Abstract
Genome-wide association studies have associated clusterin (CLU) variants with Alzheimer’s disease (AD). However the role of CLU on AD pathogenesis is not totally understood. We used CSF and plasma CLU levels as endophenotypes for genetic studies to understand the role of CLU in AD. CSF, but not plasma, CLU levels were significantly associated with AD status and CSF tau/Aβ ratio, and highly correlated with CSF apolipoprotein E (APOE) levels. Several loci showed almost genome-wide significant associations including LINC00917 (p=3.98×10−7) and interleukin 6 (IL6, p=9.94×10−6, in the entire dataset and in the APOE ε4- individuals p=7.40×10−8). Gene-ontology analyses suggest that CSF CLU levels may be associated with wound healing and immune response which supports previous functional studies that demonstrated an association between CLU and IL6. CLU may play a role in AD by influencing immune system changes that have been observed in AD or by disrupting healing after neurodegeneration.
Keywords: Alzheimer’s disease, APOE, clusterin, cerebrospinal fluid, gene ontology, immune response
1. Introduction
Clusterin (CLU), a multifunctional glycoprotein also known as apolipoprotein J, plays a role in several cellular processes including apoptosis, proliferation, and clearance of misfolded proteins (Kim and Choi, 2011,Wang, et al., 2014,Wyatt, et al., 2011). CLU is ubiquitous and is highly expressed in the brain by astrocytes (de Silva, et al., 1990). CLU is a secreted protein, but some isoforms have been discovered in the cytoplasm and nucleus (Kimura, et al., 1997,Leskov, et al., 2003).
CLU was first associated with Alzheimer’s disease (AD) in 1990 when it was found to be increased in the hippocampi of AD patients (May, et al., 1990). Two independent genome-wide association studies (GWAS) in 2009 found rs11136000, an intronic variant in CLU, associated with AD (Lambert et al, p=3.7×10−9 and Harold et al, p=8.5×10−10) (Harold, et al., 2009,Lambert, et al., 2009). In a meta-analysis of the two stage GWAS by Harold et al another single nucleotide polymorphism (SNP) in strong linkage disequilibrium (LD) with rs11136000 was also genome-wide significant, rs7982 (r2=0.95). This SNP has three alleles resulting in either a synonymous or a missense variant located in exon 5 of CLU (p=8×10−10) (Harold, et al., 2009). In a meta-analysis of >74,000 individuals in 2013, rs9331896, located in a CLU intron and also in strong LD with rs11136000 (r2=0.925) and rs7982 (r2=0.889), was also associated with AD (p=2.8×10−25) (Lambert, et al., 2013).
Increased cerebrospinal fluid (CSF) CLU levels combined with increased amyloid-beta (Aβ) levels were associated with increased entorhinal atrophy (Desikan, et al., 2014) and elevated CSF CLU levels have been observed in AD patients (Nilselid, et al., 2006). It has been suggested that CLU may play a role in AD, by interacting with apolipoprotein E (APOE) or alone, by affecting Aβ clearance and/or aggregation. APOE−/− and CLU−/− mice have similar Aβ levels, but in an APOE/CLU double knock-out mouse model Aβ was significantly increased in CSF and brain interstitial fluid, suggesting that the APOE and CLU effects on Aβ levels are additive and not completely independent (DeMattos, et al., 2004). CLU has high affinity for soluble Aβ (Ghiso, et al., 1993) and associates specifically with Aβ40 (Howlett, et al., 2013). CLU levels also appear to affect aggregation of Aβ (Oda, et al., 1995,Wilson, et al., 2008).
We hypothesized that genetic variants associated with CSF or plasma CLU levels may also play a role in AD. We performed a single-stage GWAS to look for SNPs associated with CLU levels and followed up with gene-ontology analyses to look at potential biological, cellular, and molecular categories that may be associated with plasma and CSF CLU levels. Differences in CSF CLU levels have been observed in AD patients versus controls and appear to affect Aβ levels. We hypothesized that CLU levels were also associated with CSF tau/Aβ ratio, a powerful predictor of cognitive decline that can be used to discriminate between vascular dementia and AD (de Jong, et al., 2006,Fagan, et al., 2007,Harari, et al., 2014). Animal studies suggest that CLU and APOE have synergistic effects on Aβ levels and both have been associated with AD. Given this biology we tested whether CLU and APOE levels in plasma or CSF are correlated.
2. Materials and Methods
2.1. Ethics Statement
The Institutional Review Board of all participating institutions approved the study. Written informed consent was obtained from participants or their family members.
2.2. Study Participants
There were CSF CLU, APOE, tau, and Aβ levels from 673 unrelated individuals (Table 1): 400 from the Charles F. and Joanne Knight Alzheimer’s Disease Research Center (Knight ADRC) (151 AD cases, 249 cognitively normal controls) and 273 from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (205 cases, 68 controls). There were 818 individuals with plasma analyte levels (Table 1): 312 from Knight ADRC (124 cases, 188 controls) and 506 from ADNI (434 cases, 72 controls). In the combined datasets 537 individuals had both CSF and plasma analyte levels (273 cases, 264 controls; Table 2). ADNI individuals were evaluated at the time of sample collection as described in the ADNI procedures manual (http:www.adni-info.org). Knight ADRC individuals were evaluated at the time of sample collection by Clinical Core personnel at Washington University; cases received a clinical diagnosis of AD in accordance with standard criteria and dementia severity was determined using the Clinical Dementia Rating (CDR) (Morris, 1993). Neuropsychological and clinical assessments and biological samples were collected for all participants as described previously (Cruchaga, et al., 2011,Cruchaga, et al., 2013,Shaw, et al., 2009,Toledo, et al., 2011).
Table 1.
Characteristics of plasma and CSF ADNI and Knight ADRC data
| CSF (n=673) | Plasma (n=818) | CSF & Plasma (n=537) | ||||
|---|---|---|---|---|---|---|
| ADRC | ADNI | ADRC | ADNI | ADRC | ADNI | |
| Samples | 400 | 273 | 312 | 506 | 305 | 232 |
| Age in years (mean±SD) |
73.03±6.77 | 78.05±6.71 | 73.22±7.03 | 78.30±7.40 | 73.25±6.89 | 77.84±6.97 |
| (range) | (49–91) | (60–93) | (49–91) | (55–95) | (49–91) | (60–93) |
| % AD cases | 27.50 | 69.60 | 26.92 | 86.36 | 27.54 | 81.47 |
| % female | 58.50 | 39.19 | 61.22 | 37.35 | 60.98 | 36.64 |
| % APOE E4 | 36.25 | 49.45 | 36.22 | 53.16 | 36.07 | 50.43 |
Table 2.
Characteristics of plasma and CSF data by disease status
| CSF (n=673) | Plasma (n=818) | CSF & Plasma (n=537) | ||||
|---|---|---|---|---|---|---|
| cases | controls | cases | controls | cases | controls | |
| Samples | 300 | 373 | 521 | 297 | 273 | 264 |
| Age in years (mean±SD) |
76.68±7.18 | 73.77±6.92 | 77.65±7.51 | 74.12±7.44 | 76.88±7.23 | 73.53±6.94 |
| (range) | (60–92) | (49–93) | (55–94) | (49–95) | (59–94) | (49–95) |
| % female | 40.33 | 58.98 | 38.58 | 60.27 | 39.93 | 61.36 |
| % APOE E4 | 59.00 | 27.61 | 58.93 | 25.25 | 58.97 | 25.00 |
2.3. Genotyping and Quality Control
Knight ADRC samples were genotyped with the Illumina 610 or Omniexpress chip and ADNI samples with the Illumina 610 chip. A call rate of ≤98% for SNPs and individuals was applied and SNPs not in Hardy-Weinberg equilibrium (p<1×10−6) or with MAF<0.02 were excluded. Quality control (QC) steps were applied to each genotyping array separately. X-chromosome SNPs were analyzed to verify gender identification. Duplicate and related individuals were found using pairwise genome-wide estimates of proportion identity-by-descent and eliminated from the analysis. We used EIGENSTRAT (Price, et al., 2006) to calculate principal component factors for each sample and confirm ethnicity. Imputation was performed as described before (Cruchaga, et al., 2013). Briefly the 1,000 genome data and BEAGLE v3.3.1 (Browning and Browning, 2007) software were used to impute up to 6 million SNPs. SNPs with a call rate <95% or a BEAGLE r2 ≤0.3 were removed, leaving a total of 5,970,354 imputed and genotyped variants.
2.4. Analyte Measurements and QC
Samples were measured for CLU and APOE protein levels, among others, by Rules Based Medicine, Inc (RBM) using multiplex immunoassay on the Human Discovery Multi-Analyte Profile (MAP) panel v1.0 (https://rbm.myriad.com/products-services/humanmap-services/human-discoverymap/). The Knight ADRC Biomarker Core and ADNI measured CSF Aβ42, tau, and phosphotau (ptau181) levels as described previously (Fagan, et al., 2006,Shaw, et al., 2009). Analytes with a call rate ≥90% passed QC. Before datasets were combined outliers were removed and values were normalized by log transformation then standardized by series so the mean for each analyte was equal to zero.
2.5. Statistical Analyses
SAS v9.2 for Linux (copyright © 2008 by SAS Institute Inc) was used to combine the ADNI and Knight ADRC datasets and the log-transformed, standardized values were tested for normality using the Shapiro-Wilk test. We used R v3.1.3 (Team, 2015) to perform linear regression to determine if CSF or plasma levels of CLU were influenced by age or gender. We used age, gender, and study as covariates when testing for association of CLU levels in plasma and CSF with AD status, CSF tau, CSF Aβ, and CSF tau/Aβ ratio. Pearson’s correlation was used to determine whether CLU and APOE levels in CSF and plasma were correlated.
CLU protein levels were tested for association using an additive model in PLINK v1.9 (Chang, et al., 2015)(http://www.cog-genomics.org/plink2). Covariates used were study, age, gender, and two principal component factors for population structure. Bonferroni corrected statistical significance was defined as p<5×10−8 and p<1×10−5 was considered suggestive association. The genomic inflation factor was 1, indicating no inflation due to population stratification. ANNOVAR version 2015-03-22 (Wang, et al., 2010), SNAP version 2.2 SNP dataset 1000 Genomes Pilot 1 population panel CEU (Johnson, et al., 2008) (http:www.broadinstitute.org/mpg/snap), SNPnexus (http://www.snp-nexus.org), build GRCh37/hg19 (Dayem Ullah, et al., 2012) and the NCBI Database of Single Nucleotide Polymorphisms (dbSNP) Build ID: 142 (http://www.ncbi.nlm.nih.gov/SNP/) (Sherry, et al., 2001) were used to perform SNP annotation.
2.6. Gene ontology over-representation analyses
Before analysis, we pruned GWAS SNPs using the clump function in PLINK v1.9 (Chang, et al., 2015). Significance threshold for index and clumped SNPs was 1 to include all SNPs. SNPs were clumped if they were within 1Mb and in LD with the index (r2=0.8). Index SNPs were mapped to genes using a gene map created from the Table Browser tool on the UCSC genome browser using the Feb. 2009 (GRCh37/hg19) assembly (http://www.genome.ucsc.edu/, accessed March 18, 2015) (Karolchik, et al., 2004,Kent and Haussler, 2001,Kent, et al., 2002). SNPs were mapped to a gene if they were located within 20kb of that gene; if SNPs were mapped to more than one gene, all genes were included in the analysis. Genes were only counted once regardless of how many SNPs were mapped to the gene. Using a significance threshold p<1×10−4, the CSF analysis included186 pruned SNPs in 117 genes and there were 165 SNPs in 79 genes in the plasma analysis.
Gene ontology analyses were performed using the Protein Analysis Through Evolutionary Relationships (PANTHER) statistical over-representation test v9.0 release 20150430 (http://www.go.pantherdb.org) (Mi, et al., 2013,Thomas, et al., 2006) which used data from the Gene Ontology Consortium (GOC) (http://www.geneontology.org, June 6, 2015) [33,34], and the ConsensusPathDB (CPDB) over-representation gene set analysis release 30 which used GOC version GO_201501 released January 2015 (http://www.cpdb.molgen.mpg.de) (Kamburov, et al., 2011). CPDB compares rates of GO term membership between the background and candidate sets of genes by the hypergeometric test and the resulting p-values are corrected for multiple testing using the false discovery rate method. PANTHER utilizes a binomial distribution test to calculate over-representation of candidate genes, relative to the background, for different gene ontology (GO) terms. PANTHER uses the Bonferroni model for multiple test correction which is highly conservative in this case because ontology terms include parent and child terms that are tested together in the analysis and are not independent at all. Because of this, we decided to use the PANTHER analyses without Bonferroni correction. Background gene sets came from each tool’s default which included 18,043 genes for CPDB (based on the number of HGNC gene symbols annotated to at least one GO term) and 20,814 genes for PANTHER (based on the total number of genes in human database obtained from Ensembl in April 2014). Of the 117 genes in the CSF analysis, 105 genes were assigned to at least one GO term by CPDB and PANTHER recognized 116 genes (the number of genes assigned to at least one GO term was not reported by PANTHER). Of the 79 genes in the plasma analysis, PANTHER recognized all of the genes and 77 were assigned to at least one GO term by CPDB. CPDB tested all three types of GO terms (biological, cellular, and molecular) in one analysis. PANTHER tested each GO term type separately, further separating genes that were manually assigned to that term based on experimental evidence from those assigned electronically based on bioinformatics algorithms. Categories with corrected p<0.05 for CPDB and uncorrected p<8.33×10−3 for PANTHER (based on the six separate tests) were considered significant.
Association List GO Annotator (ALIGATOR) was used to perform analysis of the non-pruned GWAS data as described previously with a few changes (Holmans, et al., 2009). A list of significant SNPs was converted into a list of genes in which the SNPs lie (between the start of the first and end of the last exon as defined by NCBI build 37.3). Each gene was counted once regardless of how many significant SNPs it contained. Replicate gene lists generated by randomly sampling SNPs (to allow for varying numbers of SNPs within genes) were used to obtain empirical enrichment p-values for each gene set. A bootstrap method was used to correct for testing multiple non-independent gene sets, and test whether the number of significantly enriched gene sets was higher than expected. Significant genes <1Mb apart and located in the same functional gene set were grouped into one signal, to correct for linkage disequilibrium between nearby genes. Gene sets were only classed as being enriched if they carried at least two signals. Only genes defined as "protein coding" by NCBI were analyzed, a total of 17,233 genes for plasma and 17,690 genes for CSF. Significance threshold for SNPs was p<1×10−4, resulting in 49 genes for plasma and 85 for CSF.
A large pathway set, covering as many areas of biology as possible, comprised GOC (downloaded July 26, 2013) (Ashburner, et al., 2000,Harris, et al., 2004), Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/, June 4, 2013) (Kanehisa, et al., 2012), PANTHER v8.1 (June 4, 2013) (Mi, et al., 2013), Mouse Genome Informatics (http://www.informatics.jax.org, August 9, 2013) (Bult, et al., 2008), Reactome pathways (http://www.reactome.org, July 27, 2013) (Croft, et al., 2014), Biocarta pathways from the Molecular Signatures Database v4.0 (http://www.broadinstitute.org/gsea/msigdb/index.jsp, July 28, 2013), and the NCI pathway interaction database (http://pid.nci.nih.gov/download.shtml, July 28, 2013) (Schaefer, et al., 2009). We restricted analysis to 9016 categories containing 10 to 1000 genes (200 for GO, given its large size relative to the other sets).
3. Results
3.1. CSF and plasma CLU levels in possible covariates
We used linear regression to determine if CSF or plasma levels of CLU were influenced by age, gender, or APOE ε4 allele. CLU levels in CSF and plasma were significantly influenced by gender but with opposite effects (p=8.21×10−5, beta=0.152 and p=1.21×10−11, beta=−0.235 respectively; Supplemental Table S1). CSF levels of CLU were strongly associated with age (p=2.88×10−9, beta=0.227), but there was no significant association between plasma CLU levels and age (p=0.142, beta=−0.052; Supplemental Table S1). CSF and plasma CLU levels were not associated with APOE genotype (p=0.296, beta=0.04 and p=0.279, beta=0.038 respectively) or APOE ε4 carrier status (p=0.091, beta=0.065 and p=0.806, beta=0.009 respectively; Supplemental Table S1).
3.2. CSF and plasma CLU levels in AD cases vs controls
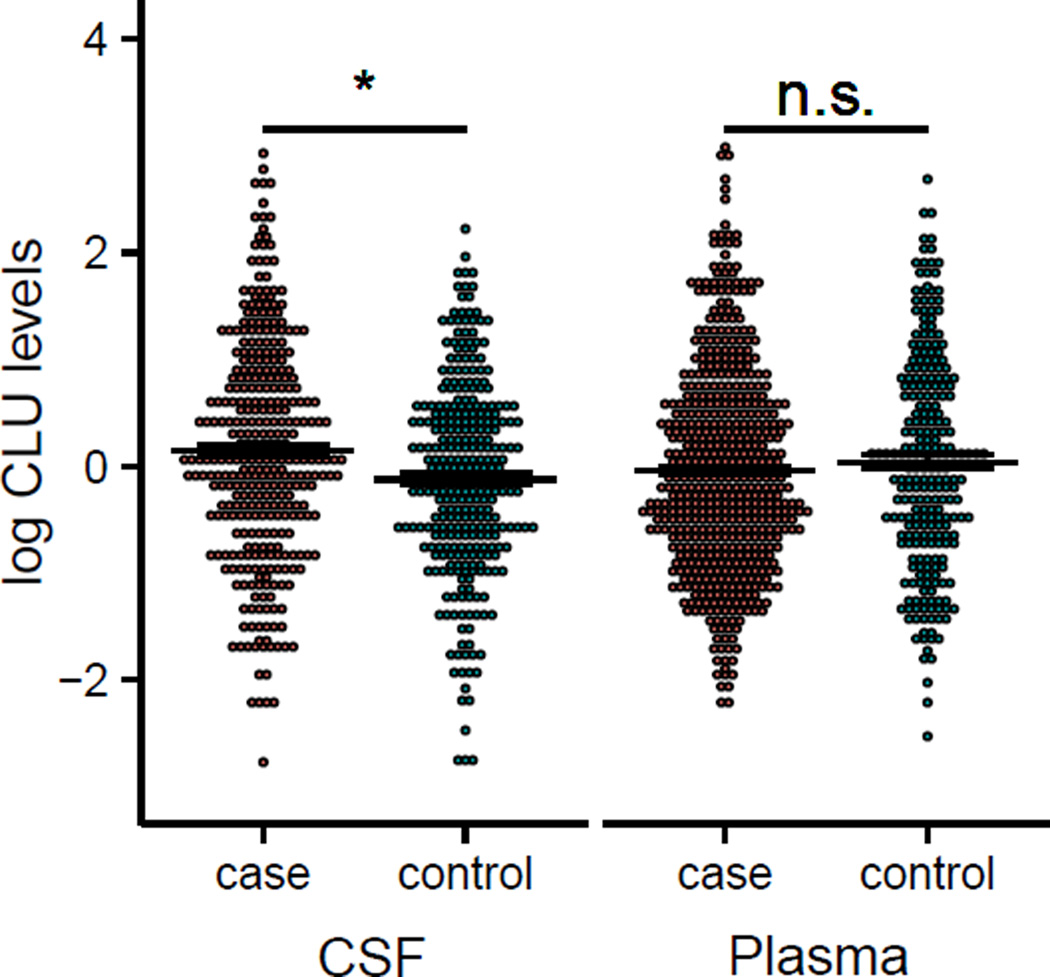
We used logistic and linear regression to determine if CSF or plasma levels of CLU were associated with AD status, CSF tau, CSF Aβ, or CSF tau/Aβ ratio. Age, gender, and study were included as covariates. CSF CLU levels were significantly higher in cases (defined by CDR>0; mean 24.74 µg/mL, SD 9.7) than controls (mean 19.92 µg/mL, SD 6.92; p=0.027, beta=−0.391, Figure 1) and there was a significant association between CSF tau/Aβ ratio and CSF CLU levels (p=3.82×10−8, beta=0.218, Supplemental Figure S1). When the dataset was stratified by APOE ε4 carrier status the association between CSF CLU levels and CSF tau/Aβ ratio was still significant (ε4+: p=1.03×10−4, beta=0.238, ε4−: p=3.66×10−4, beta=0.181, Supplemental Table S2).
Figure 1. Scatterplots of log normalized CLU levels in CSF and plasma in AD cases and controls.
Error bars (mean±sem). CSF (cases=24.74±0.56 µg/mL, controls=19.92±0.36 µg/mL, p=0.027), plasma (cases=285.05±3.12 µg/mL, controls=221.22±4.39 µg/mL, p=0.637). *p<0.05, n.s.=not significant (p>0.05)
We also compared levels of CLU between cases and controls in plasma. There was no significant difference between cases (mean 291.81 µg/mL, SD 70.54) and controls (mean 220.57 µg/mL, SD 68.70; p=0.222, beta=0.241) and there was no significant association with CSF tau/Aβ ratio (p=0.396, beta=0.035) in the whole dataset or APOE ε4 stratified datasets (Supplemental Table S2). Together these results suggest CSF CLU would be a more informative biomarker for AD than plasma CLU.
3.3. Correlation between CLU and APOE in CSF and plasma
Because previous functional studies linked CLU with APOE, we analyzed whether there was any correlation between CLU and APOE levels in CSF and plasma in the 537 individuals with both plasma and CSF levels (Supplemental Table S3). CLU levels in plasma and CSF were not correlated (r2=0.044, p=0.311), similar to what we found previously for CSF and plasma APOE (Cruchaga, et al., 2012).
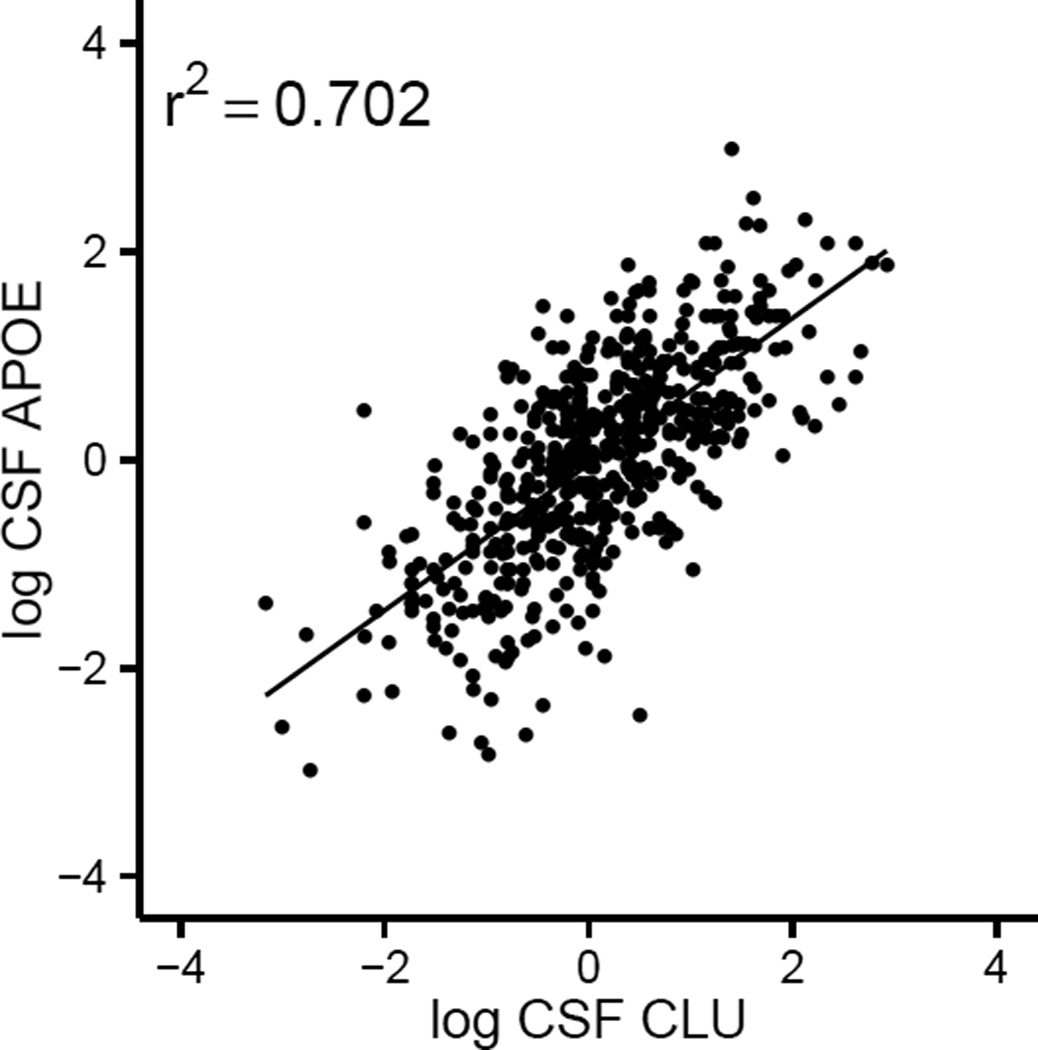
We found a strong correlation between CSF levels of CLU and APOE (r2=0.702, p<2.2×10−16, Figure 2), but the correlation in plasma was weaker (r2=0.315, p=1.22×10−13). Correlation between CSF CLU and CSF APOE was independent of AD status (cases: r2=0.708, p<2.2×10−16, controls: r2=0.721, p<2.2×10−16) and relative levels of CSF Aβ (threshold: ADNI=192 ng/mL, Knight ADRC=500 ng/mL, high Aβ: r2=0.682, p<2.2×10−16, low Aβ: r2=0.724, p<2.2×10−16). Correlation between CLU and APOE in the CSF was also independent of APOE genotype (ε4+: r2=0.703, p<2.2×10−16, ε4-: r2=0.75, p<2.2×10−16). We did not find an association between CSF and plasma CLU levels
Figure 2. Scatterplot of association of log normalized CSF levels of CLU and CSF levels APOE.
(r2=0.702, p<2.2×10−16).
3.4. GWAS for CSF and plasma CLU
In a previous GWAS we looked for genetic loci associated with CSF levels of CLU in an overall smaller dataset (n=574) by analyzing each study and performing a meta-analysis, and we found no genome-wide significant associations (Kauwe, et al., 2014). Joint analyses provide more statistical power than the power that meta-analysis approaches (Skol, et al., 2006). For this reason we performed linear regression on all 673 individuals with CSF CLU levels from ADNI and Knight ADRC. There were no genome-wide significant hits, but several loci had p-values less than the suggestive threshold p=1×10−5 (Table 2, Supplemental Figure S4). The most significant SNP was rs2581305 on chromosome 16 in an intron of Long Intergenic Non-Protein Coding RNA 917 (LINC00917) (imputed, MAF 0.052, p=3.98×10−7, beta=−0.608). The most significant genotyped SNP in LD was rs2581304 in an intron of LINC00917 (r2=0.947, D'=1, MAF 0.049, p=3.19×10−6, beta=−0.575). Testing for SNP×APOE genotype interaction showed no significant effect (p=0.287, beta=−0.346).
Because previous studies suggest an interaction with APOE and we found a strong correlation between CSF CLU levels and CSF APOE levels, we performed APOE genotype-stratified analyses. We did not find any genome-wide significant SNPs in the 270 ε4 carriers or 403 non-carriers (Table 2, Supplemental Figures S3 and S4 respectively), but in ε4-individuals one SNP almost reached genome-wide significance, rs1800795 on chromosome 7 in the promoter of interleukin 6 (IL6) (imputed, MAF 0.397, p=7.40×10−8, beta=0.375; Table 2, Supplemental Figure S6). In the overall analysis (ε4+ and ε4-combined) this SNP was below the suggestive p-value (p=9.94×10−6, beta=0.243) while in the ε4+ group there was no suggestive association (p=0.785, beta=0.024). The most significant genotyped SNP in this locus in LD with rs1800795 was rs1800797 located within 2kb of the 5’ end of IL6 (r2=0.965, D'=1, MAF 0.382, p=6.63×10−7, beta=0.241).
We did not find any genome-wide significant association in the GWAS for plasma CLU levels even though the sample size was 20% greater than in CSF (n=818, Table 3, Supplemental Figure S5). We also found no correlation between the CSF and plasma GWAS results (r2=−0.124, Supplemental Figure S6).
Table 3.
Association of top SNPs with CSF CLU levels stratified by APOE carrier status.
| All samples | APOE E4+ (n=270) | APOE E4− (n=403) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Gene | p-value | Beta | MAF | p-value | Beta | p-value | Beta |
| 16 | rs2581305 | LINC00917 | 3.98×10−7 | −0.608 | 0.052 | 5.83×10−4 | −0.615 | 4.03×10−4 | −0.577 |
| 2 | rs12470837 | 3Kb from LOC647996 | 1.16×10−6 | −0.336 | 0.167 | 1.37×10−6 | −0.468 | 1.22×10−2 | −0.243 |
| 1 | rs17507884 | MAGI3 | 1.27×10−6 | −0.466 | 0.073 | 1.45×10−3 | −0.452 | 2.72×10−4 | −0.476 |
| 7 | rs57375391 | LOC401312 | 1.58×10−6 | −0.497 | 0.073 | 1.05×10−3 | −0.539 | 1.29×10−4 | −0.514 |
| 18 | rs73431975 | SLC14A2 | 1.70×10−6 | −0.573 | 0.056 | 1.91×10−2 | −0.489 | 8.20×10−5 | −0.584 |
| 8 | rs10102274 | TMEM64 | 2.27×10−6 | 0.853 | 0.053 | 6.60×10−3 | 0.673 | 7.54×10−5 | 1.054 |
| 1 | rs7533701 | 10Kb from MAGI3 | 2.46×10−6 | −0.358 | 0.133 | 1.09×10−2 | −0.307 | 3.37×10−4 | −0.357 |
| 3 | rs417387 | VIPR1 | 2.79×10−6 | −0.353 | 0.439 | 2.73×10−3 | −0.342 | 3.40×10−4 | −0.370 |
| 6 | rs12195424 | 24Kb from DST | 2.87×10−6 | −0.350 | 0.148 | 6.11×10−3 | −0.336 | 4.07×10−4 | −0.338 |
| 8 | rs1693575 | 21Kb from SNX31 | 3.24×10−6 | 0.363 | 0.318 | 4.63×10−4 | 0.441 | 1.48×10−3 | 0.328 |
| 4 | rs1662046 | ADH1C | 3.30×10−6 | 0.451 | 0.069 | 5.11×10−3 | 0.447 | 8.68×10−5 | 0.551 |
| 6 | rs77121579 | 19Kb from DST | 3.69×10−6 | −0.384 | 0.105 | 1.68×10−2 | −0.333 | 2.69×10−4 | −0.383 |
| 14 | rs3783863 | FOXN3 | 4.53×10−6 | −0.575 | 0.043 | 2.41×10−2 | −0.496 | 8.14×10−5 | −0.608 |
| 13 | rs741668 | 12Kb from ZC3H13 | 4.64×10−6 | 0.267 | 0.299 | 2.00×10−4 | 0.302 | 7.49×10−3 | 0.225 |
| 10 | rs2456721 | SEC23IP | 4.94×10−6 | −0.266 | 0.286 | 2.60×10−3 | −0.265 | 6.67×10−4 | −0.266 |
| 13 | rs1006064 | FAM124A | 5.02×10−6 | −0.252 | 0.344 | 7.99×10−3 | −0.216 | 1.43×10−4 | −0.283 |
| 4 | rs72635116 | 43Kb from TLL1 | 5.29×10−6 | −0.313 | 0.182 | 1.13×10−1 | −0.169 | 2.32×10−5 | −0.383 |
| 15 | rs9972327 | IDH2 | 5.62×10−6 | −0.400 | 0.223 | 1.74×10−5 | −0.531 | 1.47×10−2 | −0.299 |
| 2 | rs79215379 | CTNNA2 | 7.01×10−6 | −0.490 | 0.062 | 1.32×10−2 | −0.403 | 2.46×10−4 | −0.538 |
| 10 | rs11006002 | 193Kb from IPMK | 7.49×10−6 | −0.799 | 0.060 | 1.00×10−2 | −0.753 | 3.96×10−4 | −0.815 |
| 3 | rs2442825 | SETD5 | 7.79×10−6 | −0.237 | 0.487 | 1.03×10−2 | −0.191 | 4.75×10−4 | −0.261 |
| 21 | rs9305339 | 110Kb from LINC00314 | 7.87×10−6 | 0.264 | 0.394 | 9.22×10−4 | 0.297 | 1.76×10−3 | 0.245 |
| 8 | rs17068510 | CSMD1 | 7.96×10−6 | 0.291 | 0.198 | 8.10×10−3 | 0.250 | 3.41×10−4 | 0.326 |
| 22 | rs131814 | NCAPH2 | 8.60×10−6 | −0.249 | 0.398 | 2.36×10−3 | −0.257 | 8.59×10−4 | −0.247 |
| 2 | rs6758001 | LINC00607 | 8.87×10−6 | 0.638 | 0.034 | 1.71×10−2 | 0.509 | 7.78×10−5 | 0.769 |
| 7 | rs144495862 | ABCA13 | 9.22×10−6 | −0.422 | 0.198 | 1.17×10−3 | −0.545 | 2.80×10−3 | −0.363 |
| 7 | rs1800795 | IL6 | 9.94×10−6 | 0.243 | 0.392 | 7.85×10−1 | 0.024 | 7.40×10−8 | 0.375 |
3.5. Gene ontology over-representation
In the ALIGATOR analysis of the CSF GWAS results there was a significant excess of enriched categories: 190 categories with uncorrected p<0.05 (p=0.051), 77 p<0.01 (p=0.016), and 17 p<0.001 (p=0.006). Some categories with uncorrected p<0.001 were related to wound healing and immune response (Supplemental Table S4) such as abnormal response to injury (p=5×10−5), regulation of cytokine biosynthetic process (p=8×10−5), cytokine pathway (p=1.80×10−4), abnormal wound healing (p=2.2×10−4), and delayed wound healing (p=3×10−4). ALIGATOR analysis of the plasma GWAS showed there was no significant excess of enriched categories. There were 47 categories with uncorrected p<0.05 (p=0.689), 11 p<0.01 (p=0.68), and only one p<0.001 (p=0.48). Results for the plasma CPDB and PANTHER analyses can be found in Supplemental Table S5. Due to the lack of association between plasma CLU levels and case-control status and the lack of correlation in the GWAS results between CSF and plasma, we focused our gene ontology analyses on the CSF CLU results.
In the CPDB analysis of the pruned CSF GWAS results there were 45 significant categories (Supplemental Table S6) and the PANTHER analysis resulted in 142 significant categories (Supplemental Table S7). There were 32 significant categories that overlapped in both analyses (Table 4). These categories were primarily related to lumenal side of membrane (CPDB p=1.02×10−3, PANTHER p=2.08×10−5), MHC protein complex (CPDB p=5.08×10−4, PANTHER p=2.08×10−5), wound healing (CPDB p=3.41×10−2, PANTHER p=2.88×10−4), coagulation (CPDB p=2.55×10−2, PANTHER p=1.51×10−4), hemostasis (CPDB p=2.82×10−2, PANTHER p=1.64×10−4), regulation of body fluid levels (CPDB p=2.55×10−2, PANTHER p=3×10−4), and positive regulation of immune system process (CPDB p=4.24×10−2, PANTHER p=2.12×10−4). Most of the significant categories shared between both analyses contained some of the major histocompatibility complex class I and class II genes (HLA-A, HLA-G, HLA-DPB1, HLA-DPA1) and several categories contained IL6 (including wound healing, coagulation, hemostasis, regulation of body fluid levels, and positive regulation of immune system process). IL6 was also involved in most of the categories with uncorrected p<0.001 in the ALIGATOR analysis.
Table 4.
Top SNPs (p<1×10−5) from GWAS of association with plasma CLU levels (n=818).
| Chromosome | SNP | Gene | p-value | MAF |
|---|---|---|---|---|
| 1 | rs4428865 | 928Kb from LOC102723336 | 6.53 × 10−7 | 0.224 |
| 3 | rs12492269 | LINC01014 | 2.22 × 10−6 | 0.062 |
| 12 | rs4930776 | ANO2 | 3.20 × 10−6 | 0.372 |
| 3 | rs2007029 | GRM7 | 3.94 × 10−6 | 0.169 |
| 10 | rs1575951 | 326Kb from LINC01163 | 4.01 × 10−6 | 0.116 |
| 13 | rs7995618 | LOC160824 | 4.10 × 10−6 | 0.118 |
| 12 | rs66478310 | 5Kb from LOC102723562 | 4.86 × 10−6 | 0.042 |
| 2 | rs7589728 | 32Kb from THNSL2 | 4.95 × 10−6 | 0.103 |
| 1 | rs520885 | 24Kb from MTF2 | 5.04 × 10−6 | 0.436 |
| 9 | rs11793419 | PTPRD | 5.33 × 10−6 | 0.306 |
| 6 | rs2502399 | 36Kb from LOC102724704 | 5.35 × 10−6 | 0.382 |
| 4 | rs13121109 | 78Kb from EDNRA | 6.40 × 10−6 | 0.111 |
| 8 | rs4545046 | LOC646843 | 7.12 × 10−6 | 0.379 |
| 3 | rs2029773 | 16Kb from LINC00635 | 8.12 × 10−6 | 0.473 |
| 8 | rs4637816 | 86Kb from LOC102724874 | 9.31 × 10−6 | 0.066 |
| 11 | rs58655671 | OR51E2 | 9.91 × 10−6 | 0.172 |
4. Discussion
In a previous study of 99 patients and 39 controls CSF levels of CLU were significantly higher in AD patients (p=0.002) (Nilselid, et al., 2006). In our much larger study of 356 cases and 317 controls we also found that CSF levels of CLU were significantly higher in cases than controls (p=0.03) and we found CSF CLU levels were associated with tau/Aβ ratio (p=1.06×10−7) which is highly predictive of cognitive decline. There was no difference in plasma CLU between cases and controls (p=0.114) nor any association of plasma CLU levels with CSF tau/Aβ ratio (p=0.478). In a different previous study there was no association between plasma CLU levels and incidence of AD (n=926, p=0.77), which is consistent with our findings (Schrijvers, et al., 2011). All these data suggest that CSF CLU levels would be more informative than plasma CLU levels for AD studies, although not as informative as CSF Aβ or tau.
We have previously shown that CSF APOE levels are an endophenotype for AD (Cruchaga, et al., 2012) and animal studies suggest that APOE and CLU may interact and have an additive effect on Aβ levels to play a key role in AD. A previous study demonstrated that CLU−/−/APOE−/− mice had increased amyloid deposition and significantly higher levels of soluble and insoluble Aβ40 and Aβ42 in the CSF (p<0.001) than CLU−/− or APOE−/− alone (DeMattos, et al., 2004). Additionally, Western blotting revealed that CLU levels were significantly lower in APOE−/− mice (p=1×10−4) (DeMattos, et al., 2004) suggesting that there could be an interaction between CLU and APOE. Here we report for the first time that CLU and APOE levels are strongly correlated in the CSF of humans (r2=0.702, p<2.2×10−16) which indicates that CLU and APOE may interact in the human brain as well.
Previously we successfully utilized CSF tau and phosphorylated tau levels in an endophenotype-based approach to find novel loci associated with AD (Cruchaga, et al., 2013) so we decided to look for variants associated with CLU levels since CLU has also been strongly associated with AD. In our GWAS we did not find any genome-wide significant SNPs associated with CSF or plasma CLU levels. Several SNPs had p-values<10−5, so an increase in sample size may provide enough power to observe genome-wide significant SNPs in these loci. Interestingly, the GWAS of ε4-individuals (N=403) had one SNP in IL6 of borderline genome-wide significance (p=7.58×10−8). This SNP is located in the promoter of IL6 and the - 174C allele was previously associated with higher levels of IL6 in the brain (Licastro, et al., 2003). IL6 has been associated with diffuse plaques in the brains of AD patients (Hull, et al., 1996). IL6 was also reported to induce hyperphosphorylation of tau (Quintanilla, et al., 2004). IL6 appears to play a role in modulating expression of CLU (Pucci, et al., 2009) and CLU siRNA knockdown increased IL6 baseline production in fibroblast-like synoviocytes (p<0.05) (Devauchelle, et al., 2006). Together these data suggest that IL6 may play a role in AD pathology, possibly by regulating CLU levels or by CLU modulating IL6 levels.
Additionally, our gene ontology analyses of the CSF GWAS results included categories related to wound healing and immune response indicating that CSF levels of CLU may be associated with wound healing and immune response, not only through IL6 but also other genes. IL6 is important in wound healing, not only through proinflammatory effects but also by promoting cell migration (Ebihara, et al., 2011,Nasole, et al., 2014). Previous research demonstrates that CLU may also play a role in healing, particularly in the brain. CLU improved healing after brain ischemia in wild-type mice where there was an increase in CLU mRNA in astrocytes in the peri-infarct area up to three months post-ischemia and CLU−/− mice showed a significant reduction in healing post-ischemia (Imhof, et al., 2006). Further research is necessary to explore the potential role of CLU in response to tissue damage and how this may influence the progression of AD.
It is interesting to note that the significant gene ontology categories in our CSF and plasma analyses were very different. Whereas CSF levels of CLU may be associated with response to tissue damage, plasma levels of CLU may be associated with channel and transporter activity. While there does not currently appear to be a strong association between CLU expression and channel or transporter activity, CLU was found to be important in recovery of hypofunctioning salivary glands, possibly by influencing expression of aquaporin-5 and two receptors involved in protein secretion (Mishima, et al., 2012). Aquaporin 1 was one of the genes mapped to SNPs with p<1×10−4 in the plasma GWAS results and most of the other mapped genes were receptors, channels, or subunits.
In conclusion, we have demonstrated that CSF levels of CLU are significantly associated with powerful endophenotypes for AD (tau, ptau, and Aβ42) as well as AD status and CSF tau/Aβ ratio indicating that CSF CLU levels may be a good phenotype to use when studying AD. We also found a strong correlation between CSF levels of CLU and APOE which supports previous research that indicates there may be an important interaction between these two proteins or a common pathway that influences Aβ. Our genetic analyses suggest that the role of CLU in AD may not be limited to Aβ. While there were no genome-wide significant hits in any of our GWAS, we did find several SNPs with suggestive p-values in our analyses of CSF CLU levels including one SNP in IL6 that almost reached genome-wide significance. IL6 is related to immune response, but also response to tissue damage. Our gene ontology analyses indicate CLU may also be related to wound healing as well as immune response, further research is necessary to determine this.
Supplementary Material
Table 5.
Top gene ontology categories from CPDB (p<0.05) and PANTHER (p<8.33×10−3) analyses of CLU CSF GWAS results.
| GO ID | Function | Total genes |
Candidate genes |
CPDB p- value |
PANTHER p-value |
|---|---|---|---|---|---|
| GO:0042611 | MHC protein complex | 28 | 4 | 5.08E-04 | 2.08E-05 |
| GO:0098553 | lumenal side of endoplasmic reticulum membrane | 28 | 4 | 5.08E-04 | 2.08E-05 |
| GO:0071556 | integral component of lumenal side of endoplasmic reticulum membrane |
28 | 4 | 9.96E-04 | 2.08E-05 |
| GO:0098576 | lumenal side of membrane | 28 | 4 | 1.02E-03 | 2.08E-05 |
| GO:0030662 | coated vesicle membrane | 123 | 6 | 1.88E-03 | 7.25E-05 |
| GO:0042605 | peptide antigen binding | 10 | 3 | 1.93E-03 | 2.70E-05 |
| GO:0030658 | transport vesicle membrane | 83 | 5 | 2.29E-03 | 1.12E-04 |
| GO:0030666 | endocytic vesicle membrane | 140 | 5 | 1.10E-02 | 1.19E-03 |
| GO:0051240 | positive regulation of multicellular organismal process | 517 | 12 | 2.02E-02 | 3.47E-05 |
| GO:0042613 | MHC class II protein complex | 20 | 3 | 2.24E-02 | 2.07E-04 |
| GO:0030176 | integral component of endoplasmic reticulum membrane | 121 | 4 | 2.24E-02 | 4.88E-03 |
| GO:0031227 | intrinsic component of endoplasmic reticulum membrane | 126 | 4 | 2.35E-02 | 5.62E-03 |
| GO:0050817 | coagulation | 514 | 11 | 2.55E-02 | 1.51E-04 |
| GO:0009611 | response to wounding | 726 | 13 | 2.55E-02 | 2.17E-04 |
| GO:0048518 | positive regulation of biological process | 5128 | 46 | 2.55E-02 | 2.53E-04 |
| GO:0050878 | regulation of body fluid levels | 652 | 12 | 2.55E-02 | 3.00E-04 |
| GO:0007010 | cytoskeleton organization | 830 | 12 | 2.55E-02 | 2.38E-03 |
| GO:0007596 | blood coagulation | 514 | 11 | 2.82E-02 | 1.51E-04 |
| GO:0007599 | hemostasis | 519 | 11 | 2.82E-02 | 1.64E-04 |
| GO:0030155 | regulation of cell adhesion | 593 | 11 | 2.82E-02 | 5.06E-04 |
| GO:0006996 | organelle organization | 2854 | 30 | 3.19E-02 | 3.71E-04 |
| GO:0016043 | cellular component organization | 4847 | 41 | 3.19E-02 | 2.24E-03 |
| GO:0042060 | wound healing | 649 | 12 | 3.41E-02 | 2.88E-04 |
| GO:0098552 | side of membrane | 407 | 9 | 3.76E-02 | 4.86E-04 |
| GO:0002694 | regulation of leukocyte activation | 128 | 7 | 4.24E-02 | 8.64E-06 |
| GO:0002684 | positive regulation of immune system process | 285 | 8 | 4.24E-02 | 2.12E-04 |
| GO:0001817 | regulation of cytokine production | 254 | 7 | 4.24E-02 | 5.90E-04 |
| GO:0048878 | chemical homeostasis | 826 | 13 | 4.24E-02 | 7.30E-04 |
| GO:0003281 | ventricular septum development | 44 | 3 | 4.24E-02 | 2.00E-03 |
| GO:0048522 | positive regulation of cellular process | 4400 | 38 | 4.24E-02 | 2.41E-03 |
| GO:0050865 | regulation of cell activation | 138 | 7 | 4.56E-02 | 1.40E-05 |
| GO:1902589 | single-organism organelle organization | 1895 | 21 | 4.69E-02 | 1.77E-03 |
Acknowledgments
This work was supported by Pfizer and grants from the National Institutes of Health (R01-AG044546, P01-AG003991), and the Alzheimer Association (NIRG-11-200110). This research was conducted while CC was a recipient of a New Investigator Award in Alzheimer’s disease from the American Federation for Aging Research. CC is a recipient of a BrightFocus Foundation Alzheimer's Disease Research Grant (A2013359S).
The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991, and P01 AG026276.
Some of the samples used in this study were genotyped by the ADGC and GERAD. ADGC is supported by grants from the NIH (#U01AG032984) and GERAD from the Wellcome Trust (GR082604MA) and the Medical Research Council (G0300429).
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; ; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Rev December 5, 2013 Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. Efficient multilocus association testing for whole genome association studies using localized haplotype clustering. Genet Epidemiol. 2007;31(5):365–375. doi: 10.1002/gepi.20216. [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA Mouse Genome Database, G. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, Jassal B, Jupe S, Matthews L, May B, Palatnik S, Rothfels K, Shamovsky V, Song H, Williams M, Birney E, Hermjakob H, Stein L, D'Eustachio P. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Graff C, Chiang HH, Wang J, Hinrichs AL, Spiegel N, Bertelsen S, Mayo K, Norton JB, Morris JC, Goate A. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch Neurol. 2011;68(5):581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D, Bertelsen S, Bailey M, McKean D, Shulman JM, De Jager PL, Chibnik L, Bennett DA, Arnold SE, Harold D, Sims R, Gerrish A, Williams J, Van Deerlin VM, Lee VM, Shaw LM, Trojanowski JQ, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Peskind ER, Galasko D, Fagan AM, Holtzman DM, Morris JC, Consortium, G., Alzheimer's Disease Neuroimaging, I., Alzheimer Disease Genetic, C. Goate AM. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer's disease. Neuron. 2013;78(2):256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JS, Nowotny P, Bales K, Pickering EH, Mayo K, Bertelsen S, Hinrichs A, Alzheimer's Disease Neuroimaging, I. Fagan AM, Holtzman DM, Morris JC, Goate AM. Cerebrospinal fluid APOE levels: an endophenotype for genetic studies for Alzheimer's disease. Hum Mol Genet. 2012;21(20):4558–4571. doi: 10.1093/hmg/dds296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–W70. doi: 10.1093/nar/gks364. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong D, Jansen RW, Kremer BP, Verbeek MM. Cerebrospinal fluid amyloid beta42/phosphorylated tau ratio discriminates between Alzheimer's disease and vascular dementia. J Gerontol A Biol Sci Med Sci. 2006;61(7):755–758. doi: 10.1093/gerona/61.7.755. [DOI] [PubMed] [Google Scholar]
- de Silva HV, Harmony JA, Stuart WD, Gil CM, Robbins J. Apolipoprotein J: structure and tissue distribution. Biochemistry. 1990;29(22):5380–5389. doi: 10.1021/bi00474a025. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O'Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41(2):193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Thompson WK, Holland D, Hess CP, Brewer JB, Zetterberg H, Blennow K, Andreassen OA, McEvoy LK, Hyman BT, Dale AM Alzheimer's Disease Neuroimaging Initiative, G. The role of clusterin in amyloid-beta-associated neurodegeneration. JAMA Neurol. 2014;71(2):180–187. doi: 10.1001/jamaneurol.2013.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devauchelle V, Essabbani A, De Pinieux G, Germain S, Tourneur L, Mistou S, Margottin-Goguet F, Anract P, Migaud H, Le Nen D, Lequerre T, Saraux A, Dougados M, Breban M, Fournier C, Chiocchia G. Characterization and functional consequences of underexpression of clusterin in rheumatoid arthritis. J Immunol. 2006;177(9):6471–6479. doi: 10.4049/jimmunol.177.9.6471. [DOI] [PubMed] [Google Scholar]
- Ebihara N, Matsuda A, Nakamura S, Matsuda H, Murakami A. Role of the IL-6 classic- and trans-signaling pathways in corneal sterile inflammation and wound healing. Invest Ophthalmol Vis Sci. 2011;52(12):8549–8557. doi: 10.1167/iovs.11-7956. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, LaRossa GN, Spinner ML, Klunk WE, Mathis CA, DeKosky ST, Morris JC, Holtzman DM. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Ann Neurol. 2006;59(3):512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- Fagan AM, Roe CM, Xiong C, Mintun MA, Morris JC, Holtzman DM. Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64(3):343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- Ghiso J, Matsubara E, Koudinov A, Choi-Miura NH, Tomita M, Wisniewski T, Frangione B. The cerebrospinal-fluid soluble form of Alzheimer's amyloid beta is complexed to SP-40,40 (apolipoprotein J), an inhibitor of the complement membrane-attack complex. Biochem J. 1993;293(Pt 1):27–30. doi: 10.1042/bj2930027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari O, Cruchaga C, Kauwe JS, Ainscough BJ, Bales K, Pickering EH, Bertelsen S, Fagan AM, Holtzman DM, Morris JC, Goate AM Alzheimer's Disease Neuroimaging, I. Phosphorylated tau-Abeta42 ratio as a continuous trait for biomarker discovery for early-stage Alzheimer's disease in multiplex immunoassay panels of cerebrospinal fluid. Biol Psychiatry. 2014;75(9):723–731. doi: 10.1016/j.biopsych.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R, Gene Ontology C. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Wellcome Trust Case-Control, C. Owen MJ, O'Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85(1):13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett DR, Hortobagyi T, Francis PT. Clusterin associates specifically with Abeta40 in Alzheimer's disease brain tissue. Brain Pathol. 2013;23(6):623–632. doi: 10.1111/bpa.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull M, Fiebich BL, Lieb K, Strauss S, Berger SS, Volk B, Bauer J. Interleukin-6-associated inflammatory processes in Alzheimer's disease: new therapeutic options. Neurobiol Aging. 1996;17(5):795–800. doi: 10.1016/0197-4580(96)00107-8. [DOI] [PubMed] [Google Scholar]
- Imhof A, Charnay Y, Vallet PG, Aronow B, Kovari E, French LE, Bouras C, Giannakopoulos P. Sustained astrocytic clusterin expression improves remodeling after brain ischemia. Neurobiol Dis. 2006;22(2):274–283. doi: 10.1016/j.nbd.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauwe JS, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL, Staley LA, Karch CM, Harari O, Cruchaga C, Ainscough BJ, Bales K, Pickering EH, Bertelsen S, Alzheimer's Disease Neuroimaging, I. Fagan AM, Holtzman DM, Morris JC, Goate AM. Genome-wide association study of CSF levels of 59 alzheimer's disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet. 2014;10(10):e1004758. doi: 10.1371/journal.pgen.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Haussler D. Assembly of the working draft of the human genome with GigAssembler. Genome Res. 2001;11(9):1541–1548. doi: 10.1101/gr.183201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Choi WS. Proapoptotic role of nuclear clusterin in brain. Anat Cell Biol. 2011;44(3):169–175. doi: 10.5115/acb.2011.44.3.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Asami K, Yamamoto M. Effect of heat shock treatment on the production of variant testosterone-repressed prostate message-2 (TRPM-2) mRNA in culture cells. Cell Biochem Funct. 1997;15(4):251–257. doi: 10.1002/(SICI)1099-0844(199712)15:4<251::AID-CBF748>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, European Alzheimer's Disease Initiative, I. de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer's Disease, I., Genetic, Environmental Risk in Alzheimer's, D., Alzheimer's Disease Genetic, C., Cohorts for H., Aging Research in Genomic, E. Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O'Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskov KS, Klokov DY, Li J, Kinsella TJ, Boothman DA. Synthesis and functional analyses of nuclear clusterin, a cell death protein. J Biol Chem. 2003;278(13):11590–11600. doi: 10.1074/jbc.M209233200. [DOI] [PubMed] [Google Scholar]
- Licastro F, Grimaldi LM, Bonafe M, Martina C, Olivieri F, Cavallone L, Giovanietti S, Masliah E, Franceschi C. Interleukin-6 gene alleles affect the risk of Alzheimer's disease and levels of the cytokine in blood and brain. Neurobiol Aging. 2003;24(7):921–926. doi: 10.1016/s0197-4580(03)00013-7. [DOI] [PubMed] [Google Scholar]
- May PC, Lampert-Etchells M, Johnson SA, Poirier J, Masters JN, Finch CE. Dynamics of gene expression for a hippocampal glycoprotein elevated in Alzheimer's disease and in response to experimental lesions in rat. Neuron. 1990;5(6):831–839. doi: 10.1016/0896-6273(90)90342-d. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima K, Inoue H, Nishiyama T, Mabuchi Y, Amano Y, Ide F, Matsui M, Yamada H, Yamamoto G, Tanaka J, Yasuhara R, Sakurai T, Lee MC, Chiba K, Sumimoto H, Kawakami Y, Matsuzaki Y, Tsubota K, Saito I. Transplantation of side population cells restores the function of damaged exocrine glands through clusterin. Stem Cells. 2012;30(9):1925–1937. doi: 10.1002/stem.1173. [DOI] [PubMed] [Google Scholar]
- Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43(11):2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- Nasole E, Nicoletti C, Yang ZJ, Girelli A, Rubini A, Giuffreda F, Di Tano A, Camporesi E, Bosco G. Effects of alpha lipoic acid and its R+ enantiomer supplemented to hyperbaric oxygen therapy on interleukin-6, TNF-alpha and EGF production in chronic leg wound healing. J Enzyme Inhib Med Chem. 2014;29(2):297–302. doi: 10.3109/14756366.2012.759951. [DOI] [PubMed] [Google Scholar]
- Nilselid AM, Davidsson P, Nagga K, Andreasen N, Fredman P, Blennow K. Clusterin in cerebrospinal fluid: analysis of carbohydrates and quantification of native and glycosylated forms. Neurochem Int. 2006;48(8):718–728. doi: 10.1016/j.neuint.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Oda T, Wals P, Osterburg HH, Johnson SA, Pasinetti GM, Morgan TE, Rozovsky I, Stine WB, Snyder SW, Holzman TF, et al. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1–42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp Neurol. 1995;136(1):22–31. doi: 10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pucci S, Mazzarelli P, Sesti F, Boothman DA, Spagnoli LG. Interleukin-6 affects cell death escaping mechanisms acting on Bax-Ku70-Clusterin interactions in human colon cancer progression. Cell Cycle. 2009;8(3):473–481. doi: 10.4161/cc.8.3.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla RA, Orellana DI, Gonzalez-Billault C, Maccioni RB. Interleukin-6 induces Alzheimer-type phosphorylation of tau protein by deregulating the cdk5/p35 pathway. Exp Cell Res. 2004;295(1):245–257. doi: 10.1016/j.yexcr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrijvers EM, Koudstaal PJ, Hofman A, Breteler MM. Plasma clusterin and the risk of Alzheimer disease. JAMA. 2011;305(13):1322–1326. doi: 10.1001/jama.2011.381. [DOI] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM, Trojanowski JQ Alzheimer's Disease Neuroimaging, I. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65(4):403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replicationbased analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo JB, Vanderstichele H, Figurski M, Aisen PS, Petersen RC, Weiner MW, Jack CR, Jr, Jagust W, Decarli C, Toga AW, Toledo E, Xie SX, Lee VM, Trojanowski JQ, Shaw LM Alzheimer's Disease Neuroimaging, I. Factors affecting Abeta plasma levels and their utility as biomarkers in ADNI. Acta Neuropathol. 2011;122(4):401–413. doi: 10.1007/s00401-011-0861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Cao W, Su Q, Liu Z, Zhang L. Clusterin silencing inhibits proliferation and reduces invasion in human laryngeal squamous carcinoma cells. World J Surg Oncol. 2014;12:124. doi: 10.1186/1477-7819-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Yerbury JJ, Poon S. Potential roles of abundant extracellular chaperones in the control of amyloid formation and toxicity. Mol Biosyst. 2008;4(1):42–52. doi: 10.1039/b712728f. [DOI] [PubMed] [Google Scholar]
- Wyatt AR, Yerbury JJ, Berghofer P, Greguric I, Katsifis A, Dobson CM, Wilson MR. Clusterin facilitates in vivo clearance of extracellular misfolded proteins. Cell Mol Life Sci. 2011;68(23):3919–3931. doi: 10.1007/s00018-011-0684-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.