Abstract
The Retinal homeobox (rax) gene is expressed in vertebrate retinal progenitor and stem cells and is essential for retinal development. In frogs, rax is expressed in the ciliary marginal zone (CMZ), a region containing retinal progenitor and stem cells at the anterior of the eye. Little is known regarding regulation of rax transcription and regulation of transcription of rax targets. We found that three ultra-conserved genomic elements (UCEs) flanking the rax coding region regulate expression of a rax promoter-GFP transgene in Xenopus tadpoles. One of these elements, UCE1, regulates expression of the transgene in the dorsal CMZ. UCE1 contains a Rax binding site, PCE-1. We demonstrate that rax regulates expression of the transgene through the PCE-1 site found in UCE1. Therefore, rax transcription in the CMZ is controlled, in part, by autoregulatory mechanisms.
Keywords: retinal homeobox gene, ciliary marginal zone, retinal progenitor cells, genomic element, Xenopus
INTRODUCTION
The retinal homeobox (rax) gene is essential for vertebrate retinal development [reviewed in (Bailey et al. 2004)]. rax is among the first genes to be expressed in the prospective retina at the anterior neural plate (Casarosa et al. 1997; Mathers et al. 1997; Zuber et al. 2003). Its expression continues in undifferentiated retinal neuroepithelium and is generally down-regulated as retinal neurons differentiate, with expression persisting in photoreceptors (Wang and Harris 2005; Pan et al. 2010). In fish and amphibian tadpoles, rax expression persists in retinal progenitor cells residing in the ciliary marginal or germinal zone (CMZ or CGZ, respectively) of the mature retina (Chuang et al. 1999; Wang and Harris 2005; Pan et al. 2010). Rax is an eye field transcription factor (EFTF), expressed early in eye field specification and development (Zuber et al. 2003). Many EFTFs can activate expression of other EFTFs.
The molecular mechanisms by which rax expression is regulated are not completely worked out. We previously reported the identification and partial characterization of the Xenopus laevis rax.S and rax.L (Rx1A and Rx2A) and Xenopus tropicalis rax gene promoters (Zhang et al. 2003; Martinez-de Luna et al. 2010; Pan et al. 2010). The rax transcriptional unit is flanked by highly conserved genomic elements that we refer to as ultra-conserved elements (UCEs) (Danno et al. 2008; Pan et al. 2010). Two UCEs are found in the 5’-flank of the rax transcribed region. Here, we describe the identification and preliminary characterization of UCE1, located in the 3’-flank of the rax transcribed region. It has been demonstrated that Sox2 and Otx2 are sufficient to initiate expression of rax through UCE3 in naïve Xenopus ectoderm (Danno et al. 2008). Outside of Otx2 and Sox2, trans-acting factors regulating rax expression are largely unknown. Here, we report that rax is capable of autoregulation through UCE1, resulting in regulation of rax transcriptional activity in the dorsal portion of the CMZ.
MATERIALS AND METHODS
Animals
We have complied with or the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978) in the use of animals for the studies described here.
Identification and isolation of UCE1
UCE1 was identified as a highly conserved genomic region adjacent to the Xenopus tropicalis rax transcribed region by analysis of the rax locus using the UCSC Genome Browser (http://genome.ucsc.edu/), spanning scaffold 228:557470–557770 [Aug. 2005 (JGI 4.1/xenTro2) Assembly]. UCE1 was amplified from X. tropicalis genomic DNA using gene-specific primers F: 5’ – CGCGTTGTTGGGATCCCAGTAAATATGC – 3’; R: 5’ – GATCAGATCTCTATCCAGTCCGACCCTGTAGGAATAC – 3’. The R primer contains a BglII restriction enzyme recognition sequence (underlined). Amplified DNA was cloned into pCRII Topo (TOPO TA Cloning Kit, Invitrogen) to make pCRII-UCE1 and verified by DNA sequencing.
Plasmids
UCE1 was liberated from the pCRII TOPO using BamHI and BglII and inserted into the BamHI site of Tg(rax:GFP)Hme (previuously known as tRx3000-GFP) (Pan et al. 2010) to make Tg(UCE1,rax:GFP)Hme. The BglII site was included in the R amplification primer as discussed above. The BamHI site was within the amplified genomic region.
The PCE-1 sites or Nkx sites of UCE1 (pCRII-UCE1) were mutagenized by site-directed mutagenesis using the QuickChange XL Kit (Stratagene) and the following oligonucleotides (top strands are shown; bottom strands are reverse complement oligonucleotides): PCE-1 site: 5’-CTGAGGCAGGATTATTAGGCTCAACACCTAGCTAACGAGGGCAGC-3’ Nkx site: 5’-GCGTGAGCAGCTGGATGCTGGTTAAGGAATTAATCACTAATGGG-3’. To mutate both sites, site-directed mutagenesis was carried out on the PCE-1 mutated UCE1 using Nkx mutagenic primers. Singly or doubly mutated UCE1 was then subcloned into desired plasmids as described above.
To prepare the Tg(UCE1,rax.S:GFP)Hme constructs, wild type or mutated UCE1 was liberated from the pCRII using BamHI and BglII and inserted into the BamHI site of pLITMUS28 (New England Biolabs) to make pL-UCE1. Tg(rax.S:GFP)Mjam(previously known as Rx1A-GFP), including the 3.4 kb rax.S promoter, was liberated from pBS/Rx1A-GFP (Zhang et al. 2003) using SacI and KpnI and ligated into the same sites in pL-UCE1.
Chromatin immunoprecipitation
Xenopus tropicalis embryos were generated by in vitro fertilization and injected with RNA encoding myc-tagged Rax (MT-Rax) in both dorsal animal blastomeres at the 4 cell stage (1 pg per blastomere). Abnormally developing embryos, including those exhibiting the rax gain-of-function phenotype [i.e. ectopic RPE (Mathers et al. 1997)] were discarded. Embryos were cultured to st 41 and then fixed and used for ChIP as described previously (Pan et al. 2010). ChIP products were detected by real time PCR using oligonucleotide primers spanning either UCE1 or UCE2.
Transgenesis
Transgenes were purified from agarose gels using the GeneClean Kit (Bio101) and introduced into X. laevis eggs by intra-cytosolic sperm injection (ICSI) (Sparrow et al. 2000; Kelly et al. 2005). Sperm nuclei were prepared as described previously (Kroll and Amaya 1996) but treated with digitonin instead of lysolecithin (Huang et al. 1999).
In situ hybridization
Section in situ hybridization was performed on 8 µm retinal sections processed using either digoxygenin or fluorescein-labeled antisense riboprobes as previously described (Shimamura et al. 1994; Viczian et al. 2003).
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed as described previously (Voronina et al. 2004; Pan et al. 2006). The probe used was the PCE-1 site from UCE1 (F: 5’-CTGAGGCAGGATTATTAGGCTCAACACCCAATTAACGAGGGCAGC-3’). Competitors were the UCE1 PCE-1 site, the mouse rhodopsin promoter PCE-1 site (Kimura et al. 2000), of the UCE1 Nkx site (5’-GCGTGAGCAATTAGATGCTGGTTAAGGAATTAATCACTAATGGG-3’). All were annealed to the appropriate reverse complement oligonucleotides to make double-stranded oligonucleotides.
RESULTS
The rax CDS is flanked by conserved genomic elements
We identified 3 ultra-conserved elements (UCEs) that are conserved across vertebrates and demonstrated that UCE2 and UCE3 are necessary for expression of a GFP transgene in the ciliary marginal zone (CMZ) (Pan et al. 2010). To investigate the role UCE1 might play in regulating rax promoter activity, we amplified it from X. tropicalis genomic DNA and fused it to the Tg(rax:GFP)Hme transgene (Fig 1A). We determined the expression of this transgene in Xenopus laevis tadpoles, finding that it is expressed in a portion of the CMZ. We found that Tg(UCE1,rax:GFP)Hme is expressed in the dorsal CMZ (the CMZ in the dorsal portion of the retina; Fig 1B). We previously characterized the activity of a 3.4 kb X. laevis rax.S promoter in the developing retina (Zhang et al. 2003). We found that a Tg(rax.S:GFP)Mjam transgene is not expressed in the CMZ [(Zhang et al. 2003) and Fig 1C]. Addition of UCE1 also drove expression of the resulting transgene in the dorsal CMZ (tadpoles, Fig 1D).
Figure 1.
UCE1 drives transgene expression in the dorsal CMZ. A. Schematic diagram of the rax genomic locus (top) and the Tg(UCE1,rax:GFP)Hme transgene. B. Addition of UCE1 to the Tg(rax:GFP)Hme transgene results in expression in the dorsal CMZ but not the ventral CMZ. C, D. Addition of UCE1 to the X. laevis rax.S promoter drives expression in the dorsal CMZ. C. The X. laevis Tg(rax.S:GFP)Mjam transgene is not expressed in the CMZ (open arrowheads). D. Addition of UCE1 to the Tg(rax.S:GFP)Mjam transgene results in expression in the dorsal CMZ (filled arrowhead). Transgene expression was visualized by in situ hybridization using a GFP antisense riboprobe (blue color). L – lens. Sections are oriented so that the dorsal aspect of the embryo is toward the top of the figure.
UCE1 contains two putative Rax binding sites
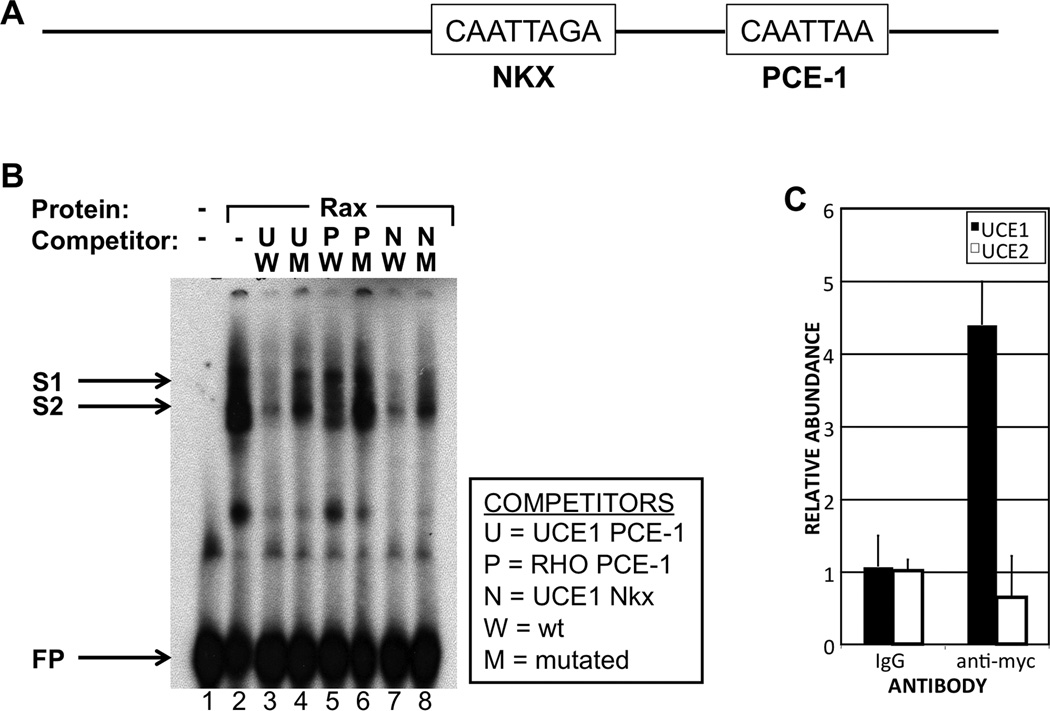
rax gene products can bind the photoreceptor conserved element 1 (PCE-1) site in vitro (Kimura et al. 2000). Analysis of UCE1 reveals that it contains two PCE-1-like sites. One of these was predicted to be a binding site for Nkx transcription factors by the UCSC Genome Browser (not shown). We refer to the two sites as the PCE-1 and Nkx sites (Fig 2A). To analyze binding of Rax to these sites, we performed electrophoretic mobility shift assays (EMSAs) using the UCE1 PCE-1 site as a probe. We observed the formation of two protein-DNA complexes, in the form of bands of shifted mobility, upon addition of Rax protein (S1 and S2, Fig 2B, lane 2). S1 band was essentially absent and the intensity of S2 was substantially reduced upon addition of a 100 X molar excess of unlabeled UCE1 PCE-1 double-stranded oligonucleotide but not a mutated version containing a disrupted version of the PCE-1 site (lanes 3, 4). This suggests that binding of Rax to the UCE1 PCE-1 site is specific. Formation of the complexes was similarly disrupted by addition of a UCE1 Nkx site competitor oligonucleotide (lanes 7, 8). Interestingly, addition of a competitor oligonucleotide spanning the rhodopsin gene promoter PCE-1 site only partially inhibited complex formation (lanes 5, 6), suggesting that Rax binds to this site with lower affinity than the UCE1 PCE-1 and Nkx sites. These data suggest that Rax can specifically bind the UCE1 PCE-1 and Nkx sites in vitro.
Figure 2.
Rax can bind to UCE1 in vitro and in vivo. A. Schematic diagram of UCE1 showing the putative PCE-1 and Nkx sites. B. Rax can bind the PCE-1 and Nkx sites in vitro. Electrophoretic mobility shift assay (EMSA) using an oligonucleotide spanning the UCE1 PCE-1 site as probe and competitor oligonucleotides spanning the same site (lanes 3, 4), the mouse rhodopsin promoter PCE-1 site (lanes 5, 6) or the UCE1 Nkx site (lanes 7, 8). For each competitor we used a wild type version (lanes 3, 5, 7) or a version in which the PCE-1 site was mutated (lanes 4, 6, 8). Addition of in vitro translated Rax protein to the probe resulted in two bands of shifted mobility (S1, S2). C. Rax can bind UCE1 in vivo. Chromatin immunoprecipitation (ChIP) performed using embryos injected with RNA encoding myc-tagged Rax (MT-Rax) and immunoprecipitated using an antibody raised agains the myc tag. UCE1 is specifically precipitated by anti-myc antibody and not control IgG. UCE1 is specifically precipitated but UCE2 is not.
Rax can interact with UCE-1 in vivo
We next investigated whether Rax binds UCE-1 in vivo by chromatin immunoprecipitation (ChIP). We have not identified an antibody that binds to Xenopus Rax, so we used an approach involving epitope-tagged Rax (Pan et al. 2010). A relatively low amount of myc-tagged Rax (MT-Rax) or myc tag alone (MT) was expressed in X. tropicalis embryos (see Materials and Methods) and ChIP was performed using an antibody raised against the myc tag. UCE-1 was enriched in ChIP from MT-Rax-expressing embryos as compared to embryos expressing MT alone (Fig 2C). Further, UCE1 was not precipitated by control IgG. Finally, UCE2 was not enriched in ChIP from MT-Rax-expressing embryos using the anti-myc antibody, even though UCE2 contains a putative PCE-1 site. Taken together, these results indicate that Rax can specifically bind to UCE1 in vivo.
The PCE-1 site is necessary for Rax-dependent regulation of UCE1-dependent reporters
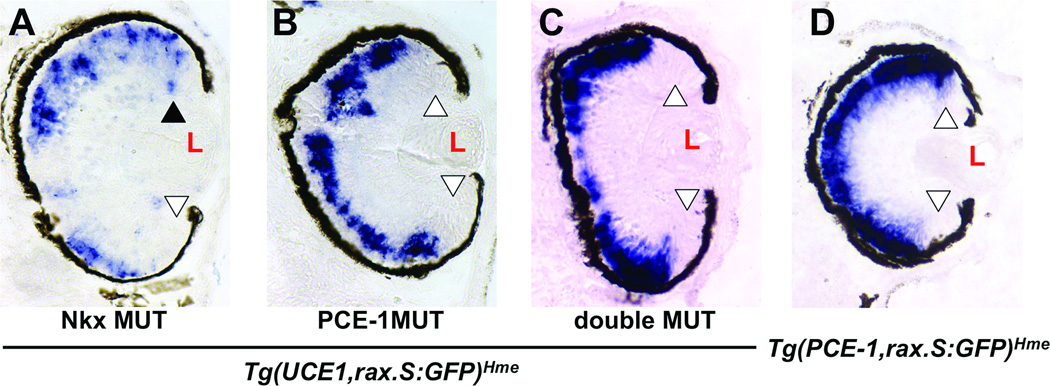
We analyzed the regulation of the UCE1 by PCE-1 in the context of retinal progenitor cells. We mutated the PCE-1 and/or Nkx sites in the context of the Tg(UCE1,rax.S:GFP)Hme transgene and used them to produce transgenic embryos. We found that mutation of the Nkx site did not affect dorsal CMZ expression (3/3 tadpoles, Fig 3A), while mutation of the PCE-1 site abolished expression of the transgene in the dorsal CMZ (5/5 tadpoles, Fig 3B). Mutation of both the Nkx and PCE-1 sites resulted in transgene expression in apparently the same pattern as Tg(rax.S:GFP)Mjam lacking UCE1 entirely (3/3 tadpoles, compare to Figures 3C to 1C), suggesting that expression of Tg(UCE1,rax.S:GFP)Hme in the dorsal CMZ was dependent on the PCE-1 site of UCE1.
Figure 3.
rax-dependent UCE1 activity is mediated through the PCE-1 site. A – C. Expression of the Tg(UCE1,rax.S:GFP)Hme transgene containing mutations in the Nkx site (A), PCE-1 site (B), or both (C). D. Expression of Tg(PCE-1,rax.S:GFP)Hme transgene. Expression is visualized by in situ hybridization of sectioned retinal tissue using a GFP antisense riboprobe. Open arrowheads indicate lack of expression in the CMZ; black arrowhead indicates expression in the CMZ. L – lens. Sections are oriented so that the dorsal aspect of the embryo is toward the top of the figure.
The PCE-1 site is not sufficient for dorsal CMZ activity of UCE1
Having determined that the PCE-1 site is necessary for transgene expression in the dorsal CMZ, we next asked if it is sufficient for transgene expression in the dorsal CMZ. We prepared the Tg(PCE-1,rax.S:GFP)Hme transgene, adding a single copy of the PCE-1 site to Tg(rax.S:GFP)Mjam and tested it in transgenic tadpoles. We found that this transgene was expressed essentially only in the photoreceptor layer and not in the CMZ, as is the Tg(rax.S:GFP)Mjam transgene (Figure 3D), suggesting that the PCE-1 site itself is not sufficient to drive transgene expression in the dorsal CMZ.
DISCUSSION
In this paper, we present evidence that rax may regulates its own transcription by binding a conserved genomic element, UCE-1. We found that Rax can bind to a specific cis-element in UCE-1 in vitro and in vivo and that this element is necessary for UCE-1 dependent transgene expression. In transgenic Xenopus tadpoles, UCE-1 directed transgene expression in the dorsal CMZ. The PCE-1 site was necessary but not sufficient for this expression. These results suggest that Rax binds the PCE-1 site and cooperates with other trans-acting factors to promote transgene expression in the dorsal CMZ. It is interesting to speculate that Rax might interact with a trans-factor that binds the Nkx site. Nkx5.3 and 5.4 (SOHo) are examples of Nkx genes that are expressed in the neural retina (Deitcher et al. 1994; Stadler and Solursh 1994; Adamska et al. 2001; Kelly and El-Hodiri 2016). Although Nkx5.3 and SOHo are expressed in the mature X. laevis neural retina, Nkx5.3 does not exhibit differential expression between dorsal and ventral CMZ and SOHo expression is not observed in the CMZ (Kelly and El-Hodiri 2016).
The PCE-1 site is best known for its conservation among a significant subset of genes expressed specifically in photoreceptors (Kikuchi et al. 1993; Batni et al. 1996; Boatright et al. 1997; Ma et al. 2001; Mani et al. 2001; Moritz et al. 2002). It is clear that Rax binds the PCE-1 site and regulates gene expression of genes containing a PCE-1 site (Kimura et al. 2000; Pan et al. 2010). rax expression is photoreceptors is diminished in rax knockdown embryos (Pan et al. 2010), supporting the proposition that rax expression is, at least to some extent, self-regulating. Even though addition of UCE1 to the X. tropicalis rax regulatory region seemed to decrease transgene in photoreceptors (Figure 1B), it is not clear that this effect was observed when UCE1 was added to the X. laevis rax.s regulatory region (compare Figure 1C and D). In any case, the knockdown results suggest that Rax is required for regulating its own expression in photoreceptors while the data presented here suggests that, in the case of UCE1-regulated transgene, Rax inhibits photoreceptor expression of the UCE1-containing transgene. Regulation of rax expression in the context of the endogenous genetic locus may be significantly different than in the context of the transgene, as evidenced by expression in the CMZ: in the knockdown, CMZ expression is lost, while expression of the transgene results in dorsal CMZ-specific expression.
It is interesting that UCE-1 drives transgene expression in the dorsal CMZ and represses it in the ventral CMZ. rax is expressed throughout the CMZ, so this represents only a portion of the rax expression domain. The dorsal and ventral portions of the CMZ develop with different timing. After the initial formation of the CMZ, the dorsal CMZ continues to develop rapidly whereas ventral CMZ cells exhibit decreased proliferation until somewhat later in development, when it gives rise to the post-metamorphic retina. This process is controlled by expression of type III deiodinase (Marsh-Armstrong et al. 1999), which is expressed exclusively in the dorsal retina as early as st 36. Other genes, such as bmp4, also exhibit dorsal CMZ expression (Hocking and McFarlane 2007), although their role in regulating dorsal-ventral differences in CMZ development have not been reported. In any case, it seems likely that Rax cooperates with other factors that interact with UCE1 to regulate its own differential expression in the CMZ.
Acknowledgments
The authors thank Carlos Alvarez for help and inspiration with comparative genomic analysis and Andy Fischer for helpful and thoughtful discussions. We also thank members of the Fischer and El-Hodiri labs for helpful critical comments. Thanks also to Andy Fischer for critical reading of the manuscript.
Funding: This work is funded by NIH/NEI grant EY015480 to HME. The funding source had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
LITERATURE CITED
- Adamska M, Wolff A, Kreusler M, Wittbrodt J, Braun T, Bober E. Five Nkx5 genes show differential expression patterns in anlagen of sensory organs in medaka: insight into the evolution of the gene family. Development genes and evolution. 2001;211(7):338–349. doi: 10.1007/s004270100162. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol. 2004;48(8–9):761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- Batni S, Scalzetti L, Moody SA, Knox BE. Characterization of the Xenopus rhodopsin gene. J Biol Chem. 1996;271(6):3179–3186. doi: 10.1074/jbc.271.6.3179. [DOI] [PubMed] [Google Scholar]
- Boatright JH, Borst DE, Peoples JW, Bruno J, Edwards CL, Si JS, Nickerson JM. A major cis activator of the IRBP gene contains CRX-binding and Ret-1/PCE-I elements. Mol Vis. 1997;3:15. [PubMed] [Google Scholar]
- Casarosa S, Andreazzoli M, Simeone A, Barsacchi G. Xrx1, a novel Xenopus homeobox gene expressed during eye and pineal gland development. Mech Dev. 1997;61(1–2):187–198. doi: 10.1016/s0925-4773(96)00640-5. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Mathers PH, Raymond PA. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84(1–2):195–198. doi: 10.1016/s0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- Danno H, Michiue T, Hitachi K, Yukita A, Ishiura S, Asashima M. Molecular links among the causative genes for ocular malformation: Otx2 and Sox2 coregulate Rax expression. Proc Natl Acad Sci U S A. 2008;105(14):5408–5413. doi: 10.1073/pnas.0710954105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitcher DL, Fekete DM, Cepko CL. Asymmetric expression of a novel homeobox gene in vertebrate sensory organs. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1994;14(2):486–498. doi: 10.1523/JNEUROSCI.14-02-00486.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking JC, McFarlane S. Expression of Bmp ligands and receptors in the developing Xenopus retina. The International journal of developmental biology. 2007;51(2):161–165. doi: 10.1387/ijdb.062185jh. [DOI] [PubMed] [Google Scholar]
- Huang H, Marsh-Armstrong N, Brown DD. Metamorphosis is inhibited in transgenic Xenopus laevis tadpoles that overexpress type III deiodinase. Proc Natl Acad Sci U S A. 1999;96(3):962–967. doi: 10.1073/pnas.96.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LE, Davy BE, Berbari NF, Robinson ML, El-Hodiri HM. Recombineered Xenopus tropicalis BAC expresses a GFP reporter under the control of Arx transcriptional regulatory elements in transgenic Xenopus laevis embryos. Genesis. 2005;41(4):185–191. doi: 10.1002/gene.20113. [DOI] [PubMed] [Google Scholar]
- Kelly LE, El-Hodiri HM. Xenopus laevis Nkx5.3 and sensory organ homeobox (SOHo) are expressed in developing sensory organs and ganglia of the head and anterior trunk. Dev Genes Evol. 2016 doi: 10.1007/s00427-016-0555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi T, Raju K, Breitman ML, Shinohara T. The proximal promoter of the mouse arrestin gene directs gene expression in photoreceptor cells and contains an evolutionarily conserved retinal factor-binding site. Mol Cell Biol. 1993;13(7):4400–4408. doi: 10.1128/mcb.13.7.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem. 2000;275(2):1152–1160. doi: 10.1074/jbc.275.2.1152. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122(10):3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Ma GC, Wang TM, Su CY, Wang YL, Chen S, Tsai HJ. Retina-specific cis-elements and binding nuclear proteins of carp rhodopsin gene. FEBS Lett. 2001;508(2):265–271. doi: 10.1016/s0014-5793(01)03058-7. [DOI] [PubMed] [Google Scholar]
- Mani SS, Batni S, Whitaker L, Chen S, Engbretson G, Knox BE. Xenopus rhodopsin promoter. Identification of immediate upstream sequences necessary for high level, rod-specific transcription. J Biol Chem. 2001;276(39):36557–36565. doi: 10.1074/jbc.M101685200. [DOI] [PubMed] [Google Scholar]
- Marsh-Armstrong N, Huang H, Remo BF, Liu TT, Brown DD. Asymmetric growth and development of the Xenopus laevis retina during metamorphosis is controlled by type III deiodinase. Neuron. 1999;24(4):871–878. doi: 10.1016/s0896-6273(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Martinez-de Luna RI, Moose HE, Kelly LE, Nekkalapudi S, El-Hodiri HM. Regulation of retinal homeobox gene transcription by cooperative activity among cis-elements. Gene. 2010;467(1–2):13–24. doi: 10.1016/j.gene.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387(6633):603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Moritz OL, Peck A, Tam BM. Xenopus laevis red cone opsin and Prph2 promoters allow transgene expression in amphibian cones, or both rods and cones. Gene. 2002;298(2):173–182. doi: 10.1016/s0378-1119(02)00923-x. [DOI] [PubMed] [Google Scholar]
- Pan Y, Martinez-De Luna RI, Lou CH, Nekkalapudi S, Kelly LE, Sater AK, El-Hodiri HM. Regulation of photoreceptor gene expression by the retinal homeobox (Rx) gene product. Dev Biol. 2010;339(2):494–506. doi: 10.1016/j.ydbio.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Nekkalapudi S, Kelly LE, El-Hodiri HM. The Rx-like homeobox gene (Rx-L) is necessary for normal photoreceptor development. Invest Ophthalmol Vis Sci. 2006;47(10):4245–4253. doi: 10.1167/iovs.06-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura K, Hirano S, McMahon AP, Takeichi M. Wnt-1-dependent regulation of local E-cadherin and alpha N-catenin expression in the embryonic mouse brain. Development. 1994;120(8):2225–2234. doi: 10.1242/dev.120.8.2225. [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Latinkic B, Mohun TJ. A simplified method of generating transgenic Xenopus. Nucleic Acids Res. 2000;28(4):E12. doi: 10.1093/nar/28.4.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler HS, Solursh M. Characterization of the homeobox-containing gene GH6 identifies novel regions of homeobox gene expression in the developing chick embryo. Developmental biology. 1994;161(1):251–262. doi: 10.1006/dbio.1994.1025. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130(7):1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Voronina VA, Kozhemyakina EA, O'Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13(3):315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- Wang JC, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285(1):101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Zhang L, El-Hodiri HM, Ma HF, Zhang X, Servetnick M, Wensel TG, Jamrich M. Targeted expression of the dominant-negative FGFR4a in the eye using Xrx1A regulatory sequences interferes with normal retinal development. Development. 2003;130(17):4177–4186. doi: 10.1242/dev.00626. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130(21):5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]