Abstract
Background
Nonalcoholic fatty liver disease (NAFLD) is a complex, multifactorial disease affected by diet, lifestyle and genetics. Proinflammatory cytokines like IL-1β and IL-6 have been shown to be elevated in nonalcoholic steatohepatitis (NASH). The goal of this study was to investigate the relationship between IL1B and IL6 gene polymorphisms and histologic features of NAFLD in the NASH CRN cohort.
Methods
604 adult (≥18 yrs) non-Hispanic Caucasians with biopsy-proven NAFLD were genotyped for the following SNPs: IL1B, rs16944, rs1143634; IL6, rs1800795, rs10499563. Logistic regression was used to examine the relationship between genotype and a definitive diagnosis and advanced histological features of NASH after controlling for the following variables selected a priori: age, sex, diabetes, obesity and HOMA-IR level.
Results
The IL6 rs10499563 C allele was independently associated with the presence of definitive NASH, and increased ballooning and Mallory bodies. The IL1B rs1143634 TT genotype was associated with advanced fibrosis and increased Mallory bodies. The IL6 rs1800795 C allele was associated with increased risk for severe steatosis, >66% but also decreased risk for advanced fibrosis and lobular inflammation and Mallory body formation.
Conclusions
These results suggest that common variants in the IL6 and IL1B genes may increase susceptibility for NASH and confer a higher risk of hepatic parenchymal damage including increased ballooning, increased Mallory bodies, and bridging fibrosis or cirrhosis. In contrast, the IL6 rs1800795 C allele may confer a higher risk for steatosis, but less parenchymal damage. Our findings support the development of therapeutics aimed at IL-1β and IL-6 suppression.
Keywords: NAFLD, Steatohepatitis, NASH, cytokines, polymorphism, inflammasome
Introduction
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease in most Westernized countries; as many as 30% of US adults may be affected [1,2]. Nonalcoholic steatohepatitis (NASH), the more severe form of the disease, is associated with hepatic inflammation, hepatocellular ballooning and may include varying degrees of fibrosis [3]. The majority of NASH patients are obese and comorbidities such as type 2 diabetes mellitus and metabolic syndrome are also common in these patients [3]. While lifestyle modifications towards a healthy diet and habitual exercise are advisable in NAFLD, as weight loss of at least 3–5% improves steatosis and weight loss of up to 10% may improve necroinflammation, and are still considered the most effective treatment, however, the combined impact of genetics, diet and lifestyle in the etiology of NASH is poorly understood [4–7]. An increasing number of potential NASH susceptibility loci have been identified in recent years including patatin-like phospholipase 3 gene (PNLPA3) [8–12], neurocan (NCAN) [11,12], protein phosphatase 1 regulatory subunit 3B (PPP1R3B) [12], apolipoprotein C3 (APOC3) [12], peroxisome proliferator-activated receptors α or γ (PPAR α or γ) [13], glucokinase regulator (GCKR) [12, 14], among others. Additionally, several recent genome-wide association study (GWAS) identified single nucleotide polymorphisms (SNPs) associated with component NAFLD traits including elevated liver enzymes [10,15–18], steatosis based on computed tomography (CT) [19] or magnetic resonance imaging (MRI) [10] or specific biopsy-proven histologic features including steatosis, lobular inflammation and fibrosis [18–20]. Together these studies underscore the multifactorial etiology of this disease, however most previous genetic studies have focused on SNPs associated with lipid metabolism. There is a paucity of studies investigating the association of inflammatory gene variants and the phenotype of NAFLD.
Levels of the proinflammatory cytokines IL6 and IL1β are elevated in NASH and obesity [21]. These cytokines are important mediators of the inflammatory response, and are both expressed in Kupffer cells, macrophages, hepatocytes and adipocytes. In addition to their role in inflammation, these cytokines may also contribute to NASH pathogenesis by promoting insulin resistance and altering lipid metabolism [21–24]. IL6 is involved in initiation of systemic inflammation and acute phase reactions in response to infection or other chronic inflammatory states such as obesity [24]. IL1β is thought to be more important in innate immune system modulation and localized tissue injury repair and is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis [24]. IL1β is mainly produced by cleavage of pro-IL1β by caspase-1 upon activation of the inflammasome, the large multi-protein complex which is assembled and activated in response to recognition of a variety of ligands including endogenous danger-associated molecular patterns (DAMPs) or pathogen-associated molecular patterns (PAMPs) [25]. DAMPs, include dsDNA and CpG-rich DNA fragments and mitochondrial components like cytochrome c and adenosine triphosphate (ATP) which are generated during tissue injury. PAMPs are generally microbial in nature such as gram negative bacterial cell wall components, including lipopolysaccharides, and various bacterial toxins.
Several common SNPs in the IL1B and IL6 genes were previously associated with increased cytokine levels; IL6-174G>C (rs1800795) [26, 27], IL6 -6331T>C (rs10499563) [28], IL1B +3954 C>T (rs1143634) [29, 30]. Moreover, IL1B and IL6 SNPs have also been associated with various NASH-related phenotypes including the presence of diabetes mellitus or insulin resistance; IL6 rs1800795 [31], IL1B -511C>T (rs16944) [32] and adiposity; IL6 rs1800795 [33], IL1B rs1143634 [34]. The IL6 rs1800795 C allele and the IL1B rs16944 T allele were recently associated with the presence of NASH in smaller Italian [35] and Japanese cohorts [36], respectively. Therefore, the aim of this study was to investigate if these 4 common SNPs in the IL1B and IL6 genes, previously reported to be associated with one or more NASH-related traits, could be associated with susceptibility for NASH or histologic features of advanced disease.
Patients and methods
Patients
A total of 604 adult (>18 yrs) non-Hispanic Caucasian subjects enrolled into the NASH CRN Database and Pioglitazone vs Vitamin E vs Placebo for Treatment of Non-Diabetic Patients With Nonalcoholic Steatohepatitis (PIVENS) therapeutic trial with biopsy-proven NAFLD and available DNA who had previously consented to genetic analysis were evaluated in the present study. Demographic information such as age, sex, ethnicity, race and medical history to identify co-morbidities and medication usage were obtained from patient interviews during screening. A physical exam including body weight and height measures was performed. Laboratory data including hepatic, hematologic, metabolic and lipid measurements were collected within 6 months of the liver biopsy. Alcohol consumption was determined from the AUDIT-C questionnaires completed during study visits closest to the time of biopsy [37]. The prevalence of metabolic syndrome in this cohort was defined using the World Health Organization criteria. All subjects gave written informed consent and the study was approved by the institutional review board at each local site of the NASH CRN.
Plasma cytokine levels
Serum tumor necrosis factor α (TNFα), IL6 and IL1β levels, determined using Luminex technology and the human cytokine LINCOplex kit (Catalog number HCYTO-60K, Millipore, St. Charles, MO), were available in 63%, 57% and 39% of non-Hispanic Caucasian subjects, respectively. The lower limit of detection for these assays was 0.66, 0.79 and 0.19 pg/mL, respectively. Serum soluble IL1 receptor 1 (sIL1R) and soluble IL6 receptor (sIL6R) were each available in 63% of non-Hispanic Caucasian subjects (Catalog number HSCR-32K, Millipore, St. Charles, MO). The lower limit of detection for these assays was 7.9 and 5.8 pg/mL, respectively.
Genotyping
Genotyping for the following SNPs in the IL1B and IL6 genes was performed using a real time PCR genotyping assay: IL1B rs16944, IL1B rs1143634, IL6 rs1800795, IL6 rs10499563. Briefly, 10 ng of genomic DNA was plated into 384 well plates using a Beckman Coulter Biomek FX robotic workstation. Each 5 μl reaction containing DNA, fluorescently labeled MGB-Eclipse probes and primers (Epoch Bioscience, Bothell WA) and 0.3 u JumpStart Taq (Sigma-Aldrich, St. Louis, MO, USA) were analyzed on an ABI HT7900. Primer/probe sequences are shown in Supplemental Table 1. The percentage of missing genotypes for each SNP was ≤ 0.38%. Details of the SNPs including type, location, genetic model used and rare allele frequency (RAF; also known as minor allele frequency) were analyzed in the current work and compared to publicly available allele frequency estimates.
Histological assessment
All liver biopsies were centrally read by the NASH CRN Pathology Committee according to the NASH CRN scoring system [38]. H&E and Masson’s Trichrome stains were evaluated for each case. Steatosis, lobular inflammation, fibrosis, ballooning, portal inflammation, acidophil bodies, megamitochondria, and Mallory-Denk bodies were assessed by a semi-quantitative method according to the NASH CRN scoring system [38]. The presence of definite steatohepatitis was determined by consensus upon central review by the study pathologists based on pattern recognition. We observed that of the patients that had definite NASH, all had >5% steatosis and lobular inflammation and all but two biopsies had ballooning (99.6%). Many ballooning hepatocytes were defined as more than just few or rare ballooning hepatocytes, as defined by the NASH-CRN pathology committee and the biopsies were reviewed at the consensus of the NASH-CRN pathology committee. Likewise, many Mallory-Denk bodies were defined as more than just few or rare Mallory-Denk bodies, as defined by the NASH-CRN pathology committee and the biopsies were reviewed at the consensus of the NASH-CRN pathology committee.
Statistical analysis
Demographic, clinical, and laboratory characteristics and NASH diagnosis were recorded as number and percentage for categorical data and means and standard deviation for continuous data. Categorical data was analyzed using Fisher’s exact test or Chi square, where appropriate. Continuous variables including laboratory measures were not normally distributed and were analyzed using the Wilcoxon rank sum test. For association of SNPs with parameters of histological scores or a definitive diagnosis of NASH, three genetic models were assessed for each SNP: additive, dominant, and recessive. The model which best fit the data by maximizing the significance of the test statistic for the majority of variables was chosen. Multivariable forward stepwise logistic regression (cutoff p<0.20) was used to investigate the independent association of each genetic model to individual advanced histological features including: grade 3 steatosis, stage 3–4 fibrosis, grade 2–3 lobular inflammation and increased ballooning, portal inflammation, acidophil bodies, Mallory bodies and megamitochondria. The effect of each SNP was simultaneously modeled and adjusted for potential confounding variables defined a priori including sex, age, obesity, HOMA-IR level and presence of type 2 diabetes mellitus. Because our candidate genotyping approach targeted just 4 SNPs, each with a high posterior probability of being true positives given a prior association to NAFLD or a related phenotype, a two-sided p-value <0.05 was used as the threshold for statistical significance. All analyses were performed using STATA (version 12, College Station, TX, USA).
Results
Candidate IL6 and IL1B genotyping
Details of the SNPs including type, location, genetic model used and rare allele frequency (RAF; also known as minor allele frequency) from this study compared to publically available allele frequency estimates (i.e., HapMap-CEU) are shown in Table 1. IL1B rs1143634 was a coding region synonymous substitution located in exon 5, while the other 3 SNPs were present in the promoter regions of each gene. The RAFs of the 4 candidate SNPs in white non-Hispanics were similar to the HapMap-CEU. Each of the SNP allele frequencies were in Hardy–Weinberg equilibrium.
Table 1.
Single nucleotide polymorphism (SNP) details
| SNP ID | Common Name | SNP Type | HapMap-CEU* RAF |
Study RAF |
Location | Model |
|---|---|---|---|---|---|---|
| rs16944 | IL1B -511C>T | promoter | 0.36 | 0.33 | chr. 7 113594867 |
Recessive |
| rs1143634 | IL1B +3954C>T | coding; synonymous, exon 5 | 0.21 | 0.24 | chr. 7 113590390 |
Recessive |
| rs1800795 | IL6 -174G>C | promoter; MEF site | 0.47 | 0.41 | chr. 7 22766645 |
Dominant |
| rs10499563 | IL6 -6331T>C | promoter; Oct-1 binding site | 0.19 | 0.21 | chr. 7 22760488 |
Dominant |
RAF=Rare Allele Frequency in white, non-hispanics
Utah residents with Northern and Western European ancestry from the CEPH collection
Patient characteristics
Characteristics of the study cohort are shown in Table 2. The mean age of the study cohort was 46.2 ± 12 yrs. The majority of subjects were female (65%) and obese (75%). The mean BMI was 34.7 ± 6.4. Nearly a third of the cohort had a history of diabetes mellitus (28%) and 72% met the criteria for metabolic syndrome. Two thirds of the cohort had a definitive diagnosis of NASH and one third had advanced (stage 3 or 4) fibrosis. Thirteen and five percent of the cohort was homozygous for the IL1B rs16944 and IL1B rs1143634 rare T alleles, respectively (Table 3). The combined homozygous and heterozygous genotypes for the dominant model IL6 SNPs, IL6 rs1800795 and IL6 rs10499563, comprised 58% and 38% of the cohort, respectively. Subjects homozygous for the IL1B rs16944 T allele were significantly younger than the other combined IL1B rs16944 genotypes (Table 3; p=0.025). Type 2 diabetes was significantly more prevalent among subjects with the IL1B rs1143634 TT genotype, compared to the combined CC and CT IL1B rs1143634 genotypes (Table 3; p=0.016). There were no significant differences in sex, waist/hip ratio, BMI, or presence of the metabolic syndrome between these genotypes.
Table 2.
Patient Characteristics.
| Characteristic | Total Cohort |
|---|---|
| Number, n(%) | 604(100) |
| Age, (yrs) | 46.2±12.0 |
| BMI, (kg/m2) | 34.7±6.4 |
| Waist-hip ratio | 0.94±0.07 |
| Female, n(%) | 392(65) |
| Obese, (≥30 BMI) | 453(75) |
| Diabetes mellitus, n(%) | 170(28) |
| Metabolic syndrome, n(%) | 429(72) |
| Current smoker, n(%) | 57(9.5) |
| NSAID usage, n(%) | 475(79) |
| Antihyperlipidemic medication usage, n(%) | 231(39) |
| Definitive NASH diagnosis | 360(60) |
| Advanced fibrosis (stage 3–4) | 182(30) |
Values are mean±SD or n(%)
NASH- nonalcoholic steatohepatitis
NSAID-nonsteroidal anti inflammatory drugs
BMI- body mass index
Table 3.
Clinical data and laboratory values in non-Hispanic Caucasian patients with nonalcoholic fatty liver disease.
| Characteristic | IL1B rs16944 | IL1B rs1143634 | IL6 rs1800795 | IL6rs10499563 | ||||
|---|---|---|---|---|---|---|---|---|
| CC + CT (n=540) | TT (n=62) | CC + CT (n=570) | TT (n=32) | GG (n=211) | GC + CC (n=391) | TT (n=375) | CT + CC (n=228) | |
| Age, (yrs) | 49.7±10.8 | 46.4±12.0* | 49.4±11.1 | 48.7±7.2 | 49.6±10.7 | 49.2±11.1 | 49.4±11.1 | 49.3±10.6 |
| BMI, (kg/m2) | 34.7±6.5 | 35.3±5.4 | 34.7±6.4 | 35.8±7.1 | 34.6±6.7 | 34.8±6.3 | 34.9±6.7 | 34.5±5.9 |
| Waist-hip ratio | 0.93±0.08 | 0.94±0.08 | 0.93±0.08 | 0.95±0.08 | 0.93±0.08 | 0.93±0.08 | 0.93±0.08 | 0.93±0.08 |
| Female, n(%) | 355(66) | 37(60) | 370(65) | 21(66) | 133(63) | 258(66) | 245(65) | 146(64) |
| Obese, (≥30 BMI) | 402(75) | 50(81) | 428(75) | 24(75) | 152(72) | 300(76) | 277(74) | 175(77) |
| Diabetes mellitus, n(%) | 152(28) | 17(27) | 155(27) | 15(47)* | 58(27) | 112(29) | 111(30) | 59(26) |
| Metabolic Synd. n(%) | 383(72) | 44(71) | 407(72) | 21(66) | 150(71) | 277(72) | 263(71) | 165(73) |
| Laboratory values‡ | ||||||||
| ALT (U/L) | 69.0±47.3 | 70.0±46.7 | 69.1±47.4 | 67.9±44.8 | 74.7±51.0 | 65.1±44.0* | 66.5±42.8 | 72.6±52.9 |
| AST (U/L) | 51.5±35.6 | 54.0±30.8 | 51.4±34.2 | 55.7±48.7 | 54.4±35.0 | 49.7±35.1* | 51.1±32.1 | 52.6±39.3 |
| AST/ALT | 0.83±0.40 | 0.87±0.34 | 0.84±0.40 | 0.87±0.32 | 0.82±0.32 | 0.85±0.43 | 0.86±0.43 | 0.81±0.31 |
| Total bilirubin (mg/dL) | 0.75±0.46 | 0.67±0.42 | 0.74±0.47 | 0.71±0.31 | 0.76±0.56 | 0.73±0.40 | 0.75±0.50 | 0.73±0.40 |
| GGT | 72.4±89.2 | 60.0±41.2 | 68.0±74.0 | 124.0±197.8 | 69.4±77.9 | 71.8±89.3 | 70.7±94.3 | 71.5±69.1* |
| Total cholesterol (mg/dL) | 196.0±41.3 | 192.5±45.1 | 195.1±41.8 | 201.8±38.9 | 196.8±42.6 | 195.0±41.2 | 194.4±39.6 | 197.5±44.8 |
| Triglycerides (mg/dL) | 181.8±119.0 | 189.1±138.2 | 182.4±120.3 | 176.0±110.8 | 195.5±127.2 | 174.5±114.7 | 180.0±124.6 | 184.6±111.0 |
| LDL (mg/dL) | 115.6±38.2 | 113.3±42.5 | 114.9±38.5 | 121.1±39.6 | 114.1±38.8 | 116±38.6 | 114.1±37.0 | 117.4±41.0 |
| HDL (mg/dL) | 44.4±11.6 | 44.2±12.0 | 44.2±38.5 | 46.0±14.2 | 43.7±11.3 | 44.7±11.8 | 44.8±11.8 | 43.6±11.4 |
| Glucose (mg/dL) | 104.9±31.4 | 108.7±38.9 | 105.2±32.5 | 108.7±28.7 | 104.7±33.1 | 105.7±31.9 | 103.7±27.7 | 108.0±38.3 |
| Fasting insulin (μU/mL) | 21.5±19.5 | 22.2±13.9 | 21.5±19.2 | 23.0±14.8 | 24.4±21.9 | 20.3±17.5* | 21.3±20.6 | 22.0±16.1 |
| HOMA-IR | 5.9±6.6 | 6.3±5.3 | 5.9±6.6 | 6.4±4.7 | 6.9±8.4 | 5.4±5.2 | 5.7±6.1 | 6.3±7.1 |
| HbA1c (%) | 6.0±1.1 | 6.0±1.1 | 6.0±1.1 | 6.3±1.1* | 6.0±1.1 | 6.0±1.1 | 5.9±1.1 | 6.0±1.1 |
| Cytokine levels | ||||||||
| IL1β (pg/ml) | 0.53±0.71 | 0.57±0.53 | 0.51±0.56 | 0.82±1.76 | 0.49±0.61 | 0.57±0.73 | 0.60±0.83 | 0.45±0.39 |
| IL6 (pg/ml) | 12.4±50.3 | 7.9±5.8 | 11.8±48.8 | 14.8±16.7 | 11.5±21.0 | 12.3±58.2 | 13.9±59.5 | 9.1±18.6 |
| sIL1R1(pg/ml) | 33.0±21.4 | 42.5±42.7 | 33.7±24.5 | 37.5± 26.3 | 34.7±30.0 | 33.4±21.0 | 33.9±26.8 | 33.8±20.5 |
| sIL6R (ng/ml) | 20.0±5.3 | 20.5±5.4 | 20.1±5.3 | 19.0±4.9 | 19.4±5.1 | 20.4±5.4 | 19.9±5.4 | 20.3±5.1 |
| TNFα (pg/ml) | 8.6±13.0 | 7.2±3.4 | 8.4±12.7 | 8.7±4.6 | 7.5±3.4 | 9.0±15.3 | 9.1±15.7 | 7.3±3.2 |
Values are mean±SD or n(%)
P-value <0.05 according to genetic model for each SNP.
Only laboratory values collected within 6 months of liver biopsy included.
BMI-Body Mass Index, sIL1R- Serum soluble IL1 receptor 1, sIL6R- soluble IL6 receptor, ALT- Alanine Amino Transferase, AST-Aspartate Amino Transferase
GGT-Gamma Glutamyl Transferase, HDL-High density lipoprotein, LDL-low density liplipoprotein, Hb1AC- Glycated Hemoglobin
Relationship between IL1B and IL6 genotype, serum cytokine levels and laboratory data
As shown in Table 3, non-Hispanic Caucasian subjects with an IL6 rs1800795 dominant C allele had significantly lower serum aminotransferase and fasting insulin levels (p<0.03). Except for gamma-glutamyl transferase (GGT) levels among different IL6 rs10499563 genotypes and percent of glycated hemoglobin (HbA1c) among the different IL6 rs1143634 genotypes, no other differences in laboratory tests between alleles at any loci were found. Plasma levels of IL1β, IL6, TNFα and sIL1R and sIL6R were available in 39–64% of non-Hispanic Caucasian subjects. There were no significant differences in cytokine plasma levels between alleles at any loci among non-Hispanic Caucasians, although several significant differences did exist in non-Caucasians (data not shown).
Increased risk of nonalcoholic steatohepatitis among carriers of IL6 rs10499563 C allele
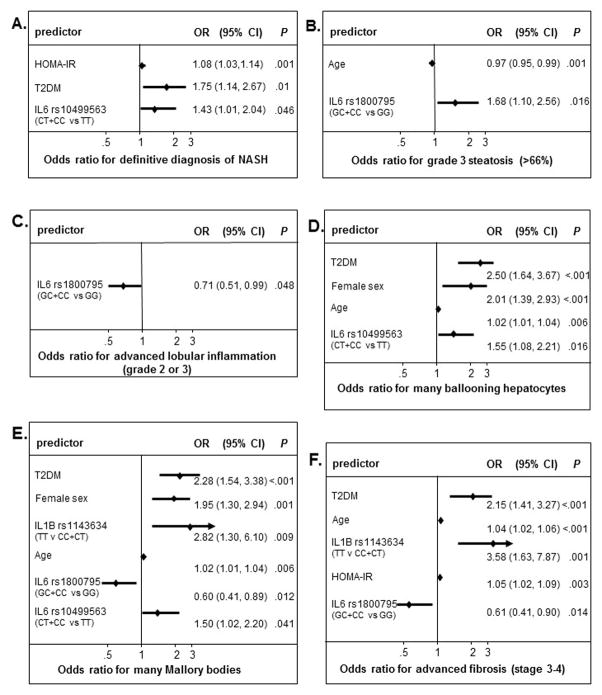
Non-Hispanic Caucasian carriers of the IL6 rs10499563 C allele were more likely to have a definitive diagnosis of NASH compared to IL6 rs10499563 TT genotype patients (65% vs 57%; Chi2 4.1, p=0.042). Moreover, the IL6 rs10499563 C allele independently predicted the presence of definitive NASH, after adjusting for sex, age, presence of diabetes, obesity and HOMA-IR (Homeostatic model assessment-Insulin resistance) levels (Fig. 1A, OR 1.43, 95% CI 1.01–2.04, p=0.046). No other genotype was associated with the presence of definitive NASH in this cohort, however increased HOMA-IR level and presence of diabetes were each associated with a definitive diagnosis of NASH (p<0.05).
Fig. 1. Multivariable logistic regression analysis for advanced histologic features of NASH in white non-Hispanic patients.
Stepwise multivariable logistic regression analysis (cutoff p<0.2) was used to model the independent risk of the presence (yes versus no) of a definitive diagnosis of nonalcoholic steatohepatitis (NASH) (Panel A) and advanced histological disease features such as presence of grade 3 steatosis (66%) (Panel B), advanced lobular inflammation-grade 2 or 3 (Panel C), presence of many ballooning hepatocytes (Panel D), presence of many Mallory Denk bodies (Panel E) and presence of advanced fibrosis (stage 3–4) (Panel F) for IL1β and IL-6 single nucleotide polymorphisms (SNPs) simultaneously and including potential confounding variables such as age, sex, diabetes, obesity and Homeostatic model assessment-Insulin resistance (HOMA-IR) level.
IL1B and IL6 SNPs are associated with advanced histological disease features
To investigate if each IL1B and IL6 polymorphism could independently confer susceptibility to histologically severe disease such as advanced fibrosis among non-Hispanic Caucasians, we performed stepwise multivariable logistic regression analysis, which included each SNP model and the following possible contributory variables chosen apriori: sex, age, presence of diabetes, obesity and HOMA-IR levels (Fig. 1). The combined IL6 rs1800795 GC and CC genotypes showed an increased risk for severe steatosis, >66% (Fig 1B; OR 1.65, 95%CI 1.08, 2.51, p=0.020), but was associated with less advanced lobular inflammation (Fig 1C; OR 0.71, 95%CI 0.51, 0.99, p=0.048), fewer Mallory bodies (Fig 1E; OR 0.60, 95%CI 0.41, 0.89, p=0.012) and less advanced fibrosis (Fig 1F; OR 0.61, 95%CI 0.41, 0.90, p=0.014). Interestingly, while older age was associated with parenchymal damage, younger age was associated with the highest grade of steatosis (>66%), which could reflect an earlier temporal pattern of onset for steatosis, relative to hepatic damage which may require addition insults (i.e., inflammation, oxidative stress, mitochondrial dysfunction etc). The presence of diabetes as well as older age were both associated with the three histological features indicative of parenchymal damage; increased ballooning hepatocytes, increased Mallory bodies and advanced fibrosis (stage 3 or 4) (Fig. 1D–F). The IL6 rs10499563 C allele and female sex were both strongly associated with increased ballooning and Mallory bodies (Fig. 1D–E). The IL1B rs1143634 TT genotype had the highest increased risk of Mallory bodies (Fig. 1E; OR 3.52, 95%CI 1.76–7.07, p<0.001) and advanced fibrosis (Fig. 1F; OR 3.58, 95%CI 1.63, 7.87, p=0.001) of all variables and thus may have the greatest impact on disease progression of all the genotypes in this study. No loci were associated with increased portal inflammation, acidophil bodies or megamitochondria.
Discussion
Proinflammatory cytokines are important mediators of NASH pathology, however, there is a paucity of studies investigating the potential of cytokine gene polymorphisms as NASH susceptibility loci, despite the fact that several common SNPs in these genes have been associated with several NASH-related traits such as diabetes, insulin resistance and adiposity. Though the pathogenesis of disease progression from NAFLD to NASH has not been fully understood, immunological mechanism like defects in innate immunity, adaptive immunity, Toll-like receptor (TLR) signaling and gut-liver axis have been increasingly recognized to be involved in the disease progression [39]. It is thought that proinflammatory mediators such as lipopolysaccharides (LPS) or interferon-γ (INFγ) induce M1 polarized macrophages and inhibit M2 polarized macrophages leading to increased secretion of proinflammatory cytokines like TNFα, IL-6, IL-23, IL-17 and decreased secretion of anti-inflammatory cytokines like IL-10, resulting in progression of NAFLD to NASH [40,41]. Activation of Kupffer cells and neutrophils in acute or chronic liver diseases, lipotoxicity and increased reactive oxygen species (ROS) production may induce production of proinflammatory cytokines leading to activation of T cells, oxidative damage to hepatic cells and hepatocyte apoptosis resulting in progression to NASH [42]. It has been shown that reduction of NK cell activity, which has an anti-fibrotic effect in the liver, may increase susceptibility to liver cirrhosis in obese subjects postulating the role of NK cells in development of NASH [42]. Few studies on experimental models have shown that TNFα, IL-6, IL-1α, IL-1β, IL-17 could have a role in the progression to NASH. TNFα produced by kupffer cells may suppress apoptotic activity of hepatic stellate cells by inducing expression of tissue inhibitor of metalloproteinase 1 (TIMP-1) mRNA resulting in liver fibrosis [43]. In hepatocytes, it may induce sterol regulatory element binding protein-1c (SREBP-1c) and inhibitors of cytokine signaling (SOCS), aggravate ROS formation resulting in liver steatosis, decreased insulin signaling and hepatic cell death. IL-18 could be involved in the development of insulin resistance-mediated NAFLD [43]. It has been recently demonstrated that NLRP3 inflammasome, an intracellular multiprotein complex involved in the production of mature IL-1β could play a crucial role in progression of NASH [44].
Association between IL-1β and type 2 diabetes in obese patients has been demonstrated in various studies [32, 34]. Recently, it has been recognized that type 2 diabetes is due to chronic inflammation resulting in dysfunction of pancreatic islets. IL-1β production and secretion has been reported from pancreatic islets; insulin producing β-cells are specifically prone to IL-1β-induced destruction and loss of function [45]. There is increased evidence that an imbalance between IL-1β and its naturally occurring antagonist, IL-1R antagonist results in recruitment and activation of IL-1β producing macrophages. This is known to mediate pancreatic islet inflammation and lead to insulitis. A randomized study done on 34 patients by de Mello et al. demonstrated a decrease in IL-1 receptor antagonist expression after weight loss and a strong correlation between the decrease in IL-1β expression and the increase in insulin sensitivity suggesting their contribution to insulin resistance in obesity and metabolic syndrome [46]. In addition, use of anakinra, a recombinant IL-1 receptor antagonist, in individuals with type 2 diabetes resulted in decreased blood glucose concentrations, improved pancreatic β cell function, and reduced circulating inflammatory markers [47]. We also found that type 2 diabetes was significantly more prevalent among subjects with the IL1B rs1143634 TT genotype, indicating an association between this IL-1β SNP and type 2 diabetes.
In the present study we found that SNPs in the IL6 and IL1B genes were independently associated with advanced histologic features of NASH among non-Hispanic Caucasians, including advanced fibrosis, severe steatosis, and increased ballooning and Mallory bodies. Specifically, this is the first report we are aware of that the IL1B rs1143634 and IL6 rs10499563 are associated with features of hepatic parenchymal damage in liver disease of any kind. In addition, the IL6 rs10499563 C allele was associated with a definitive diagnosis of NASH. Although 3 of the 4 SNPs we investigated were located in promoter regions, we did not find significant differences in plasma cytokine levels between different genotypes. We speculate that in this patient population, potential polymorphic effects may be too subtle to reflect significant differences in plasma cytokine levels, given the high obesity rate and chronic inflammatory nature of NAFLD which would increase baseline cytokine values regardless of genotype. We also cannot rule out the effects of localized differences in tissue cytokine levels within the liver.
In this study the IL6 rs10499563 C allele was associated with definitive NASH and increased ballooning and Mallory bodies. In contrast to the other three SNPs we investigated, which have been widely studied in numerous diseases, very little is known regarding the potential pathologic effects of IL6 rs10499563. Smith et al., have shown that the IL6 rs10499563 CC genotype had lower levels of IL6 compared to the TT genotype, in response to an acute inflammatory state in separate cohorts, 6–24 hours after coronary bypass grafting surgery or intensive periodontal treatment [28]. This effect was mediated by increased binding of the transcription factor Oct-1 to the T allele [28]. However, it is unclear how the acutely increased IL6 levels (levels were normalized by 7 days) seen in the TT genotype in the study by Smith et al., would relate to the chronic inflammatory state of the obese NAFLD patients our study. Additional studies are warranted to define a potential mechanism for the association of the IL6 rs10499563 C allele and worsened NAFLD disease, and to determine if this SNP may play a pathogenic role in other diseases.
Homozygosity for the IL1B rs1143634 T allele was independently associated with advanced fibrosis and Mallory bodies after controlling for sex, age, obesity, HOMA-IR and presence of diabetes in multivariable analyses. Subjects with this risk genotype were more likely to be diabetic. Although IL1β is known to contribute to the pathogenesis of diabetes through glucose and fatty acid stimulated IL1β production in pancreatic β-cells, resulting in subsequent β-cell loss 17, a review of the literature failed to identify an association between this polymorphism and Type 2 diabetes. Neither were there any reports for this SNP identifying the type of tissue damage we observed (i.e., advanced fibrosis, ballooning, and increased Mallory bodies) in other liver diseases. In one large Swedish study of 18–20 year old men the IL1B rs1143634 rare allele T was associated with decreased fat mass based on dual-energy x-ray absorptiometry (DEXA) scan measurements [34]. In another study of coronary heart disease patients from Western Australia the IL1B rs1143634 TT genotype was associated with increased waist circumference [48]. We did not observe an association to any measure of adiposity in our study for this polymorphism.
The effect of IL6 rs1800795 in the etiology of numerous diseases, including several liver diseases, has been widely studied for many years. Most of the studies of liver disease are in agreement with our results that the IL6 rs1800795 C variant is associated with a less severe disease phenotype when compared to the G variant, including among patients with chronic hepatitis C virus infection and patients with hepatocellular carcinoma [49]. One notable exception is a study of 59 biopsy-proven Italian NAFLD subjects, which found that the C allele was more prevalent among subjects with NASH compared to those without NASH 30. This study showed that this allele was an independent predictor of both NAFLD and NASH, when 79 additional healthy blood donors with normal liver transaminase levels were included as controls, but also failed to find an association between genotype and any histological disease features. The lack of healthy controls in our study and the fact that our regression models included both IL1B and IL6 SNPs, could explain the differences in these studies. In agreement with our study this group also did not find differences in plasma IL6 levels between genotypes.
An interesting finding of our study was that IL6 rs1800795 C allele carriers had lower serum transaminases and fasting insulin levels and were less likely to have advanced lobular inflammation, advanced fibrosis and Mallory bodies, but conversely were at an increased risk for developing severe steatosis. Increased hepatic IL6 gene expression is associated with NASH in humans and positively correlates with degree of inflammation and stage of fibrosis [50]. Additionally, luciferase reporter constructs containing the IL6 rs1800795 C allele show reduced expression compared to the G allele when transiently transfected into HeLa cells [26]. Thus, our findings of decreased risk of lobular inflammation, advanced fibrosis and Mallory bodies among IL6 rs1800795 C allele carriers agree with the concept that this allele may lead to less IL6 expression in the liver. Furthermore, several studies have shown that IL6-dependant activation of signal transducer and activator of transcription 3 (STAT3) reduces steatosis via inhibition of SREBP-1c mediated lipogenic gene activation as well as upregulation of fatty acid β oxidation genes [51–53]. Together these studies suggest that lower levels of IL6 could be associated with less inflammation and fibrosis but also more steatosis via depression of STAT3-mediated SREBP-1c inhibition. However, the pathogenic effects of IL6 induced STAT3 activation appears to be cell type specific and could vary depending on the degree and type of inflammatory infiltrate [51].
Recently, MR 16-1, an IL-6 receptor antibody, was effective in reducing hepatic steatosis in a mouse model of NAFLD [54]. Further, the utilization of IL-1 Receptor antagonist: IL-IRa (which binds to the IL-1 receptor and prevents IL-1 signal transduction); and the IL-1 receptor knockout mice showed a reduction in steatosis in a diet-induced model of NAFLD 41. A large study, CANTOS, is underway to determine if IL-1 neutralization is effective against cardiovascular diseases [55]. Favorable outcomes from such a study might pave the way for the use of IL-1 suppression in NASH [55].
In conclusion, we have made a novel observation that SNPs IL6 rs10499563 and IL1B rs1143634 may have a significant impact on the development of parenchymal cell damage, such as ballooning, Mallory bodies, bridging fibrosis and cirrhosis, in non-Hispanic Caucasian patients with NAFLD. The IL6 rs10499563 C allele, present in 38% of cohort, was independently associated with the presence of definitive NASH. The IL6 rs1800795 C allele could contribute to the development of steatosis but was also associated with a decreased risk of having increased Mallory bodies or an advanced disease grade or stage. Further studies are necessary to define the mechanisms for the effects of these polymorphisms in NAFLD and to determine the role of these SNPs in other liver diseases.
Supplementary Material
Acknowledgments
The Nonalcoholic Steatohepatitis Clinical Research Network (NASH CRN) is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (grants U01DK061718, U01DK061728, U01DK061731, U01DK061732, U01DK061734, U01DK061737, U01DK061738, U01DK061730, U01DK061713), and the National Institute of Child Health and Human Development (NICHD).
Several clinical centers use support from the National Center for Advancing Translational Sciences (NCATS) in conduct of NASH CRN Studies (grants UL1TR000439, UL1TR000077, UL1TR000436, UL1TR000150, UL1TR000424, UL1TR000006, UL1TR000448, UL1TR000040, UL1TR000100, UL1TR000004, UL1TR000423, UL1TR000058, UL1TR000067, UL1TR000454). This work was supported in part by the Intramural Research Program of the National Cancer Institute.
The authors would like to acknowledge the support of the BRI Genotyping Core facility and Dr. Karen Cerosaletti and Ami Charmley. The authors thank Dr. Jim Tonascia and Dr. Mark Van Atta for helpful discussions. This work was supported by NIH grants R01DK087696 and K24DK002957 to KVK.
Abbreviations
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- NAS
NAFLD activity score
- IL
interleukin
Members of the Nonalcoholic Steatohepatitis Clinical Research Network
Baylor College of Medicine, Houston, TX: Stephanie H. Abrams, MD, MS; Ryan Himes, MD; Rajesh Krisnamurthy, MD; Leanel Maldonado, RN (2007–2012); Beverly Morris
Case Western Reserve University Clinical Centers
-
•
MetroHealth Medical Center, Cleveland, OH: Patricia Brandt; Srinivasan Dasarathy, MD; Jaividhya Dasarathy, MD; Carol Hawkins, RN; Arthur J. McCullough, MD
-
•
Cleveland Clinic Foundation, Cleveland, OH: Srinivasan Dasarathy, MD; Arthur J. McCullough, MD; Mangesh Pagadala, MD; Rish Pai, MD; Ruth Sargent, LPN; Shetal Shah, MD; Claudia Zein, MD
Cincinnati Children’s Hospital Medical Center, Cincinnati, OH: Kimberlee Bernstein, BS, CCRP; Kim Cecil, PhD; Stephanie DeVore, MSPH (2009–2011); Rohit Kohli, MD; Kathleen Lake, MSW (2009–2012); Daniel Podberesky, MD; Crystal Slaughter, BA, CCRP; Stavra Xanthakos, MD
Columbia University, New York, NY: Gerald Behr, MD; Joel E. Lavine, MD, PhD; Ali Mencin, MD; Nadia Ovchinsky, MD; Elena Reynoso, MD
Duke University Medical Center, Durham, NC: Manal F. Abdelmalek, MD; Mustafa Bashir, MD; Stephanie Buie; Anna Mae Diehl, MD; Cynthia Guy, MD; Christopher Kigongo; Yi-Ping Pan; Dawn Piercy, FNP (2004–2012); Melissa Wagner
Emory University, Atlanta, GA: Adina Alazraki, MD; Rebecca Cleeton, MPH; Saul Karpen, MD, PhD; Nicholas Raviele; Miriam Vos, MD, MSPH
Indiana University School of Medicine, Indianapolis, IN: Elizabeth Byam, RN; Naga Chalasani, MD; Oscar W. Cummings, MD; Cynthia Fleming, RN, MSN; Marwan Ghabril, MD; Ann Klipsch, RN; Smitha Marri, MD; Jean P. Molleston, MD; Linda Ragozzino, RN; Kumar Sandrasegaran, MD; Girish Subbarao, MD; Raj Vuppalanchi, MD
Johns Hopkins Hospital, Baltimore, MD: Kimberly Pfeifer, RN; Ann Scheimann, MD; Michael Torbenson, MD
Mount Sinai Kravis Children’s Hospital, New York, NY: Ronen Arnon, MD; Mariel Boyd, CCRP
Northwestern University Feinberg School of Medicine/Ann & Robert H. Lurie Children’s Hospital of Chicago: Katie Amsden, Mark H. Fishbein, MD; Elizabeth Kirwan, RN; Saeed Mohammad, MD; Ann Quinn, RD (2010–2012); Cynthia Rigsby, MD; Peter F. Whitington, MD
Saint Louis University, St Louis, MO: Sarah Barlow, MD (2002–2007); Jose Derdoy, MD (2007–2012); Ajay Jain MD; Debra King, RN; Pat Osmack; Joan Siegner, RN; Susan Stewart, RN; Brent A. Neuschwander-Tetri, MD; Dana Romo
University of California San Diego, San Diego, CA: Brandon Ang; Sandra Arroyo; Cynthia Behling, MD, PhD; Archana Bhatt; Jennifer Collins; Iliana Doycheva, MD; Janis Durelle; Tarek Hassanein, MD (2004–2009); Joel E. Lavine, MD PhD (2002–2010); Rohit Loomba, MD, MHSc; Michael Middleton, MD, PhD; Kimberly Newton, MD; Phirum Nguyen; Mazen Noureddin, MD; Melissa Paiz; Heather Patton, MD; Jeffrey B. Schwimmer, MD; Claude Sirlin, MD; Patricia Ugalde-Nicalo
University of California San Francisco, San Francisco, CA: Bradley Aouizerat, PhD; Nathan M. Bass, MD, PhD (2002–2011); Danielle Brandman, MD; Linda D. Ferrell, MD; Shannon Fleck; Ryan Gill, MD, PhD; Bilal Hameed, MD; Alexander Ko; Camille Langlois; Emily Rothbaum Perito, MD; Aliya Qayyum, MD; Philip Rosenthal, MD; Norah Terrault, MD, MPH; Patrika Tsai, MD
University of California San Francisco- Fresno, Fresno, CA: PradeepAtla, MD; Cathy Hurtado; Rebekah Garcia; Sonia Garcia; Muhammad Sheikh, MD; Mandeep Singh, MD
University of Washington Medical Center and Seattle Children’s Hospital, Seattle, WA: Kara Cooper; Simon Horslen, MB ChB; Evelyn Hsu, MD; Karen Murray, MD; Randolph Otto, MD; Deana Rich; Matthew Yeh, MD, PhD; Melissa Young
Virginia Commonwealth University, Richmond, VA: Sherry Boyett, RN, BSN; Laura Carucci, MD; Melissa J. Contos, MD; Michael Fuchs, MD; Amy Jones; Kenneth Kraft, PhD; Velimir AC Luketic, MD; Kimberly Noble; Puneet Puri, MD; Bimalijit Sandhu, MD (2007–2009); Arun J. Sanyal, MD; Carol Sargeant, RN, BSN, MPH (2004–2012); Jolene Schlosser; Mohhamad S. Siddiqui, MD; Ben Wolford; Melanie White, RN, BSN (2006–2009)
Virginia Mason Medical Center, Seattle, WA: Sarah Ackermann; Shannon Cooney; David Coy, MD, PhD; Katie Gelinas; Maximillian Lee, MD, MPH; Tracey Pierce; Jody Mooney, MS; James E. Nelson, PhD; Lacey Siekas; Cheryl Shaw, MPH; Asma Siddique, MD; Chia Wang, MD
Swedish Medical Center, Seattle, WA: Kris V. Kowdley, MD, Priya Handa, PhD
Washington University, St. Louis, MO: Elizabeth M. Brunt, MD; Kathryn Fowler, MD
Resource Centers
National Cancer Institute, Bethesda, MD: David E. Kleiner, MD, PhD
National Institute of Child Health and Human Development, Bethesda, MD: Gilman D. Grave, MD
National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD: Edward C. Doo, MD; Jay H. Hoofnagle, MD; Patricia R. Robuck, PhD, MPH (2002–2011); Averell Sherker, MD
Johns Hopkins University, Bloomberg School of Public Health (Data Coordinating Center), Baltimore, MD: Patricia Belt, BS; Jeanne M. Clark, MD, MPH; Erin Corless, MHS; Michele Donithan, MHS; Milana Isaacson, BS; Kevin P. May, MS; Laura Miriel, BS; Alice Sternberg, ScM; James Tonascia, PhD; Aynur Ünalp-Arida, MD, PhD; Mark Van Natta, MHS; Ivana Vaughn, MPH; Laura Wilson, ScM; Katherine Yates, ScM
Footnotes
Disclosures: No conflicts of interest exist
Author contributions: study concept and design (JEN, KVK); acquisition of data (JEN, BA, LW, MMY); analysis and interpretation of data (JEN, BA, LW, MMY, KVK); drafting of the manuscript JEN, PH, BA, MMY); critical revision of the manuscript for important intellectual content (JEN, PH, BA, LW, LAV, KVK); statistical analysis; (JEN, BA, LW); obtained funding (JEN, BA, KVK).
References
- 1.Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 2.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34(3):274–85. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masarone M, Federico A, Abenavoli L, et al. Non alcoholic fatty liver: epidemiology and natural history. Rev Recent Clin Trials. 2014;9:126–133. doi: 10.2174/1574887109666141216111143. [DOI] [PubMed] [Google Scholar]
- 5.European Association for the Study of the Liver (EASL) Electronic address: easloffice@easloffice.eu; European Association for the Study of Diabetes (EASD); European Association for the Study of Obesity (EASO). EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64(6):1388–402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Am J Gastroenterol. 2012;107(6):811–26. doi: 10.1038/ajg.2012.128. [DOI] [PubMed] [Google Scholar]
- 7.Loria P, Lonardo A, Carulli L, et al. Review article: the metabolic syndrome and non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2005;22(Suppl 2):31–6. doi: 10.1111/j.1365-2036.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 8.Speliotes EK, Butler JL, Palmer CD, et al. GIANT Consortium, MIGen Consortium, NASH CRN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 10.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorden A, Yang R, Yerges-Armstrong LM, et al. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75:34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernaez R, McLean J, Lazo M, et al. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013;11:1183–1190. doi: 10.1016/j.cgh.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen KF, Dufour S, Hariri A, et al. Apoliporpotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362:1082–1089. doi: 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dongiovanni P, Valenti L. Peroxisome proliferator-activated receptor genetic polymorphisms and nonalcoholic Fatty liver disease: any role in disease susceptibility? PPAR Res. 2013;2013:452061. doi: 10.1155/2013/452061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan HL, Zain SM, Mohamed R, et al. Association of glucokinase regulatory gene polymorphisms with risk and severity of non-alcoholic fatty liver disease: an interaction study with adiponutrin gene. J Gastroenterol. 2014;49:1056–1064. doi: 10.1007/s00535-013-0850-x. [DOI] [PubMed] [Google Scholar]
- 16.Chambers JC, Zhang W, Sehmi J, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan X, Waterworth D, Perry JR, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalasani N, Guo X, Loomba R, et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology. 2010;139:1567–1576. doi: 10.1053/j.gastro.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen QR, Braun R, Hu Y, et al. Multi-SNP Analysis of GWAS Data Identifies Pathways Associated with Nonalcoholic Fatty Liver Disease. PLoS One. 2013;8:e65982. doi: 10.1371/journal.pone.0065982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tilg H, Moschen AR. Insulin resistance, inflammation, and non-alcoholic fatty liver disease. Trends Endocrinol Metab. 2008;19:371–379. doi: 10.1016/j.tem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Dinarello CA, Donath MY, Mandrup-Poulsen T. Role of IL-1beta in type 2 diabetes. Curr Opin Endocrinol Diabetes Obes. 2010;17:314–321. doi: 10.1097/MED.0b013e32833bf6dc. [DOI] [PubMed] [Google Scholar]
- 23.Miura K, Kodama Y, Inokuchi S, et al. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology. 2010;139:323–334. doi: 10.1053/j.gastro.2010.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woods A, Brull DJ, Humphries SE, et al. Genetics of inflammation and risk of coronary artery disease: the central role of interleukin-6. European Heart Journal. 2000;21:1574–1583. doi: 10.1053/euhj.1999.2207. [DOI] [PubMed] [Google Scholar]
- 25.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 26.Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 102:1369–1376. doi: 10.1172/JCI2629. 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens JW, Hurel SJ, Lowe GD, et al. Association between plasma IL-6, the IL6 -174G>C gene variant and the metabolic syndrome in type 2 diabetes mellitus. Mol Genet Metab. 2007;90:422–428. doi: 10.1016/j.ymgme.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Smith AJ, D’Aiuto F, Palmen J, et al. Association of serum interleukin-6 concentration with a functional IL6 -6331T>C polymorphism. Clin Chem. 2008;54:841–850. doi: 10.1373/clinchem.2007.098608. [DOI] [PubMed] [Google Scholar]
- 29.Pociot F, Mølvig J, Wogensen L, et al. TaqI polymorphism in the human interleukin-1beta (IL-1 beta) gene correlates with IL-1 beta secretion in vitro. Eur J Clin Invest. 1992;22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- 30.Licastro F, Pedrini S, Ferri C, et al. Gene polymorphism affecting alpha1-antichymotrypsin and interleukin-1 plasma levels increases Alzheimer’s disease risk. Ann Neurol. 2000;48:388–391. [PubMed] [Google Scholar]
- 31.Huth C, Heid IM, Vollmert C, et al. IL6 gene promoter polymorphisms and type 2 diabetes: joint analysis of individual participants’ data from 21 studies. Diabetes. 2006;55:2915–2921. doi: 10.2337/db06-0600. [DOI] [PubMed] [Google Scholar]
- 32.Luotola K, Pääkkönen R, Alanne M, et al. Association of variation in the interleukin-1 gene family with diabetes and glucose homeostasis. J Clin Endocrinol Metab. 2009;94:4575–4583. doi: 10.1210/jc.2009-0666. [DOI] [PubMed] [Google Scholar]
- 33.Strandberg L, Mellström D, Ljunggren O, et al. IL6 and IL1B polymorphisms are associated with fat mass in older men: the MrOS Study Sweden. Obesity (Silver Spring) 2008;16:710–713. doi: 10.1038/oby.2007.95. [DOI] [PubMed] [Google Scholar]
- 34.Strandberg L, Lorentzon M, Hellqvist A, et al. Interleukin-1 system gene polymorphisms are associated with fat mass in young men. J Clin Endocrinol Metab. 2006;91:2749–2754. doi: 10.1210/jc.2005-2786. [DOI] [PubMed] [Google Scholar]
- 35.Carulli L, Canedi I, Rondinella S, et al. Genetic polymorphisms in non-alcoholic fatty liver disease: interleukin-6-174G/C polymorphism is associated with non-alcoholic steatohepatitis. Dig Liver Dis. 2009;41:823–828. doi: 10.1016/j.dld.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Nozaki Y, Saibara T, Nemoto Y, et al. Polymorphisms of interleukin-1 beta and beta 3-adrenergic receptor in Japanese patients with nonalcoholic steatohepatitis. Alcohol Clin Exp Res. 2004;28:106S–110S. doi: 10.1111/j.1530-0277.2004.tb03226.x. [DOI] [PubMed] [Google Scholar]
- 37.Dawson DA, Grant BF, Stinson FS, et al. Effectiveness of the derived Alcohol Use Disorders Identification Test (AUDIT-C) in screening for alcohol use disorders and risk drinking in the US general population. Alcohol Clin Exp Res. 2005;29:844–854. doi: 10.1097/01.alc.0000164374.32229.a2. [DOI] [PubMed] [Google Scholar]
- 38.Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 39.Ajmera V, Perito ER, Bass NM, et al. Novel Plasma Biomarkers Associated with Liver Disease Severity in Adults with Nonalcoholic Fatty Liver Disease. Hepatology. 2016 doi: 10.1002/hep.28776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vonghia L, Magrone T, Verrijken A, et al. Peripheral and Hepatic Vein Cytokine Levels in Correlation with Non-Alcoholic Fatty Liver Disease (NAFLD)-Related Metabolic, Histological, and Haemodynamic Features. PLoS One. 2015;10(11):e0143380. doi: 10.1371/journal.pone.0143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jarrar MH, Baranova A, Collantes R, et al. Adipokines and cytokines in non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2008;27(5):412–21. doi: 10.1111/j.1365-2036.2007.03586.x. [DOI] [PubMed] [Google Scholar]
- 42.Abd El-Kader SM, El-Den Ashmawy EM. Non-alcoholic fatty liver disease: The diagnosis and management. World J Hepatol. 2015;7(6):846–58. doi: 10.4254/wjh.v7.i6.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stojsavljević S, Gomerčić Palčić M, Virović Jukić L, et al. Adipokines and proinflammatory cytokines, the key mediators in the pathogenesis of nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20(48):18070–91. doi: 10.3748/wjg.v20.i48.18070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan X, Xu C, Yu C, et al. Role of NLRP3 Inflammasome in the Progression of NAFLD to NASH. Can J Gastroenterol Hepatol. 2016;2016:6489012. doi: 10.1155/2016/6489012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maedler K, Dharmadhikari G, Schumann DM, et al. Interleukin-targeted therapy for metabolic syndrome and type 2 diabetes. Handb Exp Pharmacol. 2011;(203):257–78. doi: 10.1007/978-3-642-17214-4_11. [DOI] [PubMed] [Google Scholar]
- 46.de Mello VD, Kolehmainen M, Schwab U, et al. Effect of weight loss on cytokine messenger RNA expression in peripheral blood mononuclear cells of obese subjects with the metabolic syndrome. Metabolism. 2008;57(2):192–9. doi: 10.1016/j.metabol.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 47.Odegaard JI, Chawla A. Connecting type 1 and type 2 diabetes through innate immunity. Cold Spring Harb Perspect Med. 2012;2(3):a007724. doi: 10.1101/cshperspect.a007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter KW, Hung J, Powell BL, et al. Association of Interleukin-1 gene polymorphisms with central obesity and metabolic syndrome in a coronary heart disease population. Hum Genet. 2008;124:199–206. doi: 10.1007/s00439-008-0540-6. [DOI] [PubMed] [Google Scholar]
- 49.Giannitrapani L1, Soresi M, Balasus D, et al. Genetic association of interleukin-6 polymorphism (-174 G/C) with chronic liver diseases and hepatocellular carcinoma. World J Gastroenterol. 2013;19:2449–2455. doi: 10.3748/wjg.v19.i16.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wieckowska A, Papouchado BG, Li Z, et al. Increased hepatic and circulating interleukin-6 levels in human nonalcoholic steatohepatitis. Am J Gastroenterol. 2008;103(6):1372–1379. doi: 10.1111/j.1572-0241.2007.01774.x. [DOI] [PubMed] [Google Scholar]
- 51.Horiguchi N, Wang L, Mukhopadhyay P, et al. Cell type-dependent pro- and anti-inflammatory role of signal transducer and activator of transcription 3 in alcoholic liver injury. Gastroenterology. 2008;134:1148–1158. doi: 10.1053/j.gastro.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue H, Ogawa W, Ozaki M, et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 53.Kroy DC, Beraza N, Tschaharganeh DF, et al. Lack of interleukin-6/glycoprotein 130/signal transducers and activators of transcription-3 signaling in hepatocytes predisposes to liver steatosis and injury in mice. Hepatology. 2010;51:463–473. doi: 10.1002/hep.23322. [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi K, Nishimura T, Ishiba H, et al. Blockade of interleukin 6 signalling ameliorates systemic insulin resistance through upregulation of glucose uptake in skeletal muscle and improves hepatic steatosis in high-fat diet fed mice. Liver Int. 2015;35:550–61. doi: 10.1111/liv.12645. [DOI] [PubMed] [Google Scholar]
- 55.Tilg H, Moschen AR, Szabo G. Interleukin-1 and inflammasomes in ALD/AAH and NAFLD/NASH. Hepatology. 2016 doi: 10.1002/hep.28456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.