Abstract
Transcriptomic and proteomic approaches have separately proven effective at identifying novel mechanisms affecting addiction-related behavior; however, it is difficult to prioritize the many promising leads from each approach. A convergent secondary analysis of proteomic and transcriptomic results can glean additional information to help prioritize promising leads. The current study is a secondary analysis of the convergence of recently published separate transcriptomic and proteomic analyses of nucleus accumbens (NAc) tissue from rats subjected to environmental enrichment vs. isolation and cocaine self-administration vs. saline. Multiple bioinformatics approaches (e.g. Gene Ontology (GO) analysis, Ingenuity Pathway Analysis (IPA), and Gene Set Enrichment Analysis (GSEA)) were used to interrogate these rich data sets. Although there was little correspondence between mRNA vs. protein at the individual target level, good correspondence was found at the level of gene/protein sets, particularly for the environmental enrichment manipulation. These data identify gene sets where there is a positive relationship between changes in mRNA and protein (e.g. glycolysis, ATP synthesis, translation elongation factor activity, etc.) and gene sets where there is an inverse relationship (e.g. ribosomes, Rho GTPase signaling, protein ubiquitination, etc.). Overall environmental enrichment produced better correspondence than cocaine self-administration. The individual targets contributing to mRNA and protein effects were largely not overlapping. As a whole, these results confirm that robust transcriptomic and proteomic data sets can provide similar results at the gene/protein set level even when there is little correspondence at the individual target level and little overlap in the targets contributing to the effects.
Keywords: proteomics, transcriptomics, environmental enrichment, cocaine, self-administration, addiction
INTRODUCTION
As a naturally derived psychostimulant, cocaine has robust adverse effects on both the body and the mind. Despite the high abuse liability of cocaine, most individuals who use cocaine do not become addicted. Approximately 6% of individuals who used cocaine were dependent 24 months after their first use (O’Brien and Anthony, 2005). Therefore, understanding the individual differences in vulnerability to cocaine addiction is necessary to discover novel therapeutic targets. The environmental enrichment paradigm produces a protective addiction phenotype in rats as a non-drug, non-surgical, and non-genetic manipulation (Bardo et al., 2001; Green et al., 2002, 2010; Chauvet et al., 2009; Solinas et al., 2010). Animals reared in an enriched condition (EC) are exposed to exercise, social interaction, and novelty (access to toys that are changed daily). Compared with rats reared in an isolated condition (IC) or a pair-housed standard condition (SC), EC rats exhibit decreased drug-taking and -seeking behavior in the acquisition, maintenance, extinction, and reinstatement phases of self-administration of ethanol, cocaine, amphetamine, methylphenidate, etc. (Bardo et al., 2001; Green et al., 2002, 2010; Deehan et al., 2007).
To investigate the molecular mechanisms of the protective addiction phenotype, a transcriptomic study using high-throughput RNA sequencing and a proteomic study using high-performance liquid chromatography with tandem mass spectrometry (nanoLC–MS/MS) were conducted to explore the differential responses to cocaine in EC and IC rats at both the mRNA and protein level (Lichi et al., 2014; Zhang et al., 2016). More than 14,000 transcripts and 1900 proteins were identified and subsequently quantified in these studies.
Much was learned from the mRNA and protein analyses alone; the current secondary analyses spotlight additional information that can be gleaned from a combined analysis of the two data sets. This analysis is somewhat unique, as it is rare to have mRNA and protein data from the same animals.
EXPERIMENTAL PROCEDURES
Animals and behavior
Male Sprague–Dawley rats (Harlan, Houston) arrived at 21 days of age and were randomly assigned to an EC condition, 12 per cage (70 × 70 × 70 cm) with hard plastic children’s toys rearranged daily, or an IC condition, singly housed in standard polycarbonate cages. The rats continued in these conditions throughout behavioral testing, which commenced at 51 days of age. The facility at UTMB is an AAALAC-accredited facility. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and conform to The NIH Guide for the Care and Use of Laboratory Animals, 2011.
The overall behavioral component was a 2 × 2 design comparing EC vs. IC rats self-administering cocaine vs. saline. To eliminate differences in cocaine intake between EC and IC groups, rats were first food regulated down to 85% of free-feeding body weight and trained to bar press for 45 mg banana-flavored sucrose pellets. Rats were given access to sucrose pellets for 15 min daily, with the first session being a fixed ratio 1 (FR1) schedule. The ratio was incremented daily for four additional sessions. After the FR5 session, rats were allowed to free-feed for one week prior to catheter surgery.
For surgery, rats were anesthetized with 100 mg/kg ketamine and 10 mg/kg xylazine. Silastic catheters were constructed in-house. The jugular vein was isolated and the catheter inserted 4.5 cm. The catheter exited through the skin on the upper back. Catheters were flushed daily with 0.1 ml of a sterile saline solution containing heparin (30.0 U/ml) and Ticarcillin (250,000 U/ml). Rats were allowed one week for recovery.
For self-administration, rats were placed in an operant chamber (Med-Associated, St. Albans, VT, USA) and allowed to self-administer 0.5 mg/kg/infusion cocaine hydrochloride (NIDA Drug Supply Program, Research Triangle) for 2 h daily for a total of 14 days on an FR1 schedule of reinforcement. Rats were able to self-administer up to 30 infusions per session and were removed from the chambers at the end of the session (2 h) or when 30 infusions were delivered. The 30-infusion limit was used to eliminate EC/IC differences in intake, which could skew the results. Tissue from rats that did not acquire self-administration was not harvested. Rats were sacrificed 3 h after the beginning of the self-administration session on the last day, where the nucleus accumbens (NAc) tissue was dissected and flash-frozen for later processing. The left side of the NAc of each rat was used for quantitative RNA sequencing while the right was used for quantitative proteomics analysis.
Quantitative transcriptomics study
Detailed methods can be found in (Zhang et al., 2016). Briefly, RNA from the left NAc was purified with the RNeasy kit (Qiagen Valencia, CA, USA) and reverse transcribed into RNA. Adapters were ligated to “a-tailed” ends. RNA was sequenced using an Illumina HiSeq 1000 system and individual reads were mapped to each rat using Tophat (v2.0.4) and Bowtie2 (v2.0.0.6) software packages with a reference genome (RN4). The quality of mapping was checked with FastQC (v0.9.1). The “count” data were analyzed using the log-transformed “trimmed mean for M-values” (TMM) for normalization and tagwise dispersion with EdgeR (v3.0.8). A likelihood ratio F-test was used to analyze the main effect of environmental enrichment and cocaine, as well as the environment × cocaine interaction.
The quantitative transcriptomic data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE88736 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE88736).
Quantitative proteomics study
A full description of methods can be found in (Lichti et al., 2014). Briefly, the right NAc from two rats in the same group were pooled randomly for protein extraction. Tissue samples were washed in ice cold Tris-buffered saline (TBS), homogenized in ice cold buffer (TBS, pH 7.4, 1% Igepal-CA630 (NP-40), 1× protease inhibitor cocktail, 20 mM NaF, 1 mM Na3VO4, 10 mM DTT, and 5 mM EDTA) and centrifuged (750×g, 2 min, 4 °C). The pellet with the nuclear fraction was set aside while the top fraction was removed and centrifuged separately (20,000×g, 20 min, 4 °C). Methanol and chloroform (v/v 1:4:1) was added to the resulting supernatant and kept at 24 °C for 15–30, vortexing every 5 min, and centrifuged (16,000×g, 20 min, 4 °C). Acetone was added (500 μL) to remove methanol and chloroform. The membrane fraction (20,000×g pellet) was treated similarly to remove lipids (i.e. membrane fraction). All fractions were finally dissolved in urea buffer (6 M urea, 1% NP-40, 20 mM Tris–HCl pH 7.4, 1x protease inhibitor cocktail, 10 mM DTT and 5 mM EDTA) and stored at −80° C until preparation for LC–MS.
For nanoLC–MS, protein extracts were digested with trypsin and incubated at 37 °C overnight. The peptide mixture was extracted with a C18 Zip Tip (Pierce, Thermo Fisher Scientific, Waltham, MA, USA), eluted with 90% acetonitrile in 0.1% TFA, dried and resuspended with 5 mM Tris, pH 7.4 (25 μL). Peptide mixtures from each group were randomized and analyzed by nanoLC–MS/MS with a nanoLC chromatography system (Eksigent, Sciex, Framingham, MA, USA), coupled on-line to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) through a nanospray ion source. Peptide samples were injected into the trap column and eluted by gradient elution. LC–MS/MS data were acquired with XCalibur (v2.1.0, Thermo). Total run time was 104 min. MS files were imported into Progenesis LC–MS (v4.1, Nonlinear Dynamics) for m/z and retention time alignment. The top five spectra for each feature were exported and database searched in PEAKS (v6, Bioinformatics Solutions) against a combined UniProtKB/SwissProt rat-mouse database (v. Sept 2013), which was appended with the common Repository of Adventitious Proteins (cRAP) contaminant database. A filtered peptide intensity list was log2 transformed in Dante-R and combined with protein abundances (RRollup). Proteins were quantified by a two-way ANOVA, and p-value adjustment performed based on the Benjamini and Hochberg method (Benjamini and Hochberg, 1995). LC–MS data files are available via ProteomeXchange with identifier PXD000990.
Secondary bioinformatic analyses
The Perseus software package was first used to annotate individual targets with Gene Ontology (GO) annotations, aggregate scores within each gene set, and analyze gene sets using a 2D annotation enrichment analysis (Cox and Mann, 2012). A gene set was deemed significant at p<0.05. Next, an IPA Comparison Analysis was performed for lists of transcripts and proteins significantly regulated at the p<0.05 cutoff level of significance for the individual primary analyses. The IPA analysis uses Fisher’s exact test to generate p-values for curated transcript/protein sets separately grouped by canonical pathways, upstream regulators, and biological functions and diseases.
Venn diagrams were used to depict significantly-regulated individual mRNA/protein targets (Figs. 2A and 6A) in specific IPA canonical pathways and significantly regulated gene sets (Figs. 2B–D and 6B–D) at the p<0.05 level of significance.
To analyze the data without p-value cutoffs, Gene Set Enrichment Analyses (GSEA) were used to analyze which curated gene sets showed corresponding and statistically significant changes in mRNA or protein between environmentally enriched versus isolated animals or animals that self-administered cocaine versus saline. GSEA results in an enrichment score: a running-sum statistic based on whether the genes in the set are enriched at the top of all the ranked genes (positive value) or at the bottom (negative value). In order to compare across gene sets, the enrichment scores are converted into normalized enrichment scores (NESs) calculated by dividing the original enrichment score by the average of enrichment scores against all permutations of the dataset (from the GSEA user manual).
Scatterplot data were visualized using the Tableau software package (http://www.tableau.com/).
RESULTS
The primary analyses of tissue from NAc of enriched or isolated animals that self-administered cocaine or saline determined that environmental enrichment regulated 3393 of 14,309 transcripts (p<0.05; (Zhang et al., 2016) and 117 of 1917 proteins (Lichti et al., 2014). Cocaine regulated 1274 transcripts and 52 proteins. The current study is a secondary analysis of these previous proteomic and transcriptomic analyses to gain a more thorough understanding of the overall effects of environmental enrichment and cocaine.
Environmental enrichment
The mRNA and protein data were compared using the Perseus software package for the enriched versus isolated animals. Fig. 1A shows the comparison of mRNA and protein data for three large Gene Ontology (GO) categories: Biological Processes (GOBP), Molecular Functions (GOMF), and Cellular Compartment (GOCC). Each category contains many gene sets or groups of genes that are segregated based on a specific biological process, molecular function, or cellular compartment. A larger circle indicates a higher −log (p) value for the GO gene set. We found that for environmental enrichment, mRNA and protein Gene Ontology data show a positive correlation at the gene set level, indicated by the regression line (R2=0.109, p<0.0001, Fig. 1A).
Fig. 1.
Comparison of mRNA and protein regulation by environmental enrichment at the gene set level via Perseus. (A) Correlation of all gene sets comparing mRNA and protein data. Larger symbol indicates higher −log(p) value for the gene set. Colors of symbols correspond to colors in panels (B–F) of this figure. (B) Members of the Gene Ontology Molecular Function Ribonucleoprotein Complex gene set overlaid on all genes identified with mRNA and protein analyses. (C) Members of the Gene Ontology Molecular Function Synaptic Membrane gene set highlighted. (D) Members of the Gene Ontology Molecular Function Adherens Junction gene set highlighted. (E) Members of the Gene Ontology Biological Process Glycolysis gene set highlighted. (F) Members of the Gene Ontology Biological Process Purine Nucleoside Triphosphate Biosynthetic Process gene set highlighted. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In addition to an overall correlation in Gene Ontology, we also found that specific gene sets with similar function or cellular compartment tended to show coordinated regulation at the mRNA and protein levels. For example, the orange squares in Fig. 1A are clustered in the top right quadrant, indicating mRNA and protein levels were increased in the enriched animals. These data points are all related to mitochondria and include the gene sets: mitochondrial part (GOCC; p=5.4E−6), mitochondrion organization (GOBP; p=0.02), mitochondrial membrane part (GOCC; 7.5E−6), and mitochondrial membrane part (GOMF; 1.4E−4). Gene sets related to glycolysis (dark green) and gene sets related to the synthesis of ATP (purple) also showed increases in mRNA and increases in protein (Fig. 1A). The individual genes from the glycolysis (GOBP; p=1.9E−3) gene set are highlighted in Panel E while the genes from the purine nucleoside triphosphate biosynthetic process (GOBP; p=1.6E−5) gene set are highlighted in Panel F. Not only are the similar gene sets segregated into specific quadrants related to increases in mRNA and increases in protein, but individual genes in the gene sets are also showing correspondence in mRNA and protein. These results indicate environmentally enriched animals have higher mRNA and protein related to mitochondria, ATP, and glycolysis than isolated animals.
Gene sets related to synapses and adherens junctions in Fig. 1A show decreases in mRNA and corresponding decreases in protein in enriched animals. The gene sets synaptic membrane (GOMF; p=3.2E−4) and adherens junction (GOMF; p=2.4E−3) are shown in Panels C and D, respectively. These individual gene sets also show that individual genes are decreased by environmental enrichment at both the mRNA and protein levels.
Gene sets related to ribosomes, translation, and splicing are found in the lower right quadrant, indicating enriched animals have an increase at the mRNA level but a decrease at the protein level (Fig. 1A, light blue). The gene set ribonucleoprotein complex (GOMF; p=3.0E−9) is shown in Panel B and the majority of the genes in this gene set show opposite regulation by enrichment in mRNA and protein.
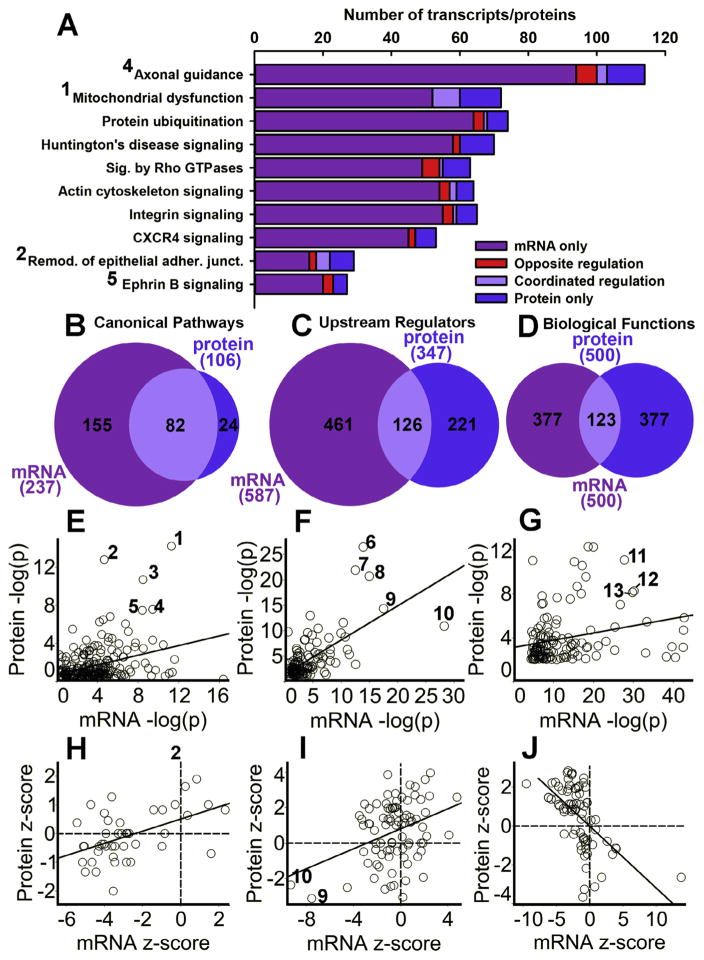
In order to examine the similarities and differences in the biological functions and pathways regulated by environmental enrichment, we used Ingenuity Pathway Analysis (IPA) to compare mRNA and protein data. Fig. 2A depicts several top-ranked canonical pathways for both protein and mRNA regulated by environmental enrichment. The bar Venn diagrams illustrate the significantly regulated transcripts that were identified in the transcriptomic study only (purple), the proteins identified in the proteomics study only (blue), and the transcripts/proteins which were identified with both and showed coordinated regulation (increased in both mRNA and protein, or decreased in both mRNA and protein; light purple) and those that showed opposite regulation (increased in one analysis, decreased in the other analysis; red) for the environmental enrichment main effect. Overall, many more transcripts were quantified than proteins, leading to a greater number of regulated targets; however, there seems to be little coordinated regulation of a given target by environmental enrichment at both the mRNA and protein level (light purple), with the exception of Mitochondrial Dysfunction.
Fig. 2.
Environmental enrichment-induced protein vs. mRNA regulation at the gene set level via Ingenuity Pathway Analysis. (A) Venn diagrams showing degree of overlap of target gene regulation for some top-regulated Canonical Pathways by environmental enrichment. Dark purple or left-most part of diagram denotes number of targets regulated exclusively at the mRNA level and blue or right-most sections represent exclusive regulation at the protein level. Light purple in the middle denotes coordinated regulation of both protein and mRNA, and red in the middle represents number of oppositely regulated targets by environmental enrichment. (B–D) Venn diagrams showing the degree of overlap of the number of significant Canonical Pathways (B), Upstream Regulators (C), and Biological Functions (D) between the mRNA and protein analyses. Negative log (p) scores (E–G) and Z-score correlations (H–J) for mRNA and proteins of individual Canonical Pathways (E, H), Upstream Regulators (F, I), and Biological Functions (G, J). Numbers in (A–I) denote Canonical Pathways: Mitochondrial Dysfunction (1), Remodeling of Epithelial Adherens Junctions (2), Oxidative Phosphorylation (3), Axonal Guidance Signaling (4), and Ephrin Receptor Signaling (5); the Upstream Regulators MAPT (6), APP (7), PSEN1 (8), CD437 (9), and RICTOR (10); and the Biological Functions Movement Disorders (11), Organization of Cytoplasm (12), and Organization of Cytoskeleton (13). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Despite a lack of coordinated regulation of individual targets, there was a good correspondence for significantly regulated canonical pathways, upstream regulators, and biological functions, (Panels B–D, respectively) for the environmental enrichment main effect. For example, 237 canonical pathways were significantly regulated by enrichment in the transcriptomic analysis, 106 were statistically significant in the proteomic analysis, and 82 canonical pathways were significantly regulated in both data sets (Fig. 2B). To identify top-regulated sets for both mRNA and protein, the −log (p) values and the z-scores for the canonical pathways, upstream regulators, and biological functions analyses were plotted for enrichment. All three IPA analyses showed significant correlations in −log (p) values and z-scores for environmental enrichment (Fig. 2 E–J, p<0.05). Top canonical pathways from Fig. 2E included (1) Mitochondrial Dysfunction, (2) Remodeling of Epithelial Adherens Junctions, (3) Oxidative Phosphorylation, (4) Axonal Guidance Signaling, and (5) Ephrin Receptor Signaling. Top upstream regulators included (6) MAPT (microtubuleassociated protein tau), (7) APP (amyloid precursor protein), (8) PSEN1 (presenilin 1), (9) the retinoic acid receptor gamma agonist CD437, and (10) RICTOR (rapamycin-insensitive companion of mTOR, see also Fig. 3E–F). Top biological functions included (11) Movement Disorders, (12) Organization of Cytoplasm, and (13) Organization of Cytoskeleton.
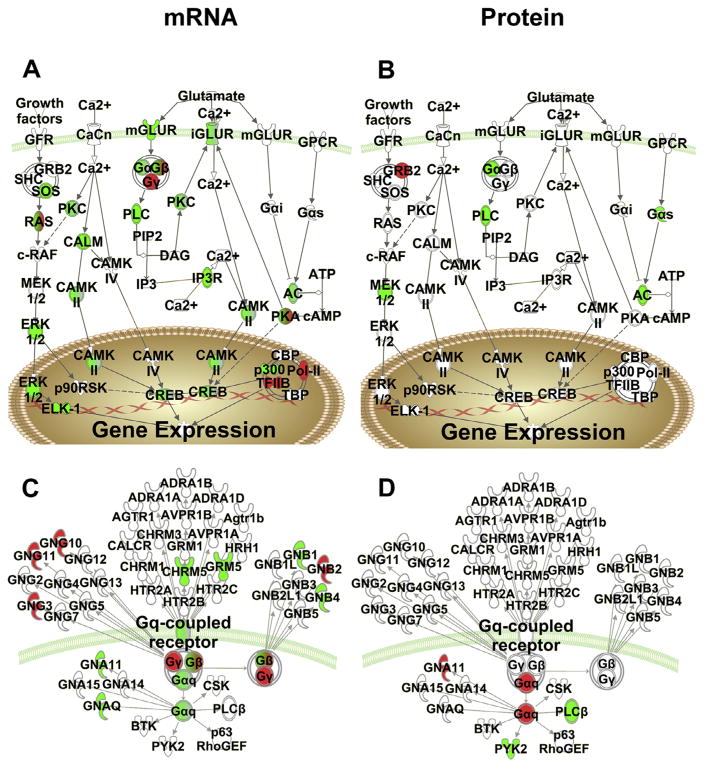
Fig. 3.
Environmental enrichment regulation of specific Canonical Pathways at the mRNA level (left) and protein level (right). (A, B) Comparison of regulated targets for the CREB signaling pathway. Red denotes upregulation by environmental enrichment and green denotes down-regulation. (C, D) Comparison of regulated targets for G-protein coupled receptor signaling. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Closer inspection of additional addiction-related canonical pathways significant in both mRNA and protein data sets for the environmental enrichment main effect underscore the lack of correspondence at the individual target level. For example, the cAMP response element binding protein (CREB) Signaling Pathway (mRNA −log (p)=6.93; protein −log (p)=3.32; Fig. 3A, B), and the Gαq Signaling Pathway (mRNA −log (p)=2.42; protein −log (p)=1.69; Fig. 3C, D) were significantly regulated by environmental enrichment, but none of the individual targets were coordinately regulated in protein and mRNA (for AC, PLC and Gα, different members of these groups were regulated at the protein and mRNA levels). This is illustrated in Fig. 3 where the colored symbols (green for downregulated and red for upregulated) are not the same between the mRNA (panels A and C) and the protein (panels B and D) for environmental enrichment. If the symbol is white it means it was not significantly regulated by environmental enrichment. These data are further showing that the significantly regulated individual mRNAs and proteins do not overlap for environmental enrichment but some of the same pathways are identified in both analyses.
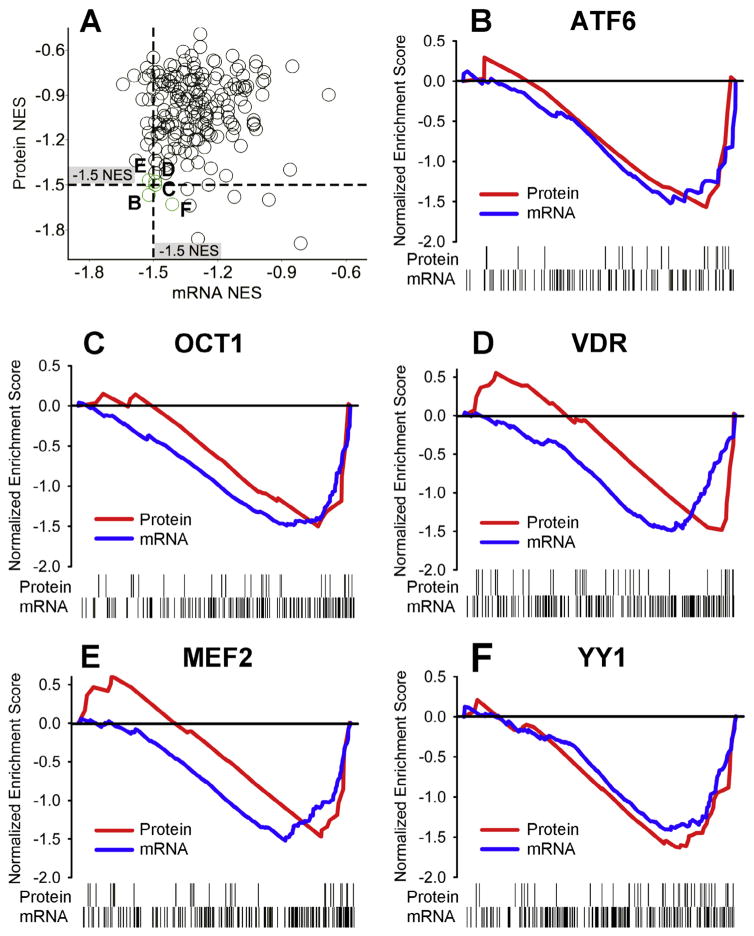
We analyzed the NES of conserved Transcription Factor Targets (TFT) gene sets from the Molecular Signature Database (MSigDB) (Subramanian et al., 2005) regulated by environmental enrichment. We were particularly interested in coordinated regulation of transcription factor targets because several transcription factors have been previously identified as involved in the protective addiction phenotype of environmental enrichment (Green et al., 2010; Pavlovsky et al., 2013; Zhang et al., 2014). When we compared the NESs for the mRNA vs. protein (Fig. 4A), we identified several coordinately-regulated transcription factor target sets for the environmental enrichment main effect (shown in Panels B–F). Specifically, the transcription factors activating transcription factor 6 (ATF6) (mRNA NES=−1.52, p=0.02; protein NES=−1.57, p=0.03), octamer-binding transcription factor 1 (OCT1) (mRNA NES=−1.49, p=0.006; protein NES=−1.5, p=0.03), vitamin D receptor (VDR) (mRNA NES=−1.48, p=0.004; protein NES=−1.48, p=0.04), myocyte enhancer factor 2 (MEF2) (mRNA NES=−1.52, p=0.008; protein NES=−1.47, p=0.04), and yin and yang 1 transcription factor (YY1) (mRNA NES=−1.42, p=0.08; protein NES=−1.63, p=0.01) showed coordinated regulation of protein and mRNA by environmental enrichment.
Fig. 4.
Environmental enrichment-induced regulation of target genes of specific transcription factors. (A) Gene Set Enrichment Analysis correlation of normalized enrichment scores (NES) for top-regulated transcription factor target genes. The labels (B–F) on Panel A label the green circles that correspond to the transcription factor target genes shown in Panels B–F of this figure. (B–F) Running sum NESs for ranked targets that are conserved targets of ATF6, OCT1, VDR, MEF2, and YY1, respectively. Downward deflection of the graph indicates the targets of these transcription factors are downregulated in enriched rats in the protein (red) or mRNA (blue) data. Vertical ticks along the bottom of each panel indicate where the gene targets appear in the ranked list of genes for the protein and the mRNA data. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Cocaine
Using the Perseus software package we compared the mRNA and protein GO data for the animals that self-administered cocaine vs. saline. The correspondence between the mRNA and protein data for three large GO categories (Biological Processes (GOBP), Molecular Functions (GOMF), and Cellular Compartment (GOCC)) is shown in Fig. 5A for the cocaine main effect. We found a positive correlation between the mRNA and protein data for the main effect of cocaine (R2=0.02; p<0.0001); however, the correlation for cocaine is less than that of environmental enrichment (R2=0.109, p<0.0001, Fig. 1A). This likely reflects the greater number of targets regulated by enrichment (mRNA=3393; protein=117) vs. cocaine (mRNA=1274; protein=52).
Fig. 5.
Comparison of mRNA and protein regulation by cocaine at the gene set level via Perseus. (A) Correlation of all gene sets comparing mRNA and protein. Larger symbol indicates higher −log(p) value for the gene set. Colors of symbols correspond to colors in panels B–F of this figure. (B) Members of the Gene Ontology Cellular Compartment Ribosome gene set highlighted overlaid on all genes identified with mRNA and protein analyses. (C) Members of the Gene Ontology Cellular Compartment Presynaptic Membrane gene set highlighted. (D) Members of the Gene Ontology Biological Process Biological Adhesion gene set highlighted. (E) Members of the Gene Ontology Cellular Compartment Mitochondrial Membrane Part gene set highlighted. (F) Members of the Gene Ontology Biological Process ATP Biosynthetic Process gene set highlighted. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As with environmental enrichment, we found clusters of the same categories of gene sets including gene sets involved in ribosomes, synapses, adhesion, mitochondrial, and ATP synthesis for the cocaine main effect. Ribosomal gene sets for cocaine are found to have increased mRNA levels but decreased protein levels similar to environmental enrichment. The GOCC gene set for Ribosome is shown in Fig. 5B (p=6.2E−6). Cocaine increased mRNA and protein targets in mitochondria and ATP synthesis gene sets Mitochondrial membrane part (GOCC; p=5.5E−6) and GOBP ATP biosynthetic process (p=6.3E−4; Fig. 5E, F, respectively). Gene sets related to synapses (e.g. Presynaptic membrane, p=8.2E−3; Fig. 5C) and adhesion (e.g. Biological adhesion, p=0.019; Fig. 5D) show coordinated decreases inmRNAand protein for the cocaine main effect.
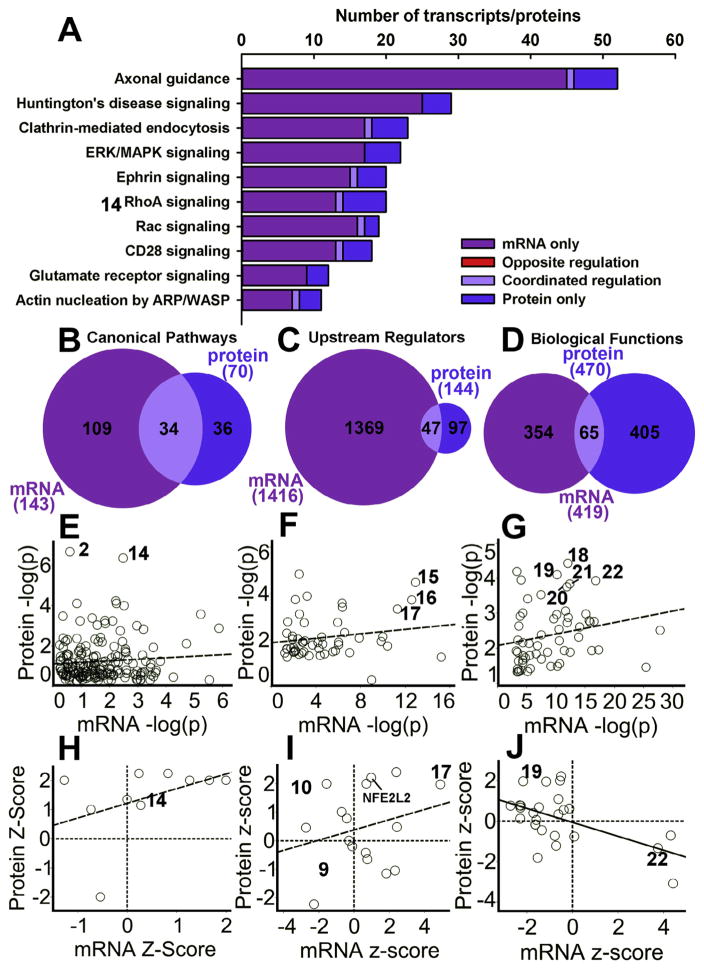
We explored the similarities and differences in the biological functions and pathways regulated by cocaine in the mRNA and protein data using IPA. Compared to the environmental enrichment data, there was much less opposite regulation between mRNA and protein in top-ranked canonical pathways (Fig. 6A). We found as with enrichment that the individual mRNAs and proteins within a specific pathway showed little correspondence, but there was more overlap at the canonical pathway level (34 of 70 significant protein pathways; Fig. 6B). There was considerably less overlap for upstream regulators and biological functions (Fig. 6 panels C, D). The canonical pathway RhoA signaling was a top-scoring canonical pathway (Fig. 6E, number 14). Top upstream regulators for cocaine included (15) huntingtin (HTT), (16) APP and (17) the cAMP activator forskolin (see also Fig. 7E, F). Top biological functions for cocaine included (18) Disorders of the basal ganglia, (19) Size of body, (20) Huntington’s disease, (21) Neuromuscular disease, and 22) Movement disorders. Analyzing the −log (p) values for protein vs. mRNA for Canonical Pathways, Upstream Regulators, and Diseases and Biological Functions did not produce significant correlations for −log (p) values from cocaine regulation (Fig. 6E–G), and only the Diseases and Biological Functions analysis produced a significant z-score correlation (Fig. 6J).
Fig. 6.
Cocaine-induced protein vs. mRNA regulation at the gene set level via Ingenuity Pathway Analysis. (A) Venn diagrams showing degree of overlap of target gene regulation for top-regulated Canonical Pathways by the cocaine main effect. Dark purple or left-most part of diagram denotes number of targets regulated exclusively at the mRNA level and blue or right-most sections represent exclusive regulation at the protein level. Light purple denotes coordinated regulation of both protein and mRNA, and red represents number of oppositely regulated targets. (B–D) Venn diagrams showing the degree of overlap of the number of significant Canonical Pathways (B), Upstream Regulators (C), and Biological Functions (D) between the mRNA and protein analyses for the cocaine main effect. Negative log(p) scores (E–G) and Z-score correlations (H–J) for mRNA and proteins of individual Canonical Pathways (E, H), Upstream Regulators (F, I), and Biological Functions (G, J). Numbers in A–J denote Canonical Pathways: Remodeling of Epithelial Adherens Junctions (2), and RhoA Signaling (14); the Upstream Regulators CD437 (9), RICTOR (10), NFE2L2, HTT (15), APP (16), and Forskolin (17); and the Biological Functions Disorder of Basal Ganglia (18), Size of Body (19), Huntington’s Disease (20), Neuromuscular Disease (21), and Movement Disorders (22). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
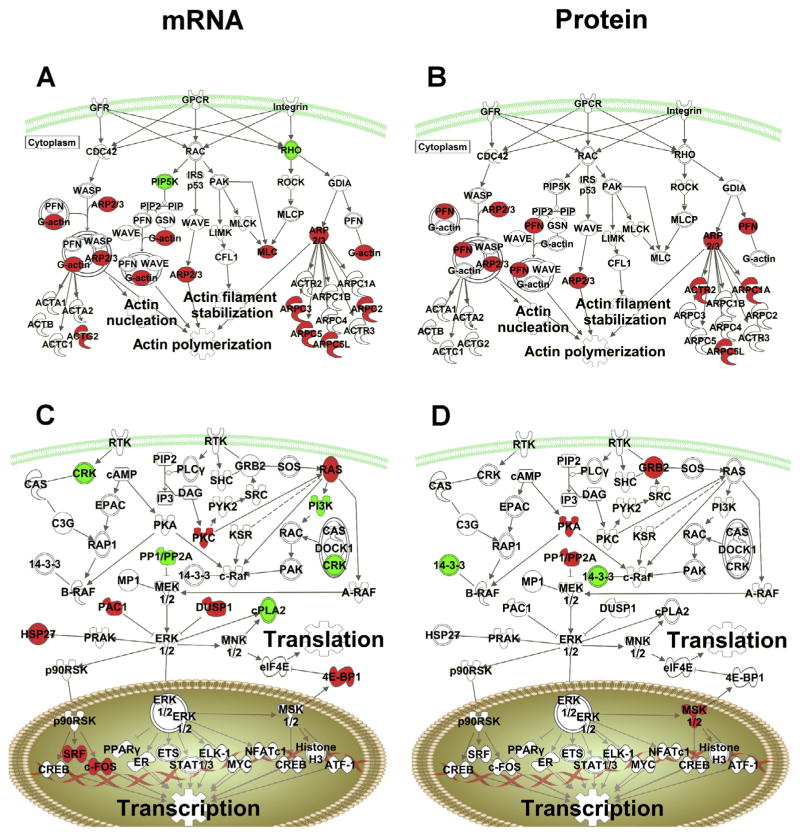
Cocaine regulation of specific Canonical Pathways at the mRNA level (left) and protein level (right). (A and B) Comparison of cocaine regulated targets for the Actin pathway. Red denotes upregulation by cocaine and green denotes down-regulation. (C and D) Comparison of regulated targets for the ERK/MAPK signaling pathway by cocaine. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We examined pathways from IPA that were significantly regulated by cocaine at the mRNA and the protein level, and we found that the significant canonical pathway for Regulation of actin-based motility by Rho was significant for protein (−log (p)=3.38, Z-score= 2.0) and mRNA ((−log (p)=1.41, Z-score= 1.7) yet had only ARPC5L in common (Fig. 7A, B). The ERK/MAPK signaling pathway had no overlap at the individual target level for the cocaine main effect (protein (−log (p)=3.12, mRNA (−log (p)=1.87; Fig. 7C, D). These data suggest that the mRNA and protein analyses are not identifying the same individual genes as contributing to the cocaine or the environmental enrichment effects but both levels of analysis are pointing toward similar pathways and gene sets.
DISCUSSION
These results show clearly that complementary proteomic and transcriptomic studies can identify similar gene sets, canonical pathways, and upstream regulators even when there is little correspondence of regulation among individual transcripts vs. proteins. This underscores the robustness of the bioinformatic approaches in the face of the high Type I and Type II error inherent to such analyses. However, these results also demonstrate that manipulations producing more robust regulation (in this case, environmental enrichment regulated 3393 transcripts and 117 proteins) result in greater correspondence among pathways than less robust manipulations (cocaine only regulated 52 proteins and 1274 transcripts), as one might expect. Overall more transcripts were identified than proteins, which is most likely due to factors idiosyncratic to both the next generation RNA sequencing and the nanoLC–MS/MS. However, even though (1) more mRNA transcripts were included in the pathway analysis than protein, and (2) genes are differentially regulated in mRNA and protein levels, the comparison analysis shows that the results concur at the gene set level despite these differences.
The advantage of assessing both the proteome and transcriptome from the same set of animals is that the gene sets significantly regulated at both levels are most likely to be substantive. This is most evident when looking at cocaine effects, as so much is already known about this drug. Several of the top pathways regulated at both the mRNA and protein levels have been identified previously as being important for cocaine neuroplasticity, including ERK/MAPK signaling (Berhow et al., 1996; Pan et al., 2011), RhoA signaling (Kim et al., 2009), RAC signaling (Dietz et al., 2012), Ephrin signaling (Yue et al., 1999), and glutamate signaling (Garcia-Keller et al., 2015). Additionally, the upstream regulator forskolin was identified by both mRNA and protein analyses. Forskolin is an activator of the transcription factor CREB, which has been shown to be an important mediator of cocaine self-administration (Green et al., 2010). Because the cocaine data identify so many previously validated mechanisms, we can have utmost confidence in the environmental enrichment results.
Because environmental enrichment produces a protective addiction phenotype (Green et al., 2002, 2010; Thiel et al., 2009; Chauvet et al., 2012), identifying molecular mechanisms contributing to this phenotype will likely provide new mechanisms for the prevention of and resilience to addiction behavior. For example, the Upstream Regulator analysis identified the retinoic acid receptor agonist CD437, implicating low retinoic acid signaling in the environmental enrichment phenotype; our recent data validate this signaling pathway as a likely mediator of the enrichment phenotype by showing the inverse: increasing retinoic acid signaling potentiates cocaine-taking and -seeking behavior (manuscript in review). The current data suggest several additional avenues for possible future therapeutic development. Several of the pathways regulated at the protein and mRNA level for environmental enrichment fall into the category of cell to cell cytoskeletal organization, including Axonal guidance, Actin cytoskeleton signaling, Remodeling of epithelial adherens junctions, and Ephrin B signaling. Several significant gene sub-sets within axonal guidance underscore its importance, including Signaling by Rho GTPases, CXCR4 signaling, and ephrinB signaling. Energy metabolism was a recurring theme, with Mitochondrial dysfunction and Oxidative phosphorylation pathways being regulated. Other pathways are involved with intracellular signaling cascades, particularly G-α12–13 signaling, G-β/γ signaling, rhoA signaling and CREB signaling. These pathways have many “druggable” targets for pharmacotherapeutic development.
It is interesting that environmental enrichment and cocaine have similar effects at the gene set level. For example, enrichment (Fig. 1A) and cocaine (Fig. 5A) both increase mRNA and protein for energy metabolism constituents. Both manipulations decrease mRNA and protein for adherens junctions and synaptic constituents. Additionally, both manipulations increase mRNA expression but at the same time decrease protein expression of constituents of ribonucleoprotein complexes. Using heat maps of expression to draw conclusions, we interpret these results as a reflection of the IC saline group being qualitatively different from the other three groups. Thus, environmental enrichment and cocaine self-administration both tax the idle brain in similar ways.
This idea supports our hypothesis that environmental enrichment is a very mild form of stress that inoculates the brain against addiction (Crofton et al., 2015). Inoculation stress is the process of developing resilience to future stressful events by first being exposed to mildly stressful experiences early in life. Environmental enrichment constitutes a chronic mild stress situation as the animals live in a complex environment, which is changed daily, and they interact non-aggressively with conspecifics. This experience early in life (P21–P51) inoculates enriched rats against subsequent stressors and/or drugs of abuse, creating a protective addiction phenotype that is a non-drug, non-surgical, and non-genetic manipulation (Crofton et al., 2015).
For this experiment, enriched cocaine rats were housed with enriched saline rats, so we cannot preclude the possibility of a “rub-off” effect between cocaine and saline rats. However, the return of the rats to the homecage after self-administration was coincident with the daily toy change when saline and cocaine rats all exhibit extremely high locomotor activity.
CONCLUSION
This paper demonstrates a convergence in results from parallel transcriptomic and proteomic analyses, and additionally demonstrates that the combined analysis of these data can help to narrow the many exciting leads from robust manipulations such as environmental enrichment and cocaine self-administration. These new leads are currently being evaluated as novel therapeutic targets for the treatment and/or prevention of addiction.
Acknowledgments
FUNDING
This work was supported by the National Institute on Drug Abuse grants DA029091 (Thomas A. Green) and T32 DA007287, and supported by the Clinical and Translational Science Award (CTSA) to The University of Texas Medical Branch UL1TR01439.
We would like to thank the Molecular Genomics Core at UTMB for performing the next generation RNA sequencing and the Mass Spectrometry Core of the Biomolecular Resource Facility at UTMB. EJC and YZ contributed equally and are co-first authors. EJC, YZ, and TAG designed the study, collected the data, analyzed the data, performed the analyses, and wrote the manuscript. XF, DL, FK, MS, BAL, HMS, CFL collected the data and analyzed the data. BAL, HMS, CFL also edited the manuscript and aided in interpretation.
Abbreviations
- APP
amyloid precursor protein
- ATF
activating transcription factor 6
- CREB
cAMP response element binding protein
- EC
enriched condition
- EDTA
ethylenediaminetetraacetic acid
- ERK
extracellular signal regulated kinase
- FR1
fixed ratio 1
- GO
Gene Ontology
- GOBP
Gene Ontology Biological Processes
- GOCC
Gene Ontology Cellular Compartment
- GOMF
Gene Ontology Molecular Functions
- GSEA
Gene Set Enrichment Analysis
- IC
isolated condition
- IPA
Ingenuity Pathway Analysis
- MAPK
mitogen-activated protein kinase
- MAPT
microtubule-associated protein tau
- MEF2
myocyte enhancer factor 2
- NAc
nucleus accumbens
- NES
normalized enrichment score
- NFE2L2
nuclear factor, erthroid 2-like 2
- OCT1
octamer-binding transcription factor 1
- PSEN1
presenlin 1
- RICTOR
rapamycin-insensitive companion of mTOR
- TBS
tris-buffered saline
- TFT
Transcription Factor Targets
- VDR
vitamin D receptor
- YY1
yin and yang 1 transcription factor
References
- Bardo MT, Klebaur JE, Valone JM, Deaton C. Environmental enrichment decreases intravenous self-administration of amphetamine in female and male rats. Psychopharmacology. 2001;155:278–284. doi: 10.1007/s002130100720. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995:289–300. [Google Scholar]
- Berhow MT, Hiroi N, Nestler EJ. Regulation of ERK (extracellular signal regulated kinase), part of the neurotrophin signal transduction cascade, in the rat mesolimbic dopamine system by chronic exposure to morphine or cocaine. J Neurosci. 1996;16:4707–4715. doi: 10.1523/JNEUROSCI.16-15-04707.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet C, Lardeux V, Goldberg SR, Jaber M, Solinas M. Environmental enrichment reduces cocaine seeking and reinstatement induced by cues and stress but not by cocaine. Neuropsychopharmacology. 2009;34:2767–2778. doi: 10.1038/npp.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet C, Goldberg SR, Jaber M, Solinas M. Effects of environmental enrichment on the incubation of cocaine craving. Neuropharmacology. 2012;63:635–641. doi: 10.1016/j.neuropharm.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012;13(Suppl 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofton EJ, Zhang Y, Green TA. Inoculation stress hypothesis of environmental enrichment. Neurosci Biobehav Rev. 2015;49c:19–31. doi: 10.1016/j.neubiorev.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deehan GA, Cain ME, Kiefer SW. Differential rearing conditions alter operant responding for ethanol in outbred rats. Alcohol Clin Exp Res. 2007;31:1692–1698. doi: 10.1111/j.1530-0277.2007.00466.x. [DOI] [PubMed] [Google Scholar]
- Dietz DM, Sun H, Lobo MK, Cahill ME, Chadwick B, Gao V, Koo JW, Mazei-Robison MS, Dias C, Maze I, Damez-Werno D, Dietz KC, Scobie KN, Ferguson D, Christoffel D, Ohnishi Y, Hodes GE, Zheng Y, Neve RL, Hahn KM, Russo SJ, Nestler EJ. Rac1 is essential in cocaine-induced structural plasticity of nucleus accumbens neurons. Nat Neurosci. 2012;15:891–896. doi: 10.1038/nn.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Keller C, Kupchik YM, Gipson CD, Brown RM, Spencer S, Bollati F, Esparza MA, Roberts-Wolfe DJ, Heinsbroek JA, Bobadilla AC, Cancela LM, Kalivas PW. Glutamatergic mechanisms of comorbidity between acute stress and cocaine self-administration. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green TA, Gehrke BJ, Bardo MT. Environmental enrichment decreases intravenous amphetamine self-administration in rats: dose–response functions for fixed- and progressive-ratio schedules. Psychopharmacology. 2002;162:373–378. doi: 10.1007/s00213-002-1134-y. [DOI] [PubMed] [Google Scholar]
- Green TA, Alibhai IN, Roybal CN, Winstanley CA, Theobald DE, Birnbaum SG, Graham AR, Unterberg S, Graham DL, Vialou V, Bass CE, Terwilliger EF, Bardo MT, Nestler EJ. Environmental enrichment produces a behavioral phenotype mediated by low cyclic adenosine monophosphate response element binding (CREB) activity in the nucleus accumbens. Biol Psychiatry. 2010;67:28–35. doi: 10.1016/j.biopsych.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Shin SR, Kim S, Jeon S, Kim JH. Cocaine regulates ezrin-radixin-moesin proteins and RhoA signaling in the nucleus accumbens. Neuroscience. 2009;163:501–505. doi: 10.1016/j.neuroscience.2009.06.067. [DOI] [PubMed] [Google Scholar]
- Lichti CF, Fan X, English RD, Zhang Y, Li D, Kong F, Sinha M, Andersen CR, Spratt H, Luxon BA, Green TA. Environmental enrichment alters protein expression as well as the proteomic response to cocaine in rat nucleus accumbens. Front Behav Neurosci. 2014;8:246. doi: 10.3389/fnbeh.2014.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien MS, Anthony JC. Risk of becoming cocaine dependent: epidemiological estimates for the United States, 2000–2001. Neuropsychopharmacology. 2005;30:1006–1018. doi: 10.1038/sj.npp.1300681. [DOI] [PubMed] [Google Scholar]
- Pan B, Zhong P, Sun D, Liu QS. Extracellular signal-regulated kinase signaling in the ventral tegmental area mediates cocaine-induced synaptic plasticity and rewarding effects. J Neurosci. 2011;31:11244–11255. doi: 10.1523/JNEUROSCI.1040-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovsky AA, Boehning D, Li D, Zhang Y, Fan X, Green TA. Psychological stress, cocaine and natural reward each induce endoplasmic reticulum stress genes in rat brain. Neuroscience. 2013;246:160–169. doi: 10.1016/j.neuroscience.2013.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas M, Thiriet N, Chauvet C, Jaber M. Prevention and treatment of drug addiction by environmental enrichment. Prog Neurobiol. 2010;92:572–592. doi: 10.1016/j.pneurobio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel KJ, Sanabria F, Pentkowski NS, Neisewander JL. Anti-craving effects of environmental enrichment. Int J Neuropsychopharmacol. 2009;12:1151–1156. doi: 10.1017/S1461145709990472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Widmer DA, Halladay AK, Cerretti DP, Wagner GC, Dreyer JL, Zhou R. Specification of distinct dopaminergic neural pathways: roles of the Eph family receptor EphB1 and ligand ephrin-B2. J Neurosci. 1999;19:2090–2101. doi: 10.1523/JNEUROSCI.19-06-02090.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Crofton EJ, Li D, Lobo MK, Fan X, Nestler EJ, Green TA. Overexpression of DeltaFosB in nucleus accumbens mimics the protective addiction phenotype, but not the protective depression phenotype of environmental enrichment. Front Behav Neurosci. 2014;8:297. doi: 10.3389/fnbeh.2014.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kong F, Crofton EJ, Dragosljvich SN, Sinha M, Li D, Fan X, Koshy S, Hommel JD, Spratt HM, Luxon BA, Green TA. Transcriptomics of environmental enrichment reveals a role for retinoic acid signaling in addiction. 2016 doi: 10.3389/fnmol.2016.00119. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]