Figure 3.
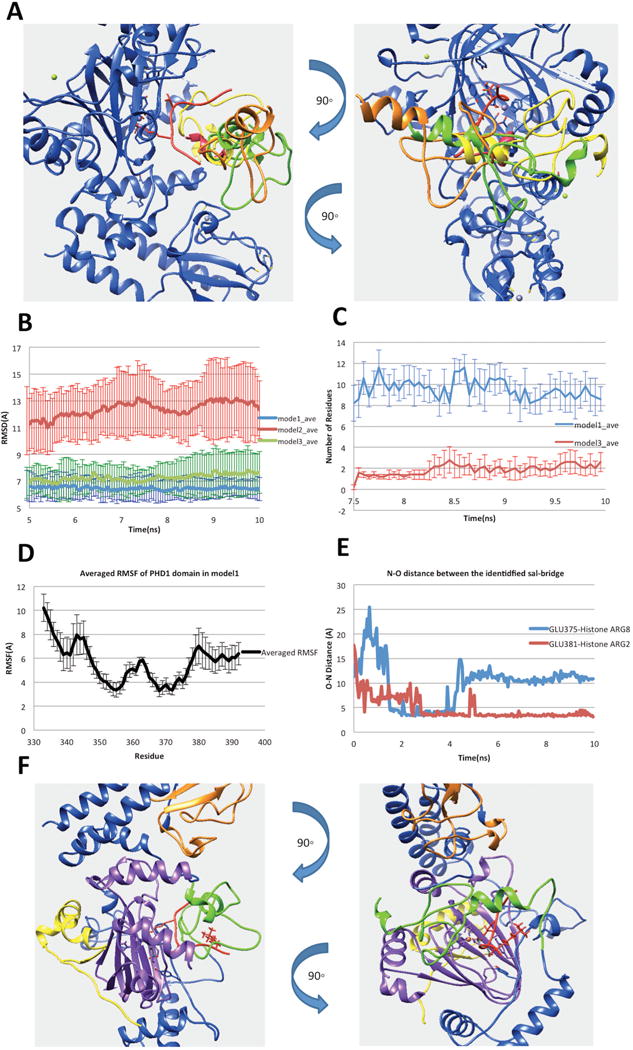
(A) Three possible binding modes after applying the constraint of linker length. (B) RMSDs of the complex of PHD1and JmjC domains. (C) Number of residues in PHD1 domain, which have any atom within 6Å of H3K9me3 in the last 2.5ns simulation time. (D) The N–O distance of identified salt bridges (Glu375-H3R8 and Glu381-H3R2) between PHD1 and substrate peptide. (E) Averaged RMSF of PHD1 domain residues calculated from model1 MD simulations. (F) Finalized model of PHD1 domain bound to KDM5C catalytic core including JmjN, PHD1, JmjC, ZF domains, inter domain region and histone peptide marked with yellow, green, purple, orange, blue and red, respectively
