Abstract
Photoacoustic (PA) imaging is a rapidly emerging biomedical imaging modality that is capable of visualizing cellular and molecular functions with high detection sensitivity and spatial resolution in deep tissue. Great efforts and progress have been made on the development of various PA imaging technologies with improved resolution and sensitivity over the past two decades. Various PA probes with high contrast have also been extensively developed, with many important biomedical applications. In comparison with chemical dyes and nanoparticles, genetically encoded probes offer easier labeling of defined cells within tissues or proteins of interest within a cell, have higher stability in vivo, and eliminate the need for delivery of exogenous substances. Genetically encoded probes have thus attracted increasing attention from researchers in engineering and biomedicine. In this review, we aim to provide an overview of the existing PA imaging technologies and genetically encoded PA probes, and describe further improvements in PA imaging techniques and the near-infrared photochromic protein BphP1, the most sensitive genetically encoded probe thus far, as well as the potential biomedical applications of BphP1-based PA imaging in vivo.
Keywords: Molecular Imaging, Photoacoustic Imaging, Photoacoustic Probe, Reporter Gene, Genetically Encoded Probe, Photoswitchable Protein.
1. Introduction
Non-invasive monitoring of physiological and pathological processes in deep tissue requires both new imaging techniques and advanced probes 1. One emerging method is photoacoustic (PA) tomography, an in vivo hybrid imaging technology that uniquely combines the absorption contrast of light with ultrasound resolution 2. PA imaging relies on the conversion of photon energy into ultrasound waves generated by the thermoelastic expansion of tissue following absorption of short laser pulses 3. The key advantages of PA imaging include 1) great imaging depth (up to 7 cm) in intact biological tissues 4, thereby breaking through the optical diffusion limit (~1 mm depth in the skin) 5; 2) highly scalable penetration and spatial resolution6, enabling multiscale imaging with a PA probe; 3) no speckle artifacts in PA images 7, in contrast to optical coherence tomography (OCT) and ultrasound imaging; 4) no need for ionizing radiation 6, reducing health hazard. Therefore, PA imaging has become a powerful tool for basic research in life sciences, preclinical applications and drug discovery 2, 8, 9.
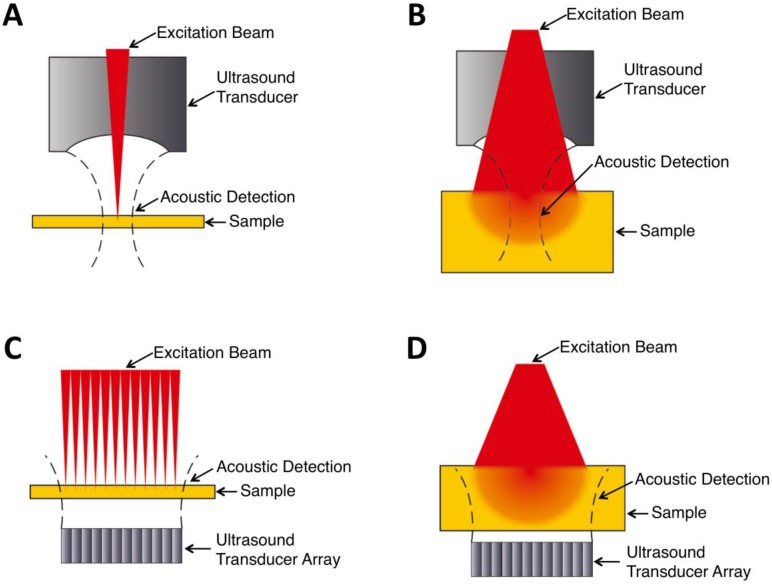
In general, there are four types of PA imaging systems 10: optical-resolution photoacoustic microscopy (OR-PAM), acoustic-resolution photoacoustic microscopy (AR-PAM), optical-resolution photoacoustic computed tomography (OR-PACT), and acoustic-resolution photoacoustic computed tomography (AR-PACT) (Figure 1). Depending on whether the laser beam or ultrasonic detection is more tightly focused, PA imaging can be divided into OR and AR with different lateral resolution. Based on the number of ultrasound detectors and whether a reconstruction algorithm is used, PA imaging can be classified into PAM and PACT. The sensitivity and resolution of these imaging systems vary depending on their optical excitation and acoustic detection configurations, as well as other design considerations of the system.
Figure 1.
Schematic cartoons showing principles of different PA imaging systems. (A) OR-PAM. (B) AR-PAM. (C) OR-PACT. (D) AR-PACT.
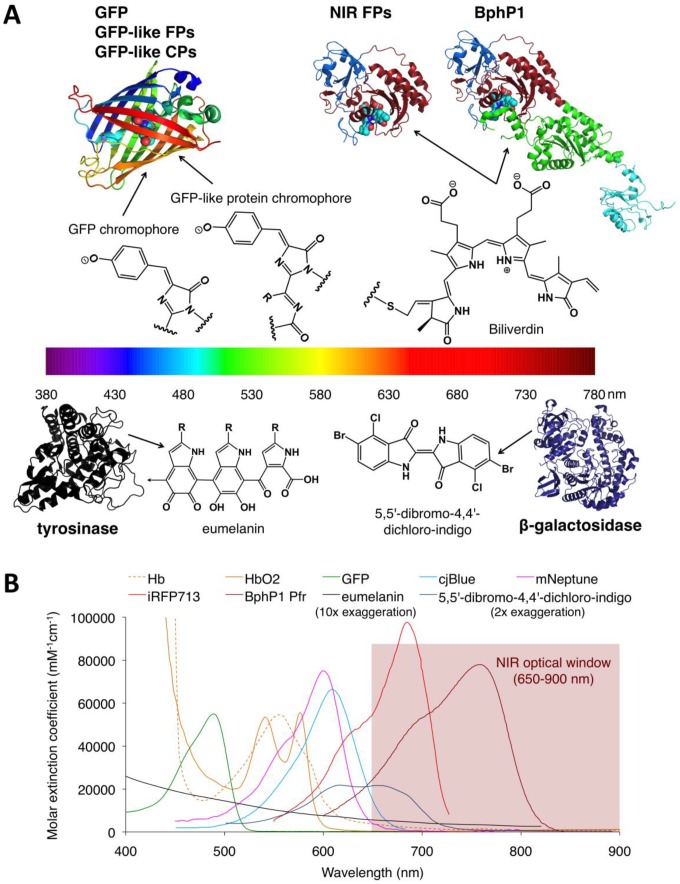
PA probes, also referred to PA contrast agents, are critical to determine the sensitivity and specificity of PA imaging 11, and have been used to track specific cell populations within an animal when used alone 12, 13 and visualize specific biochemical processes in vivo when engineered into biosensors 14, 15. An ideal PA probe for deep-tissue imaging should meet the following criteria: strong absorption in the near-infrared (NIR) optical window (650-900 nm), low fluorescence quantum yield (QY), high specificity, easy labeling of specific cells and proteins, easy development of functional biosensors, high photostability in vivo, high intracellular stability, no toxicity to mammalian cells, and no need for exogenous substances. Endogenous tissue chromophores, such as hemoglobin and eumelanin, which were widely used as cell markers in earlier studies 16-18, cannot label proteins within a cell, and thus can not be used in PA imaging of the dynamics of proteins of interest apart from hemoglobin in vivo. Moreover, hemoglobin cannot be produced in mammalian cells other than blood cells, limiting its usefulness to only blood vessel-related studies. Chemical dyes and nanoparticles overcome the labeling issues of endogenous chromophores to some extent by coupling cell- or protein-specific peptides or antibodies 19-21. However, these molecules are not very stable in living cells and are quickly cleared away from the body in vivo 22, resulting in a low effective concentration of probes in cells and difficulty in longitudinal imaging. By contrast, genetically encoded probes, particularly NIR probes, meet all the above criteria. More importantly, genetically encoded probes can be easily engineered into biosensors to sense various signaling molecules in cells, making them useful for tracking not only cell fates but also molecular functions in vivo.
In the present review, we will first describe two classes of PA imaging technologies with optical or acoustic resolution, including their imaging system setups and corresponding applications, then present and discuss genetically encoded PA probes that have been developed so far. Finally, we will give an outlook for further improvements in PA imaging techniques and genetically encoded probes, with a discussion of important applications made possible using the newly developed near-infrared photochromic protein BphP1-based differential PA imaging technique.
2. Photoacoustic Imaging Systems
2.1 Optical-resolution PA Systems
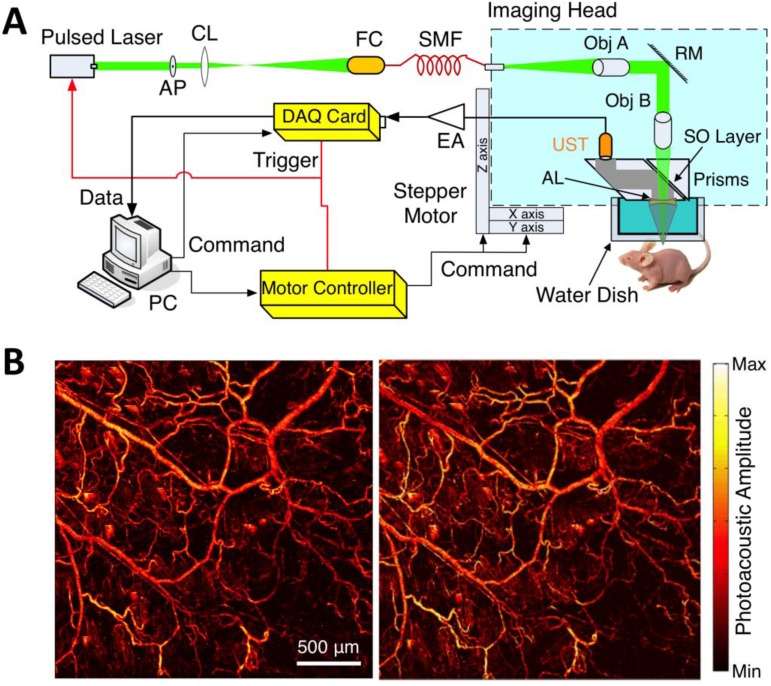
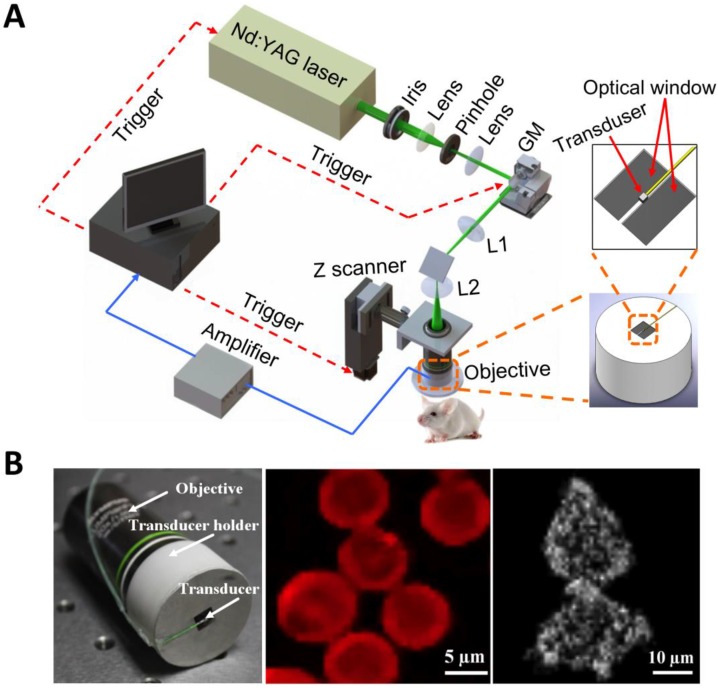
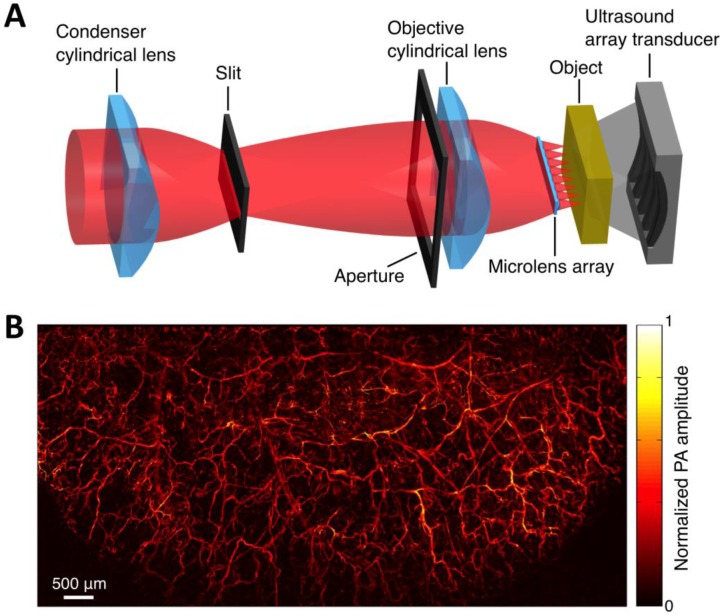
Within the optical ballistic propagation regime in biological tissue, tight optical focusing can be achieved by restricting the optical excitation to a much more confined area than that of ultrasound detection (Figure 1a, 1c) 23. Optical-resolution PA systems exploit this tight focusing to obtain a lateral resolution that can be as fine as the optical diffraction limit. The lateral resolution of optical-resolution PA systems can vary from several microns to submicrons (Table 1), depending on the numerical aperture of the optical objective lens 4. We have reported the development of blind-deconvolution OR-PAM to achieve both fine spatial resolution and extended depth of focus, and further utilized the system to visualize the mouse ear microvasculature in vivo (Figure 2) 24. By applying an optical objective lens with a much higher numerical aperture and a novel design for PA signal detection, a reflection-mode in vivo PA microscope with sub-wavelength lateral resolution was developed that is capable of imaging sub-cellular structure of red blood cells and melanoma cells (Figure 3) 17. Because OR-PAM typically uses a single-element ultrasonic transducer combined with mechanical or optical scanning to acquire the signal, its imaging speed is relatively slow (Table 1). To increase the imaging speed of OR-PAM, Song et al. developed a multifocal OR-PACT with a microlens array for optical illumination in conjunction with an ultrasound array for PA detection 25 (Figure 4A). With this new system, hemoglobin concentration in individual microvessels in vivo was monitored at a much higher speed than the single-focus OR-PAM (Figure 4B).
Table 1.
Comparison of different types of PA imaging systems
| OR-PAM | OR-PACT | AR-PAM | AR-PACT | |
|---|---|---|---|---|
| Lateral Resolution | several hundred nms to several μmsa | several μmsa | tens of μms to hundreds of μmsb | hundreds of μmsc |
| Axial Resolutiond | tens of μms | tens of μms | tens of μms to hundreds of μms | hundreds of μms |
| Imaging Depth | <1 mme | <1 mme | 1-20 mmf | 10-70 mmf |
| Imaging Speed | tens of minutes | < 1 minute | tens of minutes | < 1 minute |
| Biomedical Applications | Brain [31, 32] | Brain [33] | Tumor [26, 34] | Tumor [1, 30] |
| Tumor angiogenesis [35] | Blood vessel [25, 36] | SLNg [37] | SLNg [38, 39] | |
| Oxygen saturation [32, 40] | Brain [41, 42] | Brain [43, 44] | ||
| Oxygen metabolism [45] | Whole body [46] | Whole body [28, 47] | ||
| Hemodynamics [48] | ||||
| Subcellular imaging [17, 49] |
aOptical dependent, determined by NA (numerical aperture) of optical lens and optical wavelength
bAcoustic dependent, determined by NA of acoustic lens and acoustic central frequency
cAcoustic dependent, determined by acoustic central frequency
dAcoustic dependent, determined by acoustic frequency bandwidth
eOptical ballistic regime
fOptical diffusive regime
gSLN: Sentinel Lymph Node
Figure 2.
OR-PAM imaging of microvasculature in a mouse ear imaged in vivo. (A) Schematic diagram of the OR-PAM system. AP, aperture; CL, convex lens; FC, fiber coupler; SMF, single mode fiber; Obj, objective; RM, reflection mirror; UST ultrasonic transducer; AL, acoustic lens; SO, silicone oil; EA, electrical amplifier; DAQ, data acquisition; PC, personal computer. (B) PA images of the microvasculature in a mouse ear with OR-PAM. Left: raw data; right: after blind deconvolution. Figure adapted with permission from 24.
Figure 3.
Sub-wavelength resolution OR-PAM imaging of single cells ex vivo. (A) Schematic diagram of the reflection-mode sub-wavelength resolution PAM system. GM: galvanometer scanner; L1: scan lens; L2: tube lens. (B) Photograph of the PA signal detection probe (left); sub-wavelength resolution PA image of red blood cells (middle) and melanoma cells (right). Figure adapted with permission from 17.
Figure 4.
Multifocal OR-PACT imaging of microvasculature in a mouse ear. (A) Schematic diagram of the multifocal OR-PACT system. (B) In vivo PA MAP (maximum projection amplitude) image of the mouse ear microvasculature. Figure adapted with permission from 25.
2.2 Acoustic-resolution PA Systems
As light travels deeper into the tissue, tight optical focusing is no longer achievable due to multiple scattering events of photons 23. Correspondingly, for an acoustic-resolution PA system, weakly “focused” light is usually used to excite a significantly larger volume of the sample than that in an optical-resolution PA system (Figure 1b, 1d). However, as acoustic waves are much less scattered than photons in biological tissue, acoustic focusing is superior to optical focusing beyond a specific imaging depth and will yield a lateral resolution that is determined by the ultrasound focusing 4. The lateral resolution of acoustic-resolution PA systems is typically within hundreds of microns to tens of microns (Table 1), which is scalable with the centre frequency of the ultrasound transducer. Benefitting from the large imaging depth, the acoustic-resolution PA system is more commonly used in whole-body imaging applications (Table 1). Wang et al. applied AR-PAM system in conjunction with a novel targeted indocyanine green-doped nanoprobe to image breast cancer in mice (Figure 5) 26. For AR-PACT systems, three types of detection geometries have been reported: planar 27, circular 28 and spherical 29, 30. Wang et al. presented a three-dimensional volumetric PA imaging system built on a two-dimensional planar matrix array ultrasound probe and demonstrated the clinical potential of the system to identify sentinel lymph nodes for cancer staging purposes (Figure 6) 27. To perform whole-body imaging, Xia et al. reported a novel small animal whole-body PA imaging system with a confocal design of free-space ring-shaped light illumination and 512-element full-ring ultrasonic array signal detection (Figure 7) 28. Kruger et al. developed a dedicated human breast PA mammography system with a spherical detector capable of spiral scanning (Figure 8) 29, 30. As mentioned in the optical-resolution systems, the imaging speed of AR-PACT is much higher than that of AR-PAM because no mechanical raster scanning is needed (Table 2).
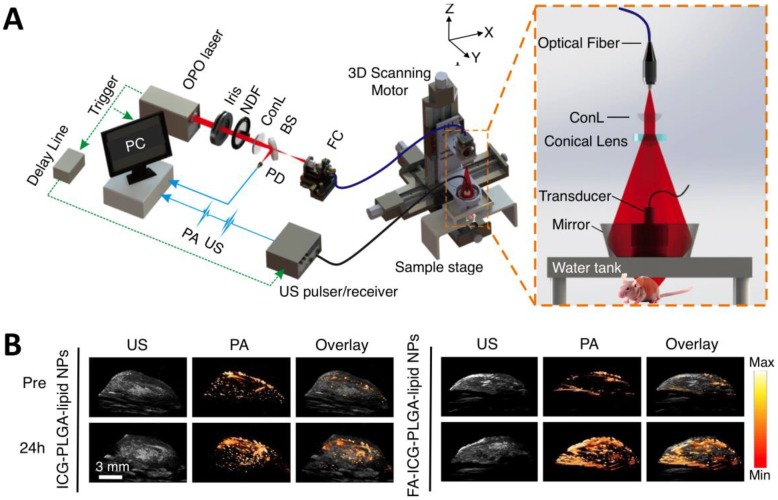
Figure 5.
AR-PAM imaging of breast cancer in vivo. (A) Schematic diagram of the AR-PAM system. OPO: optical parametric oscillator; NDF: neutral density filter; BS: beam splitter; PD: photodiode; FC: optical fiber coupler; PA: photoacoustic; US: ultrasound; ConL: convex lens. (B) Three-dimensional ultrasound (US), PA, and their overlay images of the tumor region before and 24 hours after the injection of non-targeted ICG-PLGA-lipid nanoparticles and targeted FA-ICG-PLGA-lipid nanoparticles, respectively. Figure adapted with permission from 26.
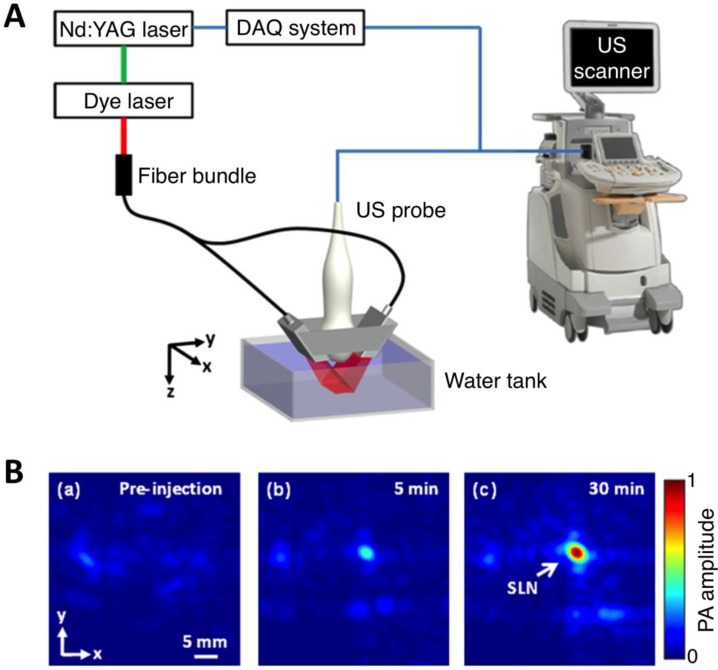
Figure 6.
Three-dimensional volumetric AR-PACT imaging of sentinel lymph node in vivo. (A) Schematic diagram of a volumetric PA imaging system with a matrix probe. DAQ: data acquisition; US: ultrasound. (B) PA images of the rat axillary region obtained (a) before methylene blue dye injection and (b) 5 min and (c) 30 min post injection, indicating the uptake of dye in the sentinel lymph node (SLN). Figure adapted with permission from 27.
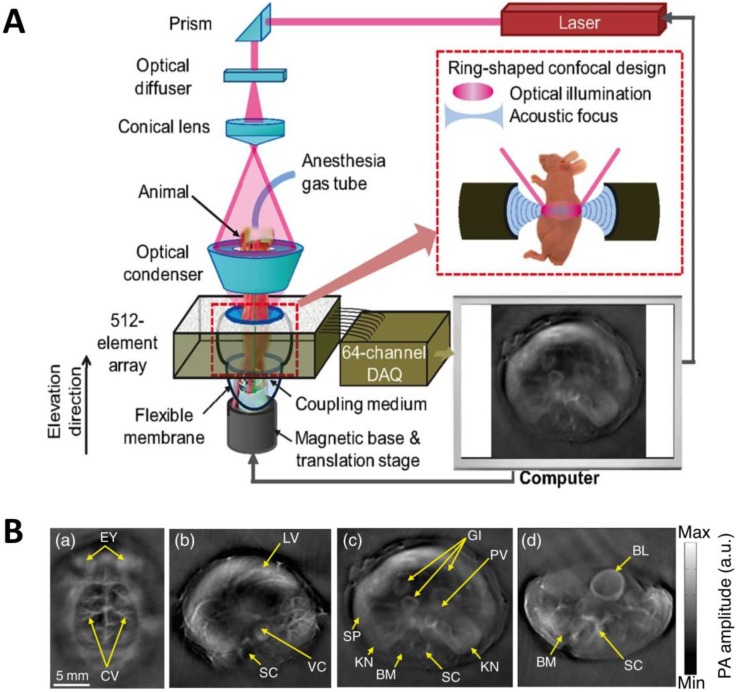
Figure 7.
Whole-body AR-PACT imaging of a mouse. (A) Schematic diagram of the whole-body PA imaging system. (B) In vivo PA images of a mouse acquired noninvasively at various anatomical locations: brain (a); liver (b); kidney (c); and bladder (d). BL, bladder; BM, backbone muscle; CV, cortical vessels; EY, eyes; GI, gastrointestinal tract; KN, kidney; LV, liver; PV, portal vein; SC, spinal cord; SP, spleen; VC, vena cava. Figure adapted with permission from 28.
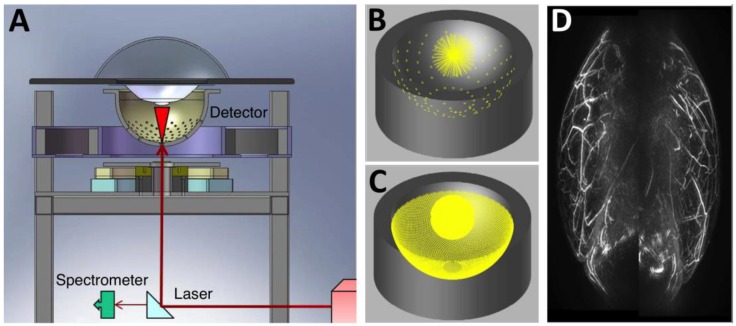
Figure 8.
Three-dimensional AR-PACT imaging of human breast. (A) Schematic diagram of a human breast PA mammography system, showing the laser beam entering the bottom of the detector bowl and illuminating the sample immersed in water and suspended above the bowl by a transparent tray. A beam splitter allows a portion of the laser beam to be monitored by a spectrometer for detection of laser power and wavelength. (B) The 128 radial projections captured by the detector geometry when data is collected for a single-bowl angular position. (C) The increased density of projections available as the bowl is rotated through 360 degrees. (D) Human breast PA maximum intensity projection image of a healthy volunteer (back-to-back image: left breast on right, right breast on left). Figure adapted with permission from 29, 30.
Table 2.
Properties of selected AFPs and CPs used in PA imaging.
| Protein | Absa | ε at peakb | ε at 650 nmc | Φd | Ref.e |
|---|---|---|---|---|---|
| EGFP | 488 | 53 | 0.2 | 0.6 | [69] |
| DsRed | 558 | 52 | 1.2 | 0.68 | [70] |
| E2-Crimson NFf, g | 583 | NDh | NDh | NDh | [67] |
| mCherry | 587 | 72 | 0.1 | 0.22 | [71] |
| Ultramarinei | 587 | 64 | 2.3 | 0.001 | [72] |
| dUltramarine2g | 587 | 82 | ~3 | <0.0001 | [73] |
| tdUltramarine2j, g | 587 | 203 | ~7.3 | <0.0001 | [73] |
| mKate | 588 | 45 | 0.2 | 0.33 | [74] |
| AQ143k | 595 | 90 | 9.7 | 0.04 | [62] |
| aeCP597i | 597 | 110 | 12 | NDh | [62] |
| mRaspberry | 598 | 86 | 1.9 | 0.15 | [75] |
| mNeptune | 600 | 67 | 2.2 | 0.2 | [76] |
| E2-Crimson | 605 | 59 | 4.6 | 0.12 | [77] |
| cjBluei | 610 | 67 | 3.6 | <0.0001 | [78] |
| cjBlue2g | 603 | 57 | <3.1 | <0.0001 | [73] |
| TagRFP657 | 611 | 34 | 4.6 | 0.1 | [79] |
aAbsorbance maximum in nm
bExtinction coefficient at peak in units of mM-1cm-1determined by alkali denaturation
cExtinction coefficient at 650 nm in units of mM-1cm-1 calculated as a product of ε at peak and excitation or absorption efficiency at 650 nm. Excitation or absorption efficiency is defined as the ratio of excitation or absorption at a given wavelength to that at peak.
dFluorescence quantum yield
eSource of data
fE2-Crimson's non-fluorescent variant
gNaturally occurring chromoproteins
hNot determined
iEngineered chromoproteins
jA tandem fusion of two copies of dUltramarine2
kaeCP597's bright variant
3. Genetically Encoded Probes for PA Imaging
3.1 Pigment-producing Enzymes
The metabolic enzymes β-galactosidase (encoded by lacZ) and tyrosinase can produce the light-absorbing pigments with broad absorption, 5,5'-dibromo-4,4'-dichloro-indigo (blue) and eumelanin (dark brown or black), respectively (Figure 9). Despite a low peak extinction coefficient (EC <10 mM-1cm-1), naturally occurring pigmented cells, such as melanoma cells, exhibited relatively strong PA signal because the intrinsic signal amplification of these enzymes produce very high concentrations of pigments in living cells 50, 51. Mammalian cells that do not naturally contain pigments become light absorber by introducing either β-galactosidase or tyrosinase alone and can be visualized in the presence of substrates using PA imaging 52, 53. The Wang Lab first established lacZ gene-expressing gliosarcoma tumor cells, imaged lacZ gene expression in tumor at a depth of up to 5 cm with ~1-mm lateral spatial resolution, and obtained well-separated PA images of lacZ and blood vessels 53-55, indicating the utility of β-galactosidase as a PA probe in non-pigmented mammalian cells. However, imaging lacZ expression requires the injection of exogenous X-gal, potentially complicating delivery and metabolism in vivo 53. Moreover, X-gal can cause skin irritation occasionally 53.
Figure 9.
Structures and absorbance spectra of genetically encoded PA probes. (A) Diversity of genetically encoded PA probes with distinct chromophores/pigments. The upper and lower parts of the spectrum color bar show the crystal structures of PA probes and the chemical structures of corresponding chromophores or pigments. The chromophores of the probes in the upper part are shown as spheres. In the BphP-based probes (NIR-FPs and BphP1), the PAS domain is shown in blue, the GAF domain in dark red, the PHY domain in green and the output transducing module (OTM) in cyan. Eumelanin is a polymer comprising numerous cross-linked 5,6-dihydroxyindole (DHI) and 5,6-dihydroxyindole-2-carboxylic acid (DHICA). The arrow in eumelanin denotes where the polymer continues. (B) Absorbance spectra of deoxygenated hemoglobin (Hb), oxygenated hemoglobin (HbO2), GFP, cjBlue (GFP-like CP), mNeptune (GFP-like FP), iRFP713 (NIR FP), BphP1 Pfr (BphP1 in Pfr state), eumelanin and 5,5'-dibromo-4,4'-dichloro-indigo. Eumelanin and 5,5'-dibromo-4,4'-dichloro-indigo were artificially augmented 10-fold and 2-fold, respectively, to render them visible in the figure. The spectra data for Hb, HbO2 and eumelanin are from http://omlc.ogi.edu, GFP from http://www.tsienlab.ucsd.edu/Documents.htm, cjBlue and iRFP713 and 5,5'-dibromo-4,4'-dichloro-indigo from 62, 63 and 53, respectively.
In contrast to β-galactosidase, tyrosinase, first found in melanocytes 56, utilizes endogenous and ubiquitous tyrosine in cells as a substrate to produce eumelanin. Thus, tyrosinase is more suited for in vivo PA imaging. Transfected cells with human tyrosinase appear darkly pigmented and can be spectrally resolved from hemoglobin in vivo using the OR-PAM system 52. However, with the PACT system, in vivo image contrast has been modest 12, 57, 58. To improve contrast, Jathoul et al. developed a new deep-tissue PA imaging system with a Fabry-Pérot-based optical ultrasound detector, and demonstrated three-dimensional images of tumors at depths up to 8 mm and obtained with a spatial resolution of ~100 μm (Figure 10) 13. Moreover, longitudinal visualization of the growth of human tyrosinase-expressing cells over long periods (>7 weeks) in vivo has been successfully achieved 13, providing a powerful tool for tracking cell fates over a long time period in vivo. Unfortunately, human tyrosinase does not function in bacterial, presumably owing to the difference in the cell membrane of mammalian cells and bacteria. melA, a bacterial tyrosinase homologue, has recently been reported as a suitable PA reporter gene for imaging E. coli 59, thus providing a complementary method to visualize the infection processes of pathogenic bacteria and monitor bacteria-based cancer treatment in vivo. It has been reported that high expression of tyrosinase in mammalian cells is correlated with cytotoxicity as transfected cells do not form melanosome where the cytotoxic semiquinone intermediates generated during eumelanin synthesis are sequestered in melanocytes 60, 61. Therefore, to minimize the cytotoxicity, the expression level of tyrosinase should be critically controlled in cells 51.
Figure 10.
In vivo PA imaging of tyrosinase-expressing tumor cells after subcutaneous injection into a nude mouse. (A) Volume-rendered image at day 0 post injection. (B-E) Horizontal (x-y, B and C) and vertical (y-z, D and E) maximum amplitude projection (MAP) images of the tumor cell injection area before (B or D) and after (C or E) cell injection. The dotted circles in (C) and (E) indicate the location of the injection. (F) Serial longitudinal in vivo MAP images acquired in the same mouse at different time points post innoculation (from 0 to 52 days). Figure adapted with permission from 13.
3.2 Fluorescent Proteins and Chromoproteins
3.2.1 Autofluorescent GFP and GFP-like Proteins
Autofluorescent proteins (AFPs) were first isolated from the jellyfish Aequorea Victoria (green fluorescent protein, GFP) and subsequently from other marine organisms (GFP-like FPs) 64. Interestingly, all AFPs share a common structure consisting of an eleven-stranded β-barrel containing an autocatalytically formed chromophore (Figure 9A). AFPs have been widely used as fluorescent markers to monitor protein dynamics in living cells or organisms because they do not require any cofactors or substrates to fluoresce and are non-toxic to mammalian cells 65. However, AFPs were first used in PA imaging only a few years ago. In 2009, Razansky et al. first demonstrated that tissue-specific expression of bright AFPs (GFP and mCherry) several millimeters deep within tissues can be detected using multispectral opto-acoustic tomography (MSOT) with higher spatial resolution than fluorescence (38 μm vs. 1 mm) (Figure 11) 66. However, due to relatively the high QYs (>0.2, Table 2) of GFP and mCherry and low absorption in the NIR region (EC at 650 nm ≤0.2, Table 2), bright AFP-based PA imaging was limited to relatively transparent zebrafish and small fruit fly pupae 66. Compared to GFP and red FPs, far-red FPs have red-shifted absorption spectra and lower QYs, and thus could offer stronger PA signals in vivo 67, 68. However, far-red FPs still exhibit low PA signals due to fluorescence emission and ground-state depopulation effects 67, 68. Moreover, most far-red FPs can be easily photobleached when exposed to ns excitation pulses typically used in PA imaging 67. Therefore, photostable and non-fluorescent proteins with strong absorption are required.
Figure 11.
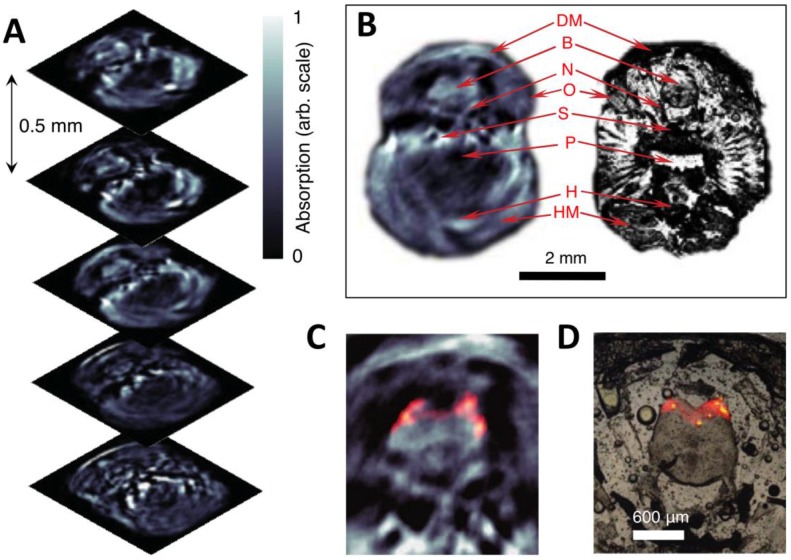
Three-dimensional in vivo imaging through the brain of an adult mCherry-expressing transgenic zebrafish. (A) Five transverse PA imaging slices through the hindbrain area of a living zebrafish. (B) Example of an imaged slice (left) and its corresponding histological section (right); DM, dorsal fin musculature; B, hindbrain; N, lateral line nerve; O, operculum; S, skull bones; P, pharynx; H, heart; HM, hypobranchial musculature. (C) Multispectral PA tomography image of the brain with mCherry expression shown in color; (D) Corresponding fluorescent histology of a dissected fish at the hindbrain level. Figure adapted with permission from 66.
3.2.2 Non-fluorescent GFP-like Chromoproteins
Among GFP-like proteins, some non-fluorescent proteins, called chromoproteins (CPs), absorb orange-red light with high ECs (>55 mM-1cm-1) and almost negligible QY (≤0.001, Table 2) and are thus better suited as PA probes for in vivo PA imaging than far-red FPs. Not surprisingly, compared to far-red FPs with low QYs, two naturally occurring CPs (aeCP597 and cjBlue) exhibit twofold stronger PA signals in vitro due to the lack of radiative relaxation, and are more resistant to repeated laser irradiation 67, presumably because of limited oxygen access to the chromophore caused by protein dimerization 80, 81. To further improve the performance of CPs in PA imaging, several improved cjBlue and Ultramarine variants with 2- to 4-fold increases in PA signals and enhanced photostability were developed using a custom PA screening system 73. Using a strategy similar to that employed in engineering fluorescence resonance energy transfer (FRET) biosensors, researchers developed CP-based PA sensors for caspase-3 and successfully visualized caspase-3 activity during staurosporine-induced cell apoptosis in cultured cells 73. This study was the first to report use of genetically encoded PA biosensors in PA imaging of molecular functions. However, despite these promising results, the low absorption (EC at 650 nm ≤12 mM-1cm-1, Table 2) of CPs in the NIR optical window, where the absorption and scattering of light by tissues are minimal, limits their application in highly light-scattering organisms, such as small animals.
3.2.3 Bacteriophytochrome-based NIR FPs
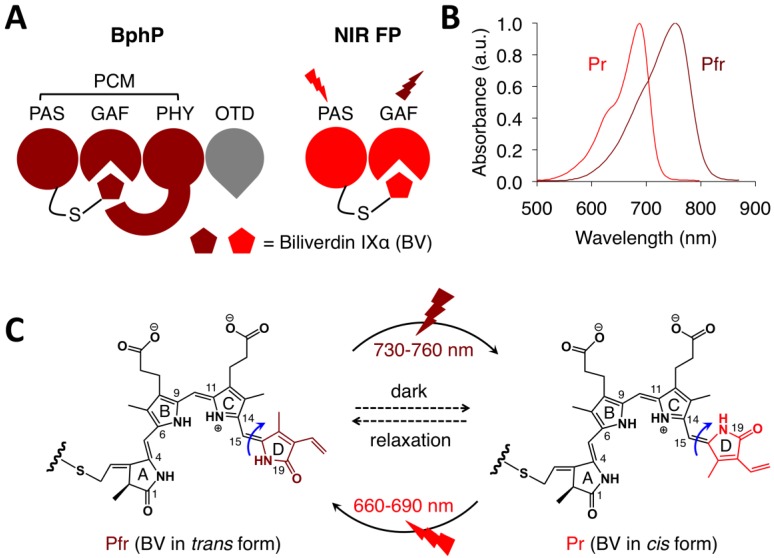
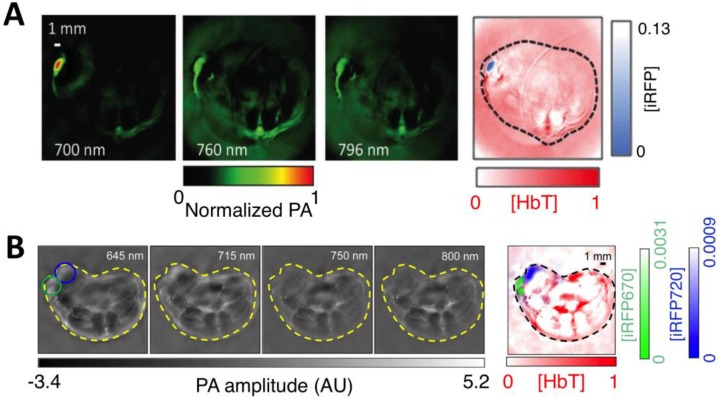
Bacteriophytochromes (BphPs) are bacterial photoreceptors that utilize biliverdin IXα (BV) available in mammalian cells as a chromophore 82. A typical BphP consists of a photosensory core module (PCM) possessing PAS, GAF and PHY domains and an output transducing domain (OTD) (Figure 12A). NIR BphPs, a large subset of BphPs, can convert from the far red-absorbing (Pfr, BV in trans form) state to the red-absorbing (Pr, BV in cis form) state upon ~750-nm illumination, and back upon ~690-nm illumination and vice versa (Figure 12B, C). NIR FPs can be achieved by truncating the PHY and effector domains of BphPs and by subsequently introducing specific mutations surrounding BV (Figure 12A and Figure 9A). In contrast to bright AFPs, most BphP-based NIR FPs have low quantum yields (<0.10) and high ECs (>90 mM-1cm-1) in the NIR optical window (Table 3), and are thus expected to be superior to their visible counterparts. In 2012, Filonov et al. first reported that NIR FP iRFP713 exhibits 2-fold higher PA signals than lysed oxygenated blood and far-red FPs under 680-nm excitation and imaged subcutaneous tumors overlaid by 4-mm-thick tissue at a 280-μm lateral resolution with the deep-tissue PAM imaging (Figure 13A) 1, suggesting that NIR FPs are promising probes for deep-tissue PA imaging. Imaging multiple biological events enables several cell populations to be tracked or the interactions between cells and their microenvironment to be monitored. Two new NIR FPs, iRFP670 and iRFP720, were recently developed, and two tumors labeled with iRFP670 and iRFP720 in the presence of blood vessels were simultaneously visualized with submillimeter resolution at a depth of up to 8 mm (Figure 13B) 83. Up to now, more than fifteen BphP-based NIR FPs with desired properties have been developed (Table 3) and can be extended to functional applications. For example, monomeric mIFP 84 might be used as a protein tag to monitor gene expression of proteins of interest in vivo using PA imaging.
Figure 12.
Structure and optical properties of bacteriophytochromes (BphPs). (A) Domain organization of BphP. In BphP, BV is secured in the chromophore-binding pocket of the GAF domain and binds covalently to the conserved cysteine residue in the N-terminus of the PAS domain. The chromophore binding module (PAS and GAF domains) can easily be engineered into NIR FPs. PCM: photosensory core module. OTD: output transducing domain. (B) Absorbance spectra of a typical NIR BphP. (C) Conversion between the Pfr state and the Pr state and vice versa is induced by far-red or red light, respectively, owing to trans-cis photoisomerization of the D-ring of the BV chromophore around the C15=C16 double bond (blue arrows). In the dark, the photoconverted state spontaneously reverts back to the ground state (dashed arrows).
Table 3.
Properties of BphP-based NIR FPs.
| NIR FP | Exa | Emb | ε at peakc | Φd | BphP | Ref.e |
|---|---|---|---|---|---|---|
| IFP1.4 | 684 | 708 | 92 | 0.077 | DrBphP | [86] |
| Wi-Phy | 701 | 719 | 93 | 0.047 | DrBphP | [87] |
| IFP1.4rev | 685 | 708 | 131 | 0.087 | DrBphP | [88] |
| IFP2.0 | 690 | 711 | 98 | 0.081 | DrBphP | [89] |
| iRFP713 | 690 | 713 | 98 | 0.063 | RpBphP2 | [63] |
| iRFP720 | 702 | 720 | 96 | 0.06 | RpBphP2 | [90] |
| iRFP713/V256Cf | 662 | 680 | 94 | 0.145 | RpBphP2 | [91] |
| iRFP682 | 663 | 682 | 90 | 0.113 | RpBphP6 | [90] |
| iRFP702 | 673 | 702 | 93 | 0.082 | RpBphP6 | [90] |
| iRFP670f | 643 | 670 | 114 | 0.111 | RpBphP6 | [90] |
| mIFP | 683 | 704 | 82 | 0.084 | BrBphP | [84] |
| iBlueberryf | 644 | 667 | 80 | 0.08 | BrBphP | [92] |
| GAF-FPg | 635 | 670 | 50 | 0.073 | RpBphP1 | [93] |
| BphP1-FP/C20Sf | 639 | 670 | 82 | 0.139 | RpBphP1 | [94] |
| AphB variants | ~695 | ~720 | ~80 | ~0.06 | AphB | [95] |
aExcitation maximum in nm
bEmission maximum in nm
cExtinction coefficient at peak in units of mM-1cm-1 calculated based on BV absorbance
dFluorescence quantum yield
eSource of data
fBV covalently binds to Cys256 in the GAF domain (the numbering is based on iRFP713)
gGAF domain only
Figure 13.
In vivo PA imaging of tumors expressing iRFP. (A) PA images of mammary gland tumor expressing iRFP713. The three images on the left were acquired at different wavelengths as indicated in the figure; the right-most image is a spectrally separated image with the iRFP713 signal in blue and the blood signal in red. (B) PA images of two different tumors labeled with iRFP670 and iRFP720. The four images on the left were acquired at different wavelengths as indicated in the figure; the right-most image is the spectrally separated image with the iRFP720 signal in blue, the iRFP670 signal in green, and the blood signal in red. Figure adapted with permission from 83.
Like other imaging modalities, the contrast of PA imaging in vivo is determined not only by probe' performance but also by tissue background 85. In PA imaging, the background mainly originates from the tissue absorber hemoglobin in blood through visible and NIR regions. Thus, even with high PA generation efficiency and strong absorption in the NIR region, BphP-based NIR FPs offer relatively low PA contrast over blood in vivo (~2), limiting visualization of a small number of probes or cells expressing probes in the vasculature. One way to suppress the background is using the difference of PA images corresponding to different locations with specially designed magnetic nanoparticles 85, which suggests that any absorption change in a given imaging area can significantly increase PA contrast. Therefore, a genetically encoded PA probe with optical absorption change should be able to dramatically enhance the imaging contrast in vivo.
3.3 Reversibly Photoswitchable Proteins
Reversibly photoswitchable proteins (rPSPs) can switch back and forth between two states with well-separated absorption peaks 96, making them useful for high-contrast PA imaging. In rPSP-based PA imaging, PA signals from a rPSP in two light-absorbing states under the illumination of same wavelength light are first recorded, and then pixel-wise subtraction of the images in low light-absorbing state from the images in high light-absorbing state is performed to generate a differential image. Up to now, two classes of rPSPs have been exploited to achieve high-contrast PA images 43, 97.
Two reversibly switchable green FPs, Dronpa and its fast-switching variant Dronpa-M159T (both are GFP-like AFPs) were first applied to PA imaging. By photoswitching the two Dronpa variants with a programmed optical illumination schedule, two FPs can be distinguished from each other and from the strong background signals with an increase in signal-to-noise ratio (SNR) by a factor of 5 using the temporally unmixed MSOT technique. However, as discussed above, Dronpa and Dronpa-M159T have very high fluorescence QY (~0.85) and low absorption in the NIR region and thus are not suitable for deep tissue PA imaging.
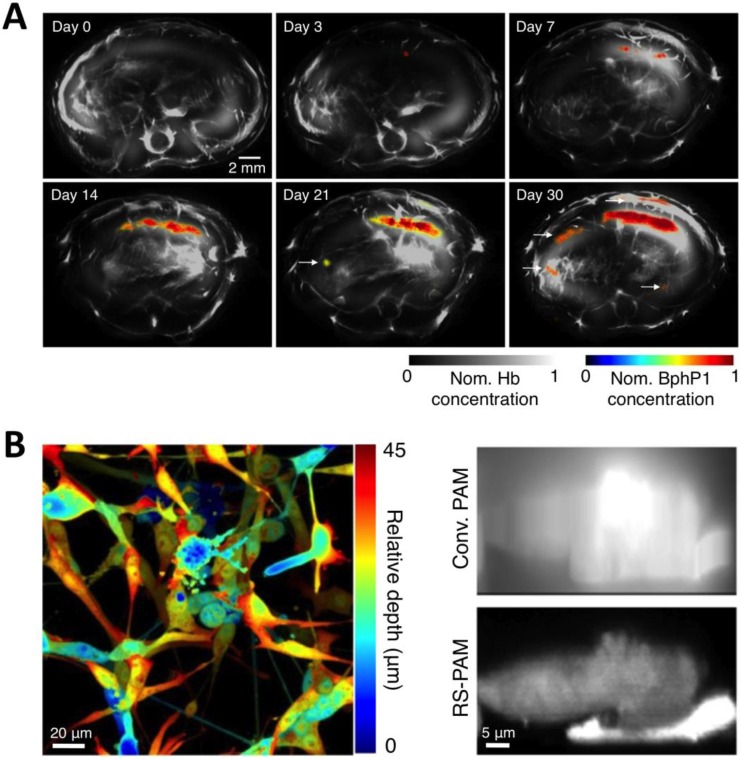
To overcome the limitations of Dronpa, very recently, Yao et al. reported a novel high-contrast imaging approach that combines deep tissue PA imaging with the NIR photochromic protein BphP1, the full-length RpBphP1 bacteriophytochrome from Rhodopseudomonas palustris (Figure 12A) 43. BphP1 adopts the Pfr state in the dark and can switch off (Pr state) and on (Pfr state) upon 780-nm and 630-nm light illumination (Figure 12B, C), respectively. Surprisingly, the contrast-to-noise ratio (CNR) for the differential image was 21 while that of the Pfr-state image was ~1 at a 10-mm depth in the scattering medium, indicating that background reduction with rPSPs can significantly increase the PA imaging contrast. To demonstrate the ability of BphP1 to visualize weak signals deep in tissues, the growth and metastasis of tumor expressing BphP1 in mouse livers were monitored over a long time period (30 days). Lateral resolution of ~100 μm was obtained with an average CNR of ~15 (Figure 14A). In addition, a sub-diffraction spatial resolution of 140 nm on individual cancer cells was achieved using RS-PAM (reversible photoswitching PAM), a two-fold improvement over that of traditional PAM (Figure 14B), enabling multiscale PA imaging using the BphP1-based single-wavelength differential method.
Figure 14.
Multiscale PA imaging of photoswitchable BphP1 with high contrast and resolution. (A) In vivo longitudinal PA monitoring of cancer metastasis in mouse liver for 30 days after the injection of tumor cells expressing BphP1 into the right liver lobe. Differential signals (in color) are overlaid on top of the structure signals from blood (in gray scale). The white arrows in the images for day 21 and 30 indicate secondary tumors due to metastasis. (B) Super-resolution RS-PAM imaging of U87 cells expressing BphP1. Left: depth-encoded RS-PAM image of a multilayered BphP1-expressing fixed U87 cells; Right: x-z cross-sectional images of two stacked U87 cells expressing BphP1, acquired with conventional PAM (Conv. PAM) and RS-PAM, demonstrating the substantially finer axial resolution of RS-PAM. Figure adapted with permission from 43.
The BphP1-based single-wavelength differential-imaging method has several advantages over the conventional method with non-photoswitchable probes. First, BphP1 is the most red-shifted of all reported BphPs, with an absorption peak at 760 nm, thus enabling deep-tissue PA imaging. Second, BphP1 resists photobleaching during photoconversion and imaging, enabling longitudinal imaging. Third, BphP1 photoconverts much more easily in vivo than photoswitchable AFPs, such as rsTagRFP because red and NIR light attenuate much less than blue and orange light do in vivo. In addition, even under daylight, a portion of BphP1 can undergo the Pr-Pfr transition. Fourth, the formation of the chromophore BV in BphP1 is oxygen independent, whereas GFP-like proteins require oxygen. Thus, BphP1 is more suitable for hypoxic tissues. Finally, compared to conventional multispectral methods, single-wavelength imaging at 780 nm greatly reduces the unknown influence of local light fluence. Therefore, the single-wavelength differential-imaging method combined with the photoswitchable BphP1 is the most promising approach for detection of weak signals from molecules and cells deep in living subjects.
4. Concluding Remarks and Outlook
Over the past two decades, different PA imaging technologies have been rapidly developed to circumvent the limited penetration depth and spatial resolution of conventional optical imaging in vivo. Deep-tissue PA imaging using genetically encoded reporter genes, particularly photoswitchable BphP1, offers a powerful tool for tracking cell fates and molecular functions in deep tissue within light-scattering living organisms. Although, unlike chemical dyes or nanoparticles, the use of BphP1 in humans remains challenging due to the requirement of introduction of exogenous genetic material 20 and potential safety issues 98, PA imaging with BphP1 provides unprecedented detection sensitivity and penetration depth and is thus useful for both basic research such as cancer, developmental biology and neurosciences and preclinical applications in small animals 10, 99, 100.
Currently, PA imaging has found broad applications both in preclinical laboratory research and clinical studies and trials. However, more work is needed to further mature this imaging technology and accelerate its clinical translation. 1) To fully explore the multi-scale capability of PA imaging, a novel system design that can achieve multiple optical resolutions by easily switching optical objectives, achieve multiple acoustic resolutions by easily switching ultrasonic transducers, and, finally, achieve full-scale PA imaging by integrating optical- and acoustic-resolution capabilities within a single PA system should be proposed. 2) Accurate quantitative PA imaging methodologies must be developed by considering both the optical and acoustic heterogeneities of biological tissues and by establishing a more sophisticated spectral unmixing model. 3) Multimodality imaging integrating PA with other imaging techniques such as ultrasound or fluorescence imaging should be further developed to provide complementary information on tissue properties in vivo 101. 4) PA systems with new imaging setups and more advanced lasers should be built to further improve imaging speed. For example, the current PACT system for BphP1 has relatively low temporal resolution: 11 min for 20 averaging switching cycles. However, many biological processes in living cells occur on a timescale of nanoseconds to seconds 102. The imaging speed can be improved using a laser with a higher repetition rate and a 512-channel data-acquisition system, to avoid data multiplexing and thus reduce the imaging time 43. 5) With the advances in laser technologies, and continuous enhancements of the performance of PA imaging systems, more clinical trials and studies are expected to fully demonstrate the potential of this technology.
Because BphP-based single-wavelength differential PA imaging offers the best performance among all genetically encoded PA probes, future studies should focus on its performance enhancement and development of BphP1-based PA biosensors. 1) Detection sensitivity can be improved by employing superior BphPs with low unconverted efficiency (uCE) and strong absorption in cells. As a differential imaging method, detection is based on the difference between the Pr state and Pfr state, and theoretically, the detection sensitivity is inversely proportional to uCE. The uCE of BphP1 is relatively high, reaching ~20% 43. In addition, BphP1-expressing cells have relatively weak absorption presumably due to the short lifetime or low BV-binding affinity in cells, or poor maturation and folding of BphP1 or a low EC. New BphPs with strong absorption and low uCE values may be obtained by directed protein evolution of BphP1 or by seeking better ones in nature. 2) Development of BphP1-based PA biosensors. Very few PA studies have sought to visualize molecular function in vivo due to the lack of biosensors for PA imaging 73, 103, 104. By contrast, numerous FP-based fluorescent biosensors are available and can be classified into three major groups with different design strategies: two FP-based FRET 105, and single FP-based circular permutation 105 and fluorescence complementation 106. BphP-based NIR FPs have recently been engineered as biosensors to sense protease activity (FRET and circular permutation) 107, 108 and protein-protein interactions (fluorescence complementation) in vivo 109. Since NIR FPs are derived from BphPs, researchers may develop BphP1-based PA biosensors by adopting similar strategies and visualize the spatio-temporal dynamics of biological molecules in vivo.
Until recently, most applications based on genetically encoded PA probes have been developed as proof-of-concepts. By utilizing BphP-based differential PA imaging combined with biological models, some long-standing biological questions may be addressed. For example, how do neurons connect with each other during development? 110, and how do key proteins in living cells interact with one another under physiological and pathological conditions in vivo? 111. In summary, the potential capabilities of the BphP1-based single-wavelength differential method are only beginning to be recognized and will be fully explored in the future.
Acknowledgments
The authors gratefully acknowledge the following funds: 1. NSFC grants 61405234 and 81430038 and 973 program grant 2015CB755500 to C. L.; 2. Shenzhen Science and Technology Innovation grant JCYJ20150521144320987 and the 'Hundred Talents' Program Award (Y64401) of the Chinese Academy of Sciences and the National Natural Science Foundation of China grant 1431670872 to J.C.; 3. National Natural Science Foundation of China (NSFC) grants 81522024 and 81427804, National Key Basic Research (973) Program of China grant 2014CB744503, International Science and Technology Cooperation Program of China (MOST) grant 2014DFG32800, and Guangdong Science and Technology grants 2013B091500090 and 2014B050505013 to L. S.; 4. NSFC grant 61475182 and Shenzhen Science and Technology Innovation grant JCYJ20150521144321005 to X. G.; NSFC grant 81471778 to Z. W. We apologize to those whose work could not be cited due to space limitations. Emily Wang (Harvard University) and Dr. Michael Lin (Stanford University) for critical reading of the manuscript, and Dr. Gang Han (University of Massachusetts Medical School) for his kind invitation and encouragement.
Abbreviations
- PA
photoacoustic
- NIR
near-infrared
- OCT
optical coherence tomography
- OR-PAM
optical-resolution photoacoustic microscopy
- AR-PAM
acoustic-resolution photoacoustic microscopy, OR-PACT: optical-resolution photoacoustic computed tomography
- AR-PACT
acoustic-resolution photoacoustic computed tomography
- QY
quantum yield
- EC
Extinction coefficient
- NA
numerical aperture
- Hb
deoxygenated hemoglobin
- HbO2
oxygenated hemoglobin
- GFP
green fluorescent protein
- AFP
autofluorescent proteins
- FP
fluorescent protein
- MSOT
multispectral opto-acoustic tomography
- CP
chromoprotein
- FRET
fluorescence resonance energy transfer
- BphP
bacteriophytochrome
- BV
biliverdin Ixα
- PCM
photosensory core module
- OTD
output transducing domain
- rPSP
reversibly photoswitchable protein
- SNR
signal-to-noise ratio
- CNR
contrast-to-noise ratio
- uCE
unconverted efficiency
- MAP
maximum projection amplitude
- RS-PAM
reversible photoswitching PAM
References
- 1.Filonov GS, Krumholz A, Xia J, Yao J, Wang LV, Verkhusha VV. Deep-tissue photoacoustic tomography of a genetically encoded near-infrared fluorescent probe. Angew Chem Int Ed Engl. 2012;51:1448–51. doi: 10.1002/anie.201107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C, Wang LV. Photoacoustic tomography and sensing in biomedicine. Phys Med Biol. 2009;54:R59–97. doi: 10.1088/0031-9155/54/19/R01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grootendorst DJ, Jose J, Wouters MW, van Boven H, Van der Hage J, Van Leeuwen TG. et al. First experiences of photoacoustic imaging for detection of melanoma metastases in resected human lymph nodes. Lasers Surg Med. 2012;44:541–9. doi: 10.1002/lsm.22058. [DOI] [PubMed] [Google Scholar]
- 4.Wang LV, Hu S. Photoacoustic tomography: in vivo imaging from organelles to organs. Science. 2012;335:1458–62. doi: 10.1126/science.1216210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao J, Song L, Wang LV. Photoacoustic microscopy: superdepth, superresolution, and superb contrast. IEEE Pulse. 2015;6:34–7. doi: 10.1109/MPUL.2015.2409100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang LV. Multiscale photoacoustic microscopy and computed tomography. Nat Photonics. 2009;3:503–9. doi: 10.1038/nphoton.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Z, Li L, Wang LV. On the speckle-free nature of photoacoustic tomography. Med Phys. 2009;36:4084–8. doi: 10.1118/1.3187231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Razansky D, Deliolanis NC, Vinegoni C, Ntziachristos V. Deep tissue optical and optoacoustic molecular imaging technologies for pre-clinical research and drug discovery. Curr Pharm Biotechnol. 2012;13:504–22. doi: 10.2174/138920112799436258. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Nie L, Chen X. Photoacoustic Molecular Imaging: From Multiscale Biomedical Applications Towards Early-Stage Theranostics. Trends Biotechnol. 2016;34:420–33. doi: 10.1016/j.tibtech.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao J, Wang LV. Photoacoustic Brain Imaging: from Microscopic to Macroscopic Scales. Neurophotonics; 2014. p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LV. Prospects of photoacoustic tomography. Med Phys. 2008;35:5758–67. doi: 10.1118/1.3013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stritzker J, Kirscher L, Scadeng M, Deliolanis NC, Morscher S, Symvoulidis P. et al. Vaccinia virus-mediated melanin production allows MR and optoacoustic deep tissue imaging and laser-induced thermotherapy of cancer. Proc Natl Acad Sci U S A. 2013;110:3316–20. doi: 10.1073/pnas.1216916110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jathoul AP, Laufer J, Ogunlade O, Treeby B, Cox B, Zhang E. et al. Deep in vivo photoacoustic imaging of mammalian tissues using a tyrosinase-based genetic reporter. Nature Photonics. 2015;9:8. [Google Scholar]
- 14.Dragulescu-Andrasi A, Kothapalli SR, Tikhomirov GA, Rao J, Gambhir SS. Activatable oligomerizable imaging agents for photoacoustic imaging of furin-like activity in living subjects. J Am Chem Soc. 2013;135:11015–22. doi: 10.1021/ja4010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Q, Cui H, Cai S, Yang X, Forrest ML. In vivo photoacoustic imaging of chemotherapy-induced apoptosis in squamous cell carcinoma using a near-infrared caspase-9 probe. J Biomed Opt. 2011;16:116026. doi: 10.1117/1.3650240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu S, Maslov K, Wang LV. Second-generation optical-resolution photoacoustic microscopy with improved sensitivity and speed. Opt Lett. 2011;36:1134–6. doi: 10.1364/OL.36.001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song W, Zheng W, Liu R, Lin R, Huang H, Gong X. et al. Reflection-mode in vivo photoacoustic microscopy with subwavelength lateral resolution. Biomed Opt Express. 2014;5:4235–41. doi: 10.1364/BOE.5.004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weight RM, Dale PS, Viator JA. Detection of circulating melanoma cells in human blood using photoacoustic flowmetry. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:106–9. doi: 10.1109/IEMBS.2009.5335145. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Stein EW, Ashkenazi S, Wang LV. Nanoparticles for photoacoustic imaging. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1:360–8. doi: 10.1002/wnan.42. [DOI] [PubMed] [Google Scholar]
- 20.Zackrisson S, van de Ven SM, Gambhir SS. Light in and sound out: emerging translational strategies for photoacoustic imaging. Cancer Res. 2014;74:979–1004. doi: 10.1158/0008-5472.CAN-13-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie L, Chen X. Structural and functional photoacoustic molecular tomography aided by emerging contrast agents. Chem Soc Rev. 2014;43:7132–70. doi: 10.1039/c4cs00086b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (Lond) 2008;3:703–17. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang LV, HU HI. Biomedical Optics: Principles and Imaging. Wiley-Interscience; 2007. [Google Scholar]
- 24.Chen J, Lin R, Wang H, Meng J, Zheng H, Song L. Blind-deconvolution optical-resolution photoacoustic microscopy in vivo. Opt Express. 2013;21:7316–27. doi: 10.1364/OE.21.007316. [DOI] [PubMed] [Google Scholar]
- 25.Song L, Maslov K, Wang LV. Multifocal optical-resolution photoacoustic microscopy in vivo. Opt Lett. 2011;36:1236–8. doi: 10.1364/OL.36.001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Liu C, Gong X, Hu D, Lin R, Sheng Z. et al. In vivo photoacoustic molecular imaging of breast carcinoma with folate receptor-targeted indocyanine green nanoprobes. Nanoscale. 2014;6:14270–9. doi: 10.1039/c4nr03949a. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Erpelding TN, Jankovic L, Guo Z, Robert JL, David G. et al. In vivo three-dimensional photoacoustic imaging based on a clinical matrix array ultrasound probe. J Biomed Opt. 2012;17:061208. doi: 10.1117/1.JBO.17.6.061208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia J, Chatni MR, Maslov K, Guo Z, Wang K, Anastasio M. et al. Whole-body ring-shaped confocal photoacoustic computed tomography of small animals in vivo. J Biomed Opt. 2012;17:050506. doi: 10.1117/1.JBO.17.5.050506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam RB, Kruger RA, Reinecke DR, DelRio S, Thornton MM, Picot PA. et al. Dynamic optical angiography of mouse anatomy using radial projections. Proc of SPIE. 2010;7564:7. [Google Scholar]
- 30.Kruger RA, Kuzmiak CM, Lam RB, Reinecke DR, Del Rio SP, Steed D. Dedicated 3D photoacoustic breast imaging. Med Phys. 2013;40:113301. doi: 10.1118/1.4824317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning B, Sun N, Cao R, Chen R, Kirk Shung K, Hossack JA. et al. Ultrasound-aided Multi-parametric Photoacoustic Microscopy of the Mouse Brain. Sci Rep. 2015;5:18775. doi: 10.1038/srep18775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao J, Wang L, Yang JM, Maslov KI, Wong TT, Li L. et al. High-speed label-free functional photoacoustic microscopy of mouse brain in action. Nat Methods. 2015;12:407–10. doi: 10.1038/nmeth.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia J, Li G, Wang L, Nasiriavanaki M, Maslov K, Engelbach JA. et al. Wide-field two-dimensional multifocal optical-resolution photoacoustic-computed microscopy. Opt Lett. 2013;38:5236–9. doi: 10.1364/OL.38.005236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim C, Cho EC, Chen J, Song KH, Au L, Favazza C. et al. In vivo molecular photoacoustic tomography of melanomas targeted by bioconjugated gold nanocages. ACS Nano. 2010;4:4559–64. doi: 10.1021/nn100736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin R, Chen J, Wang H, Yan M, Zheng W, Song L. Longitudinal label-free optical-resolution photoacoustic microscopy of tumor angiogenesis in vivo. Quant Imaging Med Surg. 2015;5:23–9. doi: 10.3978/j.issn.2223-4292.2014.11.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng J, Wang LV, Liang D, Song L. In vivo optical-resolution photoacoustic computed tomography with compressed sensing. Opt Lett. 2012;37:4573–5. doi: 10.1364/ol.37.004573. [DOI] [PubMed] [Google Scholar]
- 37.Kim C, Song KH, Gao F, Wang LV. Sentinel lymph nodes and lymphatic vessels: noninvasive dual-modality in vivo mapping by using indocyanine green in rats-volumetric spectroscopic photoacoustic imaging and planar fluorescence imaging. Radiology. 2010;255:442–50. doi: 10.1148/radiol.10090281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erpelding TN, Kim C, Pramanik M, Jankovic L, Maslov K, Guo Z. et al. Sentinel lymph nodes in the rat: noninvasive photoacoustic and US imaging with a clinical US system. Radiology. 2010;256:102–10. doi: 10.1148/radiol.10091772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song L, Kim C, Maslov K, Shung KK, Wang LV. High-speed dynamic 3D photoacoustic imaging of sentinel lymph node in a murine model using an ultrasound array. Med Phys. 2009;36:3724–9. doi: 10.1118/1.3168598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu S, Maslov K, Wang LV. Noninvasive label-free imaging of microhemodynamics by optical-resolution photoacoustic microscopy. Opt Express. 2009;17:7688–93. doi: 10.1364/oe.17.007688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nie L, Guo Z, Wang LV. Photoacoustic tomography of monkey brain using virtual point ultrasonic transducers. J Biomed Opt. 2011;16:076005. doi: 10.1117/1.3595842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kircher MF, de la Zerda A, Jokerst JV, Zavaleta CL, Kempen PJ, Mittra E. et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat Med. 2012;18:829–34. doi: 10.1038/nm.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao J, Kaberniuk AA, Li L, Shcherbakova DM, Zhang R, Wang L. et al. Multiscale photoacoustic tomography using reversibly switchable bacterial phytochrome as a near-infrared photochromic probe. Nat Methods. 2016;13:67–73. doi: 10.1038/nmeth.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasiriavanaki M, Xia J, Wan H, Bauer AQ, Culver JP, Wang LV. High-resolution photoacoustic tomography of resting-state functional connectivity in the mouse brain. Proc Natl Acad Sci U S A. 2014;111:21–6. doi: 10.1073/pnas.1311868111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Maslov K, Wang LV. Single-cell label-free photoacoustic flowoxigraphy in vivo. Proc Natl Acad Sci U S A. 2013;110:5759–64. doi: 10.1073/pnas.1215578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon M, Kim J, Kim C. Multiplane spectroscopic whole-body photoacoustic imaging of small animals in vivo. Med Biol Eng Comput. 2016;54:283–94. doi: 10.1007/s11517-014-1182-6. [DOI] [PubMed] [Google Scholar]
- 47.Brecht HP, Su R, Fronheiser M, Ermilov SA, Conjusteau A, Oraevsky AA. Whole-body three-dimensional optoacoustic tomography system for small animals. J Biomed Opt. 2009;14:064007. doi: 10.1117/1.3259361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao J, Maslov KI, Shi Y, Taber LA, Wang LV. In vivo photoacoustic imaging of transverse blood flow by using Doppler broadening of bandwidth. Opt Lett. 2010;35:1419–21. doi: 10.1364/OL.35.001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Maslov K, Wang LV. Subwavelength-resolution label-free photoacoustic microscopy of optical absorption in vivo. Opt Lett. 2010;35:3195–7. doi: 10.1364/OL.35.003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Brien CM, Rood KD, Bhattacharyya K, DeSouza T, Sengupta S, Gupta SK. et al. Capture of circulating tumor cells using photoacoustic flowmetry and two phase flow. J Biomed Opt. 2012;17:061221. doi: 10.1117/1.JBO.17.6.061221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paproski RJ, Forbrich AE, Wachowicz K, Hitt MM, Zemp RJ. Tyrosinase as a dual reporter gene for both photoacoustic and magnetic resonance imaging. BIOMEDICAL OPTICS EXPRESS. 2011;2:10. doi: 10.1364/BOE.2.000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krumholz A, Vanvickle-Chavez SJ, Yao J, Fleming TP, Gillanders WE, Wang LV. Photoacoustic microscopy of tyrosinase reporter gene in vivo. J Biomed Opt. 2011;16:080503. doi: 10.1117/1.3606568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, Zemp RJ, Lungu G, Stoica G, Wang LV. Photoacoustic imaging of lacZ gene expression in vivo. J Biomed Opt. 2007;12:020504. doi: 10.1117/1.2717531. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Zhang HF, Zemp RJ, Maslov K, Wang L. Simultaneous imaging of a lacZ-marked tumor and microvasculature morphology in vivo by dual-wavelength photoacoustic microscopy. J Innov Opt Health Sci. 2008;1:207–15. doi: 10.1142/S1793545808000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai X, Li L, Krumholz A, Guo Z, Erpelding TN, Zhang C. et al. Multi-scale molecular photoacoustic tomography of gene expression. PLoS One. 2012;7:e43999. doi: 10.1371/journal.pone.0043999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.del Marmol V, Beermann F. Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett. 1996;381:165–8. doi: 10.1016/0014-5793(96)00109-3. [DOI] [PubMed] [Google Scholar]
- 57.Qin C, Cheng K, Chen K, Hu X, Liu Y, Lan X. et al. Tyrosinase as a multifunctional reporter gene for Photoacoustic/MRI/PET triple modality molecular imaging. Sci Rep. 2013;3:1490. doi: 10.1038/srep01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paproski RJ, Heinmiller A, Wachowicz K, Zemp RJ. Multi-wavelength photoacoustic imaging of inducible tyrosinase reporter gene expression in xenograft tumors. Sci Rep. 2014;4:5329. doi: 10.1038/srep05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paproski RJ, Li Y, Barber Q, Lewis JD, Campbell RE, Zemp R. Validating tyrosinase homologue melA as a photoacoustic reporter gene for imaging Escherichia coli. J Biomed Opt. 2015;20:106008. doi: 10.1117/1.JBO.20.10.106008. [DOI] [PubMed] [Google Scholar]
- 60.van der Poel S, Wolthoorn J, van den Heuvel D, Egmond M, Groux-Degroote S, Neumann S. et al. Hyperacidification of trans-Golgi network and endo/lysosomes in melanocytes by glucosylceramide-dependent V-ATPase activity. Traffic. 2011;12:1634–47. doi: 10.1111/j.1600-0854.2011.01263.x. [DOI] [PubMed] [Google Scholar]
- 61.Weissleder R, Simonova M, Bogdanova A, Bredow S, Enochs WS, Bogdanov A Jr. MR imaging and scintigraphy of gene expression through melanin induction. Radiology. 1997;204:425–9. doi: 10.1148/radiology.204.2.9240530. [DOI] [PubMed] [Google Scholar]
- 62.Shkrob MA, Yanushevich YG, Chudakov DM, Gurskaya NG, Labas YA, Poponov SY. et al. Far-red fluorescent proteins evolved from a blue chromoprotein from Actinia equina. Biochem J. 2005;392:649–54. doi: 10.1042/BJ20051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–61. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guterl JK. et al. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol. 2007;25:443–5. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- 65.Mishin AS, Belousov VV, Solntsev KM, Lukyanov KA. Novel uses of fluorescent proteins. Curr Opin Chem Biol. 2015;27:1–9. doi: 10.1016/j.cbpa.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 66.Razansky D, Distel M, Vinegoni C, Ma R, Perrimon N, Koster RW. et al. Multispectral opto-acoustic tomography of deep-seated fluorescent proteins in vivo. Nature Photonics. 2009;3:6. [Google Scholar]
- 67.Laufer J, Jathoul A, Pule M, Beard P. In vitro characterization of genetically expressed absorbing proteins using photoacoustic spectroscopy. Biomed Opt Express. 2013;4:2477–90. doi: 10.1364/BOE.4.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deliolanis NC, Ale A, Morscher S, Burton NC, Schaefer K, Radrich K. et al. Deep-tissue reporter-gene imaging with fluorescence and optoacoustic tomography: a performance overview. Mol Imaging Biol. 2014;16:652–60. doi: 10.1007/s11307-014-0728-1. [DOI] [PubMed] [Google Scholar]
- 69.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73:2782–90. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–7. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 71.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 72.Pettikiriarachchi A, Gong L, Perugini MA, Devenish RJ, Prescott M. Ultramarine, a chromoprotein acceptor for Forster resonance energy transfer. PLoS One. 2012;7:e41028. doi: 10.1371/journal.pone.0041028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Y, Forbrich A, Wu J, Shao P, Campbell RE, Zemp R. Engineering Dark Chromoprotein Reporters for Photoacoustic Microscopy and FRET Imaging. Sci Rep. 2016;6:22129. doi: 10.1038/srep22129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA. et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–6. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 75.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–9. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin MZ, McKeown MR, Ng HL, Aguilera TA, Shaner NC, Campbell RE. et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–79. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shcherbo D, Shemiakina II, Ryabova AV, Luker KE, Schmidt BT, Souslova EA. et al. Near-infrared fluorescent proteins. Nat Methods. 2010;7:827–9. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan MC, Karasawa S, Mizuno H, Bosanac I, Ho D, Prive GG. et al. Structural characterization of a blue chromoprotein and its yellow mutant from the sea anemone Cnidopus japonicus. J Biol Chem. 2006;281:37813–9. doi: 10.1074/jbc.M606921200. [DOI] [PubMed] [Google Scholar]
- 79.Morozova KS, Piatkevich KD, Gould TJ, Zhang J, Bewersdorf J, Verkhusha VV. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J. 2010;99:L13–5. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chapagain PP, Regmi CK, Castillo W. Fluorescent protein barrel fluctuations and oxygen diffusion pathways in mCherry. J Chem Phys. 2011;135:235101. doi: 10.1063/1.3660197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW. et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–51. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem Soc Rev. 2013;42:3441–52. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krumholz A, Shcherbakova DM, Xia J, Wang LV, Verkhusha VV. Multicontrast photoacoustic in vivo imaging using near-infrared fluorescent proteins. Sci Rep. 2014;4:3939. doi: 10.1038/srep03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu D, Baird MA, Allen JR, Howe ES, Klassen MP, Reade A. et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Methods. 2015;12:763–5. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xia J, Pelivanov I, Wei C, Hu X, Gao X, O'Donnell M. Suppression of background signal in magnetomotive photoacoustic imaging of magnetic microspheres mimicking targeted cells. J Biomed Opt. 2012;17:061224. doi: 10.1117/1.JBO.17.6.061224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA. et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–7. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Auldridge ME, Satyshur KA, Anstrom DM, Forest KT. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J Biol Chem. 2012;287:7000–9. doi: 10.1074/jbc.M111.295121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bhattacharya S, Auldridge ME, Lehtivuori H, Ihalainen JA, Forest KT. Origins of fluorescence in evolved bacteriophytochromes. J Biol Chem. 2014;289:32144–52. doi: 10.1074/jbc.M114.589739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu D, Gustafson WC, Han C, Lafaye C, Noirclerc-Savoye M, Ge WP. et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat Commun. 2014;5:3626. doi: 10.1038/ncomms4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods. 2013;10:751–4. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stepanenko OV, Baloban M, Bublikov GS, Shcherbakova DM, Stepanenko OV, Turoverov KK. et al. Allosteric effects of chromophore interaction with dimeric near-infrared fluorescent proteins engineered from bacterial phytochromes. Sci Rep. 2016;6:18750. doi: 10.1038/srep18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu D, Dong Z, Gustafson WC, Ruiz-Gonzalez R, Signor L, Marzocca F. et al. Rational design of a monomeric and photostable far-red fluorescent protein for fluorescence imaging in vivo. Protein Sci. 2016;25:308–15. doi: 10.1002/pro.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rumyantsev KA, Shcherbakova DM, Zakharova NI, Emelyanov AV, Turoverov KK, Verkhusha VV. Minimal domain of bacterial phytochrome required for chromophore binding and fluorescence. Sci Rep. 2015;5:18348. doi: 10.1038/srep18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shcherbakova DM, Baloban M, Pletnev S, Malashkevich VN, Xiao H, Dauter Z. et al. Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins. Chem Biol. 2015;22:1540–51. doi: 10.1016/j.chembiol.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuan C, Li HZ, Tang K, Gartner W, Scheer H, Zhou M, Near infrared fluorescent biliproteins generated from bacteriophytochrome AphB of Nostoc sp. PCC 7120. Photochem Photobiol Sci; 2016. [DOI] [PubMed] [Google Scholar]
- 96.Adam V, Berardozzi R, Byrdin M, Bourgeois D. Phototransformable fluorescent proteins: Future challenges. Curr Opin Chem Biol. 2014;20:92–102. doi: 10.1016/j.cbpa.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 97.Stiel AC, Dean-Ben XL, Jiang Y, Ntziachristos V, Razansky D, Westmeyer GG. High-contrast imaging of reversibly switchable fluorescent proteins via temporally unmixed multispectral optoacoustic tomography. Opt Lett. 2015;40:367–70. doi: 10.1364/OL.40.000367. [DOI] [PubMed] [Google Scholar]
- 98.Amer MH. Gene therapy for cancer: present status and future perspective. Mol Cell Ther. 2014;2:27. doi: 10.1186/2052-8426-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bayer CL, Luke GP, Emelianov SY. Photoacoustic Imaging for Medical Diagnostics. Acoust Today. 2012;8:15–23. doi: 10.1121/1.4788648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laufer J, Norris F, Cleary J, Zhang E, Treeby B, Cox B. et al. In vivo photoacoustic imaging of mouse embryos. J Biomed Opt. 2012;17:061220. doi: 10.1117/1.JBO.17.6.061220. [DOI] [PubMed] [Google Scholar]
- 101.Liu Y, Kang N, Lv J, Zhou Z, Zhao Q, Ma L, Deep Photoacoustic/Luminescence/Magnetic Resonance Multimodal Imaging in Living Subjects Using High-Efficiency Upconversion Nanocomposites. Advanced Materials; 2016. [DOI] [PubMed] [Google Scholar]
- 102.Flores SC, Bernauer J, Shin S, Zhou R, Huang X. Multiscale modeling of macromolecular biosystems. Brief Bioinform. 2012;13:395–405. doi: 10.1093/bib/bbr077. [DOI] [PubMed] [Google Scholar]
- 103.Wang Y, Xia J, Wang LV. Deep-tissue photoacoustic tomography of Forster resonance energy transfer. J Biomed Opt. 2013;18:101316. doi: 10.1117/1.JBO.18.10.101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y, Wang LV. Forster resonance energy transfer photoacoustic microscopy. J Biomed Opt. 2012;17:086007. doi: 10.1117/1.JBO.17.8.086007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sample V, Mehta S, Zhang J. Genetically encoded molecular probes to visualize and perturb signaling dynamics in living biological systems. J Cell Sci. 2014;127:1151–60. doi: 10.1242/jcs.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kerppola TK. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev. 2009;38:2876–86. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.To TL, Piggott BJ, Makhijani K, Yu D, Jan YN, Shu X. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc Natl Acad Sci U S A. 2015;112:3338–43. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zlobovskaya OA, Sergeeva TF, Shirmanova MV, Dudenkova VV, Sharonov GV, Zagaynova EV. et al. Genetically encoded far-red fluorescent sensors for caspase-3 activity. Biotechniques. 2016;60:62–8. doi: 10.2144/000114377. [DOI] [PubMed] [Google Scholar]
- 109.Filonov GS, Verkhusha VV. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol. 2013;20:1078–86. doi: 10.1016/j.chembiol.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Devor A, Bandettini PA, Boas DA, Bower JM, Buxton RB, Cohen LB. et al. The challenge of connecting the dots in the B.R.A.I.N. Neuron. 2013;80:270–4. doi: 10.1016/j.neuron.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou M, Li Q, Wang R. Current Experimental Methods for Characterizing Protein-Protein Interactions. ChemMedChem; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]