Abstract
Giardiasis is a prevailing intestinal disease in children. This study aimed to determine molecular prevalence of Giardia intestinalis in children attending Cairo University Pediatrics Hospitals, using copro-PCR assays, conventional methods and to evaluate diagnostic effectiveness of used tests. 229 fecal samples were collected from children suffering from gastrointestinal symptoms and examined for Giardia by microscopy, Immuno-chromatographic test (ICT), copro-DNA using two PCR assays targeting tpi [nested-PCR (nPCR)] and 18S [conventional-PCR (cPCR)] genes. Out of 229 samples assessed, Giardia was diagnosed in 13.9, 17, 17.9, 4.8 % of cases using microscopy, ICT, nPCR (tpi) and cPCR (18S), respectively. Nominating both PCR assays as composite reference standard, microscopy and ICT were of reliable specificity (100 and 96.9 %) and accuracy (95.6 and 93.6 %) but of limited sensitivity (78.6 and 76.2 %). Kappa agreement showed, there was substantial agreement of ICT (0.776) and almost perfect agreement of microscopy (0.839) with PCR assays. Giardia showed a molecular prevalence of 18.3 % (42/229). ICT assay for Giardia surpassed microscopy but both couldn’t be used as a consistent single detection method due to their lowered sensitivities. nPCR targeting tpi is a reliable diagnostic test aiding to determine true prevalence of Giardia.
Keywords: Giardia intestinalis, Molecular prevalence, Copro-DNA, PCR
Introduction
Giardia intestinalis is one of the commonest causes of gastrointestinal infections in children. It is the most important drinking water contaminant and re-emerging as cause of food-borne disease (Farthing et al. 1986; Levy et al. 1998; Thompson et al. 2000; Al-Saeed and Issa 2010). Laboratory diagnosis of Giardia is usually done by microscopy on fresh and/or concentrated faecal samples. However, this method has many limitations beside its poor sensitivity (Mank et al. 1997; Ignatius et al. 2014). Alternatively copro-antigen detection assays have been accepted as diagnostic methods for G. intestinalis. They are easy to perform and less time-consuming but still miss a number of infection (Helmy et al. 2014). In order to improve sensitivity and estimate the true prevalence of the disease, molecular methods based on polymerase chain reaction (PCR) have been developed to characterize Giardia in stool even (Amar et al. 2002). Giardiasis is very common in developing countries (Haque 2007). The regional prevalence of the disease in African and Eastern Mediterranean area differs and may be over 30 % in children (Thompson and Smith 2011). The actual prevalence of Giardia infection may be underestimated due to the use of microscopic examination of stool in epidemiological studies of Giardia (Verweij et al. 2004). Therefore, in order to determine the true prevalence of Giardia among Egyptian children, we used two PCR assays as a molecular method in addition to conventional diagnostic procedures (microscopy and immunoassay).
Materials and methods
Study design and populations This is a cross sectional study included 229 children attending Cairo University Pediatrics Hospitals (CUPH) and suffering from GIT symptoms. Consent guidelines All patients included were informed verbally about the purpose of the study and collection of samples was performed after obtaining their parents’ consent. Collection and processing of samples Single stool sample at least was collected from all patients and divided into three parts, one for coproscopic examination by direct wet mount before and after Formalin-ethyl acetate concentration using saline and Lugol’s iodine to detect Giardia lamblia and other parasites using 10×, 40× objectives and the other parts were freshly frozen at −20 °C. Copro-antigen detection of Giardia part of frozen samples was subjected to in vitro detection of copro-antigens of Cryptosporidium/Giardia/Entamoeba Combi (N1722) using RIDA® QUICK Immuno-chromatographic test (ICT) Test (R-Biopharm AG, Germany) according to manufacturer’s instructions. Copro-PCR assay Genomic DNA extraction was done using Favor Prep stool DNA isolation Kit (Favorgen Biotech corporation ping-Tung 908, Taiwan, Cat. No. FASTI001), extraction was performed according to the manufacturer’s instruction with modifications in the form of thermal treatment of samples by using liquid nitrogen for 5 min then water bath (95 °C for 5 min) (repeated for 10 cycles). PCR amplification of ~480 bp DNA fragment of the 18S gene was amplified using the forward primer G 18S2: 5′-TCCGGTYGATTCTGCC-3′ and the reverse primer G 18S3: 5′-CTGGAATTACCGCGGCTGCT-3′ (Monis et al. 1999; Amjad et al. 2009). The reaction mixture consisted of 1 μl of each primer (200 nM), 5 μl of template DNA, 12.5 μl of Dream Taq Green PCR Master Mix (Product No. K1081, Thermo Scientific, USA) and molecular grade water to a total volume of 25 µl. Nested PCR was done using two sets of primers targeting tpi gene: AL3543: 5′-AAATIATGCCTGCTCGTCG-3′ and the reverse primer AL3546: 5′-CAAACCTTITCCGCAAACC-3′ for the primary reaction to amplify ~605 bp DNA and a fragment of ~530 bp for the secondary reaction using AL3544: 5′-CCCTTCATCGGIGGTAACTT-3′ and the reverse primer AL3545: 5′-GTGGCCACCACICCCGTGCC-3′ (Sulaiman et al. 2003). The amplified products were visualized with 1.5 % agarose gel electrophoresis after ethidium bromide staining.
Statistical analysis
Statistical analysis was done using the statistical package SPSS version 17 (Chicago, IL, USA). Sensitivity, specificity, PPV, NPV, accuracy were calculated to test the diagnostic yield and kappa agreement was done to test validity of microscopy and ICT in relation to PCR results considering it a nominated gold standard. According to composite reference standard (CRS) of Alonzo and Pepe (1999) that suppose the present gold standard test (microscopy) is highly specific but not very sensitive was used to define true positive cases. The present study considered tpi nested-PCR (nPCR) as a reference standard and 18S cPCR as a resolver. The CRS being both highly specific and quite sensitive that defines a subject as positive if either of the tests is positive and as negative only if both tests are negative.
Results
Out of the 229 samples screened for G. intestinalis, PCR targeting 18S and tpi genes was able to detect Giardia copro-DNA in 42 (18.3 %) samples, followed by ICT assay (39/229) and microscopy (32/229) (Tables 1, 2; Fig. 1). Considering PCR as a nominated CRS, ICT assay showed superior sensitivity followed by microscopy (Table 3). Of the 197 Giardia-negative samples detected by microscopy, PCR products of the expected size were generated in 9 and 1 samples, using primers for tpi and 18S genes respectively. Of the 190 Giardia-negative samples detected by ICT, PCR products of the expected size for tpi and 18S fragments were observed in 8 and 2 samples, respectively. Concerning the diagnostic yield, microscopy was the most efficient test with the highest accuracy (95.6 %), specificity (100 %) and Kappa agreement showed almost perfect agreement (0.839) with copro-PCR findings. While, ICT assay showed less accuracy (93.6) and specificity (96.9) than microscopy and Kappa agreement showed a substantial agreement (0.776) with copro-PCR findings (Table 3).
Table 1.
Results of microscopy, ICT and amplification of tpi and 18s Giardia genes from 229 stool samples
| Test | Copro PCR (n = 42) | |||||
|---|---|---|---|---|---|---|
| tpi | 18s | |||||
| +ve | −ve | Total | +ve | −ve | Total | |
| Microscopy 32 | ||||||
| Positive | 32 (13.9 %) | 0 (0 %) | 32 (13.9 %) | 10 (4.4 %) | 22 (9.6 %) | 32 (13.9 %) |
| Negative | 9 (3.9 %) | 188 (82 %) | 197 (86 %) | 1 (0.4 %) | 196 (85.6 %) | 197 (86 %) |
| Total | 41 (17.9 %) | 188 (82 %) | 229 (100 %) | 11 (4.8 %) | 218 (95.2 %) | 229 (100 %) |
| ICT 39 | ||||||
| Positive | 33 (14.4 %) | 6 (2.6 %) | 39 (17 %) | 9 (3.9 %) | 30 (13.1 %) | 39 (17 %) |
| Negative | 8 (3.4 %) | 182 (79.4 %) | 190 (83 %) | 2 (0.9 %) | 188 (82 %) | 190 (83 %) |
| Total | 41 (17.9 %) | 188 (82 %) | 229 (100 %) | 11 (4.8 %) | 218 (95.2 %) | 229 (100 %) |
Table 2.
Results of tpi and 18S amplification
| Giardia copro-PCR (tpi/18S) | |||||
|---|---|---|---|---|---|
| Positive | Negative by both | Total | |||
| Both TPI and 18S | tpi only | 18S only | Total | ||
| 10 | 31 | 1 | 42 | 187 | 229 |
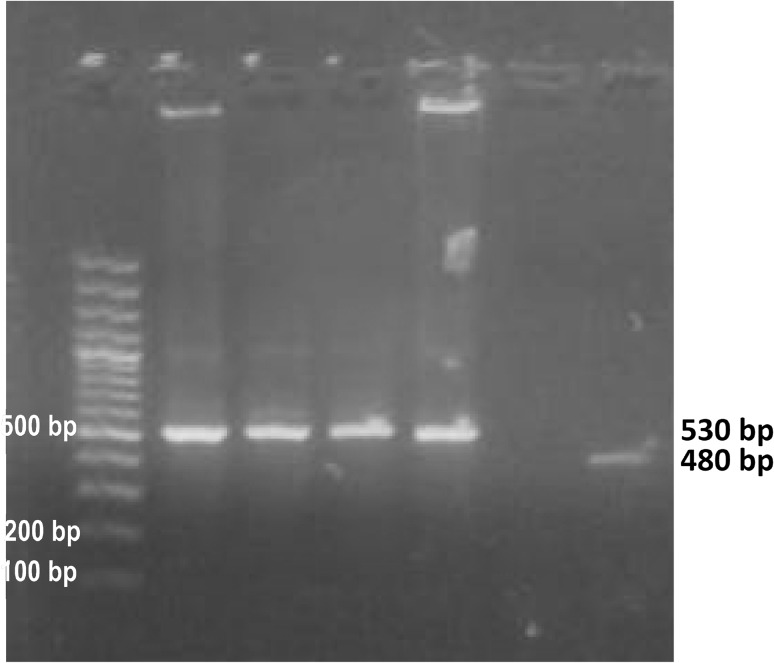
Fig. 1.
Agarose gel stained with ethidium bromide corresponding to PCR, Line 1 molecular weight marker (100 bp), lines 2–5 DNA for the products of the nPCR targeting tpi gene (530 bp), line 6 negative sample, line 7 the product of the PCR targeting 18S gene (480 bp)
Table 3.
Diagnostic effectiveness of microscopy and copro-Ag detection by ICT
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | Kappaa | |
|---|---|---|---|---|---|---|
| Microscopy | 76.2 | 100 | 100 | 94.9 | 95.6 | 0.839 |
| ICT | 78.6 | 96.9 | 84.6 | 95.4 | 93.6 | 0.776 |
| Kappa agreement interpretationa | |
|---|---|
| <0 | Poor agreement |
| 0.01–0.20 | Slight agreement |
| 0.21–0.40 | Fair agreement |
| 0.41–0.60 | Moderate agreement |
| 0.61–0.80 | Substantial agreement |
| 0.81–1.00 | Almost perfect agreement |
Discussion
Giardia is a significant worldwide cause of diarrhea and nutritional disorders in children. In our study, the molecular prevalence of Giardia among the studied group was 18.3 % (42/229), which was lower than Foronda et al. (2008), reported a prevalence of 34.6 % in Egypt and Helmy et al. (2009) who reported the amplification of the tpi gene from 42.3 % of patients all of them complaining of diarrhea. Differences in prevalence of Giardia in Egypt may be related to different geographical distribution among different Governorates as found by Fawzi et al. (2004) who detected Giardia in 24.7 % of fecal samples in Behera Governorate while a rate of 10.4 % in El-Prince (Alexandria), both in Egypt. In addition, Sadek et al. (2013) reported a Prevalence of 30 % in Menoufiya Governorate and 28.4 % in Sharkiya Governorate. Low prevalence rates may be attributed to the fact that the examined patients might have better living conditions in some Governorates (Sadek et al. 2013). Much lower prevalence rates 0.7 % (Norhayati et al. 2003), 2.0 % (Natividad et al. 2008) were reported in Malaysia and Philippines, respectively.
In this study, ICT showed higher sensitivity (78.6 %) than microscopy (76.2 %). Similar results were reported by Goñi et al. (2012) and Elsafi et al. (2013) in a study at Saudi Arabia by comparing microscopy, immunoassay (ImmunoCard STAT) and real-time (PCR) detecting the 18S rRNA gene of G. lamblia, they reported lower sensitivity of microscopy and Ignatius et al. (2014) showed higher sensitivity of immunoassays. ICT provide adequate sensitivities and specificities in outbreak situations, screening proposals (Goñi et al. 2012) especially when a single stool sample is used (Helmy et al. 2014). However it still misses number of the Giardia infections detected by PCR in hyperendemic area (Ignatius et al. 2014). in the present study, out of the 229 DNA amplified samples, since not all samples were amplified by both genes amplification of tpi and 18S gene fragments were observed respectively in 41 (17.9 %) and 11 (4.8 %) samples. The good performance of tpi gene in this work was coincide with Bertrand et al. (2005), Nahavandi et al. (2011), Zheng et al. (2014). The tpi gene is better adapted for efficient discrimination between the two major assemblages of Giardia. Thus, detection methods targeting loci with high degree of polymorphism such as tpi can be extremely useful (Bertrand et al. 2005). Some studies showed differences in the performance of commonly used genes (tpi, gdh and 18S rRNA) in DNA amplification of Giardia (Gelanew et al. 2007; Volotão et al. 2007). The cause of this difference is not yet known. However Lalle et al. (2009) mentioned that, despite the gene primers are designed to bind “conserved” regions in genes, mismatches in primer sequences could be too long to allow successful PCR amplification of some fecal isolates. Sensitivity of molecular detection of Giardia in stool was proved to be superior to microscopic examination in detecting low number of cysts (Amar et al. 2002).
Acknowledgments
We are grateful to the Scientific Research Developing Unit, Bani-suef University for granting and funding the study.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no reported conflicts of interest.
Author contribution
All manuscript authors contributed to every activity of it; idea of paper, study design, collection of materials, methodology, writing the paper and revising it.
References
- Alonzo TA, Pepe MS. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat Med. 1999;18:2987–3003. doi: 10.1002/(SICI)1097-0258(19991130)18:22<2987::AID-SIM205>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Al-Saeed AT, Issa SH. Detection of Giardia lamblia antigen in stool specimens using enzyme-linked immunosorbent assay. East Med Health J. 2010;16(4):362–364. [PubMed] [Google Scholar]
- Amar CF, Dear PH, Pedaza-Diaz S, Looker N, Linnane E, McLauchlin J. Sensitive PCR-RFLP assay for detection and genotyping of Giardia duodenalis in human feces. Clin Microbiol J. 2002;40(2):446–452. doi: 10.1128/JCM.40.2.446-452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amjad IA, Hussein AB, Yamaguchi T, Nakamoto K, Motohiro Iseki M, Tokoro M. Multiple-subgenotype infections of Giardia intestinalis detected in Palestinian clinical cases using a subcloning approach. Parasitol Internat. 2009;58:258–262. doi: 10.1016/j.parint.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Bertrand I, Albertini L, Schwartzbord A. Comparison of two target genes for detection and genotyping of Giardia lamblia in human feces by PCR and PCR-restriction fragment length polymorphism. J Clin Microbiol. 2005;43(12):5940–5944. doi: 10.1128/JCM.43.12.5940-5944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsafi SH, Al-Maqati TN, Hussein MI, Adam AA, Hassan MM, Al Zahrani EM. Comparison of microscopy, rapid immunoassay, and molecular techniques for the detection of Giardia lamblia and Cryptosporidium parvum. Parasitol Res. 2013;112(4):1641–1646. doi: 10.1007/s00436-013-3319-1. [DOI] [PubMed] [Google Scholar]
- Farthing MJ, Mata L, Urrutia JJ, Kronmal RA. Natural history of Giardia infection of infants and children in rural Guatemala and its impact on physical growth. Am J Clin Nutr. 1986;43(3):395–405. doi: 10.1093/ajcn/43.3.395. [DOI] [PubMed] [Google Scholar]
- Fawzi M, El-Sahn AA, Ibrahim HF, Shehata AI. Vegetable transmitted parasites among inhabitants of El-Prince, Alexandria and its relation to housewives’ knowledge and practices. J Egypt Public Health Assoc. 2004;79:13–29. [PubMed] [Google Scholar]
- Foronda PM, Argues N, Abreu-Acosta MV, Periago MA, Valero BV, Mas-coma S. Identification of genotypes of Giardia intestinalis of human isolates in Egypt. Parasitol Res. 2008;103:1177–1181. doi: 10.1007/s00436-008-1113-2. [DOI] [PubMed] [Google Scholar]
- Gelanew T, Lalle M, Hailu A, Pozio E, Caccio SM. Molecular characterization of human isolates of Giardia duodenalis from Ethiopia. Acta Trop. 2007;102:92–99. doi: 10.1016/j.actatropica.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Goñi P, Martín B, Villacampa M, García A, Seral C, Castillo FJ, Clavel A. Evaluation of an immunochromatographic dip strip test for simultaneous detection of Cryptosporidium spp, Giardia duodenalis, and Entamoeba histolytica antigens in human faecal samples. Eur J Clin Microbiol Infect Dis. 2012;31(8):2077–2082. doi: 10.1007/s10096-012-1544-7. [DOI] [PubMed] [Google Scholar]
- Haque R. Human intestinal parasites. J Health Popul Nutr. 2007;25:387–391. [PMC free article] [PubMed] [Google Scholar]
- Helmy MM, Abdel-Fattah HS, Rashed L. Realtime PCR-RFLP assay to detect Giardia intestinalis genotypes in human isolates with diarrhea in Egypt. J Parasitol. 2009;95(4):1–5. doi: 10.1645/GE-1670.1. [DOI] [PubMed] [Google Scholar]
- Helmy YA, Krücken J, Nöckler K, Samson-Himmelstjerna GV, Zessin KH. Comparison between two commercially available serological tests and polymerase chain reaction in the diagnosis of Cryptosporidium in animals and diarrhoeic children. Parasitol Res. 2014;113(1):211–216. doi: 10.1007/s00436-013-3645-3. [DOI] [PubMed] [Google Scholar]
- Ignatius R, Gahutu JB, Klotz C, Musemakweri A, Aebischer T, Mockenhaupt FP (2014) Detection of Giardia duodenalis assemblage A and B isolates by immunochromatography in stool samples from Rwandan children. Clin Microbiol Infect 1111(10)/1469-0691 [DOI] [PubMed]
- Lalle M, Bruschi F, Castagna B, Campa M, Pozio E, Cacciò SM. High genetic polymorphism among Giardia duodenalis isolates from Sahrawi children. Trans R Soc Trop Med Hyg. 2009;103(8):834–838. doi: 10.1016/j.trstmh.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Levy DA, Bens MS, Craun GF, Calderon RL, Herwaldt BL. Surveillance for waterborne-disease outbreaks United States, 1995–1996. CDC Surveill Summ MMWR. 1998;47(5):1–34. [PubMed] [Google Scholar]
- Mank TG, Zaat JM, Deelder AM, Eijk JThMV, Polderman AM. Sensitivity of microscopy versus enzyme immunoassay in the laboratory diagnosis of Giardiasis. Eur J Clin Microbiol Infect. 1997;16:615–619. doi: 10.1007/BF02447929. [DOI] [PubMed] [Google Scholar]
- Monis PT, Andrews RH, Mayrhofer G, Ey PL. Molecular systematic of the parasitic protozoan Giardia intestinalis. Mol Biol Evol. 1999;16:1135–1144. doi: 10.1093/oxfordjournals.molbev.a026204. [DOI] [PubMed] [Google Scholar]
- Nahavandi KH, Fallah E, Asgharzadeh M, Mirsamadi N, Mahdavipour B. Glutamate dehydrogenase and triose-phosphate-isomerase coding genes for detection and genetic characterization of Giardia lamblia in human feces by PCR and PCR-RFLP. Turk J Med Sci. 2011;41(2):283–289. [Google Scholar]
- Natividad FF, Buerano CC, Lago CB, Mapua CA, de Guzman BB, Seraspe EB, Samentar LP, Endo T. Prevalence rates of Giardia and Cryptosporidium among diarrheic patients in the Philippines. Southeast Asian J Trop. 2008;39(6):991–999. [PubMed] [Google Scholar]
- Norhayati M, Fatmah MS, Yusof S. Intestinal parasitic infections in man. A review. Med J Malays. 2003;58:2–10. [PubMed] [Google Scholar]
- Sadek GS, El-Settawy MA, Soha A, Nasr SA. Genotypic characterization of Giardia duodenalis in children in Menoufiya and Sharkiya governorates. Egypt Life Sci J. 2013;10(1):4006–4015. [Google Scholar]
- Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9(11):1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RC, Smith A. Zoonotic enteric protozoa. Vet Parasitol. 2011;182:70–78. doi: 10.1016/j.vetpar.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Thompson RC, Hopkins RM, Homan WL. Nomenclature and genetic groupings of Giardia infecting mammals. Parasitol Today. 2000;16:210–213. doi: 10.1016/S0169-4758(99)01624-5. [DOI] [PubMed] [Google Scholar]
- Verweij JJ, Blangé RA, Templeton K, Schinkel J, Brienen EA, Van Rooyen MA. Simultaneous detection of Entamoeba histolytica Giardia lamblia and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42(3):1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volotão AC, Costa-Macedo LM, Haddad FS, Brandão A, Peralta JM, Fernades O. Genotyping of Giardia duodenalis from human and animal samples from Brazil using β-giardin gene a phylogenetic analysis. Acta Trop. 2007;102(1):258–262. doi: 10.1016/j.actatropica.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Zheng G, Alsarakibi M, Liu Y, Hu W, Luo Q, Tan L, Li G. Genotyping of Giardia duodenalis isolates from dogs in Guangdong, China based on multi-locus sequence. Korean J Parasitol. 2014;52(3):299–304. doi: 10.3347/kjp.2014.52.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]