Abstract
The free-living legume symbiont Rhizobium leguminosarum is of significant economic value because of its ability to provide fixed nitrogen to globally important leguminous food crops, such as peas and lentils. Discovery based research into the genetics and physiology of R. leguminosarum provides the foundational knowledge necessary for understanding the bacterium's complex lifestyle, necessary for augmenting its use in an agricultural setting. Transposon insertion sequencing (INSeq) facilitates high-throughput forward genetic screening at a genomic scale to identify individual genes required for growth in a specific environment. In this study we applied INSeq to screen the genome of R. leguminosarum bv. viciae strain 3841 (RLV3841) for genes required for growth on minimal mannitol containing medium. Results from this study were contrasted with a prior INSeq experiment screened on peptide rich media to identify a common set of functional genes necessary for basic physiology. Contrasting the two growth conditions indicated that approximately 10% of the chromosome was required for growth, under both growth conditions. Specific genes that were essential to singular growth conditions were also identified. Data from INSeq screening on mannitol as a sole carbon source were used to reconstruct a metabolic map summarizing growth impaired phenotypes observed in the Embden-Meyerhof-Parnas pathway, Entner-Doudoroff pathway, pentose phosphate pathway, and tricarboxylic acid cycle. This revealed the presence of mannitol dependent and independent metabolic pathways required for growth, along with identifying metabolic steps with isozymes or possible carbon flux by-passes. Additionally, genes were identified on plasmids pRL11 and pRL12 that are likely to encode functional activities important to the central physiology of RLV3841.
Keywords: Rhizobium, INSeq, Tn-Seq, core functional genome, metabolism, mannitol
Introduction
Rhizobium leguminosarum is a Gram-negative soil and rhizosphere colonizing bacterium that is also capable of engaging in endosymbiosis with specific leguminous plant genera. The host specificity of rhizobial infection is dependent upon the exchange of specific chemical signals between the infecting bacterium and host plant (Oldroyd et al., 2011), and R. leguminosarum is often divided into biovars based on infectious host range. The biovar viciae is indicative of Rhizobia capable of infecting leguminous plants such as peas (Pisum sativum), lentils (Lens culinaris), and vetch (Vicia spp.). When in the endosymbiotic state, Rhizobium bacteroids reduce atmospheric nitrogen N2 to ammonia, which is then exported to the plant for assimilation. In return, the plant host provides fixed carbon and other micro-nutrients to the bacteroids to sustain the symbiosis (Wielbo, 2012; Udvardi and Poole, 2013). The availability of symbiotically supplied nitrogen enables leguminous plants to satisfy their high nitrogen demands and, in part, contributed to the evolutionary success and diversification of the Leguminosae (Doyle and Luckow, 2003).
R. leguminosarum bv. viciae 3841(RLV3841) (Johnston and Beringer, 1975), has long been considered a model organism for Rhizobium research and was one of the first rhizobial strains with a published complete genome sequence (Young et al., 2006). Aside from the overarching agricultural context of studying RLV3841, the model organism provides other interesting avenues of research because of its complex genomic structure (Young et al., 2006) and versatility of physiology and lifestyle. RLV3841 has a relatively large bacterial genome comprised of a single 5.1 Mb chromosome and 6 large, stably maintained, plasmids ranging in size from 0.87 to 0.15 Mb. The RLV3841 genome is predicted to contain approximately 7346 genes, a substantial percentage of these genes (25.2%) are annotated as hypothetical genes of unknown function, warranting further investigation. The use of high-throughput experimental approaches may allow prioritization of the study of individual genes within this large functionally unknown group.
The development of next-generation sequencing technologies has resulted in high-throughput methods of transposon (Tn) mutagenesis to study gene function at a genome scale (Gawronski et al., 2009; Goodman et al., 2009; Langridge et al., 2009; van Opijnen et al., 2009). For example, INSeq was developed through the introduction of a type II restriction enzyme site within the IS element of the himar1C9 mariner Tn allowing for the specific capture and PCR amplification of genomic DNA adjacent to the Tn insertion site (Goodman et al., 2011). Next generation sequencing of PCR amplicons derived from DNA isolated from these Tn-mutant libraries allows for the sequencing of millions of Tn insertion tags which can be mapped to the genome sequence and used to enumerate the relative abundance of individual Tn mutants within a mutant population (Barquist et al., 2013; van Opijnen and Camilli, 2013). INSeq and similar high-throughput Tn mutagenesis methods have been used to study the genetic basis of bacterial physiology (Griffin et al., 2011; Brutinel and Gralnick, 2012; Kuehl et al., 2014; Yang et al., 2014; Le Breton et al., 2015; Lee et al., 2015; Meeske et al., 2015; Pechter et al., 2015; Rubin et al., 2015; Hooven et al., 2016; Troy et al., 2016), bacterial resistance to biotic and abiotic factors (Gallagher et al., 2011; Khatiwara et al., 2012; Phan et al., 2013; Byrne et al., 2014; Murray et al., 2015; Shan et al., 2015; Yung et al., 2015; Tran et al., 2016), and colonization of hosts or specific environments (Gawronski et al., 2009; Dong et al., 2013; Kamp et al., 2013; Skurnik et al., 2013; Bishop et al., 2014; Johnson et al., 2014; Verhagen et al., 2014; Wang et al., 2014; Gutierrez et al., 2015; Moule et al., 2015; Turner et al., 2015; Capel et al., 2016). Recently, INSeq was adapted for use in the Rhizobiaceae, and was demonstrated to be a suitable tool for high-throughput functional genomic screening in RLV3841 (Perry and Yost, 2014).
In this paper we used INSeq to define a core functional genome (CFG) of RLV3841 and deconstruct central carbon metabolism for growth on mannitol, a preferred carbon source of rhizobia (Vincent, 1970; Geddes and Oresnik, 2014). Comparing the genes required for growth on mannitol with those required for growth on tryptone-yeast extract media, we estimated a core set of functional genes required for optimal growth. Furthermore, the results of this study demonstrate that using INSeq and growth on minimal media is an effective approach to gain new insight into central carbon metabolism in RLV3841.
Materials and methods
Growth conditions, strains, and plasmids
R. leguminosarum bv. viciae 3841 (Johnston and Beringer, 1975) was routinely grown at 30°C using tryptone-yeast (TY) extract growth medium (5.0 g tryptone, 3.0 g yeast extract, 3.5 mM CaCl2 per liter H2O; TY) or Vincent's minimal medium (VMM) (1.0 g K2HPO4, 1.0 g KH2PO4, 0.01 g FeCl26H2O, 0.25 g MgSO47H2O, 0.1 g CaCl26H2O, 0.6 g KNO3, 0.1 mg biotin, 0.1 mg Ca-panthenoate, and 0.1 mg thiamine per liter H2O) supplemented with 1.% (w/v) mannitol as a carbon source (Vincent, 1970). The donor E. coli strain SM10λpir harboring pSAM_Rl was routinely grown on LB medium at 37°C. Antibiotic concentrations were 500 μg/mL streptomycin (Str) and 50 μg/mL neomycin (Neo) for RLV3841, and 100 μg/mL ampicillin (Amp) and 50 μg/mL kanamycin (Kan) for E. coli.
Mariner transposon mutant pool generation and mutant selection
Mutant pools were generated as described in Perry and Yost (2014) with minor modifications. Briefly, RLV3841 and E. coli SM10λpir(pSAM_Rl) were grown in broth culture until late exponential phase. 1.0 ml of donor and 0.5 ml of recipient strains were mixed in a 1.5 mL microcentrifuge tube and pelleted at 12, 000 g for 3 min. The cell mixture was then washed twice with 1000 μL 1X phosphate buffer saline (PBS), and resuspended in a final volume of approximately 100 μL 1X PBS. Six independent conjugations were spotted onto pre-warmed VMM-mannitol plates and were incubated at 30°C overnight (~18 h). Following incubation each of the 6 conjugation spots were scraped and re-suspended in 1000 μL of 1X PBS and pooled into a total volume of 6 ml, representing the RLV3841 Tn mutant library.
Selection of mutant pools on VMM-mannitol was conducted using six 245 × 245 mm2 (Corning) Neo and Str containing VMM-mannitol agar plates. For each selection plate, 500 μL of the RLV3841 mutant pool was spread plated and allowed to dry. The agar plates were incubated at 30 C for 72 hr representing between 15 and 18 generations of RLV3841 growth on minimal media. Cells from each plate were harvested by scraping the thin film of cell growth and re-suspending in 5 mL of 1X PBS, vortexed thoroughly to homogenize the cells, and then a 1000 μL aliquot of each cell suspension was used for cell pelleting and DNA isolation (Perry and Yost, 2014).
Transposon insertion sequencing
The mutant pools recovered from 2 of 6 selection plates were pooled into 3 independent technical replicates for DNA extraction and Tn insertion sequencing. The method used for library preparation and sequencing is described by Perry and Yost (2014) with modification to the adaptor sequences INSeq_Adpt_Top and INSeq_Adpt_Bottom (Table S1). The final library concentration of the 3 stock library preparations was 1.29, 1.21, and 1.39 ng/μL after size selection. DNA sequencing was performed on an Ion Torrent PGM using 200 bp sequencing chemistry and a 316v2 sequencing chip. The raw sequence output for the 3 technical replicates was 1.4, 1.1, and 1.1 M reads, respectively, and can be found under SRA deposit number: SRR3400585-7 (TY datasets are deposited under SRR3400588-90). Sequencing data from the 3 technical replicates were pooled for a combined total of 3.6 million to achieve sufficient read depth for hidden Markov model analysis (DeJesus and Ioerger, 2013). Raw sequencing data was processed, aligned, and analyzed as previously described (Perry and Yost, 2014). Briefly, raw reads were clipped at the end of the pSAM_Rl mariner IR element, and clipped on the 3′ end at the beginning of the INSeq_Adpt sequence. The resulting reads were screened for the presence of a 5′-TA insertion site, and a length ≥15 bp. Trimmed reads were aligned to the RLV3841 reference genome using bowtie, with the option to suppress reads with multiple alignments from the output file enabled. The alignment files were then converted to wig files and analyzed using the tn-hmm.py python module. The pipeline resulted in a total of 2,374,819 reads being aligned onto the RLV3841 reference genome, after quality filtering and discarding of unaligned reads. The HMM then assigned each “TA” insertion site to one of four growth states which was used to assign each gene to a specific growth phenotype (DeJesus and Ioerger, 2013).
Curation of INSeq data
Outputs from the HMM for both the TY (Perry and Yost, 2014) and VMM dataset were combined based on RLV3841 locus number. Riley functional classifications for each gene were obtained from the lab of Phillip Poole, University of Oxford (http://rhizosphere.org/lab-page/molecular-tools/genomes/rlv3841-genome) and appended to the dataset. Duplicate gene sequences were manually examined using reciprocal BLAST to the RLV3841 reference genome for all genes with <0.30 insertion density to avoid miss-classification as an essential gene due to a lack of mapped insertions as a result of the alignment penalty for multiple mapping locations. The final compiled and curated dataset is found in Supplementary File 1.
Insertional mutagenesis of RL0920 and RL3335
Two previously uncharacterized VMM-mannitol growth impaired genes (RL0920 and RL3335) were mutated to verify the INSeq growth phenotype data. Mutants were created using a single crossover mutagenesis approach with pJQ200SK (Quandt and Hynes, 1993), as described in Vanderlinde et al. (2010). Briefly, a 563 bp internal fragment of RL0920 was PCR amplified using primers RL0920_Fwd and RL0920_Rev, which introduced 5′ ApaI and 3′ SpeI restriction enzyme sites. The 563 bp amplicon was then directionally cloned into pJQ200SK using ApaI and SpeI. The new vector pJQ200SK-RL0920 was conjugated into RLV3841 wildtype using E. coli strain S17-1, and single cross over mutants were selected for on TYSmGm, and then screened for sucrose sensitivity to confirm the plasmid integration. The identical procedure was used to create a single crossover mutant in RL3335 using primers RL3335_Fwd and RL3335_Rev to generate a 603 bp internal gene fragment for cloning into pJQ200SK (Table S1). The resulting mutants in RL0920 and RL3335 were named MA0920 and MA3335.
Growth curve analysis of MA0920, MA3335, and RLV3841 wildtype
Growth curves of RLV3841, MA0920, and MA3335 were performed using a shaking head Synergy HT Microplate Reader (Biotek) with 250 μL of inoculated growth media per well in a 96-well NuncR Optical Bottom Plate (Thermo Scientific), and a 40 μL Anti-Evaporation Oil (Ibidi) overlay. Inoculated growth medium was prepared by scraping cells from freshly grown TYSm or SmGm plates, washing twice with 1XPBS, standardizing the cell suspension for an approximate initial OD600 of 0.01. Cells were grown at 30°C with 10 min of shaking followed by OD600 measurements every 30 min, for 72 h. Each growth curve was derived from the mean OD600 measurements of 7 replicates. Mean generation times (MGT) were calculated in early exponential phase (OD600 < 0.100) and late exponential phase (0.100 < OD600 < 0.200) by calculating the average time required to double the optical density of the cell cultures, within the defined growth phases.
Results
Tn insertion sequencing and transposition summary of VMM-mannitol mutant pools
The RLV3841 genome contains 140,056 potential mariner insertion sites distributed across the chromosome and 6 mega-plasmids. Insertion densities within the genome ranged from 0.65 to 0.86 across the 7 replicons, with an average insertion density of 0.80 (Table S2). The insertion densities were similar to the insertion densities previously observed in an INSeq experiment using TY medium for selection (Perry and Yost, 2014). HMM analysis of the VMM INSeq data assigned 7.2 and 2.6% of genes to essential (ES) or growth defective (GD) growth phenotypes, respectively (Table S3). Given sufficient generations of growth GD mutants would be excluded from the mutant community. Therefore, ES and GD states were pooled into a single growth impaired (GI) category (9.8% of the genes) for annotation with Riley functional groupings. 87.1% of genes were observed to have no impact on growth (NE), and 1.20% of genes with Tn insertions became over-represented within the mutant communities, and were predicted to confer a growth advantage (GA) phenotype (Table S3). To further simplify interpretation of the INSeq data by focusing exclusively on loss-of-function phenotypes the NE and GA genes were pooled into a single category termed growth neutral (GN). Within the genome 1.6% of genes had sequence duplications resulting in no information concerning their impact on growth due to the multiple Tn insertion tag mapping location penalty imposed. As well, 0.3% of genes were observed to lack a “TA” dinucleotide motif leaving them without a target site for mariner Tn insertion.
The RLV3841 CFG, VMM-growth impaired, and TY-growth impaired phenotypes
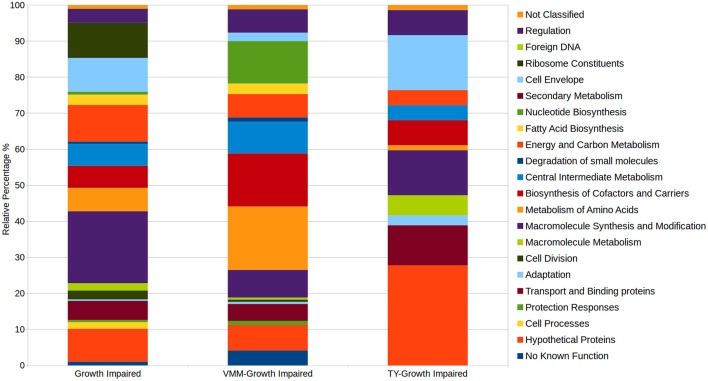
Comparison of the VMM-mannitol INSeq dataset with the TY INSeq dataset identified a set of genes that when mutated conferred a GI phenotype under both conditions; these 491 genes were assigned to the CFG (Figure 1). Whereas, 170 and 72 genes, when interrupted by Tn insertion, resulted in a condition dependent impaired ability to grow on VMM (VGI) and TY (TGI), respectively. Genes within the CFG were represented by 5 major Riley classification groups: macro-molecule synthesis and metabolism (20.0%), energy and carbon metabolism (10.2%), ribosomal constituents (9.8%), cell envelope (9.4%), and conserved hypothetical proteins (9.2%) (Figure 2). Genes that gave rise to a VGI phenotype were composed of 3 major functional groups: metabolism of amino acids (17.6%), biosynthesis of co-factors and carriers (14.7%), and nucleotide biosynthesis (11.8%). While the 4 major groups of TGI genes consisted of hypothetical proteins (27.8%), cell envelope (15.3%), macro-molecule synthesis and metabolism (12.5%), and transport and binding proteins (11.1%).
Figure 1.
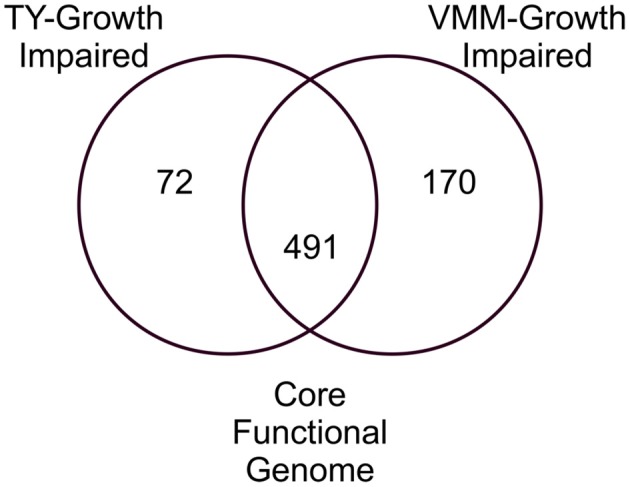
Venn diagram of growth impaired genes observed for growth on TY and VMM-mannitol. Growth impaired genes observed uniquely on TY or VMM were assigned to TY-growth impaired or VMM-growth impaired. Growth impaired genes observed in both treatments were assigned to the CFG. Genes without potential mariner insertion sites, or which had highly similar sequence redundancy in the RLV3841 genome were discounted.
Figure 2.
Relative distributions of Riley functional gene classifications within growth impaired categories. Growth impaired genes on both VMM-Mannitol and TY media were assigned to the CFG. CFG, TY-growth impaired, and VMM-mannitol growth impaired genes were assigned Riley functional classification based on Young et al. (2006). The relative abundance of genes within observed Riley functional groups were then calculated within each growth impaired category.
Open reading frames annotated as hypothetical proteins represent approximately 25.2% of the RLV3841 genome. Of these, 103 hypothetical proteins were observed to have a GI phenotype on VMM, TY, or both growth conditions (Table 1). Sequence duplication within the RLV3841 genome resulted in 20 annotated hypothetical proteins not being assayed for a growth phenotype using INSeq, due to multiple potential alignments of sequenced Tn insertion tags.
Table 1.
Growth impaired hypothetical proteins.
| VMM INSeq Data | TY INSeq Data | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Growth phenotype | N | Insertion density | Average read depth | Gene state | N | Insertion density | Average read depth | Gene state | Riley key | Annotation | |
| CORE FUNCTIONAL GENOME HYPOTHETICAL PROTEINS | ||||||||||||
| RL1763 | GI | 2 | 0 | 0 | ES | 2 | 0 | 0 | ES | 0.0.0 | Hypothetical protein | |
| RL2220 | GI | 7 | 0.57 | 29.75 | ES | 7 | 0.57 | 30.25 | ES | 0.0.0 | Hypothetical protein | |
| RL2402 | GI | 13 | 0.46 | 13.5 | GD | 13 | 0.46 | 11.33 | GD | 0.0.0 | Hypothetical protein | |
| RL2581 | GI | 6 | 0 | 0 | ES | 6 | 0.33 | 1.5 | ES | 0.0.0 | Hypothetical protein | |
| RL4714 | GI | 1 | 0 | 0 | ES | 1 | 1 | 2 | GD | 0.0.0 | Hypothetical protein | |
| RL0128 | GI | 8 | 0.13 | 1 | ES | 8 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL0393 | GI | 8 | 0.38 | 9.33 | GD | 8 | 0.5 | 4.25 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL0412 | GI | 10 | 0 | 0 | ES | 10 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL0548 | GI | 7 | 0 | 0 | ES | 7 | 0.14 | 2 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL0612 | GI | 2 | 0 | 0 | ES | 2 | 0 | 0 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL0620 | GI | 7 | 0 | 0 | GD | 7 | 0 | 0 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL1432 | GI | 5 | 0.2 | 7 | ES | 5 | 0.6 | 4.33 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL1566 | GI | 11 | 0.18 | 5 | ES | 11 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL2245 | GI | 8 | 0.13 | 22 | ES | 8 | 0.25 | 30 | GD | 0.0.1 | Conserved hypothetical protein with RimI domain, putative N-acetyltransferase | |
| RL3754 | GI | 6 | 0.17 | 8 | ES | 6 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL3757 | GI | 5 | 0.6 | 16 | GD | 5 | 0.4 | 33.5 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL3762 | GI | 14 | 0 | 0 | ES | 14 | 0.07 | 1 | ES | 0.0.1 | Conserved hypothetical exported protein | |
| RL4061 | GI | 10 | 0 | 0 | ES | 10 | 0.3 | 1.67 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL4324 | GI | 1 | 0 | 0 | ES | 1 | 0 | 0 | GD | 0.0.1 | Conserved hypothetical exported protein | |
| RL4434 | GI | 3 | 0 | 0 | ES | 3 | 0 | 0 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL4437 | GI | 8 | 0 | 0 | GD | 8 | 0.13 | 2 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL4733 | GI | 8 | 0 | 0 | ES | 8 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL0126 | GI | 8 | 0 | 0 | ES | 8 | 0.13 | 3 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL0144 | GI | 5 | 0 | 0 | ES | 5 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL0456A | GI | 4 | 0 | 0 | NE | 4 | 0 | 0 | NE | 0.0.2 | Conserved hypothetical protein | |
| RL0619 | GI | 4 | 0.25 | 1 | GD | 4 | 0.25 | 1 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL0626 | GI | 13 | 0.46 | 12.67 | GD | 13 | 0.62 | 14.25 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL0936 | GI | 30 | 0.07 | 1 | ES | 30 | 0.17 | 2 | GD | 0.0.2 | Conserved hypothetical protein (TPR repeat family) | |
| RL1094 | GI | 17 | 0.12 | 8 | ES | 17 | 0.18 | 5.33 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL1116 | GI | 9 | 0.11 | 18 | ES | 9 | 0.11 | 39 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL1569 | GI | 21 | 0.05 | 1 | ES | 21 | 0.14 | 1 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL1598 | GI | 6 | 0 | 0 | ES | 6 | 0.17 | 1 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL1706 | GI | 3 | 0 | 0 | ES | 3 | 0 | 0 | GD | 0.0.2 | Conserved hypothetical protein in NADH-ubiquinone oxidoreductase region | |
| RL2034 | GI | 8 | 0.5 | 3.75 | GD | 8 | 0.38 | 1 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL2042 | GI | 29 | 0.28 | 8.5 | ES | 29 | 0.17 | 5.4 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL2056 | GI | 5 | 0.2 | 1 | GD | 5 | 0.2 | 2 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL2207 | GI | 6 | 0.17 | 1 | ES | 6 | 0.17 | 2 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL2232 | GI | 11 | 0.09 | 1 | ES | 11 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL2584 | GI | 9 | 0 | 0 | ES | 9 | 0.11 | 2 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL2697 | GI | 5 | 0.2 | 21 | ES | 5 | 0.2 | 22 | ES | 0.0.2 | Conserved hypothetical exported protein | |
| RL3259 | GI | 1 | 0 | 0 | NE | 1 | 0 | 0 | NE | 0.0.2 | Conserved hypothetical protein | |
| RL3314 | GI | 8 | 0 | 0 | ES | 8 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL3464 | GI | 15 | 0.27 | 1.5 | GD | 15 | 0.53 | 2.63 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL3948A | GI | 31 | 0.06 | 2 | ES | 31 | 0.16 | 1 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL3967 | GI | 19 | 0 | 0 | ES | 19 | 0.53 | 3.5 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL4037 | GI | 8 | 0.5 | 3.25 | GD | 8 | 0.63 | 6.8 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL4280 | GI | 20 | 0.4 | 2.5 | GD | 20 | 0.25 | 4 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL4523 | GI | 13 | 0.23 | 7.67 | ES | 13 | 0.23 | 12.33 | ES | 0.0.2 | Conserved hypothetical exported protein | |
| RL4562 | GI | 10 | 0.1 | 3 | ES | 10 | 0.1 | 4 | ES | 0.0.2 | Conserved hypothetical exported protein | |
| RL4734 | GI | 14 | 0 | 0 | ES | 14 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein | |
| VMM-GROWTH IMPAIRED HYPOTHETICAL PROTEINS | ||||||||||||
| RL0044 | VGI | 13 | 0.62 | 22.88 | ES | 13 | 0.69 | 32.33 | NE | 0.0.0 | Hypothetical protein | |
| RL1117 | VGI | 6 | 0.5 | 15.33 | ES | 6 | 0.67 | 8 | NE | 0.0.0 | Hypothetical protein | |
| RL1628 | VGI | 39 | 0.64 | 8.32 | ES | 39 | 0.67 | 10.35 | NE | 0.0.0 | Hypothetical protein | |
| RL1946 | VGI | 13 | 0.62 | 1.75 | GD | 13 | 0.69 | 3.33 | NE | 0.0.0 | Hypothetical protein | |
| RL2117A | VGI | 33 | 0.76 | 11.36 | ES | 33 | 0.79 | 13.31 | NE | 0.0.0 | Hypothetical protein | |
| RL2154 | VGI | 16 | 0.69 | 7.55 | GD | 16 | 0.75 | 7.25 | NE | 0.0.0 | Hypothetical protein | |
| RL2611 | VGI | 10 | 0.5 | 11.6 | ES | 10 | 0.9 | 14 | NE | 0.0.0 | Hypothetical protein | |
| RL1524 | VGI | 5 | 0.6 | 3.33 | GD | 5 | 0.8 | 9.75 | NE | 0.0.1 | Conserved hypothetical protein | |
| RL3760 | VGI | 7 | 0 | 0 | ES | 7 | 0.29 | 9 | NE | 0.0.1 | Conserved hypothetical protein | |
| RL4043 | VGI | 2 | 0.5 | 2 | ES | 2 | 1 | 29 | NE | 0.0.1 | Conserved hypothetical protein | |
| RL4066 | VGI | 24 | 0.42 | 2.8 | GD | 24 | 0.79 | 9.68 | NE | 0.0.1 | Conserved hypothetical protein | |
| RL0571 | VGI | 3 | 0 | 0 | ES | 3 | 1 | 26.67 | NE | 0.0.2 | Conserved hypothetical protein | |
| RL0960 | VGI | 10 | 0.3 | 1.33 | GD | 10 | 0.5 | 2.6 | NE | 0.0.2 | Conserved hypothetical protein (Sua5 family) | |
| RL1523 | VGI | 4 | 0.25 | 2 | GD | 4 | 1 | 4.25 | NE | 0.0.2 | Conserved hypothetical protein | |
| RL2210 | VGI | 5 | 0.2 | 1 | GD | 5 | 0.4 | 1 | NE | 0.0.2 | Conserved hypothetical protein | |
| RL2289 | VGI | 3 | 0.33 | 1 | GD | 3 | 1 | 19 | NE | 0.0.2 | Conserved hypothetical protein | |
| RL2291 | VGI | 7 | 0.71 | 2 | GD | 7 | 0.86 | 20 | NE | 0.0.2 | Conserved hypothetical protein | |
| RL2527 | VGI | 13 | 0.31 | 8.5 | GD | 13 | 0.69 | 6.78 | NE | 0.0.2 | Conserved hypothetical exported protein | |
| RL4728 | VGI | 13 | 0.23 | 6 | ES | 13 | 0.69 | 6.11 | NE | 0.0.2 | Conserved hypothetical protein | |
| TY-GROWTH IMPAIRED HYPOTHETICAL PROTEINS | ||||||||||||
| RL1527 | TGI | 2 | 1 | 24 | NE | 2 | 1 | 3.5 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL1562 | TGI | 9 | 0.56 | 14.8 | NE | 9 | 0.56 | 16.4 | ES | 0.0.1 | Conserved hypothetical protein | |
| RL1618A | TGI | 17 | 1 | 9.82 | NE | 17 | 0.65 | 2.18 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL2307 | TGI | 4 | 0.75 | 5.67 | NE | 4 | 0.75 | 10 | GD | 0.0.1 | Conserved hypothetical protein | |
| RL4065 | TGI | 3 | 0.67 | 1 | NE | 3 | 0 | 0 | NE | 0.0.1 | Conserved hypothetical protein | |
| RL4716 | TGI | 18 | 1 | 37.17 | NE | 18 | 0.61 | 6.55 | GD | 0.0.1 | Conserved hypothetical exported protein | |
| RL0109 | TGI | 8 | 0.75 | 8.5 | NE | 8 | 0.25 | 2.5 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL0890 | TGI | 16 | 0.81 | 19.54 | NE | 16 | 0.5 | 13.75 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL1526 | TGI | 11 | 0.91 | 34.9 | NE | 11 | 0.64 | 5.43 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL1528 | TGI | 11 | 0.91 | 14.3 | NE | 11 | 0.45 | 7.2 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL2086 | TGI | 6 | 0.83 | 5.8 | NE | 6 | 0.5 | 1.33 | GD | 0.0.2 | Conserved hypothetical exported protein | |
| RL2142 | TGI | 17 | 0.71 | 13.33 | NE | 17 | 0.65 | 17.55 | ES | 0.0.2 | Conserved hypothetical protein | |
| RL2542 | TGI | 2 | 1 | 4.5 | NE | 2 | 0 | 0 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL2625 | TGI | 15 | 0.8 | 9.08 | NE | 15 | 0.47 | 3.71 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL2641 | TGI | 9 | 0.67 | 14 | NE | 9 | 0.56 | 5.8 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL3499 | TGI | 14 | 0.93 | 17.08 | NE | 14 | 0.5 | 2.29 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL3500 | TGI | 18 | 1 | 34.89 | NE | 18 | 0.67 | 5.75 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL3761 | TGI | 14 | 0.93 | 13.23 | NE | 14 | 0.21 | 1 | ES | 0.0.2 | Conserved hypothetical exported protein | |
| RL4016 | TGI | 5 | 0.8 | 27.5 | NE | 5 | 0 | 0 | GD | 0.0.2 | Conserved hypothetical protein | |
| RL4503 | TGI | 4 | 0.75 | 3 | NE | 4 | 1 | 4.75 | GD | 0.0.2 | Conserved hypothetical protein | |
| PLASMID GROWTH IMPAIRED HYPOTHETICAL PROTEINS | ||||||||||||
| pRL100111 | PGI | 38 | 0.45 | 3.18 | GD | 38 | 0.45 | 3.71 | ES | 0.0.0 | Hypothetical protein | |
| pRL70135 | PGI | 17 | 0.47 | 22.75 | ES | 17 | 0.65 | 23 | ES | 0.0.0 | Hypothetical protein | |
| pRL120721 | PGI | 6 | 0.17 | 1 | ES | 6 | 0.33 | 2.5 | ES | 0.0.1 | Conserved hypothetical protein | |
| pRL100012 | PGI | 3 | 0 | 0 | ES | 3 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein | |
| pRL110465 | PGI | 8 | 0.13 | 1 | GD | 8 | 0.13 | 2 | GD | 0.0.2 | Conserved hypothetical protein | |
| pRL70167 | PGI | 11 | 0.27 | 11 | ES | 11 | 0.27 | 19 | ES | 0.0.2 | Conserved hypothetical protein | |
| pRL80098 | PGI | 11 | 0.18 | 9.5 | ES | 11 | 0.64 | 8.14 | GD | 0.0.2 | Conserved hypothetical protein | |
| PLASMID/(VMM/TY)-GROWTH IMPAIRED HYPOTHETICAL PROTEINS | ||||||||||||
| pRL100010 | PVGI | 3 | 0.33 | 3 | GD | 3 | 1 | 11.67 | NE | 0.0.2 | Conserved hypothetical protein | |
| pRL100149 | PVGI | 58 | 0.83 | 14.56 | ES | 58 | 0.83 | 18.54 | NE | 0.0.2 | Conserved hypothetical protein | |
| pRL110044 | PVGI | 6 | 0.33 | 1.5 | GD | 6 | 0.83 | 4 | NE | 0.0.2 | Conserved hypothetical protein | |
| pRL110108 | PVGI | 17 | 0.76 | 9.85 | ES | 17 | 0.76 | 13.38 | NE | 0.0.2 | Conserved hypothetical protein | |
| pRL110351A | PTGI | 10 | 0.7 | 9.71 | NE | 10 | 0.6 | 14.67 | ES | 0.0.2 | Conserved hypothetical protein | |
| pRL70166 | PVGI | 9 | 0.33 | 6.33 | ES | 9 | 0.67 | 6.83 | NE | 0.0.2 | Conserved hypothetical protein | |
| pRL90280 | PVGI | 6 | 0.5 | 4.33 | ES | 6 | 0.83 | 6.6 | NE | 0.0.2 | Conserved hypothetical protein | |
| Locus | Growth phenotype | N | Insertion density | Average read depth | Gene state | N | Insertion density | Average read depth | Gene state | Riley key | Annotation | Sequence duplications |
| GROWTH IMPAIRED OR PLASMID GROWTH IMPAIRED HYPOTHETICAL PROTEINS WITH SEQUENCE DUPLICATION | ||||||||||||
| pRL70102 | PGI | 4 | 0 | 0 | ES | 4 | 0 | 0 | ES | 0.0.0 | Hypothetical protein | pRL70102, pRL110582, downstream pRL100152, upstream pRL100166 |
| RL1481 | GI | 6 | 0 | 0 | ES | 6 | 0 | 0 | ES | 0.0.0 | Hypothetical protein | RL1481, 3′-end of RL00456B, 5′-end of RL0457, 3′-end of pRL100088, 5′-end of pRL100089 |
| pRL110342 | PGI | 2 | 0 | 0 | ES | 2 | 0 | 0 | ES | 0.0.0 | Hypothetical protein | upstream pRL100046, upstream RL2679 |
| pRL100089 | PGN | 5 | 0.4 | 5.5 | NE | 5 | 0.6 | 5.33 | NE | 0.0.1 | Conserved hypothetical protein | pRL100089, RL1481 |
| pRL70033 | PGI | 8 | 0 | 0 | ES | 8 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | pRL70033, pRL70047D, pRL70147 |
| pRL70047 | PGI | 6 | 0 | 0 | ES | 6 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | pRL70142, pRL70047 |
| pRL70142 | PGI | 6 | 0 | 0 | ES | 6 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | pRL70142, pRL70047 |
| pRL70179 | PGI | 3 | 0 | 0 | ES | 3 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | pRL70169, pRL70179, PRL100095 |
| pRL120379 | PGI | 35 | 0 | 0 | ES | 35 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | pRL90004, pRL90319, pRL100469, pRL120379 |
| pRL90004 | PGI | 35 | 0 | 0 | ES | 35 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | pRL90004, pRL90319, pRL100469, pRL120379 |
| pRL120650 | PGI | 6 | 0 | 0 | ES | 6 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | RL2139, RL0835, pRL120650 |
| RL0835 | GI | 7 | 0 | 0 | ES | 7 | 0.14 | 21 | ES | 0.0.1 | Conserved hypothetical protein | RL2139, RL0835, pRL120650 |
| RL2139 | GI | 6 | 0 | 0 | ES | 6 | 0 | 0 | ES | 0.0.1 | Conserved hypothetical protein | RL2139, RL0835, pRL120650 |
| pRL110353 | PGN | 4 | 0.25 | 2 | NE | 4 | 0.25 | 3 | NE | 0.0.2 | Conserved hypothetical protein | pRL110353, RL2153 |
| RL2153 | GI | 4 | 0.25 | 3 | GD | 4 | 0.25 | 4 | GD | 0.0.2 | Conserved hypothetical protein | pRL110353, RL2153 |
| pRL100095 | PGI | 2 | 0 | 0 | NE | 2 | 0 | 0 | NE | 0.0.2 | Conserved hypothetical protein | pRL70169, pRL70179, PRL100095 |
| pRL70169 | PGI | 2 | 0 | 0 | ES | 2 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein | pRL70169, pRL70179, PRL100095 |
| pRL100469 | PGI | 35 | 0 | 0 | ES | 35 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein | pRL90004, pRL90319, pRL100469, pRL120379 |
| pRL90319 | PGI | 35 | 0 | 0 | ES | 35 | 0 | 0 | ES | 0.0.2 | Conserved hypothetical protein, possible fusion protein | pRL90004, pRL90319, pRL100469, pRL120379 |
Growth curve analysis of two predicted VMM-growth impaired mutants
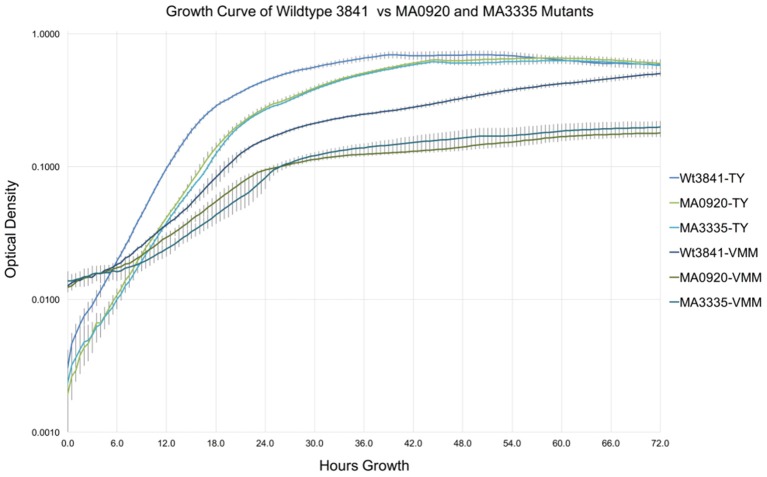
Growth curves of RLV3841, MA0920 (INSeq predicted RL0920 VGI), and MA3335 (INSeq predicted RL3335 VGI), in TY and VMM-Mannitol broth over 72 h are shown in Figure 3. After 72 h growth RLV3841, MA0920, and MA3335 reached final mean OD600 readings of 0.696, 0.657, and 0.628 on TY; and 0.588, 0.179, and 0.198 on VMM, respectively. Mean generation times for RLV3841, MA0920, and MA3335 on TY were 2.5, 3.0, and 3.0 h in early exponential phase; and 3.5, 4.5, and 4.0 h in late exponential phase. The MGTs in VMM-Mannitol were observed to be 6.0, 7.5, and 8.5 h in early exponential phase for RLV3841, MA0920, and MA3335 respectively. In late exponential phase, MA0920 and MA3335 halted growth and did not complete an additional doubling, whereas RLV3841 wildtype continued to double with a MGT of 9.5 h.
Figure 3.
Growth curve comparison of RLV3841, MA0920, and MA3335 in TY and VMM. Growth curves represent the mean OD600 of 7 replicates measured at 30 min intervals over 72 h in TY and VMM-Mannitol liquid growth media; error bars indicate the standard deviation of the mean. MA0920 and MA3335 had longer mean generation times than the wildtype in TY and VMM-mannitol. After 72 h of growth in VMM-mannitol, MA0920, and MA3335 reached cell densities that were ~1/3 of the wildtype.
The genetics of central carbon metabolism for growth on mannitol
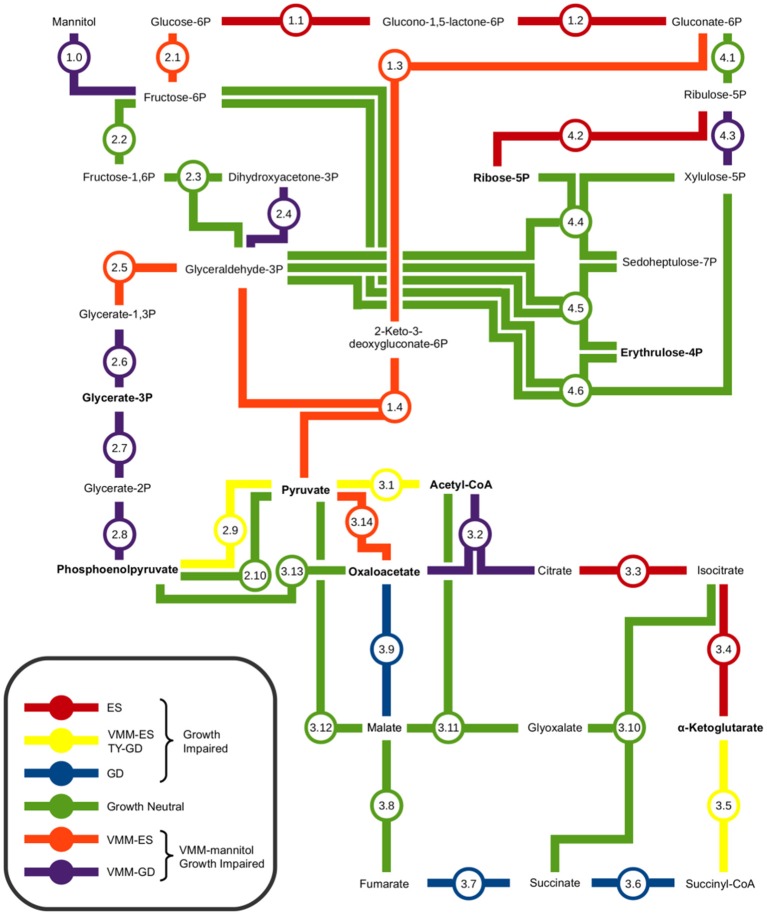
INSeq was used to identify a potential minimal central carbon metabolism pathway for growth on mannitol. Figure 4, and accompanying Table 2, provide a metabolic map illustrating the interconnections of the Embden-Meyerhof-Parnas (EMP) pathway, Entner-Doudoroff (ED) pathway, pentose phosphate (PP) pathway, and tricarboxylic acid cycle (TCA) that compose central carbon metabolism, with an overlay of the observed growth impaired phenotypes. It was observed that the genes required for mannitol uptake and conversion to fructose-6P (F6P) were VGI. Genes required for conversion of F6P to pyruvate were observed to be VGI within the ED pathway. Genes required for conversion of F6P into glyceraldehyde-3P (GA3P), via the upper EMP pathway, were observed to have no impact on growth when mutated. Genes required for the conversion of GA3P to pyruvate as part of the lower EMP pathway were found to be VGI when mutated. Assimilation of pyruvate into the TCA cycle was observed to be VGI via more than one metabolic pathway. Mutation of genes within the TCA cycle were observed to result in GI or VGI phenotypes, with the exception of a growth neutral step at the conversion between fumarate and malate (Figure 3; Table 2).
Figure 4.
Metabolic map of central carbon metabolism of Rhizobium leguminosarum bv. viciae 3841. Interconnections between the Embden-Meyerhof-Parnas pathway, Entner-Doudoroff pathway, Pentose Phosphate pathway, and Tri-Carboxylic Acid cycle were based on the proposed central carbon metabolism of Sinorhizobium meliloti and Agrobacterium tumefaciens (Fuhrer et al., 2005; Geddes and Oresnik, 2014). Amino acid precursors are indicated in bold text. The impact of mutations in each metabolic step on growth were determined by contrasting results from TY and VMM-mannitol INSeq experiments. Genes observed to be growth defective or essential exclusively on VMM-Mannitol were concluded to be involved in central carbon metabolism of mannitol. Genes observed to be GD or ES on TY and VMM-mannitol were grouped, and assumed to have roles in central carbon metabolism that were not mannitol dependent.
Table 2.
Growth phenotypes of genes involved in RLV3841 central carbon metabolism.
| VMM INSeq | TY INSeq | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolic step | RLV locus | RLV symbol | Putative product | Growth phenotype | Insertion density | Average read depth | Gene state | Insertion density | Average read depth | Gene state | Riley key | Riley functional information |
| MANNITOL UPTAKE AND CONVERSION TO FRUCTOSE | ||||||||||||
| 1.0 | RL4219 | – | Transcriptional regulator | GN | 0.86 | 16.67 | NE | 0.79 | 25.27 | NE | 6.3.10 | Putative deor family transcriptional regulator (repressor) of sorbitol/mannitol operon |
| 1.0 | RL4215 | – | Mannitol ABC transporter ATP-binding protein | VGI | 0.41 | 1.71 | GD | 0.88 | 12.6 | NE | 1.5.3 | Putative ATP-binding component of ABC transporter CUT1 mannitol transporter (S. Mel SBP homolog smc01496 induced by dulcitol, sorbitol, mannitol) |
| 1.0 | RL4216 | mtlG | Mannitol transmembrane permease component of ABC transporter | VGI | 0.56 | 3.2 | GD | 1 | 34.11 | NE | 1.5.3 | Putative permease component of ABC transporter CUT1 mannitol transporter (S. Mel SBP homolog smc01496 induced by dulcitol, sorbitol, mannitol) |
| 1.0 | RL4217 | mtlF | Mannitol transmembrane permease component of ABC transporter | VGI | 0.53 | 2.78 | GD | 0.94 | 36.06 | NE | 1.5.3 | Putative permease component of ABC transporter CUT1 mannitol transporter (S. Mel SBP homolog smc01496 induced by dulcitol, sorbitol, mannitol) |
| 1.0 | RL4218 | mtlE | Mannitol-binding component of ABC transporter | VGI | 0.48 | 2.3 | GD | 0.9 | 26.11 | NE | 1.5.3 | Putative SBP of ABC transporter CUT1 mannitol transporter (S. Mel SBP homolog smc01496 induced by dulcitol, sorbitol, mannitol) |
| 1.0 | RL4214 | mtlK | Mannitol 2-dehydrogenase | VGI | 0.12 | 7.5 | GD | 0.88 | 24.8 | NE | 3.4.3 | Putative mannitol 2-dehydrogenase |
| 1.0 | RL0098 | – | Mannitol dehydrogenase | GN | 0.93 | 17 | NE | 0.93 | 17.62 | NE | 3.4.3 | Putative mannitol dehydrogenase |
| 1.0 | RL0502 | frk | Fructokinase | VGI | 0.21 | 3 | GD | 1 | 15.71 | NE | 3.5.5 | Fructokinase |
| ENTNER–DOUDOROFF PATHWAY | ||||||||||||
| 1.1 | RL0753 | zwf1 | Putative glucose-6-phosphate 1-dehydrogenase | GI | 0.05 | 1 | ES | 0.09 | 1 | ES | 3.5.6 | Putative glucose-6-phosphate 1-dehydrogenase |
| 1.1 | RL1315 | zwf1 | Glucose-6-phosphate 1-dehydrogenase | GN | 0.93 | 18.72 | NE | 1 | 18.7 | NE | 3.5.6 | Putative glucose-6-phosphate 1-dehydrogenase |
| 1.1 | pRL120561 | – | Glucose-6-phosphate 1-dehydrogenase | PGN | 1 | 19 | NE | 1 | 14.5 | NE | 6.3.0 | Putative XRE family transcriptional regulator |
| 1.2 | RL0752 | pgl | 6-phosphogluconolactonase | GI | 0 | 0 | ES | 0 | 0 | ES | 3.5.6 | Putative 6-phosphogluconolactonase |
| 1.3 | RL0751 | edd | Phosphogluconate dehydratase | VGI | 0 | 0 | ES | 1 | 8.73 | NE | 3.3.3 | Putative phosphogluconate dehydratase |
| 1.4 | RL4162 | eda | Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase | VGI | 0.13 | 14 | ES | 0.88 | 3.14 | NE | 3.3.3 | Putative 2-dehydro-3-deoxyphosphogluconate aldolase |
| EMBDEN-MEYERHOF-PARNAS PATHWAY | ||||||||||||
| 2.1 | RL0504 | pgi | Glucose-6-phosphate isomerase | VGI | 0.06 | 30 | ES | 0.76 | 10.69 | NE | 3.5.5 | Putative glucose-6-phosphate isomerase |
| 2.2 | RL3322 | pfp | Pyrophosphate-fructose-6-phosphate 1-phosphotransferase | GN | 1 | 16.8 | NE | 0.95 | 21.79 | NE | 3.5.5 | Putative pyrophosphate–fructose 6-phosphate 1-phosphotransferase |
| 2.3 | RL4012 | Fructose-bisphosphate aldolase | GN | 0.86 | 22.42 | NE | 0.93 | 21.23 | NE | 3.5.5 | Putative fructose-bisphosphate aldolase | |
| 2.3 | pRL120027 | Aldolase | PGN | 0.86 | 23.92 | NE | 0.93 | 21.46 | NE | 3.3.15 | Putative aldolase | |
| 2.3 | pRL120196 | Fructose-1,6-bisphosphate aldolase | PGN | 0.95 | 20.67 | NE | 1 | 22.53 | NE | 3.3.9 | Putative fructose-bisphosphate aldolase | |
| 2.4 | RL2513 | tpiA | Triosephosphate isomerase | GN | 0.9 | 14.56 | NE | 0.9 | 24.22 | NE | 3.6.0 | Putative triosephosphate isomerase |
| 2.4 | pRL120209 | tpiA | Triosephosphate isomerase | PVGI | 0.38 | 2.33 | GD | 0.88 | 8.14 | NE | 3.6.0 | Putative triosephosphate isomerase |
| 2.5 | RL4007 | gap | Glyceraldehyde-3-phosphate dehydrogenase | VGI | 0 | 0 | ES | 1 | 9 | NE | 3.5.1 | Putative glyceraldehyde-3-phosphate dehydrogenase |
| 2.6 | RL4011 | pgk | Phosphoglycerate kinase | VGI | 0.25 | 3.67 | GD | 0.83 | 38.2 | NE | 3.5.1 | Putative phosphoglycerate kinase |
| 2.7 | RL0179 | gpmA | Phosphoglyceromutase | VGI | 0.36 | 1.5 | GD | 0.73 | 9.88 | NE | 3.5.5 | Putative 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (Phosphoglyceromutase) (PGAM) (BPG-dependent PGAM) (dPGM) |
| 2.7 | RL2655 | Hypothetical protein | GN | 0.88 | 10.71 | NE | 1 | 9.38 | NE | 0.0.0 | Hypothetical protein | |
| 2.7 | RL0954 | gpmB | Phosphoglycerate mutase | GN | 1 | 19.22 | NE | 0.89 | 21.38 | NE | 3.5.5 | Putative phosphoglycerate mutase |
| 2.7 | RL1010 | gpmB | Phosphoglycerate mutase | GN | 0.83 | 15.2 | NE | 0.83 | 26.2 | NE | 3.5.5 | Putative phosphoglycerate mutase |
| 2.7 | RL2997 | Phosphoglycerate mutase | GN | 1 | 34.29 | NE | 1 | 39.12 | NE | 3.5.5 | Putative phosphoglycerate mutase | |
| 2.8 | RL2239 | eno | Phosphopyruvate hydratase | VGI | 0.19 | 1 | GD | 0.81 | 4.77 | NE | 3.5.5 | Putative enolase |
| 2.9 | RL4060 | pykA | Pyruvate kinase | GI | 0.23 | 8.67 | ES | 0.92 | 4.83 | GD | 3.5.5 | Putative pyruvate kinase |
| 2.10 | RL1086 | ppdK | Pyruvate phosphate dikinase | GN | 0.84 | 21.26 | NE | 0.91 | 24.69 | NE | 3.5.5 | Pyruvate, phosphate dikinase (pyruvate, orthophosphate dikinase) |
| TRICARBOXYLIC ACID CYCLE | ||||||||||||
| 3.1 | RL2241 | pdhA | Pyruvate dehydrogenase subunit | GI | 0 | 0 | ES | 0.18 | 1 | GD | 3.5.5 | Putative pyruvate dehydrogenase subunit A |
| 3.1 | RL2242 | pdhB | Pyruvate dehydrogenase subunit beta | GI | 0 | 0 | ES | 0.22 | 1.25 | GD | 3.5.5 | Putative pyruvate dehydrogenase subunit B |
| 3.1 | RL2243 | pdhC | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex | GI | 0 | 0 | ES | 0 | 0 | GD | 3.5.5 | Putative dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (PDC) |
| 3.1 | pRL80081 | – | Hydrolase | PGN | 0.86 | 9.33 | NE | 0.86 | 9.33 | NE | 3.3.15 | Putative hydrolase |
| 3.2 | RL2234 | gltA | Type II citrate synthase | VGI | 0.7 | 4.32 | GD | 0.85 | 14.09 | NE | 3.5.8 | Putative citrate synthase |
| 3.2 | RL2508 | gltA | Citrate synthase II | GN | 0.85 | 14.82 | NE | 1 | 13.54 | NE | 3.5.8 | Putative citrate synthase II |
| 3.2 | RL2509 | citA | Citrate synthase 2 | GN | 0.8 | 19 | NE | 0.8 | 19.17 | NE | 3.5.8 | Putative citrate synthase I |
| 3.3 | RL4536 | acnA | Aconitate hydratase | GI | 0.05 | 1 | ES | 0.05 | 1 | ES | 3.5.8 | Putative aconitate hydratase |
| 3.4 | RL2631 | icd | Isocitrate dehydrogenase | GI | 0.12 | 10.5 | ES | 0.12 | 8 | ES | 3.5.8 | Putative isocitrate dehydrogenase [NADP] |
| 3.5 | RL4435 | sucA | 2-oxoglutarate dehydrogenase E1 component | GI | 0.05 | 1.5 | ES | 0.23 | 1.5 | GD | 3.5.8 | Putative 2-oxoglutarate dehydrogenase E1 component |
| 3.5 | RL4433 | citM | Dihydrolipoamide succinyltransferase | GI | 0.2 | 9 | ES | 0.3 | 4.33 | GD | 3.5.8 | Putative dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase |
| 3.6 | RL4436 | sucD | Succinyl-CoA synthetase subunit alpha | GI | 0.15 | 1 | GD | 0.23 | 1.33 | GD | 3.5.8 | Putative succinyl-coa synthetase alpha chain |
| 3.6 | RL4438 | sucC | Succinyl-CoA synthetase subunit beta | GI | 0.09 | 1 | GD | 0.36 | 1 | GD | 3.5.8 | Putative succinyl-coa synthetase beta chain |
| 3.7 | RL4443 | sdhB | Succinate dehydrogenase iron-sulfur subunit | GI | 0.13 | 3 | GD | 0.73 | 2.82 | GD | 3.5.8 | Putative succinate dehydrogenase iron-sulfur protein |
| 3.7 | RL4444 | sdhA | Succinate dehydrogenase flavoprotein subunit | GI | 0.53 | 1.5 | GD | 0.76 | 3.77 | GD | 3.5.8 | Putative succinate dehydrogenase flavoprotein subunit |
| 3.7 | RL4445 | sdhD | Succinate dehydrogenase hydrophobic membrane anchor protein | GI | 0.67 | 1.25 | GD | 0.67 | 4 | GD | 3.5.8 | Putative succinate dehydrogenase hydrophobic membrane anchor protein |
| 3.7 | RL4446 | sdhC | Succinate dehydrogenase cytochrome b556 subunit | GI | 0.46 | 1.83 | GD | 0.77 | 4.6 | GD | 3.5.8 | Putative succinate dehydrogenase cytochrome b556 subunit |
| 3.8 | RL2701 | fumC | Fumarate hydratase class II | GN | 0.94 | 18.56 | NE | 0.88 | 19.8 | NE | 3.5.8 | Putative fumarate hydratase class II |
| 3.8 | RL2703 | fumA | Fumarate hydratase class I | GN | 0.93 | 23.36 | NE | 0.96 | 20.81 | NE | 3.5.8 | Putative fumarate hydratase class I, aerobic |
| 3.9 | RL4439 | mdh | Malate dehydrogenase | GI | 0.25 | 1.33 | GD | 0.17 | 1 | GD | 3.5.8 | Putative malate dehydrogenase |
| 3.10 | RL0761 | - | Isocitrate lyase | GN | 0.91 | 17.29 | NE | 0.83 | 25.79 | NE | 3.3.5 | Putative isocitrate lyase |
| 3.11 | RL0054 | glcB | Malate synthase G | GN | 0.88 | 20 | NE | 1 | 24.06 | NE | 3.3.5 | Putative malate synthase |
| 3.12 | RL0407 | maeB | Malic enzyme | GN | 0.86 | 18.97 | NE | 0.89 | 24.74 | NE | 3.5.8 | Putative NADP-dependent malate dehydrogenase |
| 3.12 | RL2671 | maeB | Malic enzyme | GN | 0.83 | 31.47 | NE | 0.96 | 26.41 | NE | 3.5.8 | Putative NADP-dependent malic enzyme |
| 3.13 | RL0037 | pckA | Phosphoenolpyruvate carboxykinase | GN | 1 | 16.09 | NE | 0.91 | 21.55 | NE | 3.3.4 | Putative phosphoenolpyruvate carboxykinase |
| 3.14 | RL4638 | – | Pyruvate carboxylase | VGI | 0 | 0 | ES | 0.94 | 29.35 | NE | 3.5.8 | Putative pyruvate carboxylase |
| PENTOSE PHOSPHATE PATHWAY | ||||||||||||
| 4.1 | RL2807 | gnd | 6-phosphogluconate dehydrogenase | GN | 0.92 | 10.27 | NE | 0.92 | 12.27 | NE | 3.5.6 | Putative phosphogluconate dehydrogenase |
| 4.1 | RL3998 | gntZ | 6-phosphogluconate dehydrogenase | GN | 0.89 | 20.68 | NE | 1 | 21.89 | NE | 3.5.6 | Putative 6-phosphogluconate dehydrogenase,decarboxylating |
| 4.2 | RL2547 | rpiB | Ribose-5-phosphate isomerase B | GN | 0.57 | 25.75 | NE | 0.71 | 16 | NE | 3.3.9 | Putative ribose-5-phosphate isomerase B |
| 4.2 | RL2698 | rpiA | Ribose-5-phosphate isomerase A | GI | 0 | 0 | ES | 0.33 | 7 | ES | 3.3.9 | Putative ribose-5-phosphate isomerase A |
| 4.2 | pRL120210 | – | Ribose-5-phosphate isomerase B | PVGI | 0.83 | 5.2 | GD | 0.83 | 21.8 | NE | 3.3.9 | Putative ribose-5-phosphate isomerase B |
| 4.3 | RL2598 | rpe | Ribulose-phosphate 3-epimerase | VGI | 0.38 | 2.33 | GD | 0.63 | 9.2 | NE | 3.3.9 | Putative ribulose phosphate 3-epimerase |
| 4.3 | pRL120033 | – | D-allulose-6-phosphate 3-epimerase | PGN | 1 | 20.6 | NE | 1 | 21 | NE | 3.3.9 | Putative D-allulose-6-phosphate 3-epimerase |
| 4.4, 4.6 | RL2718 | – | Transketolase | GN | 1 | 16.23 | NE | 0.92 | 14.83 | NE | 3.3.15 | Putative transketolase, alpha subunit, terpenoid biosynth? |
| 4.4, 4.6 | RL2719 | – | Transketolase | GN | 0.93 | 17.31 | NE | 0.93 | 24.38 | NE | 3.3.15 | Putative transketolase, beta subunit, terpenoid biosynth? |
| 4.4, 4.6 | RL4006 | cbbT | Transketolase | GN | 0.67 | 8.38 | NE | 0.92 | 18.95 | NE | 3.5.1 | Putative transketolase |
| 4.4, 4.6 | pRL100453 | – | Transketolase | PGN | 0.86 | 17.16 | NE | 0.9 | 22.46 | NE | 3.5.6 | Putative transketolase |
| 4.5 | RL4203 | talB | Transaldolase B | GN | 0.56 | 4.2 | NE | 0.89 | 32.13 | NE | 3.3.9 | Putative transaldolase B |
Plasmid growth impaired genes
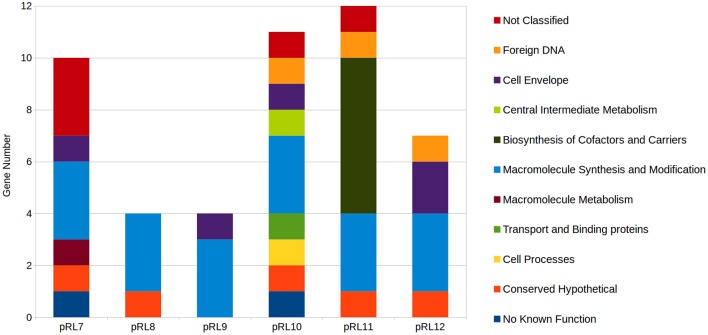
Mutation of genes encoded on RLV3841 megaplasmids that resulted in a growth impaired phenotype were assigned as plasmid growth impaired (PGI), while plasmid genes that when mutated impaired growth on VMM or TY uniquely were designated PVGI and PTGI respectively. Collectively, 48 genes distributed across the 6 megaplasmids were predicted to result in a PGI phenotype when mutated (Supplementary File 1). The 48 PGI genes included 11 Riley functional classes (Figure 5). All 6 plasmids were observed to have a set of 3 replication protein encoding genes categorized as PGI. Plasmid pRL11 was observed to carry the most PGI biosynthetic genes including 6 genes of an 8 gene cluster predicted to code for cobalamin biosynthesis (Figure 5).
Figure 5.
Distribution of plasmid growth impaired genes by Riley functional classification across RLV3841 megaplasmids. Plasmid borne genes observed to be growth defective or essential for growth on both TY and VMM-Mannitol were grouped. The contribution of growth defective or essential genes to megaplasmid stability, or growth of RLV3841 at an organism level, cannot be discriminated by INSeq analysis alone; therefore, the grouped growth defective and essential genes were assigned to plasmid growth impaired. Riley functional classifications were assigned to PGI genes for comparison of PGI profiles between megaplasmids.
Discussion
The CFG of RLV3841
Young et al. (2006) used phylogeny of conserved genes and GC% to describe a core and accessory genome within the RLV3841. The present study helps to improve the level of resolution for distinguishing between the RLV3841 core genes and accessory genes using functional genetic screening. We propose that RLV3841 has a CFG which can be defined as the core set of genes required for normal growth, independent of any specific environmental condition. In this study we approximate the CFG of RLV3841 by contrasting INSeq generated data sets from growth on complex peptide rich and minimally defined media with mannitol as the sole carbon source. Cross referencing VGI and TGI chromosomal genes identified an overlapping set of 491 genes that we putatively assigned to the CFG of RLV3841, as their loss of function resulted in a GI phenotype that appears to be independent of the growth media used. The number of CFG genes was less than that of both the TGI and VGI genes, which represented 563 and 661 genes respectively (Figure 1). This is to be expected if the CFG represents a central set of genes required for core cellular functions.
Defining a CFG in RLV3841 provides context for subsequent INSeq and classical genetic studies. For example, the described CFG for RLV3841 will help explain if a mutation resulting in a GI phenotype in a plant associated environment is the result of impairing some aspect of the RLV3841 CFG or is instead the result of a plant specific interaction. This is particularly important for genes encoding hypothetical proteins of unknown function. Summarizing the distribution of gene functions in the CFG identified 20 functional groupings (Figure 2). Five major categories accounted for over half of the total CFG (287 genes). These 5 functional categories included: macromolecule synthesis and modification (98 genes), energy and carbon metabolism (50 genes), ribosome constituents (48 genes), cell envelope (46 genes), and hypothetical proteins (50 genes) (Figure 2). Hypothetical proteins aside, these 4 categories logically compose the majority of the CFG as they represent the central genes required for the synthesis of the major cellular components, central conversion of carbon for generation of reductant and ATP, production and modification of protein synthesis machinery, and synthesis of the cell envelope.
The fact that genes encoding hypothetical proteins was one of the five major categories assigned to the CFG reinforces the broadly acknowledged observation among geneticists that the function of many genes involved in core cellular processes still remain uncharacterized. INSeq is a powerful technique that can help identify hypothetical proteins required for survival in specific growth conditions, and ultimately will advance our rate of discovery in this large and under studied category of genes. For example, in this study we identified a total of 103 hypothetical proteins observed to have an impaired growth phenotype under at least one growth condition; of which 19 were VGI, 20 were TGI, 14 were plasmid associated, and the remainder belonged to the CFG (Table 1).
Chromosomal VMM and TY specific growth impairment
Growth on minimal media requires the biosynthesis of several metabolites that can be scavenged from a complex growth media. Therefore, it was expected that there were more VGI genes compared to TGI genes, and that a substantial portion of the VGI genes are functionally classified for the biosynthesis of amino acids (30 genes), cofactors and carriers (25 genes), nucleotides (20 genes), and metabolic intermediates (15 genes) (Figure 2).
To further interrogate the VGI dataset, two previously uncharacterized genes, RL0920 a putative ATP-binding mrp family protein and RL3335 a putative lysophospholipase, were selected for targeted mutagenesis. A growth curve in liquid culture was used to characterize the generation time and growth response of the mutants. As expected, MA0920 and MA3335 were substantially growth impaired with increased generation times in VMM-mannitol. After 72 h of growth the mutant cultures were 1/3 the density of the wildtype in VMM-mannitol, whereas in TY they had achieved similar densities to the wildtype (Figure 3).
The TGI genes were composed of the smallest number of GI genes and the least functional complexity (Figure 2). Unlike the VGI genes, the general mechanism underlying the TGI genes does not hinge on metabolic biosynthesis, which is not surprising as complex media will contain many, if not all, metabolic intermediates required for growth. The largest functional categories observed in the TGI genes were hypothetical proteins (20 genes), cell envelope (11 genes), macromolecule synthesis and modification (9 genes), and transport and binding proteins (8 genes). Previous studies have identified several TGI genes, which collectively are implicated in outer membrane integrity or periplasmic function, suggesting that growth on complex media may require specific envelope traits (Gilbert et al., 2007; Vanderlinde et al., 2009, 2011; Foreman et al., 2010; Vanderlinde and Yost, 2012a,b). Additionally, we observed a putative cold shock response protein (RL2964), a LysR family transcriptional regulator, multiple transcriptional response regulators from predicted two-component systems (RL0036, RL1433, RL1729), a histidine kinase component of a two-component system (RL1382), as well as hypothetical proteins containing transmembrane domains (RL1526, RL2641), AAA–ATPase domain (RL2625), or N-terminal secretion signals (RL3761, RL1528, RL1618A, RL2086, RL4716) that were GI on TY medium (Supplementary File 1). These findings suggest that the TGI phenotype collectively may be centered around cell sensing and stress response at the cell envelope-environment interface.
Central carbon metabolism of mannitol
Using INSeq we were able to screen the 4 major conserved central carbon metabolic pathways for genes required for growth on mannitol (Figure 4; Table 2). As expected, disruption of the genes required for mannitol transport and conversion to F6P, along with glucose-6-phosphate isomerase (pgi) required for conversion of F6P into glucose-6P (Keele et al., 1969; Arias et al., 1979), resulted in a GI phenotype. Genes required for conversion of G6P into gluconate-6P (GN6P) through the ED pathway (overlapping the oxidative branch of the pentose phosphate pathway) resulted in a GI phenotype when mutated on both TY and VMM (Figure 4: reaction 1.1–2). The essential nature of these reactions may be due to several factors: (1) the NADPH generated during conversion (Spaans et al., 2015), (2) the possibly toxic accumulation of phosphorylated intermediates (Cerveñanský and Arias, 1984; Kadner et al., 1992), (3) the role of G6P in the biosynthesis of osmoprotectants (Barra et al., 2003), and (4) the need for carbon flux into the ED pathway for glycolytic growth (Arias et al., 1979; Glenn et al., 1984; Stowers, 1985). Conversion of GN6P into pyruvate through the ED pathway was determined to be VGI (Figure 4: reaction 1.3–4). Mutations in the upper EMP pathway, aside from pgi, were observed to be neutral, which is in agreement with previous work (Glenn et al., 1984). The lower EMP pathway (sometimes considered shared by the ED pathway), converts glyceraldehyde-3P (GA3P) into pyruvate, with mutants at all enzymatic steps appearing GI on mannitol, and at some steps on TY as well (Figure 4: reaction 2.5–9). The VGI nature of the lower EMP when grown on mannitol is possibly due to mutants being unable to metabolize GA3P produced from the ED pathway into the amino acid precursors glycerate-3P (G3P) or phosphoenolpyruvate (PEP), and as well catabolize GA3P into pyruvate (Finan et al., 1988) for use in the TCA cycle. The overall GI phenotype of mutants in the entire ED pathway, in contrast to the EMP pathway, suggests that it is the central pathway for glycolytic conversion of carbon into the central carbon intermediate pyruvate. This is in agreement with previous research indicating the ED pathway to be the preferred route of carbon metabolism for glycolytic growth of rhizobia and closely related genera (Stowers, 1985; Fuhrer et al., 2005; Geddes and Oresnik, 2014).
Conversion of pyruvate into the TCA cycle was observed to have 2 unique GI pathways (Figure 4: reaction 3.1–14). Conversion of pyruvate into the TCA through acetyl-CoA as an intermediate is a less direct route than the direct conversion of pyruvate into oxaloacetate, and mutants in pyruvate carboxylase (Figure 4: reaction 3.14) were observed to be more severally GI than in citrate synthase (Figure 4: reaction 3.2). Additionally, on TY media mutants in pyruvate dehydrogenase appeared GI, while mutants in pyruvate carboxylase were not. These findings are in agreement with anaplerotic production of oxaloacetate (OAA), via pyc-mediated fixation of CO2, being important for replenishing OAA pools under minimal growth conditions (Gokarn et al., 2001; Sirithanakorn et al., 2014).
When grown on mannitol as a sole carbon source, mutants in TCA cycle genes were all observed to be GI, aside from the conversion from fumarate to malate (Figure 4: reaction 3.2–3.9). Mutants in fumC and fumA (RL2701 and RL2703) were observed to be growth neutral, possibly due to functional redundancy between the two isozymes, which has been previously reported in Bradyrhizobium japonicum (Acuña et al., 1991). Future INSeq studies with prolonged exposure to selective pressure may identify which isoenzyme is dominant. Several TCA steps were observed to be GI on both VMM-mannitol and TY media, confirming that the TCA cycle is an important component of the CFG in RLV3841. Mutants in isocitrate dehydrogenase (icd) were both VGI and TGI (Figure 4: reaction 3.4). It has been previously shown that mutants in icd develop glutamate auxotrophy (McDermott and Kahn, 1992), which may explain the impaired growth phenotype on minimal medium, but does not explain growth impairment on peptide rich TY medium. Mutants in sucB and citM (sucA) were also observed to be growth impaired on both media (Figure 4: reaction 3.5). For growth on VMM-mannitol, the GI nature of these mutants is possibly due to increased α-ketoglutarate concentration due to the inability to metabolize into succinyl-CoA, and possibly perturbation of the GOGAT cycle via shunting of excess α-ketoglutarate (Bravo and Mora, 1988; Dunn, 1998).
The genes encoding enzymes for the glyoxalate by-pass, phosphoenolpyruvate carboxykinase and fructose bisphosphate aldolase, were observed to be GN on VMM-mannitol and TY media; suggesting that gluconeogenesis is not be required for growth in either condition (Kornberg, 1966; McKay et al., 1985; Stowers, 1985). This seems reasonable, as growth on mannitol is presumably glycolytic, therefore sugar conversion can be performed on metabolic intermediates generated during the breakdown of mannitol. And on TY media, many carbohydrates and polyols are likely already present in trace amounts, mitigating the need for gluconeogenesis.
Almost every gene involved in the non-oxidative branch of the PP pathways was observed to have a neutral impact on growth when mutated. The only two reactions in the PP pathway that were GI on mannitol were for the conversion of ribulose-5P into ribose-5P, or alternatively xylulose-5P (Figure 4: reaction 4.2 and 4.3). The mutation of ribose-5-phosphate isomerase A appearing GI is logical as ribose-5P is a precursor for 5-phosphoriboosyl-α-1-pyrophosphate, which is the branching point for flux of carbon in nucleotide, histidine, nicotinamide, and tryptophan biosynthesis (Kilstrup et al., 2005; Switzer, 2009). The conversion of GN6P into ribulose-5P however was observed to be GN. There are two possible explanations for mutants in this step appearing GN: (1) functional redundancy in isozymes Gnt and GntZ compensates for mutation of either (Figure 4: reaction 4.1), or (2) ribulose-5P can be replenished from xylulose-5P derived from either F6P and G3P being shunted into the PP pathway (Figure 4: reaction 4.4–6) or the phosphoketolase pathway (EC 4.1.2.9). In general, the interconnectedness of the PP pathway makes it difficult to study single gene knock-outs, as mutants may adapt to interrupted pathways by using alternative metabolic routes or isozymes (Geddes and Oresnik, 2014).
Plasmid growth impaired genes
There are unique opportunities and challenges for exploring plasmid biology when conducting INSeq experiments on bacterial species with genomes containing multiple large plasmids. Mutations that result in the loss of a plasmid from the accessory genome, due to impaired plasmid stability, will appear phenotypically identical to a GI mutant lost from the mutant pool due to a decreased growth rate. Therefore, it cannot be concluded directly from the INSeq data if a particular transposon insertion within a plasmid resulted in a GI phenotype, or instead compromised plasmid stability or replication. All plasmids contained a three gene cluster of rep genes that were observed to be essential (PGI) when mutated (Figure 5; Supplementary File 1), which in conjunction with the annotated function of these genes suggests the loss of these Tn insertion tags from the mutant population was due to impaired plasmid replication.
Beyond identifying putative rep genes, INSeq can be useful in identifying plasmid genes that provide the host cell with growth benefits under specific conditions. For example, pRL11 contains a putative 8 gene operon (pRL110625-32) predicted to be involved in cobalamin biosynthesis, that was severely PGI on VMM-mannitol, and moderately PGI on TY when mutated. Previous studies in R. etli identified similar growth phenotypes when a homologous cobalamin biosynthetic cluster on p42e was deleted (Landeta et al., 2011). Additionally, pRL120209 (putative tpiA) and pRL120210 (putative rpiB), which encode enzymes predicted to function in central carbon metabolism, were found to be PGI when grown on mannitol as a sole carbon source. Their chromosomal homologs RL2513 (tpiA) and RL2547 (rpiB) were both observed to be GN (Table 2). In a closely related R. leguminosarum bv. viciae strain VF39, the homologs of pRL120209 and pRL120210 are required for growth on erythritol as a sole carbon source (Yost et al., 2006). This suggests that pRL12 may carry genes important for the normal growth of RLV3841 under specific conditions, which is in agreement with previous studies that showed pRL12 cured strains of R. leguminosarum were unable to grow on minimal media (Hynes et al., 1989).
Technical considerations of INSeq in RLV3841
INSeq, like all high-throughput molecular techniques, is not without limitations. Genes with large regions of sequence duplication, or no mariner “TA” insertion sites, cannot be assayed using INSeq. These genes represent only 1.9% of the genome. However, the targeted library preparation method and robust statistical analysis afforded by the use of MmeI-adapted mariner transposon appears to outweigh its disadvantages. A sufficient saturation of neutral mariner insertion sites within the mutant community allows for confident identification of regions that lack insertions due to the negative selection resulting from a GI phenotype. In this and previous, work a sufficient level of neutral “TA” site saturation has been recovered to allow Bayesian methods of analysis, using a relatively modest amount of sequencing data when compared to other mariner INSeq studies. Considerations in inoculation density, the number of generations of growth during negative selection, and the ability to recover mutant populations needs to be carefully considered in order to ensure enough complexity is retained in the mutant pools post-selection to allow for statistical analysis.
Author contributions
BP and CY conceived and designed the research; BP and MA conducted the experiments; BP conducted the data analysis; BP and CY prepared and finalized the manuscript.
Funding
The presented research was conducted with support from a Discovery Grant awarded by the Natural Sciences and Engineering Council of Canada.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Barney Geddes for his helpful discussions and comments concerning RLV3841 metabolism, and Dr. Stephen Fitzgerald for his helpful criticisms of the draft manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01873/full#supplementary-material
References
- Acuña G., Ebeling S., Hennecke H. (1991). Cloning, sequencing, and mutational analysis of the Bradyrhizobium japonium fumC-like gene: evidence for the exsistence of two different fumarases. J. Gen. Microbiol. 137, 991–1000. 10.1099/00221287-137-4-991 [DOI] [PubMed] [Google Scholar]
- Arias A., Cervenansky C., Gardiol A., Martinez-Drets G. (1979). Phosphoglucose isomerase mutants of Rhizobium meliloti. J. Bacteriol. 137, 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquist L., Boinett C. J., Cain A. K. (2013). Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol. 10, 1161–1169. 10.4161/rna.24765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra L., Pica N., Gouffi K., Walker G. C., Blanco C., Trautwetter A. (2003). Glucose 6-phosphate dehydrogenase is required for sucrose and trehalose to be efficient osmoprotectants in Sinorhizobium meliloti. FEMS Microbiol. Lett. 229, 183–188. 10.1016/S0378-1097(03)00819-X [DOI] [PubMed] [Google Scholar]
- Bishop A. H., Rachwal P. A., Vaid A. (2014). Identification of genes required by Bacillus thuringiensis for survival in soil by transposon-directed insertion site sequencing. Curr. Microbiol. 68, 477–485. 10.1007/s00284-013-0502-7 [DOI] [PubMed] [Google Scholar]
- Bravo A., Mora J. (1988). Ammonium assimilation in Rhizobium phaseoli by the glutamine synthetase-glutamate synthase pathway. J. Bacteriol. 170, 980–984. 10.1128/jb.170.2.980-984.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel E. D., Gralnick J. A. (2012). Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol. Microbiol. 86, 273–283. 10.1111/j.1365-2958.2012.08196.x [DOI] [PubMed] [Google Scholar]
- Byrne R. T., Chen S. H., Wood E. A., Cabot E. L., Cox M. M. (2014). Escherichia coli genes and pathways involved in surviving extreme exposure to ionizing radiation. J. Bacteriol. 196, 3534–3545. 10.1128/JB.01589-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel E., Zomer A. L., Nussbaumer T., Bole C., Izac B., Frapy E., et al. (2016). Comprehensive identification of Meningococcal genes and small noncoding RNAs required for host cell colonization. MBio 7, e01173–e01116. 10.1128/mbio.01173-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveñanský C., Arias A. (1984). Glucose-6-phosphate dehydrogenase deficiency in pleiotropic carbohydrate-negative mutant strains of Rhizobium meliloti. J. Bacteriol. 160, 1027–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus M. A., Ioerger T. R. (2013). A hidden Markov model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14:303. 10.1186/1471-2105-14-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong T. G., Ho B. T., Yoder-Himes D. R., Mekalanos J. J. (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628. 10.1073/pnas.1222783110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Luckow M. A. (2003). The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 131, 900–910. 10.1104/pp.102.018150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn M. F. (1998). Tricarboxylic acid cycle and anaplerotic enzymes in rhizobia. FEMS Microbiol. Rev. 22, 105–123. 10.1111/j.1574-6976.1998.tb00363.x [DOI] [PubMed] [Google Scholar]
- Finan T. M., Oresnik I., Bottacin A. (1988). Mutants of Rhizobium meliloti defective in succinate metabolism. J. Bacteriol. 170, 3396–3403. 10.1128/jb.170.8.3396-3403.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman D. L., Vanderlinde E. M., Bay D. C., Yost C. K. (2010). Characterization of a gene family of outer membrane proteins (ropB) in Rhizobium leguminosarum bv. viciae VF39SM and the role of the sensor kinase ChvG in their regulation. J. Bacteriol. 192, 975–983. 10.1128/JB.01140-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T., Fischer E., Sauer U. (2005). Experimental identification and quantification of glucose metabolism in seven bacterial species. Society 187, 1581–1590. 10.1128/jb.187.5.1581-1590.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Shendure J., Manoil C. (2011). Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio 2:e00315. 10.1128/mBio.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski J. D., Wong S. M. S., Giannoukos G., Ward D. V., Akerley B. J. (2009). Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U.S.A. 106, 16422–16427. 10.1073/pnas.0906627106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes B. A., Oresnik I. J. (2014). Physiology, genetics, and biochemistry of carbon metabolism in the alphaproteobacterium Sinorhizobium meliloti. Can. J. Microbiol. 60, 491–507. 10.1139/cjm-2014-0306 [DOI] [PubMed] [Google Scholar]
- Gilbert K. B., Vanderlinde E. M., Yost C. K. (2007). Mutagenesis of the carboxy terminal protease CtpA decreases desiccation tolerance in Rhizobium leguminosarum. FEMS Microbiol. Lett. 272, 65–74. 10.1111/j.1574-6968.2007.00735.x [DOI] [PubMed] [Google Scholar]
- Glenn A. R., McKay I. A., Arwas R., Dilworth M. J. (1984). Sugar metabolism and the symbiotic properties of carbohydrate mutants of Rhizobium leguminosarum. Microbiology 130, 239–245. 10.1099/00221287-130-2-239 [DOI] [Google Scholar]
- Gokarn R. R., Evans J. D., Walker J. R., Martin S. A., Eiteman M. A., Altman E. (2001). The physiological effects and metabolic alterations caused by expression of Rhizobium etli pyruvate carboxylase in Escherichia coli. Appl. Microbiol. Biotechnol. 56, 188–195. 10.1007/s002530100661 [DOI] [PubMed] [Google Scholar]
- Goodman A. L., McNulty N. P., Zhao Y., Leip D., Mitra R. D., Lozupone C. A., et al. (2009). Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289. 10.1016/j.chom.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A. L., Wu M., Gordon J. I. (2011). Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat. Protoc. 6, 1969–1980. 10.1038/nprot.2011.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011). High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M. G., Yoder-Himes D. R., Warawa J. M. (2015). Comprehensive identification of virulence factors required for respiratory melioidosis using Tn-seq mutagenesis. Front. Cell. Infect. Microbiol. 5:78. 10.3389/fcimb.2015.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooven T. A., Catomeris A. J., Akabas L. H., Randis T. M., Maskell D. J., Peters S. E., et al. (2016). The essential genome of Streptococcus agalactiae. BMC Genomics 17:406. 10.1186/s12864-016-2741-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes M. F., Quandt J., O'Connell M. P., Pühler A. (1989). Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene 78, 111–120. 10.1016/0378-1119(89)90319-3 [DOI] [PubMed] [Google Scholar]
- Johnson J. G., Livny J., DiRita V. J. (2014). High-throughput sequencing of Campylobacter jejuni insertion mutant libraries reveals mapA as a fitness factor for chicken colonization. J. Bacteriol. 196, 1958–1967. 10.1128/JB.01395-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston A. W. B., Beringer J. E. (1975). Identification of Rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 87, 343–350. 10.1099/00221287-87-2-343 [DOI] [PubMed] [Google Scholar]
- Kadner R. J., Murphy G. P., Stephens C. M. (1992). Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J. Gen. Microbiol. 138, 2007–2014. 10.1099/00221287-138-10-2007 [DOI] [PubMed] [Google Scholar]
- Kamp H. D., Patimalla-Dipali B., Lazinski D. W., Wallace-Gadsden F., Camilli A. (2013). Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog. 9:e1003800. 10.1371/journal.ppat.1003800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B. B., Jr., Hamilton P. B., Elkan G. H. (1969). Glucose catabolism in Rhizobium japonicum. J. Bacteriol. 97, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatiwara A., Jiang T., Sung S. S., Dawoud T. (2012). Genome scanning for conditionally essential genes in Salmonella enterica serotype Typhimurium. Appl. Environ. Microbiol. 78, 3098–3107. 10.1128/aem.06865-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilstrup M., Hammer K., Ruhdal Jensen P., Martinussen J. (2005). Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 29, 555–590. 10.1016/j.fmrre.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Kornberg H. L. (1966). The role and control of the glyoxylate cycle in Escherichia coli. Biochem. J. 99, 1–11. 10.1042/bj0990001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl J. V., Price M. N., Ray J., Wetmore K. M., Esquivel Z., Kazakov A. E., et al. (2014). Functional genomics with a comprehensive library of transposon mutants for the sulfate-reducing bacterium Desulfovibrio alaskensis G20. MBio 5, 1–13. 10.1128/mBio.01041-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeta C., Dávalos A., Cevallos M. Á., Geiger O., Brom S., Romero D. (2011). Plasmids with a chromosome-like role in rhizobia. J. Bacteriol. 193, 1317–1326. 10.1128/JB.01184-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge G. C., Phan M.-D., Turner D. J., Perkins T. T., Parts L., Haase J., et al. (2009). Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19, 2308–2316. 10.1101/gr.097097.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y., Belew A. T., Valdes K. M., Islam E., Curry P., Tettelin H., et al. (2015). Essential genes in the core genome of the human pathogen Streptococcus pyogens. Sci. Rep. 5:9838. 10.1038/srep09838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. A., Gallagher L. A., Thongdee M., Staudinger B. J., Lippman S., Singh P. K., et al. (2015). General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 5189–5194. 10.1073/pnas.1422186112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott T. R., Kahn M. L. (1992). Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174, 4790–4797. 10.1128/jb.174.14.4790-4797.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay I. A., Glenn A. R., Dilworth M. J. (1985). Gluconeogenesis in Rhizobium leguminosarum MNF3841. Microbiology 131, 2067–2073. 10.1099/00221287-131-8-2067 [DOI] [Google Scholar]
- Meeske A. J., Rodrigues C. D. A., Bradly J., Lim H. C., Bernhardt T. G., Rudner D. Z. (2015). High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLoS Biol. 14:e1002341. 10.1371/journal.pbio.1002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule M. G., Spink N., Willcocks S., Lim J., Guerra-Assunção J. A., Cia F., et al. (2015). Characterization of new virulence factors involved in the intracellular growth and survival of Burkholderia pseudomallei. Infect. Immun. 84, 701–710. 10.1128/IAI.01102-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. L., Kwon T., Marcotte E. M., Whiteley M. (2015). Intrinsic antimicrobial resistance determinants in the superbug Pseudomonas aeruginosa. MBio 6, e01603–e01615. 10.1128/mBio.01603-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G. E. D., Murray J. D., Poole P. S., Downie J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. 10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- Pechter K. B., Gallagher L., Pyles H., Manoil C. S., Harwood C. S. (2015). Essential genome of the metabolically versatile Alphaproteobacterium Rhodopseudomonas palestris. J. Bacteriol. 198, 867–876. 10.1128/JB.00771-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry B. J., Yost C. K. (2014). Construction of a mariner-based transposon vector for use in insertion sequence mutagenesis in selected members of the Rhizobiaceae. BMC Microbiol. 14:298. 10.1186/s12866-014-0298-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan M. D., Peters K. M., Sarkar S., Lukowski S. W., Allsopp L. P., Gomes Moriel D., et al. (2013). The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 9:e1003834. 10.1371/journal.pgen.1003834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt J., Hynes M. F. (1993). Versatile suicide vector which allows direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. 10.1016/0378-1119(93)90611-6 [DOI] [PubMed] [Google Scholar]
- Rubin B. E., Wetmore K. M., Price M. N., Diamon S., Shultzaberger R. K., Lowe L. C., et al. (2015)., The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. U.S.A. 112, E6634–E6643. 10.1073/pnas.1519220112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Lazinski D., Rowe S., Camilli A., Lewis K. (2015). Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. MBio 6, e00078–e00015. 10.1128/mBio.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirithanakorn C., Adina-Zada A., Wallace J. C., Jitrapakdee S., Attwood P. V. (2014). Mechanisms of inhibition of Rhizobium etli pyruvate carboxylase by L-aspartate. Biochemistry 54, 7100–7106. 10.1021/bi501113u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurnik D., Roux D., Aschard H., Cattoir V., Yoder-Himes D., Lory S., et al. (2013). A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 9:e1003582. 10.1371/journal.ppat.1003582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaans S. K., Weusthuis R. A., van der Oost J., Kengen S. W. M. (2015). NADPH-generating systems in bacteria and archaea. Front. Microbiol. 6:742. 10.3389/fmicb.2015.00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers M. D. (1985). Carbon metabolism in Rhizobium species. Annu. Rev. Microbiol. 39, 89–108. 10.1146/annurev.mi.39.100185.000513 [DOI] [PubMed] [Google Scholar]
- Switzer R. L. (2009). Discoveries in bacterial nucleotide metabolism. J. Biol. Chem. 284, 6585–6594. 10.1074/jbc.X800012200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T., Ran Q., Ostrer L., Khodursky A. (2016). De novo characterization of genes that contribute to high-level ciprofloxacin resistance in Echerichia coli. Antimicrob. Agents Chemother. 60, 6353–6355. 10.1128/AAC.00889-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy E. B., Lin T., Gao L., Lazinski D. W., Lundt M., Camilli A., et al. (2016). Global Tn-seq analysis of carbohydrate utilization and vertebrate infectivity of Borrelia burgdorferi. Mol. Microbiol. 101, 1003–1023. 10.1111/mmi.13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner K. H., Wessel A. K., Palmer G. C., Murray J. L., Whiteley M. (2015). Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 112, 4110–4115. 10.1073/pnas.1419677112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M., Poole P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. 10.1146/annurev-arplant-050312-120235 [DOI] [PubMed] [Google Scholar]
- van Opijnen T., Bodi K. L., Camilli A. (2009). Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–775. 10.1038/nmeth.1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T., Camilli A. (2013). Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442. 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinde E. M., Harrison J. J., Muszynsji A., Carlson R. W., Turner R. J., Yost C. K. (2010). Identification of a novel ABC transporter required for desiccation tolerance, and biofilm formation in Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol. Ecol. 71, 327–40. 10.1111/j.1574-6941.2009.00824.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinde E. M., Magnus S. A., Tambalo D. D., Koval S. F., Yost C. K. (2011). Mutation of a broadly conserved operon (RL3499-RL3502) from Rhizobium leguminosarum biovar viciae causes defects in cell morphology and envelope integrity. J. Bacteriol. 193, 2684–2694. 10.1128/JB.01456-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinde E. M., Muszynski A., Harrison J. J., Koval S. F., Foreman D. L., Ceri H., et al. (2009). Rhizobium leguminosarum biovar viciae 3841, deficient in 27-hydroxyoctacosanoate-modified lipopolysaccharide, is impaired in desiccation tolerance, biofilm formation and motility. Microbiology 155, 3055–3069. 10.1099/mic.0.025031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinde E. M., Yost C. K. (2012a). Genetic analysis reveals links between lipid A structure and expression of the outer membrane protein gene, ropB, in Rhizobium leguminosarum. FEMS Microbiol. Lett. 335, 130–139. 10.1111/j.1574-6968.2012.02645.x [DOI] [PubMed] [Google Scholar]
- Vanderlinde E. M., Yost C. K. (2012b). Mutation of the sensor kinase chvG in Rhizobium leguminosarum negatively impacts cellular metabolism, outer membrane stability, and symbiosis. J. Bacteriol. 194, 768–777. 10.1128/JB.06357-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen L. M., de Jonge M. I., Burghout P., Schraa K., Spagnuolo L., Mennens S., et al. (2014). Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PLoS ONE 9:e89541. 10.1371/journal.pone.0089541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J. M. (1970). A Manual for the Practical Study of Root-nodule Bacteria. IBP Handbook. Oxford: Blackwell; Scientific. [Google Scholar]
- Wang N., Ozer E. A., Mandel M. J., Hauser A. R., Hauser R. (2014). Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio 5, e01163–e01114. 10.1128/mBio.01163-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielbo J. (2012). Rhizobial communities in symbiosis with legumes: genetic diversity, competition and interactions with host plants. Cent. Eur. J. Biol. 7, 363–372. 10.2478/s11535-012-0032-5 [DOI] [Google Scholar]
- Yang H., Krumholz E. W., Brutinel E. D., Palani N. P., Sadowsky M. J., Odlyzko A. M., et al. (2014). Genome-scale metabolic network validation of Shewanella oneidensis using transposon insertion frequency analysis. PLoS Comput. Biol. 10:e1003848. 10.1371/journal.pcbi.1003848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost C. K., Rath A. M., Noel T. C., Hynes M. F. (2006). Characterization of genes involved in erythritol catabolism in Rhizobium leguminosarum bv. viciae. Microbiology 152, 2061–2074. 10.1099/mic.0.28938-0 [DOI] [PubMed] [Google Scholar]
- Young J. P., Crossman L. C., Johnston A. W. (2006). The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome 7, R34. 10.1186/gb-2006-7-4-r34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung M. C., Park D. M., Overton K. W., Blow M. J., Hoover C. A., Smit J., et al. (2015). Transposon mutagenesis paired with deep sequencing of Caulobacter crescentus under uranium stress reveals genes essential for detoxification and stress tolerance. J. Bacteriol. 197, 3160–3172. 10.1128/JB.00382-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.