013), were deleted in the UV mutant SG139 (kindly provided by J. Avalos, University of Sevilla), which has lost the entire GA gene cluster and, therefore, the ability to release the volatile GA precursor ent-kaurene (Tudzynski et al., 2001). For chemical analyses the wild type and KMT6kd strains were grown on solid CM (Pontecorvo et al., 1953). For identification of a new sesquiterpene the mutant F. fujikuroi SG139 was used as background strain for overexpression of STC5-TF and concomitant expression of F. mangiferae STC5 (FmSTC5).
Cultivations for ChIP-seq and protoplasting were performed as previously described (Studt et al., 2013). For verification of KMT6kd mutants, DNA isolation and western blot analyses, strains were grown for 3 days on solid CM covered with Cellophane sheets at 28 °C in darkness. Microarray-, expression-, SM- and ChIP experiments were performed using strains grown on solid CM and ICI medium (Geissman et al., 1966), covered with cellophane for 3 days (10 days in case of volatile SMs) at 28 °C in darkness. Plate assays were carried out for 7 days at 28 °C in the dark on solid V8 agar (20% v/v vegetable juice) (Campbell Food, Puurs, Belgium), CM, PDA (Sigma-Aldrich, Germany), CD (Sigma-Aldrich, Germany), synthetic ICI and FMM (Reyes-Dominguez et al., 2012). Conidial formation was assessed on solid V8 agar inoculated with 5 mm agar plaques and incubated for two weeks at 20 °C and 12 h/12 h light/dark cycles. Cultivation of S. cerevisiae and E. coli were performed as previously described (Schumacher, 2012).
Plasmid constructions
Generation of pΔKMT6, pH3K27R, pYE-FfSTC5, pYE-FmSTC5 and FfpromFmSTC5_IL was accomplished by yeast recombinational cloning (Colot et al., 2006). In case of pΔKMT6, upstream and downstream regions of KMT6 were amplified using the primer pairs KMT6_5F and KMT6_5R as well as KMT6_3F and KMT6_3R, respectively. All primer pairs used in this study are listed in Supplementary Table S4. The hygromycin resistance cassette, hygR, driven by the A. nidulans trpC promoter sequence was amplified from pCSN44 (Christianson et al., 1992) with hphF//hphR. For amino acid exchange at histone 3 (H3K27R), H3 and 1 kb upstream region were amplified with primer pairs Histone3_Mut_1F//Histone3_Mut_K27R_1R and Histone3_Mut_K27R_2F//Histone3_Mut_2R harbouring the codon for arginine instead of lysine at position 27. The gene was followed by the terminator sequence Tgluc from Botrytis cinerea and hygR amplified with the primer pairs BcGlu-Term-F2//Tgluc-nat1-R and hphR-trpC-T2//hphF using genomic DNA of B. cinerea B05.10 and pCSN44, respectively, as templates. H3 downstream sequence was amplified with the primer pair Histone3_Mut_4F//Histone3_Mut_4R. Obtained fragments were cloned together with the EcoRI/XhoI-restricted pRS426 into S. cerevisiae FY834 yielding pΔKMT6 and pH3K27R (Fig. S2A/B). In case of over-expression, STC5 (FFUJ_11739) and STC5-TF (FFUJ_11740) were amplified together with about 150 bp terminator sequence using the primer pairs GPD-STC5-F//GPD-STC5-R and GPD-OE-TF-11740-F//GPD-OE-TF-11740-R, respectively. Obtained fragments were cloned into S. cerevisiae FY834 together with the HindIII-restricted modified pRS426 (Studt et al., 2013) containing the geneticin resistance cassette, genR, amplified from pKSgen (Bluhm et al., 2008) using the primer pair geni-OE-Prom//geni-OE-Term. The gene of interest is driven by the constitutive gpd promoter amplified from pveAgfp (Wiemann et al., 2010) with the primer pair gpd-yeast-for//gpd-yeast-rev, yielding pOE-STC5 and pOE-STC5-TF (Fig. S6A). For heterologous expression of FfSTC5 and FmSTC5, the coding regions of the genes were amplified from cDNA with heYE-FfSTC5_fwd//heYE-FfSTC5_rev and heYE-FmSTC5_fwd//heYE-FfSTC5_rev, respectively. Obtained fragments were cloned together with the EcoRI/HindIII-restricted pYE-Express (Dickschat et al., 2014) into S. cerevisiae FY834, yielding pYE-FfSTC5 and pYE-FmSTC5 (Fig. S6B). The STC5-TF over-expression construct containing the FmSTC5 gene driven by the F. fujikuroi STC5 promoter was generated by amplifying about 1 kb of the native F. fujikuroi STC5 promoter sequence using the primer pair Ffuj_prom-pRS_1F//Ffuj_prom-Fman_STC5_1R. FmSTC5 (FMAN_14887) was amplified from genomic DNA of F. mangiferae strain MRC7560 (MRC culture collection, National Research Institute for Nutritional Diseases, Tygerberg, South Africa) using the primer pair Fman_STC5-Ffuj_prom 2F//Fman_STC5-Tgluc_2R. The gene was fused to BcTgluc, followed by the nourseothricin resistance cassette, natR, itself driven by the trpC promoter. The fragment trpC-P::natR was amplified from pZPnat1 using a proof-reading polymerase and the primer pair hphF//hphR-trpC-T2 (Schumacher, 2012). The downstream region of FfSTC5 was amplified with Ffuj_term-trpC 3F//Ffuj_term-pRS 3R. S. cerevisiae FY834 was transformed with the obtained fragments together with the EcoRI/XhoI-restricted pRS426 yielding plasmid FfpromFmSTC5_IL (Fig. S6C). Knock-down of KMT6 was accomplished as previously described (Fitzgerald et al., 2004). Hairpin inverted-repeat fragments were amplified from F. fujikuroi genomic DNA using the primer pairs RNAi-Kmt6-NcoI-F1//RNAi-Kmt6-SalI-R1 (A) and RNAi-Kmt6-NotI-F2//RNAi-Kmt6-SalI-R2 (B), containing indicated restriction sites, and a proof-reading polymerase. Fragments A and B were cloned into pGEM®-T Easy (Promega, Massachusetts, USA) yielding pKMT6-RNAi_A and pKMT6-RNAi_B. Both fragments were sequenced, re-isolated from the plasmids by digestion and ligated together with the NcoI/NotI-digested pNDH-OGG (Schumacher, 2012) yielding pKMT6kd (Fig. S2C). A proof-reading polymerase was used for amplification of all plasmid parts and correct assembly of generated plasmids was verified by sequencing and/or restriction (data not shown).
Bacterial transformations and in vitro incubation experiments
Resulting plasmids were shuttled into E. coli BL21 via electroporation and transformed cells were selected and cultivated as described by (Burkhardt et al., 2016). Gene expression was induced by addition of IPTG (final concentration 400 μM). Cells were harvested by centrifugation, resuspended in 10 mL ice-cooled lysis buffer (2 mM Na2HPO4, 0.5 M NaCl, 20 mM imidazole, 1 mM MgCl2) and disrupted by sonication. The target enzyme was purified via Ni2+-NTA-affinity chromatography as described by (Burkhardt et al., 2016). Incubation experiments were done with 2 mL pure enzyme fraction and 2 mL incubation buffer (50 mM Tris-HCl, 10 mM MgCl2, 20% glycerol (v/v), pH 7) containing 0.5 mg/mL FPP. After 3 h of incubation at 28 °C the reaction mixture was extracted with hexane (0.5 mL), dried with MgSO4, and directly subjected to GC-MS analysis.
Fungal transformations
For generation of F. fujikuroi mutants, protoplasts were prepared from the wild-type strain IMI58289, FfSG139/ΔΔFFUJ_10353/12585 mutant (Brock et al., 2013) as well as from FfSG139 (Tudzynski et al., 2001) and fungal transformations were performed as previously described (Tudzynski et al., 1996). About 107 protoplasts were transformed with 10 μg of the generated constructs, i.e. pKMT6kd, pH3K27R, pOE-STC5, pOE-STC5-TF and pFfpromFmSTC5_IL. For KMT6 deletion, the knock-out fragment was amplified from pΔKMT6 with the primer pair KMT6_5F//KMT6_3R (Tab. S4) using TAKARA® polymerase according to the manufacturer's instructions. Plasmids H3K27R and FfpromFmSTC5_IL were restricted prior to transformation with PvuII and ApaI, respectively (Fig. S2B, Fig. S6C). Plasmids pKMT6kd, pOE-STC5 and pOE-STC5-TF were transformed without prior digestion (Fig. S2C, Fig. S6A). Transformed protoplasts were regenerated as described by Tudzynski et al. (1999). The medium contained 100 ppm of the appropriate resistance marker. In the case of KMT6 deletion, homologous recombination of the knock-out construct was verified using primer pairs dia_KMT6_5’//pCSN44-hph-trpC-T and dia_KMT6_3’//pCSN44-trpC-P for upstream and downstream recombination, respectively. Presence of the wild-type gene was tested using the primer pair FfWT_F//FfWT_R (Fig. S2A). Verification of homologous integration and exchange of lysine for arginine was accomplished by PCR and sequencing using the primer pairs Histone3_Mut_K9R_2F//dia_Mut H3 and Histone3_Mut_K9R_2F//hph-hi-F, respectively. Presence of the KMT6kd construct was verified with primer pairs dia_Kmt6_RNAi_I_F//dia_Kmt7_RNAi_I_R and dia_Kmt6_RNAi_II_F//dia_Kmt6_RNAi_ II_R (data not shown) and successful down-regulation of KMT6 transcript levels was analysed by RT-qPCR using the primer pair RTPCR_KMT6_RNAi_F//RT-PCR_KMT6_RNAi_R. FfSG139/ΔΔFFUJ_10353/12585 was transformed with pOE-STC5 and pOE-STC5-TF, yielding the mutant strains OE-STC5 and OE-STC5-TF, respectively. Integration of pOE-STC5 and pOE-STC5-TF was verified by diagnostic PCR using the primer pair Gpd-dia-for//gpd-TF11739-R. Successful over-expression of the respective genes was verified by RT-qPCR and primer pairs qPCR-FFUJ_11739_F//qPCR-FFUJ_11739_R (FFUJ_11739; STC5) and qPCR-FFUJ_11740_F//qPCR-FFUJ_11740_R (FFUJ_11740; STC5-TF) (Fig. S6A). For exchange of FfSTC5 for FmSTC5 under STC5-TF over-expression, the FfSG139 mutant strain was transformed with pOE-STC5-TF, yielding FfGS139/OE-STC5-TF. Next, FfSG139-OE-STC5-TF was transformed with pFfpromFmSTC5_IL. In loco integration was verified using the primer pair STC_man_inloco_dia5F//Tglucseq_R2 and successful over-expression of STC5 was verified as described above.
Standard molecular techniques
For DNA isolation lyophilised mycelium was ground to a fine powder and suspended in extraction buffer as previously described (Cenis, 1992). Isolated DNA was used for PCR amplification. PCR reactions were set up as described elsewhere (Studt et al., 2012). Phusion® polymerase (Finnzymes, Thermo Fisher Scientific, Finland) was used as a proof-reading polymerase. RNA for expression analyses was isolated from lyophilised mycelium using the RNagents total RNA isolation kit (Promega, Mannheim, Germany) according to the manufacturer's instruction. Reverse transcription PCR was performed using the Superscript II (Invitrogen, Groningen, The Netherlands) and 1 μg of template RNA according to the manufacturer's instructions. For microarray analysis RNA was extracted using TRIzol Reagent (Invitrogen™) according to manufacturer's instructions. Generated plasmids were extracted and purified from E. coli with the GeneJET™ plasmid miniprep kit (Fermentas GmbH, St. Leon-Rot, Germany). Sequencing was performed using the BigDye Terminator v3.1 cycle sequencing kit according the manufacturer's instructions. For western blot analysis mycelium from 3 day-old strains was ground to a fine powder with liquid nitrogen and proteins were extracted as described by Gacek-Matthews et al. (2015). Roughly 15 μg of proteins were used for SDS-Page and western blotting. The membrane was probed with anti-H3K27me3 primary antibody (Abcam, ab6002) and anti-rabbit (Sigma A0545) HRP conjugated secondary antibody. Chemoluminescence was detected with Clarity™ ECL Western Substrate and ChemDoc™ XRS (Bio-Rad).
Microarray analysis
Expression data analysis was carried out using the NimbleGen F. fujikuroi custom microarray (Wiemann et al., 2013). For each biological treatment the test was done in triplicate. Hybridisation of microarrays was done at Arrows Biomedical (Münster, Germany). Processing and analysis of the data was performed as previously described (Studt et al., 2013). Microarray data has been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through the GEO Series accession number GSE80479.
Chemical analysis
To identify putative novel compounds both the wild-type and the KMT6kd strains were cultivated on solid CM and synthetic ICI medium. Agar plugs were subsequently extracted with EtOAc/MeOH/CH2Cl2 (3:2:1, v/v) and the samples were characterised via HPLC-HRMS as described in Wiemann et al. (2013), with minor adjustments. A Nucleodur C18 Isis column, 150 mm × 2 mm, 5 μm, from Macherey-Nagel (Düren, Germany) was used at 40 °C with 350 μL/min. Water + 0.1% formic acid was used as solvent A, acetonitrile + 0.1% formic acid as solvent B. The gradient started at 10% B, rising up in 60 min to 100% B. These conditions were held for 5 min to rinse the column; thereafter, an equilibration step at 10% B was run for 10 min. The capillary temperature of the HRMS was set to 300 °C instead of 275 °C.
Volatile SMs emitted by agar plate cultures were trapped on charcoal filters by use of a CLSA as reported previously (Citron et al., 2012). The trapped material was extracted with 50 μL CH2Cl2 and the obtained extracts were directly subjected to GC-MS analysis. The GC-MS analyses were performed as described by (Burkhardt et al., 2016). Retention indices (I) were determined from a homologous series of n-alkanes (C8-C40).
ChIP
ChIP-seq experiments, preparation of sequencing libraries and subsequent data processing was accomplished as previously described (Wiemann et al., 2013). The ChIP-seq data are available from the NCBI Gene Expression Omnibus (GEO) under the series accession numbers GSE80479. We observed frequent reversion of the KMT6kd phenotype that occurred more readily in liquid than on solid medium. Therefore, gene-specific ChIP was performed with strains grown on solid medium to secure absence of putative suppressor mutations or other reversion phenomena, and thus appropriate execution of the experiments. Crosslinking and sample preparation was performed as described above. ChIP was performed using an anti-H3K27me3 antibody (AM39155) according to Gacek-Matthews et al. (2015). ChIP experiments were done in a biological repetition. Each ChIP experiment was set up in duplicate (technical repetition), and for each technical replicate two qPCRs were run in parallel (technical repetition).
qPCR and RT-qPCR
Verification of the microarray data was performed by RT-qPCR for STC5 genes (FFUJ_11738-44), DMATS3, PKS-NRPS1, NRPS4 and NRPS22 using primer pairs qPCR-FFUJ11738-11744_F//qPCR-FFUJ11738-11744_R, DMATS3_RTPCR_F//DMATS3_RTPCR_R, PKS1_RTPCR_F//PKS1_RTPCR_R, NRPS4_RTPCR_F//NRPS4_RTPCR_R and qPCR-BEA1_F//qPCR-BEA1_R, respectively. In all cases the primer efficiency was kept between 90-110%, annealing temperature was set to 60 °C. Expression of the wild type grown on solid ICI medium was arbitrarily set to 1, Ct values beyond 32 were taken as non-detectable, i.e. not expressed. Results were calculated according to the ΔΔCt (Pfaffl, 2001). Expression of all tested genes was normalised to the expression of actin (FRACRTPCRFW//FRACRTPCRRV), a GDP-mannose transporter (FGMTRTPCRFW//FGMTRTPCRRV) and ubiquitin (FUBRTPCRFW// FUBRTPCRRV) according to Wiemann et al. (2013).
To reveal changes in H3K27me3 at SM genes qPCR was performed using the primer pairs STC5_chipqPCR_F//STC5_chipqPCR_R, 11741_chipqPCR_F//11741_chipqPCR_R (putative P450), 11737_chipqPCR_F//11737_chipqPCR_R (putative ABC-transporter), NRPS22_chipqPCR_F//NRPS22_chipqPCR_R (FFUJ_09296), STC2_chipqPCR_F//STC2_chipqPCR_R (FFUJ_00969). Relative amounts of DNA were calculated by dividing the immunoprecipitated DNA by the input DNA. qPCR and RT-qPCR experiments were performed in technical and biological replicates. All qPCR and RT-qPCR reactions were performed with the BioRad iQ SYBR Green Supermix and the iCycler Thermal Cycler (BioRad). Sequences of F. fujikuroi ORFs were extracted from the publicly available genome sequence of F. fujikuroi (Wiemann et al., 2013).
Supplementary Material
Acknowledgements
We thank Ulrich Güldener for microarray and ChIP-seq data submission, Eduard Spitzer and Kristina M. Smith for microarray and ChIP-seq data analysis, respectively, Tobias Pöppelmann for help with the LC-HRMS experiments and Kathleen Huß for excellent technical assistance. We thank Joseph Strauss and Harald Berger for very helpful discussions. We are grateful to Brian Williamson and Joseph Strauss for critical reading of the manuscript. The work was funded by the DFG (TU101/17-3, HU 730/9-3, DI1536/7, GRK1409) and during a stay of Lena Studt at OSU by an NIH grant (GM097637) to MF.
Footnotes
We declare that no conflict of interest exists.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bargen KW, Niehaus EM, Bergander K, Brun R, Tudzynski B, Humpf HU. Structure elucidation and antimalarial activity of apicidin F: An apicidin-like compound produced by Fusarium fujikuroi. J. Nat. Prod. 2013;76:2136–2140. doi: 10.1021/np4006053. [DOI] [PubMed] [Google Scholar]
- Von Bargen KW, Niehaus E-M, Krug I, Bergander K, Würthwein E-U, Tudzynski B, Humpf H-U. Isolation and structure elucidation of fujikurins A–D: products of the PKS19 gene cluster in Fusarium fujikuroi. J. Nat. Prod. 2015;78:1809–1815. doi: 10.1021/np5008137. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bluhm BH, Kim H, Butchko R. a E., Woloshuk CP. Involvement of ZFR1 of Fusarium verticillioides in kernel colonization and the regulation of FST1, a putative sugar transporter gene required for fumonisin biosynthesis on maize kernels. Mol. Plant Pathol. 2008;9:203–211. doi: 10.1111/j.1364-3703.2007.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode HB, Bethe B, Höfs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang Y-M, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Suppl. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a Regulator of Secondary Metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA. Regulation of fungal secondary metabolism. Nat. Rev. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Brock NL, Huss K, Tudzynski B, Dickschat JS. Genetic dissection of sesquiterpene biosynthesis by Fusarium fujikuroi. ChemBioChem. 2013;14:311–315. doi: 10.1002/cbic.201200695. [DOI] [PubMed] [Google Scholar]
- Burkhardt I, Siemon T, Henrot M, Studt L, Rösler SM, Christmann M, Tudzynski B, Dickschat JS. Mechanistic Characterisation of two sesquiterpene cyclases from the plant pathogenic fungus Fusarium fujikuroi. Angew. Chem. Int. Ed. Engl. 2016;55 doi: 10.1002/anie.201603782. DOI: 10.1002/anie.201603782R1. [DOI] [PubMed] [Google Scholar]
- Cánovas D, Marcos AT, Gacek A, Ramos MS, Gutiérrez G, Reyes-Domínguez Y, Strauss J. The histone acetyltransferase GcnE (GCN5) plays a central role in the regulation of Aspergillus asexual development. Genetics. 2014;197:1175–1189. doi: 10.1534/genetics.114.165688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara M, Fanelli F, Mulè G, Logrieco AF, Pesole G, Leslie JF, Horner DS, Toomajian C. Genome sequencing of multiple isolates highlights subtelomeric genomic diversity within Fusarium fujikuroi. Genome Biol. Evol. 2015;7:3062–3069. doi: 10.1093/gbe/evv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Chujo T, Scott B. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte-plant symbiosis. Mol. Microbiol. 2014;92:413–434. doi: 10.1111/mmi.12567. [DOI] [PubMed] [Google Scholar]
- Citron CA, Rabe P, Dickschat JS. The scent of bacteria: headspace analysis for the discovery of natural products. J. Nat. Prod. 2012;75:1765–1776. doi: 10.1021/np300468h. [DOI] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-thoughput gene knockout procedure for Neurospora crassa reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. 2006;113:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly LR, Smith KM, Freitag M. The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 2013;9:e1003916. doi: 10.1371/journal.pgen.1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickschat JS, Pahirulzaman KA, Rabe P, Klapschinski TA. An improved technique for the rapid chemical characterisation of bacterial terpene cyclases. ChemBioChem. 2014;15:810–814. doi: 10.1002/cbic.201300763. [DOI] [PubMed] [Google Scholar]
- Dreyfuss M, Härri E, Hofmann H, Kobel H, Pache W, Tscherter H. Cyclosporin A and C. Eur. J. Appl. Microbiol. 1976;3:125–133. [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald A, Van Kan JAL, Plummer KM. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 2004;41:963–971. doi: 10.1016/j.fgb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Fleming A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929;10:226–236. [Google Scholar]
- Gacek A, Strauss J. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 2012;95:1389–1404. doi: 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacek-Matthews A, Noble LM, Gruber C, Berger H, Sulyok M, Marcos AT, Strauss J, Andrianopoulos A. KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans. Mol. Microbiol. 2015;96:839–860. doi: 10.1111/mmi.12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissman TA, Verbiscar AJ, Phinney BO, Cragg G. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry. 1966;5:933–947. [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJ. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bargen KW, Niehaus EM, Bergander K, Brun R, Tudzynski B, Humpf HU. Structure elucidation and antimalarial activity of apicidin F: An apicidin-like compound produced by Fusarium fujikuroi. J. Nat. Prod. 2013;76:2136–2140. doi: 10.1021/np4006053. [DOI] [PubMed] [Google Scholar]
- Von Bargen KW, Niehaus E-M, Krug I, Bergander K, Würthwein E-U, Tudzynski B, Humpf H-U. Isolation and structure elucidation of fujikurins A–D: products of the PKS19 gene cluster in Fusarium fujikuroi. J. Nat. Prod. 2015;78:1809–1815. doi: 10.1021/np5008137. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Schümann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bluhm BH, Kim H, Butchko R. a E., Woloshuk CP. Involvement of ZFR1 of Fusarium verticillioides in kernel colonization and the regulation of FST1, a putative sugar transporter gene required for fumonisin biosynthesis on maize kernels. Mol. Plant Pathol. 2008;9:203–211. doi: 10.1111/j.1364-3703.2007.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode HB, Bethe B, Höfs R, Zeeck A. Big effects from small changes: possible ways to explore nature's chemical diversity. ChemBioChem. 2002;3:619–627. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang Y-M, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Suppl. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a Regulator of Secondary Metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA. Regulation of fungal secondary metabolism. Nat. Rev. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Brock NL, Huss K, Tudzynski B, Dickschat JS. Genetic dissection of sesquiterpene biosynthesis by Fusarium fujikuroi. ChemBioChem. 2013;14:311–315. doi: 10.1002/cbic.201200695. [DOI] [PubMed] [Google Scholar]
- Burkhardt I, Siemon T, Henrot M, Studt L, Rösler SM, Christmann M, Tudzynski B, Dickschat JS. Mechanistic Characterisation of two sesquiterpene cyclases from the plant pathogenic fungus Fusarium fujikuroi. Angew. Chem. Int. Ed. Engl. 2016;55 doi: 10.1002/anie.201603782. DOI: 10.1002/anie.201603782R1. [DOI] [PubMed] [Google Scholar]
- Cánovas D, Marcos AT, Gacek A, Ramos MS, Gutiérrez G, Reyes-Domínguez Y, Strauss J. The histone acetyltransferase GcnE (GCN5) plays a central role in the regulation of Aspergillus asexual development. Genetics. 2014;197:1175–1189. doi: 10.1534/genetics.114.165688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenis JL. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 1992;20:2380. doi: 10.1093/nar/20.9.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara M, Fanelli F, Mulè G, Logrieco AF, Pesole G, Leslie JF, Horner DS, Toomajian C. Genome sequencing of multiple isolates highlights subtelomeric genomic diversity within Fusarium fujikuroi. Genome Biol. Evol. 2015;7:3062–3069. doi: 10.1093/gbe/evv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Chujo T, Scott B. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte-plant symbiosis. Mol. Microbiol. 2014;92:413–434. doi: 10.1111/mmi.12567. [DOI] [PubMed] [Google Scholar]
- Citron CA, Rabe P, Dickschat JS. The scent of bacteria: headspace analysis for the discovery of natural products. J. Nat. Prod. 2012;75:1765–1776. doi: 10.1021/np300468h. [DOI] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. A high-thoughput gene knockout procedure for Neurospora crassa reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. 2006;113:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly LR, Smith KM, Freitag M. The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 2013;9:e1003916. doi: 10.1371/journal.pgen.1003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickschat JS, Pahirulzaman KA, Rabe P, Klapschinski TA. An improved technique for the rapid chemical characterisation of bacterial terpene cyclases. ChemBioChem. 2014;15:810–814. doi: 10.1002/cbic.201300763. [DOI] [PubMed] [Google Scholar]
- Dreyfuss M, Härri E, Hofmann H, Kobel H, Pache W, Tscherter H. Cyclosporin A and C. Eur. J. Appl. Microbiol. 1976;3:125–133. [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald A, Van Kan JAL, Plummer KM. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 2004;41:963–971. doi: 10.1016/j.fgb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Fleming A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929;10:226–236. [Google Scholar]
- Gacek A, Strauss J. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 2012;95:1389–1404. doi: 10.1007/s00253-012-4208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacek-Matthews A, Noble LM, Gruber C, Berger H, Sulyok M, Marcos AT, Strauss J, Andrianopoulos A. KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans. Mol. Microbiol. 2015;96:839–860. doi: 10.1111/mmi.12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissman TA, Verbiscar AJ, Phinney BO, Cragg G. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry. 1966;5:933–947. [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJ. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- Jamieson K, Rountree MR, Lewis Z. a, Stajich JE, Selker EU. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc. Natl. Acad. Sci. U. S. A. 2013;110:6027–6032. doi: 10.1073/pnas.1303750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson K, Wiles ET, McNaught KJ, Sidoli S, Leggett N, Shao Y, Garcia BA, Selker EU. Loss of HP1 causes depletion of H3K27me3 from facultative heterochromatin and gain of H3K27me2 at constitutive heterochromatin. Genome Res. 2016;26:97–107. doi: 10.1101/gr.194555.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science (80−. ) 2015;350 doi: 10.1126/science.aac4383. aac4383–aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N, Hohn T. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- König CC, Scherlach K, Schroeckh V, Horn F, Nietzsche S, Brakhage AA, Hertweck C. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. ChemBioChem. 2013;14:938–942. doi: 10.1002/cbic.201300070. [DOI] [PubMed] [Google Scholar]
- Lachner M, Sengupta R, Schotta G, Jenuwein T. Trilogies of histone lysine methylation as epigenetic landmarks of the eukaryotic genome. Cold Spring Harb. Symp. Quant. Biol. 2004;69:209–218. doi: 10.1101/sqb.2004.69.209. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu N, Yin Y, Chen Y, Jiang J, Ma Z. Histone H3K4 methylation regulates hyphal growth, secondary metabolism and multiple stress responses in Fusarium graminearum. Environ. Microbiol. 2015;17:4615–4630. doi: 10.1111/1462-2920.12993. [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. 32 Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse CB, Studt L, Janevska S, Sieber CMK, Arndt B, Espino JJ, Humpf H-U, Güldener U, Tudzynski B. The global regulator FfSge1 is required for expression of secondary metabolite gene clusters, but not for pathogenicity in Fusarium fujikuroi. Environ. Microbiol. 2014;17:2690–2708. doi: 10.1111/1462-2920.12592. [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Niehaus EM, Von Bargen KW, Espino JJ, Pfannmüller A, Humpf HU, Tudzynski B. Characterization of the fusaric acid gene cluster in Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2014;98:1749–1762. doi: 10.1007/s00253-013-5453-1. [DOI] [PubMed] [Google Scholar]
- Niehaus E-M, Janevska S, von Bargen KW, Sieber CMK, Harrer H, Humpf H-U, Tudzynski B. Apicidin F: characterization and genetic manipulation of a new secondary metabolite gene cluster in the rice pathogen Fusarium fujikuroi. PLoS One. 2014;9:e103336. doi: 10.1371/journal.pone.0103336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus EM, Kleigrewe K, Wiemann P, Studt L, Sieber CMK, Connolly LR, Freitag M, Güldener U, Tudzynski B, Humpf HU. Genetic manipulation of the Fusarium fujikuroi fusarin gene cluster yields insight into the complex regulation and fusarin biosynthetic pathway. Chem. Biol. 2013;20:1055–1066. doi: 10.1016/j.chembiol.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Nützmann H, Reyes-dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schümann J, Hertweck C, Strauss J, Brakhage AA. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga / Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. 2011;108:14282–14287. doi: 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, MacDonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell. Mol. Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Boedi S, Sulyok M, Wiesenberger G, Stoppacher N, Krska R, Strauss J. Heterochromatin influences the secondary metabolite profile in the plant pathogen Fusarium graminearum. Fungal Genet. Biol. 2012;49:39–47. doi: 10.1016/j.fgb.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Narendja F, Berger H, Gallmetzer A, Fernandez-Martin R, Garcia I, Scazzocchio C, Strauss J. Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryot. Cell. 2008;7:656–663. doi: 10.1128/EC.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Ortiz R, Limón MC, Avalos J. Regulation of carotenogenesis and secondary metabolism by nitrogen in wild-type Fusarium fujikuroi and carotenoid- 33 overproducing mutants. Appl. Environ. Microbiol. 2009;75:405–413. doi: 10.1128/AEM.01089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler SM, Sieber CMK, Humpf H-U, Tudzynski B. Interplay between pathway-specific and global regulation of the fumonisin gene cluster in the rice pathogen Fusarium fujikuroi. Appl. Microbiol. Biotechnol. 2016 doi: 10.1007/s00253-016-7426-7. doi:10.1007/s00253–016–7426–7. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Güldener U, Mannhaupt G, Münsterkötter M, Mewes HW. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S, Takemoto D, Tapper BA, Lane GA, Fraser K, Scott B. Functional analysis of an indole-diterpene gene cluster for lolitrem B biosynthesis in the grass endosymbiont Epichloë festucae. FEBS Lett. 2012;586:2563–2569. doi: 10.1016/j.febslet.2012.06.035. [DOI] [PubMed] [Google Scholar]
- Schotanus K, Soyer JL, Connolly LR, Grandaubert J, Happel P, Smith KM, Freitag M, Stukenbrock EH. Histone modifications rather than the novel regional centromeres of Zymoseptoria tritici distinguish core and accessory chromosomes. Epigenetics Chromatin. 2015;8:41. doi: 10.1186/s13072-015-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh V, Scherlach K, Nützmann H-W, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J. Tools for Botrytis cinerea: New expression vectors make the gray mold fungus more accessible to cell biology approaches. Fungal Genet. Biol. 2012;49:483–497. doi: 10.1016/j.fgb.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Seemann M, Zhai G, de Kraker J-W, Paschall CM, Christianson DW, Cane DE. Pentalenene synthase. Analysis of active site residues by site-directed mutagenesis. J. Am. Chem. Soc. 2002;124:7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- Shaver S, Casas-Mollano JA, Cerny RL, Cerutti H. Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics. 2014;5:301–312. doi: 10.4161/epi.5.4.11608. [DOI] [PubMed] [Google Scholar]
- Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup AA, Chiang YM, Bok JW, Reyes-Dominguez Y, Oakley BR, Wang CCC, Strauss J, Keller NP. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol. 2012;86:314–330. doi: 10.1111/j.1365-2958.2012.08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt L, Janevska S, Niehaus E-M, Burkhardt I, Arndt B, Sieber CMK, Humpf H-U, Dickschat JS, Tudzynski B. Two separate key enzymes and two pathway-specific transcription factors are involved in fusaric acid biosynthesis in Fusarium fujikuroi. Environ. Microbiol. 2016;18:936–956. doi: 10.1111/1462-2920.13150. [DOI] [PubMed] [Google Scholar]
- Studt L, Schmidt FJ, Jahn L, Sieber CMK, Connolly LR, Niehaus E-M, Freitag M, Humpf H-U, Tudzynski B. Two histone deacetylases, FfHda1 and FfHda2, are important for Fusarium fujikuroi secondary metabolism and virulence. Appl. Environ. Microbiol. 2013;79:7719–7734. doi: 10.1128/AEM.01557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt L, Wiemann P, Kleigrewe K, Humpf H-U, Tudzynski B. Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl. Environ. Microbiol. 2012;78:4468–4480. doi: 10.1128/AEM.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski B, Hedden P, Carrera E, Gaskin P. The P450-4 gene of Gibberella fujikuroi encodes ent-kaurene oxidase in the gibberellin biosynthesis pathway. Appl. Environ. Microbiol. 2001;67:3514–3522. doi: 10.1128/AEM.67.8.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski B, Homann V, Feng B, Marzluf G. a. Isolation, characterization and disruption of the areA nitrogen regulatory gene of Gibberella fujikuroi. Mol. Gen. Genet. 1999;261:106–114. doi: 10.1007/s004380050947. [DOI] [PubMed] [Google Scholar]
- Tudzynski B, Mende K, Weltring K-M, Kinghorn JR, Unkles SE. The Gibberella fujikuroi nia D gene encoding nitrate reductase: isolation, sequence, homologous transformation and electrophoretic karyotype location. Microbiology. 1996;142:553–539. doi: 10.1099/13500872-142-3-533. [DOI] [PubMed] [Google Scholar]
- Wiemann P, Brown DW, Kleigrewe K, Bok JW, Keller NP, Humpf HU, Tudzynski B. FfVel1 and fflae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol. Microbiol. 2010;77:972–994. doi: 10.1111/j.1365-2958.2010.07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P, Sieber CMK, von Bargen KW, Studt L, Niehaus EM, Espino JJ, Huß K, Michielse CB, Albermann S, Wagner D, Bergner SV, Connolly LR, Fischer A, Reuter G, Kleigrewe K, Bald T, Wingfield BD, Ophir R, Freeman S, et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P, Willmann A, Straeten M, Kleigrewe K, Beyer M, Humpf HU, Tudzynski B. Biosynthesis of the red pigment bikaverin in Fusarium fujikuroi: Genes, their function and regulation. Mol. Microbiol. 2009;72:931–946. doi: 10.1111/j.1365-2958.2009.06695.x. [DOI] [PubMed] [Google Scholar]
- Williams RB, Henrikson JC, Hoover AR, Lee AE, Cichewicz RH. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- Zhang X, Tamaru H, Khan SI, Horton JR, Keefe LJ, Selker EU, Cheng X. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. Cell. 2002;111:117–127. doi: 10.1016/s0092-8674(02)00999-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.