Abstract
Calpain-5 is a calcium-activated protease expressed in the retina. Mutations in calpain-5 cause autosomal dominant neovascular inflammatory vitreoretinopathy (ADNIV, OMIM#193235). The structure of calpain-5 has not been determined, thus hindering the investigation of its proteolytic targets and pathological role in ADNIV. Herein, we report models of the proteolytic core of calpain-5 (mini-calpain-5) containing two globular domains (termed DIIa-IIb) connected by a short, flexible linker, consistent with small-angle x-ray scattering (SAXS) data. Structural modeling in the absence of calcium suggests that mini-calpain-5 adopts a more open conformation when compared to previously determined structures of other calpain cores. This open conformation, achieved by a rotation of DIIa and DIIb with respect to each other, prevents formation of the active site and constrains the enzyme in an inactivated form. The relative domain rotation of 60-100°we found for mini-calpain-5 (a non-classical calpain) is significantly greater than the largest rotation previously observed for a classical calpain (i.e., 55.0°for mini-calpain-9). Together with our prediction that, in the full-length form, a long loop in DIIb (loop C1), a few residues downstream of the inter-domain linker, likely interacts with the shorter, acidic, inactivating loop on domain-III (DIII), these structural insights illuminate the complexity of calpain regulation. Moreover, our studies argue that pursuing higher resolution structural studies are necessary to understand the complex activity regulation prevalent in the calpain family and for the design of specific calpain inhibitors.
Keywords: CAPN5, ADNIV, SAXS, protease, uveitis
Introduction
CAPN5 encodes calpain-5, a calcium-activated, signaling protease expressed by retinal photoreceptors (Mahajan et al., 2012). Coding mutations in CAPN5 cause an inherited uveitis called autosomal dominant neovascular inflammatory vitreoretinopathy (ADNIV, OMIM #602537) (Mahajan et al., 2012). ADNIV is characterized by progressive, severe intraocular inflammation, photoreceptor degeneration, retinal neovascularization, intraocular fibrosis, and retinal detachment. Otherwise, affected patients have no associated systemic conditions. ADNIV-associated mutations in calpain-5 reside in a flexible loop that gates the active site and are expected to alter enzymatic activity (Wert et al., 2014), likely leading to a gain of function (Mahajan et al., 2012). Since the natural protein targets of calpain-5 proteolysis are unknown, a functional assay for calpain-5 is not currently available. Nevertheless, in general, high calpain activity is associated with retinal and cell degeneration (Azuma and Shearer, 2008; Huang and Wang, 2001; Vanderklish and Bahr, 2000). Accordingly, expressing mutant calpain-5 alleles only in the mouse retina is sufficient to trigger disease (Wert et al., 2014), while deleting the gene (in the knockout mouse) causes no phenotype (Franz et al., 2004).
Calpain-5 is the most distant paralog of the calpain family, and a non-classical calpain, but little is known regarding its functional features and natural substrates. Although some substrates were identified for other members of the calpain family, calpain-5 is atypical: it lacks the Ca2+-binding, penta-EF-hand domains that are predicted to play a role in regulating enzymatic activity (Barnes and Hodgkin, 1996; Dear et al., 1997; Matena et al., 1998), dimerization (either homodimerization, as with calpain-3 (Ravulapalli et al., 2009), or heterodimerization, as with the large and small subunits of calpain-1 and -2 with their small subunits, calpain-S1 and calpain-S2), and interaction with the endogenous calpain inhibitor, calpastatin. Instead, calpain-5 has a C2-like domain that is structurally similar to the C2L domain implicated in transient membrane binding (reviewed in Campbell and Davies, 2012). Its closest homolog is calpain-6 (with 46% identity), a calpain without proteolytic activity. Most importantly, calpain-5 has conserved the calcium binding sites in the protease core DI-II, and is therefore expected to undergo a realignment of the active site residues for catalysis in the presence of calcium.
Investigation into proteolysis by calpain-5 has been hampered by the instability of the recombinant protein in purification strategies (Bassuk et al., 2015). Thus, the activity and regulatory mechanisms of calpain-5 remain poorly understood. Nevertheless, key clues about regulation of calpain-5 activity and target specificity were gained from structural models of the calpain catalytic core (Bassuk et al., 2015). This core sequence is highly conserved among the various calpain family members, strongly suggesting that key functions are also highly conserved. Other studies imply that calpains recognize targets via sequences in the proteolytic core (evidenced by yeast two-hybrid screens where 29 of the 37 calpain-1 interactors captured directly contact the proteolytic domain itself) (Jiang et al., 2002). This idea is supported by structural modeling that overlap the various calpains and show the most highly divergent amino acid stretches in the proteolytic domains lie in the flexible loops near the active site where they might mediate specificity (Campbell and Davies, 2012). To gain insights into the relative domain orientation which may modulate activation of the protease, in this study we purified the protease core of calpain-5 (mini-calpain-5) and determined its solution structure by small-angle x-ray scattering (SAXS). We also determined thermostability of mini-calpain-5 in presence and absence of calcium, an inhibitor (leupeptin) and a chelating agent (EGTA). We find that while mini-calpain-5 seems to adopt the typical closed form on addition of calcium, it exists in solution in a highly open form otherwise. Since this open form is quite different from the open form seen for other inactivated calpains, specific inhibitors for calpain-5 could be envisioned that lock the enzyme in this inactivated state where the active site has still not formed. Our thermostability studies show that binding to calcium alone and in presence of the inhibitor leupeptin significantly stabilizes the enzyme as expected. However, binding to leupeptin in the absence of calcium significantly destabilizes the enzyme.
Materials and Methods
Homology modeling of mini-calpain-5 based on sequence homology to calpain-9
Homology models for mini-calpain-5 were generated using MODELLER 9.14 (Webb and Sali, 2014), as described previously (Bassuk et al., 2015). Briefly, a BLAST search for mini-calpain-5 against the Protein Database (PDB) returned the structures of the catalytic core of human calpain-9 as the top hit, with a sequence identity of 42%. Other close matches were the catalytic cores of calpain-1, calpain-2, and calpain-8. Homology models of the calpain-5 catalytic core were generated using the structures of calpain-9 (PDB ID 2P0R ; calcium and leupeptin bound closed and PDB ID 1ZIV; calcium and beta-mercaptoethanol bound open), calpain-1 (PDB ID 2ARY; calcium-bound closed), calpain-2 (PDB ID 3BOW; calcium and calpstatin bound closed and PDB ID 1DF0; no calcium open), calpain-1/2 hybrid (PDB ID 1QXP; no calcium open) and calpain-8 (PDB ID 2NQA; calcium and leupeptin-bound closed) as templates. The open and closed structures provide distinct orientations of DIIa relative to DIIb and overlap poorly upon alignment (for example, rmsd of 6.6 for 304 Cα atoms for 2P0R and 1ZIV). Ten models were generated using each template, which superimposed well within each group with major variations in the N- and C-terminal regions and minor variations in the loop regions. PyMOL was used to generate all structure figures (Schrödinger Corporation, 2014). Sequence alignments were performed using ClustalW (Thompson et al., 1994).
Cloning
The catalytic core (DIIa-IIb) of human calpain-5 was cloned into a pUC57 vector with an Xho I restriction site, with an N-terminal thrombin cleavage site, and a C-terminal TEV protease cleavage site followed by a 6xHis tag, as previously described (Bassuk et al., 2015). Calpain constructs were transferred into the pMAL-C5X vector to obtain an N-terminal maltose binding protein (MBP) as a fusion partner that could be cleaved off with thrombin after purification. Sequence of the calpain flanking regions was confirmed by sequencing of constructs. Plasmids were amplified and isolated from DH5α cells and then were transformed into E. coli BL21 (DE3).
Protein purification
For structural studies, samples of mini-calpain-5 were prepared to a purity greater than 95%, as described below. E. coli BL21 (DE3) cells expressing mini-calpain-5 were grown in 1 L shake-flask cultures, at 250 rpm in Terrific Broth (Invitrogen), at 37 °C until an OD 600 of 1.0 and then induced with 0.2 mM IPTG. Cells were grown at 16 °C for 19 hrs, harvested, and centrifuged at 4000g for 25 minutes and pellet frozen at −20 °C in 25 ml of lysis buffer (20 mM Tris, 300 mM NaCl, 1 mM DTT, pH 7.5, one tablet of EDTA-free protease inhibitor (Roche), DNase (Roche)). Cells were lysed using an EmulsiFlex (Avestin) and centrifuged for 50 min. at 120,000g. Cell debris was discarded and the supernatant (30 ml) loaded onto a column packed with 12 ml (1CV) amylose resin (New England Biolabs) connected to a BioLogic DuoFlow (BioRad Laboratories Inc., CA). The column was washed with 5 to 10 CV of wash buffer (20 mM Tris, 300 mM NaCl, pH 7.5) and eluted with 6 to 8 CV of elution buffer (20 mM Tris, 300 mM NaCl, 10 mM maltose, pH 7.5). Eluted fractions were pooled and concentrated to 6 ml at 4 mg/ml (30 kDa NMWL spin concentrator; Millipore). MBP was removed with thrombin (2 units/mg of protein; thrombin from bovine plasma, T7201, Sigma-Aldrich) at 4 °C for 5 hrs and the reaction mix passed over Ni-NTA (Qiagen) resin to remove cleaved-off MBP using 6 CV wash buffer (20 mM Tris, 500 mM NaCl, 10 mM Imidazole, pH 7.5). Cleaved and remaining uncleaved mini-calpain-5 was eluted from the column with 5 CV of elution buffer (20 mM Tris, 500 mM NaCl, 200 mM Imidazole, pH 7.5). The eluted protein was concentrated with a 10 kDa NMWL spin concentrator and passed over a Superdex 75 (GE) size-exclusion column to remove residual, uncleaved fusion protein. The column was equilibrated with CD buffer (20 mM Tris-HCl, 200 mM NaCl, 2 mM DTT, pH of 7.5), DLS buffer (20 mM Tris-HCl, 300 mM NaCl, pH of 7.5) or SAXS buffer (20 mM Tris-HCl, 150 mM NaCl, 2-5 mM DTT, pH of 7.5) depending on the analysis being performed on the sample.
Circular dichroism (CD) analysis
To confirm protein secondary structure, circular dichroism (CD) spectra were collected using a Jasco J-815 spectrometer (Department of Biochemistry, University of Iowa) in the 200 to 260 nm range with a 1 mm Spectrosil (Starna) quartz cuvette at 25 °C, with a data interval of 0.1 nm and scan speed of 100 nm/min. Mini-calpain-5 samples were dialyzed against the CD buffer and diluted to a concentration of 0.48 mg/ml (11.6 μM). Ten measurements for each sample were averaged and compared after buffer subtraction. CD spectra deconvolution was performed using the Dichroweb server (Whitmore and Wallace, 2004) with the K2D method (Andrade et al, 1993).
Dynamic light scattering analysis
Purified mini-calpain-5 (0.42 mg/ml) samples were dialyzed against the DLS buffer. Sample monodispersity and hydrodynamic radius was determined at 22 °C for dialyzed samples using a DynaPro Nanostar dynamic light scattering (DLS) instrument (Wyatt Technology). The data were analyzed using the Dynamics 7.1.7 software. Thermostability of the samples was assessed by recording DLS measurements while heating the samples from 20 °C to 80 °C at 1 °C/min. Onset of protein thermal unfolding (Tonset) was determined by the sudden increase in hydrodynamic radius during the temperature ramp. Measurements were collected without additional calcium or after addition of EGTA (75 mM), Ca2+ (2-100 mM CaCl2) and Leupeptin (1-15 mM).
Small-angle X-ray scattering (SAXS) data collection and analysis
Small-angle X-ray scattering (SAXS) data were collected on the SIBYLS beamline (beamline 12.3.1) at the Advanced Light Source, Lawrence Berkeley National Laboratory. Scattering was measured by a Pilatus3 2M (Dectris) hybrid pixel array detector coaxial with the incident beam. The SAXS data were collected at 10 °C at three concentrations (0.75, 1.50, 2.25 mg/ml) in the order of lowest to highest concentration with 0.3 s exposures for 10 s resulting in 32 frames per concentration. No concentration-dependent effects were observed on comparison of the initial exposures for each concentration; however, radiation damage was generally found to occur faster at higher concentration. Data from frames before radiation damage occurred were averaged for the lowest concentration of each sample and used for modeling. Before data were collected, each sample was thoroughly dialyzed in the SAXS buffer. Scattering curves of the dialysate buffer were collected and subtracted from each protein-scattering measurement.
Scattering data were reduced using beamline scripts and analyzed using PRIMUS and related programs from the ATSAS Program package (Konarev et al., 2006). To assess aggregation or interparticle interference, all samples, at all concentrations and exposures, were assessed by analyzing the Guinier region (low-q region) of the scattering data using autorg. The program datgnom was used to compute the pair distribution function, P(r), the maximum dimension of the macromolecule, Dmax and the real space radius of gyration, Rg. Molecular weight (MWcalc) was estimated by dividing the Porod volume by 1.7. Experimental SAXS data were compared with simulated scattering curves for crystal structures of various mini-calpains and the mini-calpain-5 homology models based off representative templates of the closed and open forms using the program CRYSOL. BILBOMD (Pelikan et al., 2009) was used to sample conformations of 10 mini-calpain-5 homology models by treating DIIa and DIIb as rigid bodies, but allowing the linker region between the two domains (residues 194-196) and the N- and C-terminus (1-15 and 344-360) to remain flexible. The numbering used here includes the 4 residues post the thrombin cleavage site on the N-terminus and 14 residues containing the His-tag on the C-terminus. To generate an ensemble of models, conformational space was explored by MD simulations of the flexible regions at 1500 K. Rg was restrained to within 22-27 Å and 800 conformations per Rg were generated.
Domain motion analysis
DynDom (Lee et al., 2003) was used to calculate the rotation angles of DIIa and DIIb with respect to mini-calpain-1 (2ARY) about the flexible linker for all the structures and models.
RESULTS
Purification of mini-calpain-5
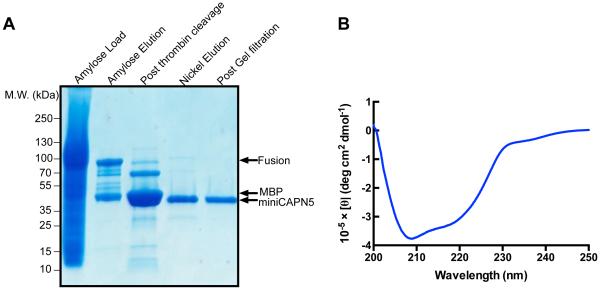
A three-step purification process yielded in high quality mini-calpain-5 suitable for structural studies. MBP-fused mini-calpain-5 was bound and eluted from an amylose resin, cleaved by thrombin to remove the MBP tag, passed over a Ni resin to isolate a clean His-tagged mini-calpain-5, and further refined by size exclusion chromatography. The result was a pure sample that gave a single band on a Coomassie stained SDS-PAGE gel (Fig.1A).
Fig. 1. Purified human mini-calpain-5 is well folded.
(A) The maltose binding protein (MBP) tag of MBP-mini-calpain-5 (83.8 kDa) was cleaved by thrombin, releasing MBP (42.3 kDa) and mini-calpain-5 (41.4 kDa). His-tagged mini-calpain-5 was separated from MBP with nickel resin. Size-exclusion chromatography then yields a very pure, mini-calpain-5 (lane 5). (B) Molar ellipticity measured using circular dichroism in the far UV range (200-250 nm) was plotted for human mini-calpain-5 wild type at 0.48 mg/ml (no added Ca2+). Mini-calpain-5 was well folded and indicative of an α/β-mixed protein.
Circular dichroism shows mini-calpain-5 has secondary structure
We confirmed that the purified mini-calpain-5 was, in fact, well-folded by investigating its secondary structure using circular dichroism (CD) spectroscopy. The CD spectrum shows a strong negative band at 208 nm and a broad shoulder from 215-222 nm indicating the protein has both alpha helical and beta sheet characteristics (Fig. 1B). Deconvolution of the CD spectrum suggests 25% alpha helical, 42% beta sheet and 33% random coil content compared to 29-32% alpha helical, 45-50% beta sheet and 21—23% random coil observed for mini-calpains 1 and 9 in the PDB.
Calcium gives mini-calpain-5 greater thermostability
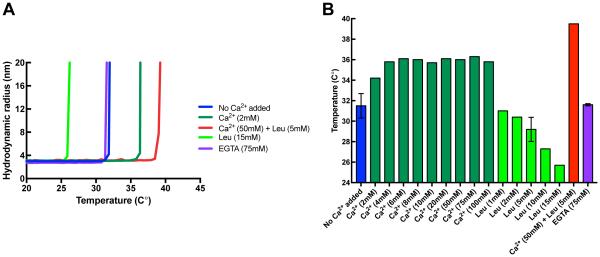
The thermostability of mini-calpain-5 was analyzed by recording dynamic light scattering (DLS) data as the protein was heated from 20 °C to 80 °C. As the temperature rises, the protein starts unfolding, causing an increase in its hydrodynamic radius, which can be measured by DLS (Fig. 2A). The temperature at which the protein starts unfolding is called the onset temperature (Tonset). Mini- calpain-5 unfolds at a temperature of 32.0±0.3°C in the absence of added calcium (Fig. 2A and 2B). With increasing amounts of Ca2+ to 100 mM, the Tonset increased until it reached a plateau of about 36 °C at 6 mM Ca2+. Adding the non-specific calpain inhibitor leupeptin (5 mM) in the presence of calcium (50 mM) further stabilized mini-calpain-5 (Tonset = 39.5 °C). Addition of 75 mM EGTA, a Ca2+ favoring chelator, to the purified mini-calpain-5 did not reduce its thermostability (Tonset = 31.6 ± 0.1 °C), suggesting that either the enzyme did not pick up any Ca2+ during expression or did not retain it during purification. None of the purification buffers had Ca2+ as a component. Interestingly, increasing amounts of leupeptin in the absence of Ca2+ progressively made mini-calpain-5 more unstable, with a Tonset of 25.7 °C for the highest amount of leupeptin (15 mM) tested. Wild type and mutant versions of mini-calpain-1 have previously been reported to have a Tonset in the 35-41 °C range (Tmelt 44-47 °C) in the absence of Ca2+, as measured by differential scanning calorimetry (Moldoveanu et al., 2004) (Tonset approximated from Fig 2A in the reference).
Fig. 2. Thermal stability of human mini-calpain-5.
(A) Representative thermal denaturation curves of human mini-calpain-5 wild type at 0.42 mg/ml with no Ca2+ added (blue) or on addition of 72 EGTA (purple), 10 mM Ca2+ (dark green), 50 mM Ca2+ with 5 mM leupeptin (red) or 5 mM leupeptin (light green) as determined by an increase in hydrodynamic radius on unfolding during a 1 °C/min temperature ramp by dynamic light scattering. (B) Concentration dependence of additives on mini-calpain-5 thermostability. Mini-calpain-5 starts unfolding at 32.0± 0.3°C, has a relatively unaltered Tonset of 31.6 ± 0.1 °C on addition of 75 mM EGTA, increasingly becomes more stable in the presence of Ca2+ (Tonset = 36.8 ± 0.1 °C; p ≤ 0.01) and is significantly more stable at 50 mM Ca2+ with 5 mM leupeptin (Tonset = 39.5 ± 0 °C; p ≤ 0.001). Increasing amounts of leupeptin alone, however, are destabilizing (Tonset = 25.7 °C at 15 mM leupeptin).
SAXS can capture conformational changes in mini-calpain-5 on addition of calcium and leupeptin
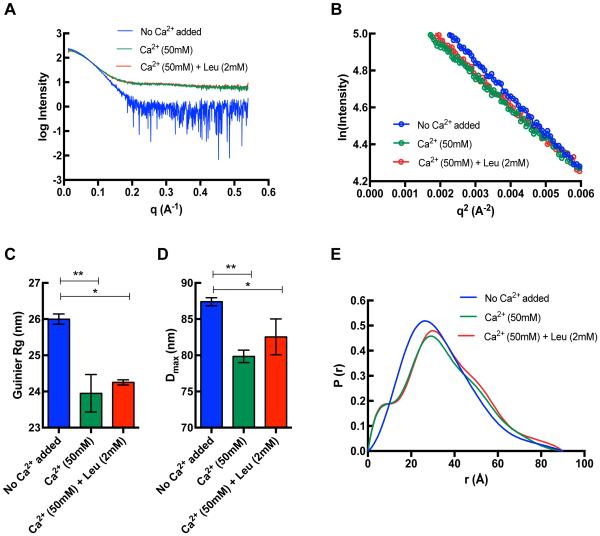
Scattering data were collected on monodisperse, homogeneous samples in three conditions: no added Ca2+, 50 mM Ca2+, and 50 mM Ca2+ with 2 mM leupeptin. Single scattering curves for each condition were derived from averaging of successive exposures that did not show radiation damage and used later for modeling. These averaged curves are shown in Fig. 3A and the linear fits to the Guinier region in Fig. 3B. The addition of 50 mM Ca2+ and 50 mM Ca2+ with 2 mM leupeptin led to a significant decrease in Guinier Rg from 26.0 ± 0.1 Å to 23.9 ± 0.5 Å and 24.3 ± 0.1 Å, respectively (Fig. 3C). A similar trend can be seen with the Dmax (from 87.4 ± 0.6 Å to 79.9 ± 0.9 Å and 82.6 ± 2.5 Å, respectively) suggesting that binding to Ca2+ or Ca2+ and leupeptin together makes mini-calpain-5 more compact (Fig. 3D). The P(r) function highlights this and also suggests that following the significant conformation change on addition of Ca2+, there is a relatively minor change on further addition of leupeptin (Fig 3E). The SAXS-derived parameters are shown in Table S1.
Fig. 3. SAXS curves for human mini-calpain-5.
(A) Buffer-subtracted averaged scattering curves for 0.75 mg/ml human mini-calpain-5 in with no added Ca2+ (blue) and in the presence of 50 mM Ca2+ (green) and 50 mM Ca2+ with 2 mM leupeptin (red). (B) The linear portion of the Guinier region in the low q region (0.0028 –0.0054 Å−1) such that q.Rg < 1.3 where Rg is 25.7 Å, 23.6 Å and 24.4 Å respectively. (C) The guinier Rg was significantly decreased in the presence of 50 mM Ca2+ (green; p ≤ 0.01) and 50 mM Ca2+ with 2 mM leupeptin (red; p = ≤ 0.05) (data shown before curve averaging). (D) A similar trend was observed where the addition of 50 mM Ca2+ and 50 mM Ca2+ with 2 mM leupeptin led to a significant decrease in the Dmax (p ≤ 0.01 and ≤ 0.05, respectively). (E) The pairwise distance distribution function P(r) of the averaged data suggests a major change in conformation of the no Ca2+ added form on addition of 50 mM Ca2+ or 50 mM Ca2+ with 2 mM leupeptin.
SAXS suggests an unusually large open form of Ca2+ free mini-calpain-5
To better understand how ADNIV disease mutations affect calpain-5 function, we need to determine the structure of the catalytic domain and the mutants, preferably interacting with a substrate. We believe that ADNIV mutations lead to increased calpain-5 proteolytic activity and specific inhibitors that lock it in its inactivated form would be desirable. Purification and high-resolution structure determination of calpains has proved to be challenging; structures of the catalytic cores of only calpain-1, 2, 8 and 9 have been solved and of calpain-1 and 2 for the full length protein (Table 1). Calcium-free inactivated form structures have been determined for full-length calpain-2 and a chimera of calpain-1 and 2, but none for just the catalytic core. We believe SAXS can be useful for identifying conformational changes on calcium, ligand and inhibitor binding for wild type and mutant forms of mini-calpain-5. In the absence of a crystal structure, we started with homology models of the calpain-5 catalytic core which we have developed and used before (Bassuk et al., 2015). Previously, calpain-9 was our template for homology modeling, because it had the highest BLAST score of the structures in the PDB database and a respectable 42% sequence identity match. A number of other known calpain catalytic core structures also match calpain-5, with similar BLAST scores and greater than 40% identity (Table S2). Nevertheless, significant differences remain in relative DIIa and DIIb orientations, depending on the calpain type, the presence or absence of activating calcium, and whether a ligand/inhibitor binds in the active site (Table 1).
Table 1.
Closed and open calpain catalytic cores.
| Protein | Typ e |
PD B ID |
Ch ain |
Catalytic calciums |
Inhibi tor |
For m |
Rota tion angl e (°) |
Rg of derived homology models |
|---|---|---|---|---|---|---|---|---|
| human-mini-calpain-1 | clas sical |
2AR Y |
A | yes | - | clo sed |
- | 23.8 ± 0.4 |
| human-mini-calpain-1 | clas sical |
1ZC M |
A | yes | ZLLY CH2F |
clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
1TL 9 |
A | yes | leupe ptin |
clo sed |
- | - |
| human-mini-calpain-8 | clas sical |
2N QA |
A | yes | leupe ptin |
clo sed |
- | 22.9 ± 0.3 |
| rat-mini-calpain-2 | clas sical |
1M DW |
A | yes | - | clo sed |
- | - |
| human-mini-calpain-9 | clas sical |
2P0 R |
A | yes | leupe ptin |
clo sed |
- | 23.4 ± 0.3 |
| rat-mini-calpain-1 | clas sical |
1KX R |
A | yes | - | clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
2R9 C |
A | yes | ZLAK- 3001 |
clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
1TL O |
A | yes | E64 | clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
2G8 E | A | yes | SNJ-1715 | clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
2N QI |
A | yes | WR13 (R,R) |
clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
2N QG |
A | yes | WR18 (S,S) |
clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
2G8 J |
A | yes | SNJ- 1945 |
clo sed |
- | - |
| rat-mini-calpain-1 | clas sical |
2R9 F |
A | yes | ZLAK- 3002 |
clo sed |
- | - |
| rat-mini-calpain-2 | clas sical |
3DF 0* |
A | yes | calpst atin |
clo sed |
- | - |
| rat-mini-calpain-2 | clas sical |
3B OW * |
A | yes | calpst atin |
clo sed |
- | 23.4 ± 0.4 |
| rat-mini-calpain-2 | clas sical |
1DF 0* |
A | no | - | ope n |
26.4 | 24.7 ± 0.2 |
| rat-mini-calpain-2 | clas sical |
1U5 I* |
A | no | - | ope n |
23.1 | - |
| rat-mini-calpain-2 | clas sical |
1KF X* |
L | no | - | ope n |
24.6 | - |
| human-mini-calpain-2 | clas sical |
1KF U* |
L | no | - | ope n |
24.5 | - |
| rat-mini-calpain-1-like | clas sical |
1Q XP* |
A | no | - | ope n |
24.9 | 23.9 ± 0.2 |
| human-mini-calpain-9 | clas sical |
1ZI V |
A | yes | - | ope n |
55.0 | 25.5 ± 0.3 |
| human-mini-calpain-5 (SAXS BILBOMD) |
non- clas sical |
- | no** | no** | ope n |
59.5- 102. 5 |
25.3 ± 0.1 |
: Full-length calpain trimmed to the start of DIII for this analysis (~ residue 360)
: Inferred from thermostability results of this study
All ligands and waters, except calciums, if present, were removed for SAXS calculations.
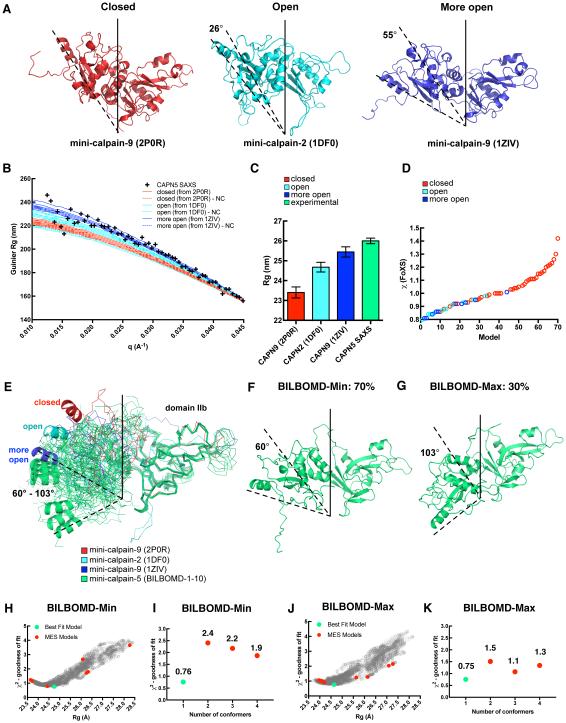
For mini-calpain-5, we investigated which of these three-dimensional structures was most consistent with our SAXS data. The sequence of our mini-calpain-5 construct (360 residues including a 6x-His tag) was modeled onto seven crystal structures of calpain-1, calpain-2, calpain-8, and calpain-9 representing 4 closed and 3 open forms, which had anywhere from 301 to 354 residues of the catalytic-core resolved in their respective electron density maps. Ten homology models were generated for each template to capture some of the conformational heterogeneity that would result from the flexible N- and C-terminus regions and the internal loop regions (Fig. S1). Of the templates chosen, the closed form represents the activated, calcium (and often ligand) bound form with the catalytic triad in position for catalysis (such as PDB ID 2P0R) (Moldoveanu et al., 2002); the open form represents the inactive, calcium-free form without the catalytic triad in position (such as PDB ID 1DF0) (Campbell and Davies, 2012); and the more open form represents the unique mini-calpain-9 structure (PDB ID 1ZIV) (Davis et al., 2007), which does not have the catalytic triad in position despite having both Ca2+bound (Fig 4A). Simulated SAXS curves in the low-q (Guinier) region for the homology models generated from 2P0R, 1DF0 and 1ZIV with and without the flexible N- and C- terminus are compared with the experimental SAXS data obtained for mini-calpain-5 (Fig 4B). The tight banding of the simulated data is more dependent on the relative orientation of the two domains and less dependent on the conformations of the flexible N- and C-terminus (or even their presence for that matter). The best fit to the SAXS data in this low-q region is the cluster of 1ZIV homology models indicating that mini-calpain-5 not only has an open form in the absence of Ca2+, but it actually may be more open than what is conventionally observed for other calpains. Theoretical Rg values calculated using CRYSOL for the homology models from 2P0R, 1DF0 and 1ZIV compared with the experimental Rg for mini-calpain-5 SAXS data also suggest that the latter is more open than 1ZIV (Fig 4C). Homology models of mini-calpin-5 generated using the 7 closed, open and more open templates indicated previously, on average, give a better fit (lower χ2) for the open and more open models against experimental SAXS data than the closed models (Fig 4D). These rigid models fit the SAXS data with χ2 ranging from 0.63 to 1.65, with better fits being models of mini-calpain-9 (PDB ID 1ZIV) based on χ2 and Rg.
Fig. 4. Fit of different mini-calpain-5 models to SAXS data.
(A) Calpain protease core structures adopt one of three conformations: closed (mini-calpain-9; PDB ID 2P0R), open (mini-calpain-2; PDB ID 1DFO), and more open (mini-calpain-9; PDB ID 1ZIV). (B) Low q (Guinier) region and fit of representative rigid homology models of closed, open and more open forms of mini-calpain-5 to SAXS data. The homology models with the floppy N- and C-terminus trimmed are labeled as “-NC”. (C) Rg calculated for the representative homology models of mini-calpain-5 compared with the Guinier Rg of SAXS data. (D) Fits of 70-homology models based off 4 closed and 3 open forms to SAXS data. (E) The 10 best-fit BILBOMD models of mini-calpain-5 starting with a homology model based off 1ZIV are shown in green. DIIb of all the structures have been superimposed to highlight the relative orientation of DIIa. Rotation angles calculated using DynDom with respect to mini-calpain-1 (2ARY) are reported. The BILBOMD models fell into two clusters, represented by BILBOMD-Min (F) and BILBOMD-Max (G) that comprised 70% and 30% of the best-fit conformations, respectively. (H) Graph representing the goodness of fit (χ2) for 4,000 conformations generated for BILBOMD-Min with their Rg values. The green and red dots represent the best-fit model and the minimal ensemble search (MES) models that were weighted, respectively. (I) MES fits for BILBOMD-Min to SAXS data indicate that the single conformer has the lowest χ2 and is the best-fit model. (J) Goodness of fit (χ2) for 4,000 conformations generated for BILBOMD-Max with their Rg values. (K) MES fits for BILBOMD-Max to SAXS data.
To sample various respective orientations of DIIa-IIb and simultaneously explore different termini conformations, the linker region between the two domains along with the N- and C-terminal regions were allowed to be flexible in BILBOMD. Each of the 10 homology models based off our best fit rigid model template (1ZIV; open form of 55°) was used as seed for an independent BILBOMD run and 4,000 models were generated per run. The single models from each run that best fit the SAXS data resulted in Rg of 25.3 ± 0.1 Å and χ2 of 0.57 ± 0.01. All these BILBOMD best-fit single models were of the open form (59.5 - 102.5°) (Fig. 4 E-G and Table S4). Compared to the starting model, in these best-fit BILBOMD models, DIIa-IIb had further rotated by about 5 - 48°, yielding an even more open form. Minimal ensemble searches of 2, 3 and 4 conformations failed to provide any mixtures of conformations that had better fits to the SAXS data than the best-fit single models (Fig. 4 H-K).
Primary sequence analysis and structural alignments
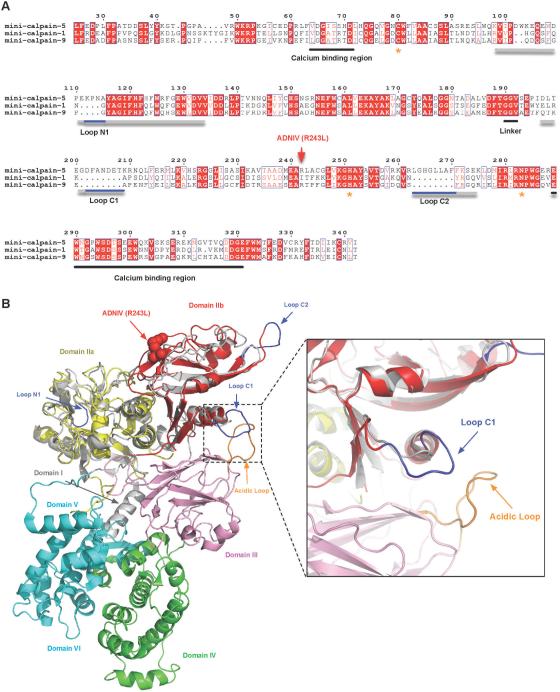
Alignment of proteolytic core sequences uncovered clear differences between calpain-1 and calpain-9 (two classical calpains with well characterized structures) and calpain-5 (a non-classical calpain). Alignment revealed that mini-calpain-5 contains three stretches of amino acids that are much longer than corresponding sequences in either mini-calpain-1 or mini-calpain-9. These comprise gap regions for calpain-1 and calpain-9, as shown in Fig. 5A, and map to loops in mini-calpain. We refer to them as N1, C1 and C2 loops, denoting their proximity to either the N- or C-terminus. Although structural alignment of mini-calpain-5 to mini-calpain-1 yields an rmsd of 6.8 Å across 307 Cα atoms, an alignment of the individual sub-domains of mini-calpain-1 and -5 (IIa and IIb) yields rmsd values of 0.55 Å (111 Cα) for IIa and 0.63 Å (93 Cα) for IIb. To predict the position of these loops relative to other domains in the full-length protein, the mini-calpain-5 subdomains were individually superimposed on the crystallographic structure for full-length calpain-2 (PDB ID: 1KFU) (Fig. 5B). Although calpain-5 does not contain a domain V or VI, this comparison highlights the relative location of loops N1, C1, and C2 with respect to DIII and DIV. Interestingly, loop C1 of mini-calpain-5 contains additional acidic residues and lies near the acidic loop of DIII, a loop believed to modulate calpain activation via calcium and phospholipid binding.
Fig. 5. Identification of loop extensions unique to calpain-5.
(A) Multiple sequence alignment of mini-calpain-1, -5, and -9. Loops N1, C1, and C2 are underlined in gray and with calpain-5 extensions, which appear as gaps in the mini-calpain-1 and -9 sequences, underlined in blue. Asterisks denote the catalytic residues. The calcium binding regions and the interdomain linker are underlined in black. The residue numbering shown is for calpain-5. (B) DIIa-IIb of a mini-calpain-5 homology model superimposed on the crystal structure of human calpain-2 (PDB ID: 1KFU). DIIa and DIIb of calpain-5 are colored yellow and red, respectively, while the corresponding calpain-1 domains are represented in gray. Extensions of the calpain-5 loops are colored in blue and correspond to the primary sequences shown in Fig. 5A. Loop C1 falls in close proximity to the calcium-binding acidic loop of DIII (inset).
Discussion
Calpains are implicated in a number of disease processes (Goll et al., 2003; Sorimachi et al., 1997; Zatz and Starling, 2005); and mutations in calpain-5 cause ADNIV, an autoimmune inflammatory retinal disease (Bassuk et al., 2015; Mahajan et al., 2012; Wert et al., 2015). Specific inhibitors of calpains (e.g., SNJ-1945) were designed and optimized for calpain-1 (Cuerrier et al., 2006); however, little structural information is available to facilitate optimization of calpain-5 inhibitors. The structure of calpain-5 is needed not only to guide rational drug design, but also to further characterize its pathogenic role in ADNIV. Our SAXS data offer new insight into the global structure of the calpain-5 proteolytic core especially of its inactivated open form.
Previously, crystallographic analysis of human calpains showed that, relative to calpain-1, the calpain-9 catalytic core (PDB ID 1ZIV) even with calcium bound could adopt a more open inactive conformation. This open conformation might mediate calpain-9 auto-inhibition because it is predicted to occlude the substrate binding site and misalign the catalytic triad (Davis et al., 2007). The presence of beta-mercaptoethanol bound to two cysteines in the crystal structure of 1ZIV could potentially have resulted in this unexpected open form. However, the calcium-bound structure of the catalytic core of calpain-1 (PDB ID 2ARY) by the same authors with beta-mercaptoethanol modifying equivalent cysteines is virtually identical to the conventional closed form seen for mini-calpains (Davis et al., 2007). This suggests there are slightly different mechanisms of inactivation/activation in the calpain family. Intriguingly, our SAXS data suggests mini-calpain-5 can adopt an even more open conformation than mini-calpain-9, and may undergo a similar mechanism of auto-inhibition. Of all the structures of mini-calpains that can be extracted from the PDB, homology models built using the open forms as templates fit our SAXS data, collected in the absence of calcium, much better than the closed forms. Interestingly, the best fits were obtained using the most open form (55°; 1ZIV) as the template. To sample even more open forms, we allowed the interdomain linker to be flexible and found the best fits to be always open and in the 60-100°range. Minimal ensemble searches failed to find a mix of open and closed forms that could fit the SAXS data any better. These largely open forms in the absence of calcium suggest that calpain-5 may require high levels of calcium for activation. Experimental data concerning the activity and calcium dependence of calpain-5 have not yet been reported.
Our thermostability studies found that on addition of the calcium favoring chelator EGTA, there was no change in the Tonset for mini-calpain-5, suggesting that under the calcium-free purification conditions no calciums had indeed bound the enzyme. Addition of calcium and the calpain inhibitor leupeptin, on the other hand, significantly made mini-calpain-5 more stable (ΔTonset= 4-7 °C). Interestingly, addition of increasing amounts of leupeptin in the absence of calcium progressively made the enzyme less stable (ΔTonset= −7 °C). It appears, that leupeptin stabilizes mini-calpain-5 when there is an available active site for it to bind to, whereas, in the absence of a specific binding pocket, non-specific interactions of leupeptin with the open inactive form destabilize the enzyme.
Analysis of the primary sequence and structural alignment of mini-calpains identified three loops in calpain-5 that are much longer than the corresponding loops in calpain-1 and -9. Loop C1 (just a few residues downstream of the inter-domain linker) likely sits near the acidic loop of DIII and may play a role in the auto-inhibition or regulating activity of calpain-5, in what is considered to be the first of the two parts of calpain activation (Khorchid and Ikura, 2002; Moldoveanu et al., 2002; Suzuki et al., 2004). The DIII acidic loop, believed to hold calcium-binding sites of its own, likely stabilizes the inactive conformation of DII (Tompa et al., 2001). However, since the crystal structure of calcium-bound calpain-2 does not show evidence of calcium binding in the DIII acidic loop, there may be a different mechanism of regulation performed by this extended C1 loop that may be elucidated by the calpain-5 crystal structure.
Interestingly, the previously described human p.R243L mutant of calpain-5 appears to be more active than the wild type and require less calcium for activation (Wert et al., 2015). Since the p.R243L mutation does not fall within loop C1, the mutation more likely affects the second part of calpain activation—calcium binding to the catalytic domain and subsequent sub-domain rearrangements that form the active site.
Despite the appealing model of an extreme conformation of a potentially inactivated form of a calpain that is offered by our analysis, there is a caveat. The presence of the other two domains in the full-length protein may impose constraints and limit some of the extreme open forms we have modeled. A high-resolution crystal structure is necessary to understand interactions at the atomic level. A crystal structure would be helpful in identifying residue-level differences at the active and regulatory sites, the effect of the ADNIV mutations on the local structure, identifying binding modes of known inhibitors such as SNJ-1945, and ultimately in designing/optimizing calpain-5 specific inhibitors to regulate the activity of this overactive disease-causing enzyme.
Supplementary Material
Fig. S1. Conformational heterogeneity in mini-calpain-5 homology models: Ten superimposed homology models of 6xHis-tag containing mini-calpain-5 generated using (A) mini-calpain-1 (2ARY; closed), (B) mini-calpain-9 (2P0R; closed), (C) mini-calpain-8 (2NQA; closed), (D) mini-calpain-2 (3BOW; closed), (E) mini-calpain-1-like (1QXP; open), (F) mini-calpain-2 (1DF0; open), and (G) mini-calpain-9 (1ZIV; open) highlighting the mobility of the loops and the N- and C-terminus regions.
Fig. S2. Fit of BILBOMD models to SAXS data: Simulated scattering curves from the 10 best-fit models from BILBOMD fit to SAXS data for human mini-calpain-5 (no Ca2+ added).
ACKNOWLEDGEMENTS
We wish to thank Melinda Smits and Diana Colgan for technical assistance. Part of this work was conducted at the Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, supported by DOE Office of Biological and Environmental Research. Additional support comes from the National Institute of Health project MINOS (R01GM105404). We thank the staff at the SIBYLS beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory, for SAXS data collection.
FUNDING
V.B.M. is supported by the National Institutes of Health [K08EY020530, R01EY016822], Doris Duke Charitable Foundation Grant #2013103, and Research to Prevent Blindness, New York, NY. G.V. is supported by the NIH [T32GM007337].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
References
- Azuma M, Shearer TR. The role of calcium-activated protease calpain in experimental retinal pathology. Surv. Ophthalmol. 2008;53:150–163. doi: 10.1016/j.survophthal.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes TM, Hodgkin J. The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 1996;15:4477–4484. [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, Yeh S, Wu S, Martin DF, Tsang SH, Gakhar L, Mahajan VB. Structural Modeling of a Novel CAPN5 Mutation that Causes Uveitis and Neovascular Retinal Detachment. PLoS One. 2015;10:e0122352. doi: 10.1371/journal.pone.0122352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RL, Davies PL. Structure-function relationships in calpains. Biochem. J. 2012;447:335–351. doi: 10.1042/BJ20120921. [DOI] [PubMed] [Google Scholar]
- Cuerrier D, Moldoveanu T, Inoue J, Davies PL, Campbell RL. Calpain inhibition by alpha-ketoamide and cyclic hemiacetal inhibitors revealed by X-ray crystallography. Biochemistry. 2006;45:7446–7452. doi: 10.1021/bi060425j. [DOI] [PubMed] [Google Scholar]
- Davis TL, Walker JR, Finerty PJ, Jr., Mackenzie F, Newman EM, Dhe-Paganon S. The crystal structures of human calpains 1 and 9 imply diverse mechanisms of action and auto-inhibition. J. Mol. Biol. 2007;366:216–229. doi: 10.1016/j.jmb.2006.11.037. [DOI] [PubMed] [Google Scholar]
- Dear N, Matena K, Vingron M, Boehm T. A new subfamily of vertebrate calpains lacking a calmodulin-like domain: implications for calpain regulation and evolution. Genomics. 1997;45:175–184. doi: 10.1006/geno.1997.4870. [DOI] [PubMed] [Google Scholar]
- Franz T, Winckler L, Boehm T, Dear TN. Capn5 is expressed in a subset of T cells and is dispensable for development. Mol. Cell. Biol. 2004;24:1649–1654. doi: 10.1128/MCB.24.4.1649-1654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wang KK. The calpain family and human disease. Trends Mol. Med. 2001;7:355–362. doi: 10.1016/s1471-4914(01)02049-4. [DOI] [PubMed] [Google Scholar]
- Jiang LQ, Wen SJ, Wang HY, Chen LY. Screening the proteins that interact with calpain in a human heart cDNA library using a yeast two-hybrid system. Hypertens. Res. 2002;25:647–652. doi: 10.1291/hypres.25.647. [DOI] [PubMed] [Google Scholar]
- Khorchid A, Ikura M. How calpain is activated by calcium. Nat. Struct. Biol. 2002;9:239–241. doi: 10.1038/nsb0402-239. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RA, Razaz M, Hayward S. The DynDom database of protein domain motions. Bioinformatics. 2003;19:1290–1291. doi: 10.1093/bioinformatics/btg137. [DOI] [PubMed] [Google Scholar]
- Mahajan VB, Skeie JM, Bassuk AG, Fingert JH, Braun TA, Daggett HT, Folk JC, Sheffield VC, Stone EM. Calpain-5 mutations cause autoimmune uveitis, retinal neovascularization, and photoreceptor degeneration. PLoS Genet. 2012;8:e1003001. doi: 10.1371/journal.pgen.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matena K, Boehm T, Dear N. Genomic organization of mouse Capn5 and Capn6 genes confirms that they are a distinct calpain subfamily. Genomics. 1998;48:117–120. doi: 10.1006/geno.1997.5133. [DOI] [PubMed] [Google Scholar]
- Moldoveanu T, Jia Z, Davies PL. Calpain activation by cooperative Ca2+ binding at two non-EF-hand sites. J. Biol. Chem. 2004;279:6106–6114. doi: 10.1074/jbc.M310460200. [DOI] [PubMed] [Google Scholar]
- Moldoveanu T, Hosfield CM, Lim D, Elce JS, Jia Z, Davies PL. A Ca(2+) switch aligns the active site of calpain. Cell. 2002;108:649–660. doi: 10.1016/s0092-8674(02)00659-1. [DOI] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravulapalli R, Campbell RL, Gauthier SY, Dhe-Paganon S, Davies PL. Distinguishing between calpain heterodimerization and homodimerization. FEBS J. 2009;276:973–982. doi: 10.1111/j.1742-4658.2008.06833.x. [DOI] [PubMed] [Google Scholar]
- Schrödinger Corporation PyMOL, New York. 2014 Accessed 2015 May 27. http://www.pymol.org/
- Sorimachi H, Ishiura S, Suzuki K. Structure and physiological function of calpains. Biochem. J. 1997;328:721–732. doi: 10.1042/bj3280721. Pt 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure, activation, and biology of calpain. Diabetes. 2004;53(Suppl 1):S12–18. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P, Emori Y, Sorimachi H, Suzuki K, Friedrich P. Domain III of calpain is a ca2+-regulated phospholipid-binding domain. Biochem. Biophys. Res. Commun. 2001;280:1333–1339. doi: 10.1006/bbrc.2001.4279. [DOI] [PubMed] [Google Scholar]
- Vanderklish PW, Bahr BA. The pathogenic activation of calpain: a marker and mediator of cellular toxicity and disease states. Int. J. Exp. Pathol. 2000;81:323–339. doi: 10.1111/j.1365-2613.2000.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2014;47:5 6 1–5 6 32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- Wert KJ, Skeie JM, Bassuk AG, Olivier AK, Tsang SH, Mahajan VB. Functional validation of a human CAPN5 exome variant by lentiviral transduction into mouse retina. Hum. Mol. Genet. 2014;23:2665–2677. doi: 10.1093/hmg/ddt661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wert KJ, Bassuk AG, Wu WH, Gakhar L, Coglan D, Mahajan M, Wu S, Yang J, Lin CS, Tsang SH, Mahajan VB. CAPN5 mutation in hereditary uveitis: the R243L mutation increases calpain catalytic activity and triggers intraocular inflammation in a mouse model. Hum. Mol. Genet. 2015;24:4584–4598. doi: 10.1093/hmg/ddv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatz M, Starling A. Calpains and disease. N. Engl. J. Med. 2005;352:2413–2423. doi: 10.1056/NEJMra043361. [DOI] [PubMed] [Google Scholar]
- Whitmore L, Wallace BA. DICHROWEB: an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Research. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Chacón P, Merelo JJ, Morán F. Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Prot. Engineering. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Conformational heterogeneity in mini-calpain-5 homology models: Ten superimposed homology models of 6xHis-tag containing mini-calpain-5 generated using (A) mini-calpain-1 (2ARY; closed), (B) mini-calpain-9 (2P0R; closed), (C) mini-calpain-8 (2NQA; closed), (D) mini-calpain-2 (3BOW; closed), (E) mini-calpain-1-like (1QXP; open), (F) mini-calpain-2 (1DF0; open), and (G) mini-calpain-9 (1ZIV; open) highlighting the mobility of the loops and the N- and C-terminus regions.
Fig. S2. Fit of BILBOMD models to SAXS data: Simulated scattering curves from the 10 best-fit models from BILBOMD fit to SAXS data for human mini-calpain-5 (no Ca2+ added).