Abstract
Biofilms enhance rates of gene exchange, access to specific nutrients, and cell survivability. Haloarchaea in Deep Lake, Antarctica, are characterized by high rates of intergenera gene exchange, metabolic specialization that promotes niche adaptation, and are exposed to high levels of UV-irradiation in summer. Halorubrum lacusprofundi from Deep Lake has previously been reported to form biofilms. Here we defined growth conditions that promoted the formation of biofilms and used microscopy and enzymatic digestion of extracellular material to characterize biofilm structures. Extracellular DNA was found to be critical to biofilms, with cell surface proteins and quorum sensing also implicated in biofilm formation. Quantitative proteomics was used to define pathways and cellular processes involved in forming biofilms; these included enhanced purine synthesis and specific cell surface proteins involved in DNA metabolism; post-translational modification of cell surface proteins; specific pathways of carbon metabolism involving acetyl-CoA; and specific responses to oxidative stress. The study provides a new level of understanding about the molecular mechanisms involved in biofilm formation of this important member of the Deep Lake community.
Halorubrum lacusprofundi is an important member of Deep Lake in Antarctica, representing ~10% of the lake population1. Deep Lake is in the Vestfold Hills, East Antarctica (68°33′36.8S, 78°11′48.7E) and is 36 m deep and perennially cold (down to −20 °C)1,2,3,4. The lake was originally a marine environment, having separated from the ocean ~3,500 years ago, and is now a closed system with salinity ~10x marine concentration1,2,3,4. Haloarchaea dominate the lake, and a high level of gene exchange occurs throughout the lake’s depth between distinct haloarchaeal genera1. The mechanisms of gene exchange have not been determined, although metaproteomic and CRISPR spacer analyses have identified viruses that infect multiple genera, thereby illustrating the potential for gene exchange to occur via transduction of cellular genes5. Transformation, conjugation, and cell fusion leading to heterodiploid formation and recombination, have also been considered as potential mechanisms for gene exchange1.
By providing high cell-density and cell-cell contact, biofilms may facilitate the exchange of genetic material6. Metaproteomics analysis of Deep Lake identified novel pili and cell surface proteins synthesized by the haloarchaea, including Hrr. lacusprofundi ACAM34, that were speculated to function in aggregation or attachment5. Laboratory studies of Hrr. lacusprofundi identified extracellular material and biofilms forming during growth7,8. The Deep Lake haloarchaea have been shown to possess distinct nutrient preferences, which possibly promotes niche adaptation1,9,10. The Antarctic haloarchaea are also exposed to high levels of UV-irradiation during the Antarctic summer3,4. Biofilms may therefore not only promote gene exchange, but enhance the survival of haloarchaea to UV-irradiation, and facilitate access to particular types of nutrients; characteristics that have previously been associated with bacterial biofilms11,12,13. Seven Deep Lake isolates (three strains of Hrr. lacusprofundi and four strains of Halohasta litchfieldiae) were assessed for their ability to adhere to plastic surfaces, and two strains (Hrr. lacusprofundi DL28 and Hht. litchfieldiae DL24) were found to strongly adhere8. The two strongly adhering strains exhibited different biofilm structures, with Hrr. lacusprofundi DL28 forming large aggregates and Hht. litchfieldiae DL24 forming carpet-like, multilayered biofilms containing macrocolonies. Using staining methods, the biofilms were shown to contain extracellular material consisting of extracellular DNA and glycoconjugates8.
Other than these Antarctic haloarchaea, the best characterized biofilm structures for cold-adapted Archaea are for SM1 Euryarchaeon that grows in sulfurous marsh waters at ~10 °C14,15. Forming macroscopic structures (e.g. 3 mm in diameter), the archaeon synthesizes unique appendages (hami) and appears to synthesize a polysaccharide matrix in which it also encases a specific species of Thiothrix sipK4 or IMB1 Epsilonproteobacteria15,16,17. The biofilm formed is thought to facilitate nutrient exchange between the two species, enabling syntrophic anaerobic sulfur metabolism. In general for Archaea, biofilm development has not been well studied, and the composition of extracellular material present in biofilms has been reported to be variable8,18. The process of cell signaling, or quorum sensing, is often important in biofilm development, but while it has been linked to bacterial biofilms19,20,21, few reports exist for equivalent analyses in Archaea; the presence of quorum sensing molecules has been described in haloarchaea22,23 and a methanogen24. A limited number of proteome-based studies have been used to assess archaeal biofilm development, with studies of Ferroplasma acidarmanus25 and Sulfolobus sp.26 identifying specific metabolic and morphological characteristics of cells in biofilms.
In view of Hrr. lacusprofundi producing extracellular material and forming biofilms, and the potential ecological importance of this capacity, here we used strain ACAM34 to study cell morphology, the composition of extracellular material and quorum sensing associated with biofilms, and used quantitative iTRAQ proteomics to assess the cellular pathways and processes involved. These analyses complement other ongoing studies of this species and collectively serve to expand our understanding of the ecophysiology of cold adapted Archaea27.
Results
Growth conditions leading to biofilm formation
The ability of media composition to promote biofilm formation of Hrr. lacusprofundi ACAM34 was identified during studies aimed at assessing the ability of the strain to utilize urea. Hrr. lacusprofundi ACAM34 was previously reported to have weak growth in DBCM2 minimal medium supplemented with low concentrations of peptone (0.025% w/v) and yeast extract (0.005% w/v) plus pyruvate (10 mM) as a carbon source and urea (10 mM) as a nitrogen source9. However, the strain was also found to be urease negative with the genome lacking identifiable genes for urea transport or catabolism9. To gain further understanding about the capacity of Hrr. lacusprofundi to utilize urea, cells were examined throughout the growth phase in medium containing 5 mM NH4Cl (medium A) or 5 mM urea (medium B), plus varying concentrations of peptone and yeast extract (Fig. 1, Fig. S1). Growth in the presence of urea, and absence or low concentrations of peptone (2.5 × 10−4% w/v) and yeast extract (5 × 10−5% w/v), produced essentially no growth over 30 d indicating that urea cannot be used as a sole source of nitrogen by Hrr. lacusprofundi ACAM34 (Fig. S1A). In contrast, growth in the presence of urea but with higher concentrations of peptone (0.025% w/v) and yeast extract (0.005% w/v) (i.e. 1 x concentration of medium B) enabled cells to reach OD600 > 0.2 (Fig. S1A). When ammonium was substituted for urea (1 x concentration of medium A), a higher growth rate and final OD600 (>0.3) was achieved (Fig. S1B), illustrating that, unlike urea, ammonium supports growth of this strain.
Figure 1.
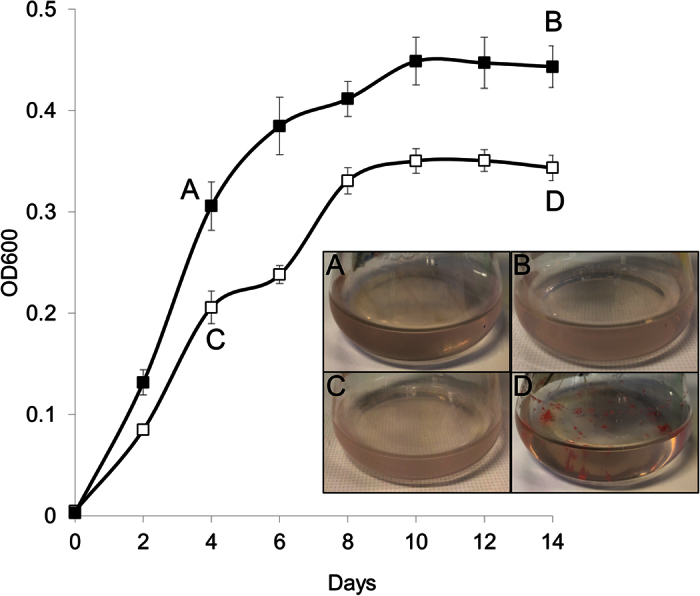
Growth response of Hrr. lacusprofundi in medium containing or lacking ammonium. Cultures were grown in 50 mL medium A containing 5 mM NH4Cl (full symbols) or medium B lacking NH4Cl (open symbols), plus peptone (0.025% w/v) and yeast extract (0.005% w/v) in 250 mL flasks at 30 °C. Error bars represent the standard error of the mean of four experiments. Cells were harvested at various time points for microscopy (see Figs 2 and 3) and proteomics (labelled A–D). Inset: images of planktonic cells (A–C) and biofilms (D) in flasks corresponding to the labels on the growth curves.
These growth studies revealed that the cell aggregation state developed differently in the two types of media. With ammonium added to the medium (medium A), cells remained planktonic throughout the 14 d growth phase, but without the addition of ammonium (medium B), aggregates (floating clumps) formed and then attached to flask walls late in the growth phase (Fig. 1). Aggregates were first visible around day 6, increased in size by day 8, and despite cultures being shaken at 120 rpm, aggregates attached to the walls of the flasks by day 10. The attached aggregates could only be removed by being physically pried off the wall of the flask (e.g. using a wire loop). Between day 10 and 14, planktonic cells, floating aggregates and attached aggregates all remained at similar levels. The floating or attached aggregates are herein referred to as biofilms, based on the definition of biofilms by Costerton and colleagues28.
Cell morphology, extracellular material, and quorum sensing associated with biofilms
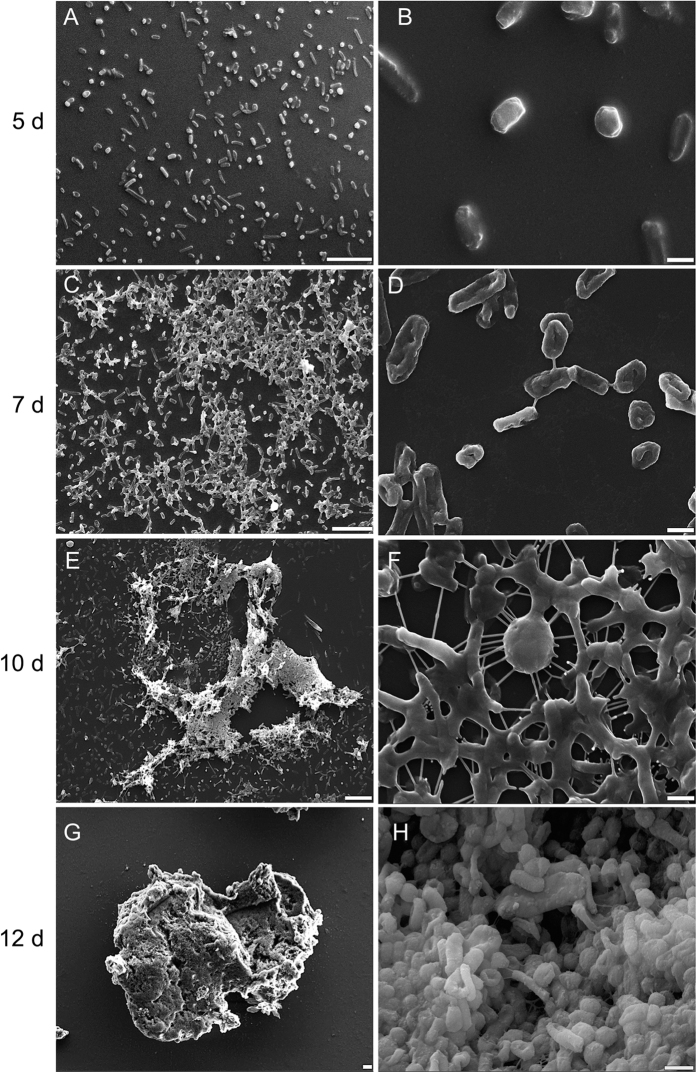
To learn about the mechanisms involved in forming biofilms, cell morphology and the presence of extracellular material were assessed using scanning electron microscopy, differential interference contrast microscopy and fluorescence microscopy of cells grown in the absence of added ammonium. Scanning electron microscopy revealed that Hrr. lacusprofundi cell shape was pleomorphic, with rods and cocci present (Fig. 2A), consistent with the original description of the organism29. Scanning electron microscopy performed on cells from 5, 7, 10 and 12 d revealed an increasing extent of extracellular structures connecting cells (Fig. 2). Cell morphology changed from discrete, relatively smooth cellular units (Fig. 2A,B), to include stringy protuberances (Fig. 2D,F,H) and heavy or rough cell surfaces (Fig. 2H), with sheets or rafts of extracellular material also present (Fig. 2C,E,G).
Figure 2.
Scanning electron microscopy images of Hrr. lacusprofundi biofilm development. Temporal progression (5, 7, 10, 12 d) shown (top to bottom) with images recorded at two different magnifications. (A,C,E,G) scale bar, 10 μm; (B,D,F,H) scale bar, 1 μm. (A,B) Planktonic cells at day 5; individual cells with smooth surfaces. (C,D) Cells aggregate and begin to attach day 7; stringy connections visible between cells. (E,F) Biofilms grow in size in solution and on the walls of flasks day 10 (images are for cells from biomass scraped of flask walls plus those in solution); protuberances and stringy connections between cells, with cells developing thick layers. (G,H) Thick biofilms form day 12–14; day 12 shown; cells with rough surfaces, and large connecting rafts of extracellular material.
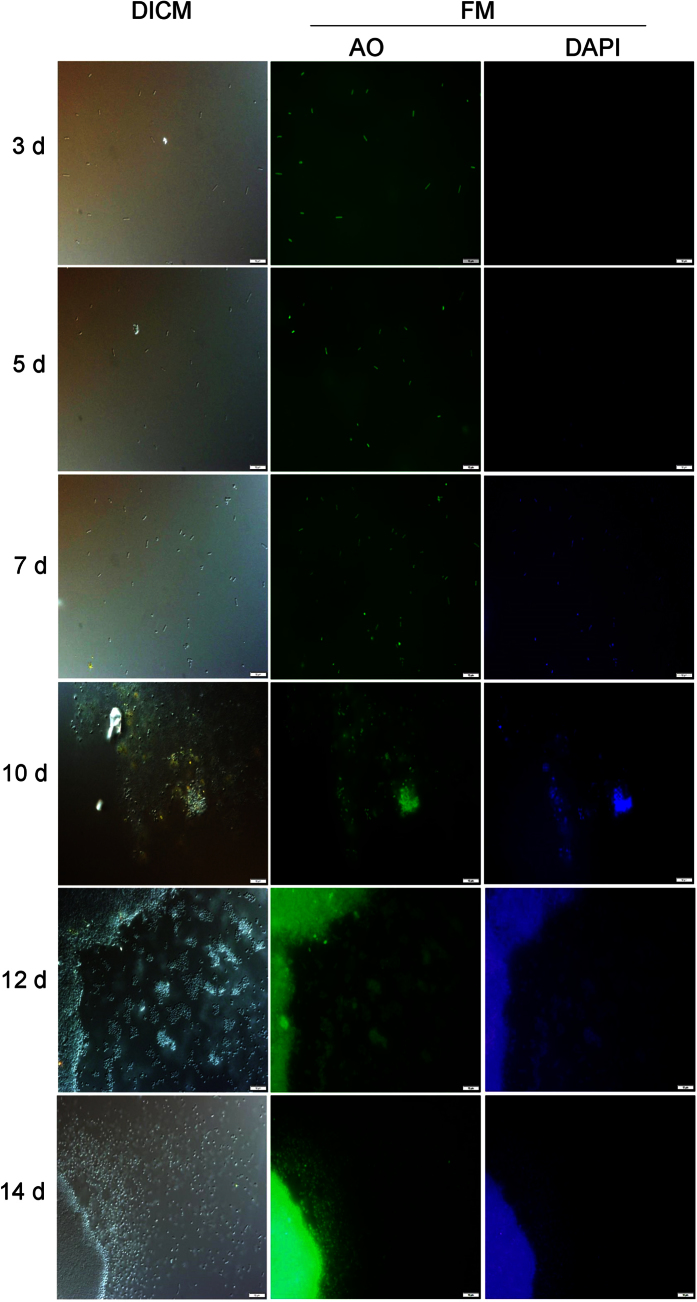
Cells were stained with acridine orange to highlight cellular and extracellular DNA or 4′,6-diamidino-2-phenylindole to highlight just extracellular DNA8,30, and fluorescence microscopy images were compared to differential interference contrast microscopy images of unstained cells (Fig. 3). 4′,6-diamidino-2-phenylindole was previously shown to preferentially stain extracellular DNA in haloarchaea and has been used for staining extracellular DNA in haloarchaeal biofilms8,30. In contrast to planktonic cells (3 and 5 d), 4′,6-diamidino-2-phenylindole signals were higher for biofilm cells (7, 10, 12 and 14 d), indicating that elevated levels of extracellular DNA were present in biofilms (Fig. 3).
Figure 3.
Fluorescence microscopy of Hrr. lacusprofundi biofilm development. Images of cells stained with acridine orange (cellular plus extracellular DNA) or 4′,6-diamidino-2-phenylindole (extracellular DNA) compared to differential interference contrast microscopy images. Extracellular DNA in biofilms commenced around day 7 and continued to grow through day 14. The scale bar represents 10 μm. Abbreviations: DICM, differential interference contrast microscopy; FM, fluorescence microscopy; AO, acridine orange; DAPI, 4′,6-diamidino-2-phenylindole.
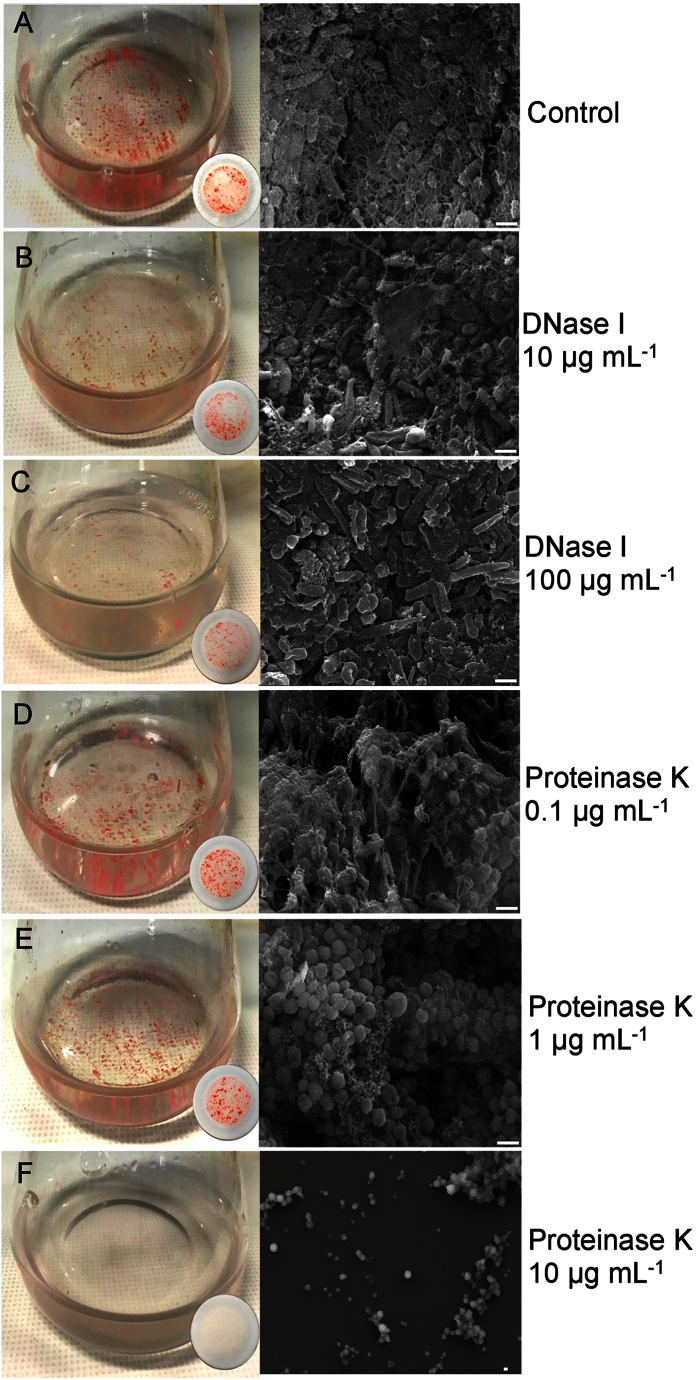
To further assess the roles of DNA and protein in extracellular material and biofilm formation, cultures were treated with DNase I or proteinase K. DNase I31,32,33 and proteinase K34,35,36 have previously been used to study the structure of biofilms. In order to assess the effects of the enzymatic activity on the formation or disruption of biofilms, the nuclease or protease was added to media before inoculation with cells, or to preformed biofilms, respectively. Quantitation of biofilms was performed by crystal violet staining or bicinchoninic acid assay, with total DNA quantitated by acridine orange staining (Table 1), and cells and extracellular material visualized by scanning electron microscopy (Fig. 4).
Table 1. Effect of DNase I and proteinase K on biofilm formation of Hrr. lacusprofundi.
| Treatment | Biofilm biomass | Total intra- and extra-cellular DNA | |
|---|---|---|---|
| Bicinchoninic acid assay (total protein μg) | Crystal violet assay (OD600) | Acridine orange assay [(LAU-B) mm−2] | |
| Control | 1400 ± 30 | 1.0 ± 0.1 | 101000 ± 1100 |
| DNase I 10 μg mL−1 | 1100 ± 60* | 0.75 ± 0.08** | 76000 ± 710** |
| DNase I 100 μg mL−1 | 600 ± 40** | 0.50 ± 0.05*** | 26800 ± 270*** |
| Proteinase K 0.1 μg mL−1 | 1900 ± 50* | 1.7 ± 0.1*** | 181000 ± 9700* |
| Proteinase K 1 μg mL−1 | 1500 ± 60 | 1.1 ± 0.09 | 257000 ± 12000** |
| Proteinase K 10 μg mL−1 | 17 ± 6*** | 0.03 ± 0.01**** | 3190 ± 130*** |
Bicinchoninic acid, crystal violet and acridine orange assays were performed at 10 d. The results are the means ± standard errors from three biological replicates. Asterisks indicate the significance of the difference compared to the value for the control, calculated using a paired t-test: *p < 0.05, **p < 0.01, ***p < 0.001; ****p < 0.0001.
Figure 4.
The effects of DNase I or proteinase K on the ability of Hrr. lacusprofundi to form biofilms. Cells grown in the absence of DNase I or proteinase K (A); 10 μg mL−1 DNase I (B); 100 μg mL−1 DNase I (C); 0.1 μg mL−1 proteinase K (D); 1 μg mL−1 proteinase K (E); 10 μg mL−1 proteinase K (F). Biofilms captured on filter papers for each enzyme treatment (25 mL medium, 100 mL flask) are shown beside their respective flasks. The scanning electron microscopy scale bar represents 1 μm. (A,B,C) Increasing the concentration of DNase I reduced the extent of extracellular lattice network present between cells. (D,E,F) Increasing the concentration of proteinase K led to collapse of the integrity of extracellular material, and changes in cell morphology and number of viable cells.
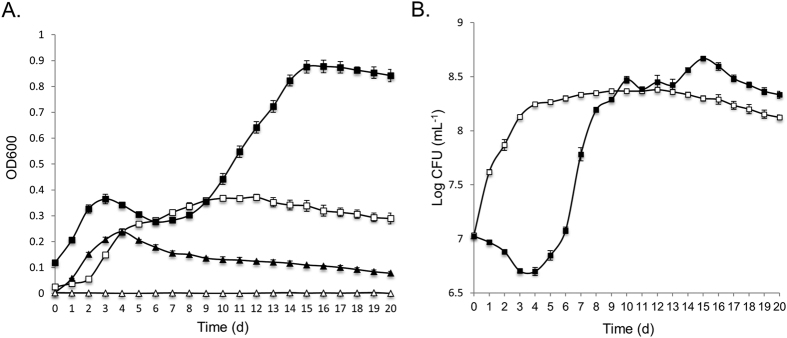
Treatment with DNase I showed a concentration-dependent (0, 10 and 100 μg mL−1) reduction in the development of biofilms and total DNA (Table 1, Fig. 4). Scanning electron microscopy revealed a reduction in the extent of lattice-like extracellular material surrounding cells, while the morphology of individual cells remained largely unchanged (Fig. 4). Higher concentrations of DNase I (1 mg mL−1) completely inhibited biofilm formation, and led to a large increase in final OD600, consistent with the growth of only planktonic cells (Fig. 5A). The high concentration of DNase I (1 mg mL−1) produced an early spike in OD600 (for cultures and uninoculated controls) (Fig. 5A) consistent with some of the enzyme aggregating in the high salt, and this coincided with a period of no increase in colony forming units (Fig. 5B). However, DNase I activity (assessed by in vitro digestion of Hrr. lacusprofundi DNA) was retained throughout the 14 d incubation period (Fig. S2). Moreover, the replacement of DNase I with an equivalent concentration of bovine serum albumin (up to 1 mg mL−1) led to a spike in OD600 (later in the incubation period) (Fig. S3), but had no effect on biofilm formation throughout the 20 d period (data not shown). Collectively the data for DNase I treatments illustrate that the extracellular material contains DNA that is important for biofilm formation.
Figure 5.
Growth response of Hrr. lacusprofundi in medium containing DNase I. Hrr. lacusprofundi were grown in 50 mL medium (250 mL flasks) with or without 1 mg mL−1 DNase I and OD600 (A) and viability (B) determined. (A) OD600 for cultures without DNase I (open squares); cultures with 1 mg mL−1 DNase I (full squares); uninoculated medium without DNase I (open triangles); uninoculated medium with 1 mg mL−1 DNase I (full triangles). (B) Colony forming units for cultures without DNase I (open squares); cultures with 1 mg mL−1 DNase I (full squares). For uninoculated medium with 1 mg mL−1 DNase I, the number of cells per mL was below the detection limit (100 cells mL−1) (data not shown). Error bars represent the standard error of the mean of three experiments.
Proteinase K treatment (0.1 and 1 μg mL−1) disrupted lattice-like extracellular material and led to changes in cellular morphology (small spherical cells), with 10 μg mL−1 greatly reducing the rate of cell growth, viability, biomass formation and the production of biofilms (Table 1, Fig. 4). Increasing the proteinase K concentration to 100 μg mL−1 completely prevented cell growth (data not shown). Similar to DNase I, activity of proteinase K was retained in the growth medium throughout 14 d of incubation (data not shown). While the structure of the extracellular material lost its integrity with relatively low concentrations (0.1 and 1 μg mL−1) of proteinase K (Fig. 4), the yield of filtered biofilm biomass and total DNA increased relative to the control (Table 1). These findings are consistent with proteinase K causing the release of DNA from cells, with DNA accumulating in extracellular material and biomass, and all of it being captured on filters.
The biomass of biofilms was unchanged after 10 d-old cultures with preformed biofilms were incubated for a further 4 d with a high concentration of DNase I (1 mg mL−1) or proteinase K (10 μg mL−1) (data not shown), indicating enzyme activity primarily affected growing cells. The findings are overall consistent with cell growth leading to DNA being released from the cell by active export or cell lysis, with the DNA fulfilling a structural role in the extracellular material involved in biofilm formation. Because proteinase K appeared to cause release of DNA and destroy extracellular material lattice structure, it is likely that cell surface proteins were proteolytically degraded during growth of the cells thereby compromising cell structural integrity and possibly protein structures involved in linkages to extracellular material.
The presence of N-acyl homoserine lactone-like quorum sensing molecules was assessed using an Escherichia coli green fluorescent protein reporter assay37 with the supernatant fraction of planktonic cells (4 d growth) or biofilms (14 d growth) (Fig. 6). Both quantitative measurements that were normalized to protein concentration (Fig. 6) and fluorescence images (Fig. S4) showed markedly higher green fluorescent protein fluorescence in biofilm cells, indicating that N-acyl homoserine lactone-like compounds may be involved in biofilm formation.
Figure 6. Quorum sensing-like activity of Hrr. lacusprofundi associated with biofilm cells.
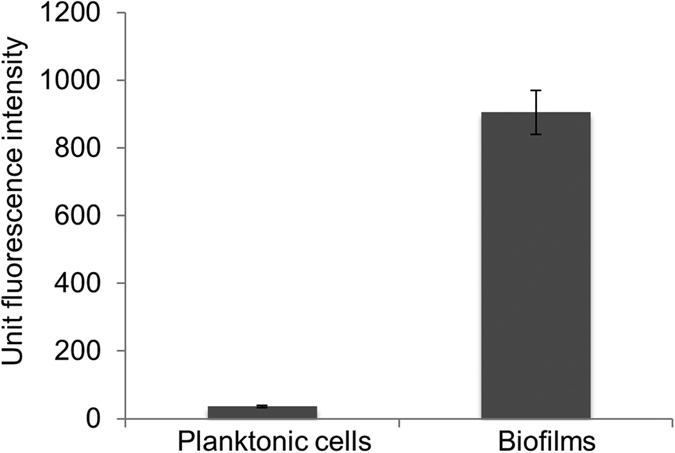
N-acyl homoserine lactone-like quorum sensing was assessed using the supernatant fraction of planktonic cells (4 d growth) or biofilms (14 d growth). Relative fluorescence units from the E. coli MT102 biosensor were determined by subtracting the background fluorescence (media blank), and unit fluorescence intensity was shown normalized to protein concentration of whole cell extracts. Error bars represent the standard error of the mean of three experiments. Fluorescence microscopy images of samples are shown in Fig. S4.
Proteomics of biofilm formation
Proteomics was performed on whole cells and the extracellular fraction (supernatant) of Hrr. lacusprofundi ACAM34 to learn about global gene expression during biofilm formation and from that data infer pathways and cellular processes involved in forming biofilms. A total of 1996 proteins were detected, accounting for ~54% of the 3665 protein-coding genes in Hrr. lacusprofundi ACAM34. An 8-plex iTRAQ labelling approach38 (Table S1) was used for assessments of differential abundance, enabling multiple growth conditions to be simultaneously compared. A total of 109 proteins had significant abundance differences (≥1.5-fold) between log phase (4 d growth) and stationary phase (14 d growth) in medium lacking ammonium (Table S2); 165 were differentially abundant between stationary phase cultures from media containing or lacking ammonium (Table S2); and 56 proteins were common to both assessments and represented proteins important for biofilms under both the growth phase and growth medium conditions that were tested (Table 2). The core set of 56 proteins included 36 with higher abundance and 20 with lower abundance (Table 2), and described specific cellular processes (Table 2, Fig. 7). A total of 13 additional proteins were added to the core set because they had ≥1.5-fold differential abundance in one assessment (e.g. log vs stationary) and 1.2–1.5 fold differential abundance in the other assessment (e.g. plus or minus ammonium) and were considered relevant to biofilm formation because they belonged to the functional processes already defined by the 56 core proteins (Table 2, Fig. 7). All original iTRAQ datasets for two iTRAQ labelling experiments, which included four biological replicates for each growth condition, are provided in Tables S3–S10.
Table 2. Proteins from proteomics linked to Hrr. lacusprofundi biofilms.
| Locus_tag | Annotation | Differential abundance (biofilm vs planktonic) | |||
|---|---|---|---|---|---|
| Medium B | Stationary phase | ||||
| Stationary vs log | Medium B vs A | ||||
| WCF | SF | WCF | SF | ||
| Secreted, membrane, and cell surface proteins | |||||
| Hlac_0389 | signal peptide; no identifiable domains | ns | 1.8 | ns | 1.8 |
| Hlac_1583 | signal peptide; peptidase M14 carboxypeptidase A family (zinc-dependent)-possible S-layer modulation | ns | 1.6 | ns | 4.3 |
| Hlac_1867 | signal peptide; predicted lipoprotein; similarity to bacterial and archaeal DNA (including eDNA) metabolism proteins | — | 1.6 | — | 1.5 |
| Hlac_2426 | preprotein translocase, SecY subunit (9 TMDs) | 0.76 | — | 0.65 | — |
| Hlac_2472 | signal peptide; PGF-CTERM archaeal protein-sorting signal; 1 TMD | ns | 1.5 | ns | 1.3 |
| Hlac_3146 | TAT signal; no identifiable domains | — | 2.1 | — | 3.2 |
| Transport | |||||
| Hlac_0674 | ABC transporter oligopeptide/dipeptide-binding protein | 1.7 | ns | 2.2 | ns |
| Hlac_1191 | ABC transporter zinc-binding lipoprotein | 2.0 | 3.9 | 1.4 | 1.3 |
| Hlac_1631 | electron transport protein SCO1/SenC (signal peptide) | ns | 1.8 | ns | 1.9 |
| Hlac_2057 | ABC transporter molybdate-binding lipoprotein | 0.44 | 1.6 | 0.69 | 1.6 |
| Hlac_2623 | Rh family protein/ammonium transporter (Amt) | 3.3 | ns | 8.3 | ns |
| Nitrogen metabolism | |||||
| Hlac_2285 | acetamidase/formamidase | 3.3 | 1.6 | 4.5 | 2.1 |
| Hlac_2374 | glutamine synthetase, type I (GlnA) | 1.5 | 2.2 | 2.7 | 3.1 |
| Central carbon metabolism | |||||
| Hlac_0890 | pyruvate ferredoxin: oxidoreductase, beta subunit (PorB) | 1.2 | ns | 1.7 | ns |
| Hlac_0891 | pyruvate:ferredoxin oxidoreductase alpha subunit (PorA) | 1.2 | ns | 1.9 | ns |
| Hlac_1306 | acetate:CoA ligase (Acs) | 3.3 | ns | 2.3 | ns |
| Hlac_2153 | isocitrate lyase (AceA) | 3.9 | ns | 2.5 | ns |
| Hlac_2311 | phosphoenolpyruvate carboxylase (Ppc) | 1.5 | ns | 2.0 | ns |
| Hlac_3040 | aconitate hydratase | 1.3 | ns | 1.7 | ns |
| One carbon metabolism | |||||
| Hlac_1238 | formate dehydrogenase, alpha subunit (FdhA) | 1.5 | — | 2.0 | — |
| Carbohydrate metabolism | |||||
| Hlac_1300 | gluconate dehydratase (GnaD) | 1.6 | ns | 2.3 | ns |
| Hlac_1672 | glyceraldehyde-3-phosphate dehydrogenase, type II (Gap2) | 0.4 | ns | 0.6 | ns |
| Hlac_1891 | NAD-dependent epimerase/dehydratase (possible glycosylation of S-layer protein) | 1.7 | ns | 1.8 | ns |
| Hlac_2371 | glyceraldehyde-3-phosphate dehydrogenase, type I (Gap) | 1.6 | ns | 2.2 | ns |
| Glycerol metabolism | |||||
| Hlac_1122 | glycerol kinase (GlpK) | 1.7 | ns | 1.4 | ns |
| Hlac_1124 | glycerol 3-phosphate dehydrogenase subunit B (GlpB) | 1.4 | — | 1.5 | — |
| Hlac_1458 | dihydroxyacetone kinase, K subunit (DhaK) | 1.3 | ns | 1.6 | ns |
| Hlac_2109 | glycerol 2-dehydrogenase (NAD+) (GldA) | 2.0 | — | 2.3 | — |
| Amino acid metabolism | |||||
| Hlac_0452 | phosphoserine phosphatase (SerB) | 0.46 | ns | 0.52 | ns |
| Hlac_0967 | methylglyoxal synthase | 1.5 | ns | 1.6 | ns |
| Hlac_1941 | anthranilate synthase (TrpE) | 1.2 | — | 1.5 | — |
| Nucleotide metabolism | |||||
| Hlac_1250 | phosphoribosylformylglycinamidine synthase (PurS) | 1.5 | — | 1.8 | — |
| Hlac_1295 | phosphoribosylamine/glycine ligase (PurD) | 1.7 | ns | 2.1 | ns |
| Energy conservation | |||||
| Hlac_2310 | menaquinol–cytochrome-c reductase | ns | 1.5 | ns | 2.0 |
| General metabolism | |||||
| Hlac_0596 | rhodanese domain + metallo-beta-lactamase domain protein | 1.5 | — | 1.6 | — |
| Hlac_1098 | isochorismatase-like hydrolase | 1.6 | — | 1.8 | — |
| Hlac_1619 | NAD (P)-binding oxidoreductase domain | 1.5 | — | 1.5 | — |
| Hlac_1837 | alcohol dehydrogenase, zinc-dependent | 2.3 | — | 2.5 | — |
| Oxidative stress | |||||
| Hlac_0175 | FeS assembly protein SufB | 1.3 | ns | 1.6 | ns |
| Hlac_0176 | FeS assembly ATPase SufC | 1.3 | ns | 1.5 | ns |
| Hlac_1548 | catalase/peroxidase (HPI) | 0.61 | ns | 0.50 | 0.50 |
| Hlac_1677 | alkyl hydroperoxide reductase (Ahp)/peroxiredoxin | 1.5 | ns | 1.7 | ns |
| Hlac_1763 | cysteine synthase A (CysK) | 1.5 | — | 1.5 | — |
| Hlac_2298 | signal peptide; DUF541 domain; homolog of E. coli antioxidant protein YggE | — | 1.6 | — | 1.9 |
| Transcription | |||||
| Hlac_0605 | winged helix-turn-helix domain protein | — | 1.7 | — | 1.9 |
| Hlac_0693 | phosphate uptake regulator, PhoU | 1.5 | ns | 1.6 | ns |
| Hlac_1558 | putative transcriptional regulator, XRE family | 1.5 | ns | 1.8 | ns |
| DNA replication, protection and repair | |||||
| Hlac_0021 | non-histone chromosomal MC1 family protein | 1.5 | — | 2.1 | — |
| Hlac_3022 | type III restriction protein Res subunit | 1.5 | — | 1.5 | — |
| Proteolysis | |||||
| Hlac_0185 | proteasome regulatory subunit (PsmR) | 0.61 | ns | 0.67 | ns |
| Cell division | |||||
| Hlac_1716 | cell division control protein CdcH | 0.71 | ns | 0.62 | ns |
| Protein chaperones | |||||
| Hlac_0416 | group II chaperonin | 0.45 | 0.63 | 0.61 | 0.70 |
| Hlac_0682 | chaperone protein DnaK | 0.65 | ns | 0.51 | ns |
| Hlac_2662 | group II chaperonin | 0.42 | ns | 0.36 | ns |
| Translation | |||||
| Hlac_0100 | ribosomal protein S7 | 0.66 | ns | 0.67 | ns |
| Hlac_0413 | ribosomal protein L15e | 0.67 | ns | 0.65 | ns |
| Hlac_0615 | ribosomal S13S15 domain protein | 0.58 | ns | 0.62 | ns |
| Hlac_0618 | ribosomal protein S3Ae | 0.63 | ns | 0.59 | ns |
| Hlac_0827 | ribosomal protein L31e | 0.61 | ns | 0.47 | ns |
| Hlac_1816 | ribosomal protein S13 | 0.59 | ns | 0.59 | ns |
| Hlac_1842 | ribosomal protein L7Ae/L30e/S12e/Gadd45 | 0.62 | ns | 0.63 | ns |
| Hlac_2277 | ribosomal protein L21e | 0.47 | — | 0.67 | — |
| Hlac_2312 | ribosomal protein S19e | 0.63 | ns | 0.67 | ns |
| Hlac_2436 | ribosomal protein L5 | 0.65 | ns | 0.63 | ns |
| Hlac_2447 | ribosomal protein L25/L23 | 0.67 | 0.69 | 0.66 | ns |
| Hlac_2448 | ribosomal protein L4/L1e | 0.66 | ns | 0.63 | ns |
| Hlac_2534 | ribosomal protein L10 | 0.61 | ns | 0.61 | ns |
| Uncategorized proteins | |||||
| Hlac_0492 | no identifiable domains | 1.8 | ns | 1.7 | ns |
| Hlac_1516 | DUF555 domain; uncharacterised protein family UPF0212 | 1.8 | ns | 1.7 | ns |
Proteins associated with biofilms (higher differential abundance, normal font) or planktonic cells (lower differential abundance, italic font) under both the growth phase and growth medium conditions that were tested. Protein functions assigned based on manual annotation. Differential abundance values are expressed as ratios for two fractions: whole cell fraction (WCF) or extracellular supernatant fraction (SF). Core proteins (total of 56) with significant differential abundance (≥1.5-fold) in both assessments (medium B, stationary vs log phase; stationary phase, medium B vs medium A) are shown in bold font; proteins (total of 13) from functional categories represented by core proteins with ≥1.5-fold differential abundance in one assessment and 1.2–0.5 fold differential abundance in the other assessment are shown in plain font. Abbreviations: predicted transmembrane domain, TMD; twin-arginine translocation signal, TAT signal; detected in the expressed proteome, but not with significant differential abundance, ns; not detected in the expressed proteome, -.
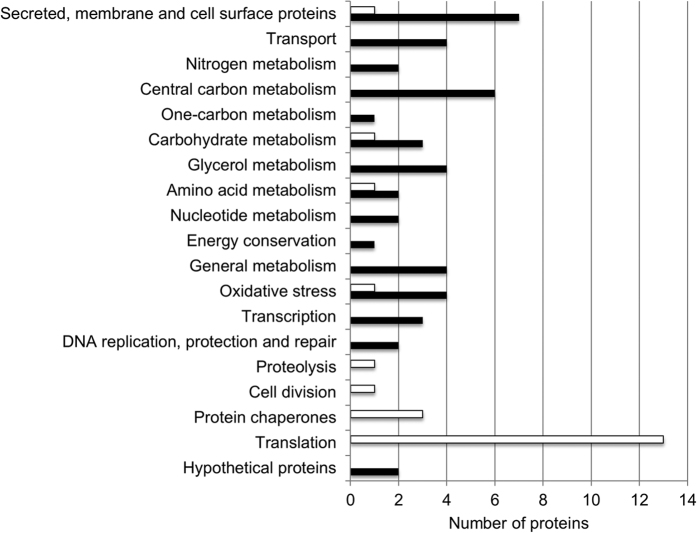
Figure 7. Protein functional categories associated with biofilms or planktonic cells.
Proteins were identified using an 8-plex iTRAQ proteomics protocol with higher abundance in biofilms (black bars) or lower abundance in biofilms (white bars) under both the growth phase and growth medium conditions that were tested, and assigned to functional categories (also see Table 2).
Proteins associated with biofilms included higher abundances of a nucleoside-diphosphate-sugar epimerase involved in glycosylation (Hlac_1891) and a range of secreted proteins (Hlac_3146, Hlac_1583, Hlac_0389, Hlac_2298, Hlac_2472, Hlac_1867). Specific transporters, and enzymes requiring the imported solutes as cofactors, had higher abundance in biofilms; these included ABC transporter lipoproteins for molybdate (Hlac_2057) and zinc (Hlac_1191), a zinc-dependent formaldehyde dehydrogenase (Hlac_1837), a zinc-dependent carboxypeptidase (Hlac_1583), and a molybdopterin-dependent formate dehydrogenase (Hlac_1238). Consistent with biofilms forming in medium lacking ammonium, proteins with higher abundance related to nitrogen metabolism included an ammonium transporter (Hlac_2623), glutamine synthetase (GS; Hlac_2374), and an acetamidase (Hlac_2285).
Cultures forming biofilms also had higher abundances of metabolic enzymes including: acetyl-CoA synthesis (Hlac_1306, Hlac_0890, Hlac_0891, Hlac_0967); the utilization of acetyl-CoA via the glyoxylate cycle (Hlac_3040; Hlac_2153); the carboxylation of phosphoenolpyruvate (PEP) to oxaloacetate (Hlac_2311); the conversion of glycerol into dihydroxyacetone phosphate (DHAP) (Hlac_2109, Hlac_1458, Hlac_1122, Hlac_1124); and purine synthesis (Hlac_1295, Hlac_1250). Proteins associated with responses to oxidative stress or DNA damage in biofilms included a higher abundance of alkyl hydroperoxide reductase (Ahp; Hlac_1677), a secreted antioxidant protein (YggE homolog; Hlac_2298), the cysteine biosynthesis protein CysK (Hlac_1763), proteins involved in the assembly of iron-sulfur (FeS) clusters (Hlac_0175, Hlac_0176), chromosomal protein MC1 (Hlac_0021), and a predicted DNA helicase (Hlac_3022), and the lower abundance of catalase/peroxidase (HPI; Hlac_1548).
Proteins that decreased in biofilms were mainly involved in protein synthesis and folding, including a large number of ribosomal proteins, and protein chaperones (Table 2, Fig. 7). Additionally, biofilms had lower levels of a proteasome protein (Hlac_0185), a protein translocase (Hlac_2426), and a cell division protein (Hlac_1716) (Table 2).
Discussion
The major metabolic pathways and cellular processes involved in biofilm formation were inferred by integrating the core proteomic data with cell morphology, extracellular material, and quorum sensing data (Fig. 8). The abundance data that were most informative about biofilm formation were the relatively large number of proteins that had elevated levels (see below). However, the proteins that decreased in biofilms, particularly those involved in protein synthesis (Table 2), informed about the overall reduced demand for cell growth; this finding is consistent with bacterial biofilm cells growing more slowly than planktonic cells39,40,41.
Figure 8.
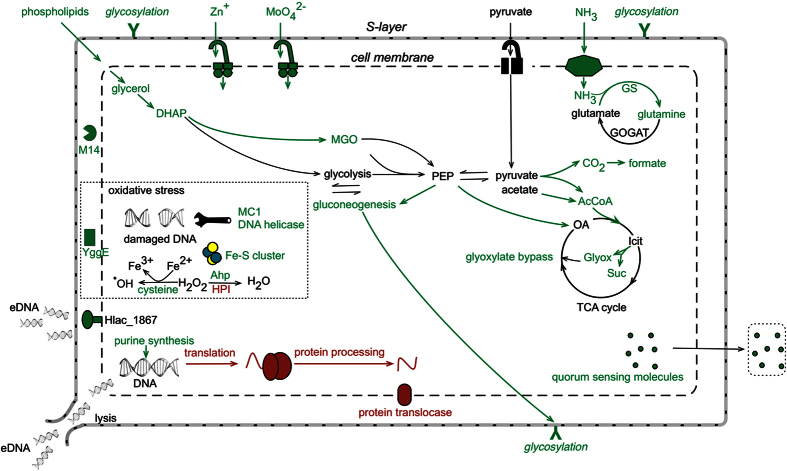
Depiction of the major metabolic pathways and cellular processes in Hrr. lacusprofundi involved in forming biofilms. The depiction represents findings for cell morphology, extracellular material, and quorum sensing integrated with the core proteomic data (Fig. 7 and Table 2). Pathways and processes linked to biofilms (green) vs planktonic cells (red) are shown, highlighting the importance of extracellular DNA (including extracellular DNA processing by Hlac_1867 and cellular release by lysis), post-translationally modified cell surface proteins (Y symbol), carbohydrate synthesis, specific responses to oxidative stress, carbon metabolism involving acetyl-CoA, ammonium assimilation and quorum sensing. Abbreviations: eDNA, extracellular DNA; DHAP, dihydroxyacetone phosphate; MGO: methyglyoxal; AcCoA, acetyl-CoA; OA, oxaloacetate; Icit, isocitrate; Glyox, glyoxylate; Suc, succinate; M14, peptidase M14 carboxypeptidase A; YggE, antioxidant protein; MC1, non-histone chromosomal MC1 family protein (Hlac_0021); Ahp, alkyl hydroperoxide reductase; HPI, catalase/peroxidase.
Modification of the cell envelope
In Archaea, the surface layer (S-layer) is composed of identical protein subunits arranged into a monolayer, forming a highly porous lattice structure42 that maintains the structural integrity of the cell envelope43,44. Biofilms appear to require post-translational modification of the S-layer. There is evidence for N-glycosylation44,45 by a nucleoside-diphosphate-sugar epimerase, and peptidolysis by Hlac_1583, which is homologous to an extracellular endopeptidase I from Lysinibacillus sphaericus that cleaves spore cortex peptidoglycan46,47. As Hlac_1583 is a zinc-dependent carboxypeptidase, the elevated levels of the zinc ABC transporter lipoprotein (Hlac_1191) may derive from an increased demand for zinc. The inferred importance of cell envelope proteins is consistent with the potent inhibitory effect of proteinase K on Hrr. lacusprofundi biofilm formation (Fig. 4).
Metabolic networks in biofilm development
Biofilms are associated with changes in carbon metabolism, particularly via acetyl-CoA (Fig. 8). Acetyl-CoA is proposed to function as a metabolic sensor of the availability of nutrients in E. coli, and accumulation of acetyl-CoA promotes biofilm production48,49. The dual action of the glyoxylate cycle (which bypasses the decarboxylation steps of the tricarboxylic acid cycle) and phosphoenolpyruvate carboxylase (which performs anaplerotic fixation of CO2) allows conservation of carbon for biosynthetic purposes (such as biosynthesis of carbohydrates). The increased expression of phosphoenolpyruvate carboxylase may relate to sugar synthesis during growth in the media used, which lacks carbohydrates. Phosphoenolpyruvate carboxylase is reported to play a critical role for extracellular material production in biofilm development in Salmonella50. Thus, it appears that carbon sources that are metabolized to acetyl-CoA and further to specific carbohydrates contribute to biofilm formation in Hrr. lacusprofundi.
The increased levels of enzymes involved in glycerol catabolism also appear to represent a route for carbon utilization. However, in this case, cells in biofilms may utilize the glycerophosphate backbones of lipids51 from neighboring lysed cells, as has been suggested for phospholipids from Desulfovibrio vulgaris biofilms52. The data are consistent with evidence of increased cell lysis within biofilms (see eDNA and regulation of biofilm formation). More broadly, these proteomic data highlight the relevance of carbohydrate metabolism in biofilm formation, and may reflect a role for carbohydrates as components of extracellular material involved in forming biofilms.
Responses to oxidative damage
The increased abundance of a YggE homolog (Hlac_2298) in biofilms was an indicator of oxidative stress as it is homologous to a periplasmic protein from E. coli that is inferred to protect cells against oxidative stress53,54. Intracellularly, cysteine serves as a reductant that drives the Fenton reaction, which generates hydroxyl radicals from iron (II) and hydrogen peroxide, thereby causing damage to DNA55. The predicted increase in cysteine biosynthesis (CysK) may therefore enhance oxidative stress. The elevated levels of proteins involved in the assembly of FeS clusters may also be in response to oxidative stress, as has been reported for some bacteria56,57. Moreover, the increased abundance of a predicted DNA helicase, and chromosomal protein MC1 which has been reported to protect DNA in Methanosarcina sp. CHTI 5558, may reflect an increased demand to protect and repair DNA damage (Fig. 8).
However, the specific enzymes directly involved in catalyzing the decomposition of hydrogen peroxide indicate that levels of this oxidant may be lower in biofilms than in planktonic cells. Ahp is associated with scavenging low levels of hydrogen peroxide, whereas HPI is associated with high levels59. The increase in abundance of Ahp and decrease of HPI in biofilms suggests that under the growth conditions that were used, hydrogen peroxide is present, but at relatively low levels. This would be consistent with biofilms growing more slowly than planktonic cells and thus generating lower levels of hydrogen peroxide. An increased abundance of Ahp was previously identified in biofilms of Sulfolobus26, several species of Bacteria60,61,62, and the fungus Candida albicans63.
Extracellular DNA and regulation of biofilm formation
Hlac_1867 is homologous to ComA from Halobacterium sp. NRC-1, with both possessing a metallo-β-lactamase domain and Lamin Tail Domain (Fig. S5A). In common with Hlac_1867, the Bacillus subtilis competence protein ComEC and Neisseria gonorrhoeae ComA64,65 possesses a metallo-β-lactamase domain (Fig. S5B), and the Haloferax volcanii extracellular DNA metabolism protein Hvo_1477, possesses a Lamin Tail Domain66 (Fig. S5A). A Lamin Tail Domain is present in some membrane associated hydrolases and likely affords membrane association67. Metallo-β-lactamase domains are often associated with nucleases68 and have been speculated to directly catalyse hydrolysis of nucleic acids69. In Hvo_1477, the metallo-β-lactamase domain is replaced by a thermonuclease domain, and the protein has been speculated to function in the Hfx. volcanii biofilm lifecycle66. Hlac_1867, Hvo_1477 and Halobacterium sp. NRC-1 ComA also possess N-terminal lipobox motifs, indicating they may be lipoproteins. The overall functional similarity of these archaeal and bacterial proteins suggests that Hlac_1867 fulfils a role in DNA metabolism in Hrr. lacusprofundi, and its elevated abundance in biofilms is consistent with the important role that extracellular DNA plays in biofilm formation (Fig. 3).
The elevated levels of proteins involved in purine biosynthesis may also be a response to the demand for extracellular DNA synthesis during biofilm formation. The requirement for purine synthesis is a characteristic of bacterial biofilms70,71,72; for example, Bacillus cereus produces extracellular material containing extracellular DNA, but mutants with defects in purine biosynthesis fail to synthesize biofilms70. For some bacterial biofilms, cell lysis and/or secretion of DNA are the main sources of extracellular DNA73,74,75. Both mechanisms seem possible for Hrr. lacusprofundi, and if active DNA export does occur, it may be facilitated by Hlac_1867. Given the implications for purine biosynthesis, the role of Hlac_1867, and the effects of DNase I on preventing the formation of biofilms (Figs 4 and 5, Table 1), extracellular DNA stands out as a particularly important component of biofilms in Hrr. lacusprofundi.
Cell signaling via N-acyl homoserine lactones has been directly linked to biofilm development, including the processes of initial surface attachment, biofilm maturation and biofilm detachment20,76. The enhanced fluorescence of quorum-sensing induced biosensors observed with Hrr. lacusprofundi biofilms (Figs 6 and S4) suggests quorum sensing may signal biofilm development, perhaps in response to changes in nutrient availability (Fig. 8). The Hrr. lacusprofundi genome does not possess genes known to be involved in N-acyl homoserine lactone quorum sensing (e.g. luxI or luxR homologues), indicating the effector compounds are likely to be mimics of N-acyl homoserine lactones.
Current knowledge of biofilm development in Archaea
A total of three global gene expression studies examining biofilms have been performed on Archaea: F. acidarmanus25, Sulfolobus spp.26, and Hrr. lacusprofundi (this study). The majority of F. acidarmanus biofilm associated proteins related to anaerobic growth, consistent with anaerobic zones occurring in some bacterial biofilms77,78,79, and the ability of F. acidarmanus to grow anaerobically25; a characteristic not shared by Sulfolobus spp. or Hrr. lacusprofundi. The only class of enzyme with increased abundance from all three studies was Ahp/peroxiredoxin (refs 25, 26 and this study), indicating that a common feature of archaeal biofilms appears to be cells mounting a specific response to relatively low levels of hydrogen peroxide. In Sulfolobus acidocaldarius biofilms, an increased abundance of extracellular material, a putative glycosyl transferase and polysaccharides on cell surfaces were reported26, which accords with the proposed role of post-translational modification of the S-layer in Hrr. lacusprofundi biofilms. In terms of quorum sensing, Sulfolobus biofilms were reported to have a decreased abundance of a FabG homolog which is proposed to be involved in the production of a quorum sensing autoinducer26, and no evidence for quorum sensing was reported for F. acidarmanus25; findings that differ from Hrr. lacusprofundi. Other than oxidative stress and the role of extracellular material, collectively, these studies do not identify general principles involved in archaeal biofilm formation. This is perhaps not surprising as while the three organisms are members of the Euryarchaeota, they represent three very different ecophysiologies: F. acidarmanus, iron-dependent chemolithotrophic acidophile25; Sulfolobus spp., sulfur-dependent heterotrophic/autotrophic thermoacidophile26; Hrr. lacusprofundi, psychrophilic salt-dependent heterotrophic halophile. Clearly additional types of studies (e.g. gene knockout), and studies of other Archaea that form biofilms are warranted.
Conclusion
By achieving high proteome coverage (~54%), analyzing two cellular fractions (whole cell extract and extracellular fraction) and comparing two distinct growth conditions (with or without ammonium, stationary phase; log vs stationary phase, no ammonium), we obtained strong evidence for specific proteins involved in Hrr. lacusprofundi biofilm development. To the best of our knowledge, this is the first quantitative proteomics report to assess biofilms from haloarchaea or Antarctic archaea. The ultrastructural analyses visually characterized the development and extent of extracellular material associated with biofilms, and demonstrated the important role of extracellular DNA in forming biofilms. Hrr. lacusprofundi is an important member of the Deep Lake community representing ~10% of the cellular population, and was identified through network analyses as supporting the highest level of intergenera exchange of high identity regions between haloarchaea in the lake1. The establishment of proteomic methods for Hrr. lacusprofundi coincides well with the development of a system for genetic manipulation of Hrr. lacusprofundi80, enabling future work to better define the mechanisms involved in formation and regulation of Hrr. lacusprofundi biofilms, and the role of biofilms in promoting gene transfer events.
Methods
Growth of Hrr. lacusprofundi
Hrr. lacusprofundi ACAM34 was grown in DBCM2 basal salt medium81 supplemented with 10 mM pyruvate and either 5 mM NH4Cl (medium A) or 5 mM urea (medium B), plus varying concentrations of peptone and yeast extract (1 x: 5 × 10−3% yeast extract and 2.5 × 10−2% peptone; 1/10 x: 5 × 10−4% yeast extract and 2.5 × 10−3% peptone; 1/100 x: 5 × 10−5% yeast extract and 2.5 × 10−4% peptone; 0 x: no yeast extract or peptone). Hrr. lacusprofundi was inoculated 1:100 from cultures grown under the same conditions either in 50 mL medium in 250 mL flasks or 25 mL medium in 100 mL flasks, at 120 rpm. Hrr. lacusprofundi grows in the laboratory across a wide range of temperatures (−1 °C to ~44 °C)7,29 and 30 °C was chosen for cultivation to expedite biomass production and growth assessments, and because the growth regime was suitable for generating both planktonic and biofilm cells; the growth temperature does not simulate natural environmental temperatures in Deep Lake which range from −20 °C at depth to a ~10 °C on the surface in summer2,3.
Microscopy and DNase I and Proteinase K treatments
Cells, including biofilms were examined by fluorescence and differential interference contrast microscopy using an Olympus BX61 microscope with DP71 camera (Olympus, Tokyo, Japan) and cellSens Standard 1.11 (Olympus, Tokyo, Japan), and scanning electron microscopy using a JEOL 7001 F field emission scanning electron microscope (JEOL, Freising, Germany), based on previously described methods8,82. Acridine orange (Ex 505 nm/Em 525 nm) was used at 10 μg mL−1 (final concentration) to stain extracellular and intracellular DNA, and 4′,6-diamidino-2-phenylindole (Ex 359 nm/Em 461 nm) was used at 5 μg mL−1 (final concentration) to stain extracellular DNA8,30. To prepare samples for imaging by scanning electron microscopy, 1 mL of cells was directly fixed with 2% glutaraldehyde at 4 °C overnight, cells were washed three times in DBCM2 salt solution to remove glutaraldehyde, and dehydrated sequentially in 30, 50, 70, 90, 100, 100, 100% ethanol, 2:1 ethanol/hexamethyldsilazane, 1:1 ethanol/hexamethyldsilazane, and 100, 100, 100% hexamethyldsilazane for 10 min each. Samples were mounted on scanning electron microscopy sample stubs and chromium coated.
DNase I31,32,33 and proteinase K34,35,36 treatments have previously been used to disrupt biofilms. Here, DNase I (Sigma-Aldrich, MO, USA) or proteinase K from Tritirachium album (Sigma-Aldrich) at specific concentrations was added to cultures (25 ml in 100 mL flasks) grown in medium B at the commencement of growth (DNase I: 10 μg mL−1, 100 μg mL−1, 1 mg mL−1; proteinase K: 0.1 μg mL−1, 1 μg mL−1, 10 μg mL−1, 100 μg mL−1), or after biofilms had formed (DNase I: 1 mg mL−1; proteinase K: 10 μg mL−1).
To quantify biofilm biomass, biofilms attached to the inner surfaces of flasks were scraped into solution, and the solution was passed through Whatman no. 54 filter paper using vacuum filtration to collect total biofilms on the filter papers and planktonic cells in the filtrate. The biomass captured on filters was washed three times with a 30% salt water solution83. Filter papers were placed in tubes in 25 mL of 30% salt water solution at 30 °C and 120 rpm overnight to release cells into the liquid phase, filters removed, cells pelleted by centrifugation for 5 min at 4,500 × g and resuspended in 2 ml 30% salt water solution. For assessments of the retention of DNase I (1 mg mL−1) activity in cultures, aliquots (100 μL) were withdrawn daily and incubated with 500 μg Hrr. lacusprofundi genomic DNA at 37 °C for 1 h and the sample analysed by agarose (1%) gel electrophoresis. Quantitative assessments of total biofilm biomass using crystal violet (Sigma-Aldrich) was performed based on previously described methods84,85 and bicinchoninic acid assays were performed using a Thermo Scientific Pierce BCA Protein Assay Kit. The crystal violet method84,85 detects bacterial peptidoglycan in cell walls, so the method was modified to make it suitable for Hrr. lacusprofundi. Additional details of methods are provided in Supplementary Information.
Quorum sensing
N-acyl homoserine lactone-like compounds were prepared by an ethyl acetate extraction procedure and quorum sensing-like responses determined using an E. coli reporter system, based upon published methods37,86,87. Hrr. lacusprofundi cells were centrifuged at 4,500 × g for 20 min at room temperature and the supernatant recovered. Ethyl acetate extractions86 were performed using 200 mL of cell free supernatant and 200 mL of medium B as a control. Extracts were redissolved in 300 μL of methanol acidified with 0.1% (v/v) formic acid, and 10 μL of the redissolved extract and 10 μL of 200 nM oxo-hexanoyl homoserine lactone (Sigma Aldrich) control were evaporated in a 96 well plate (all in triplicate). The reporter strain, E. coli MT 102 harbouring the plasmid pJBA132, was grown aerobically in LB broth supplemented with ampicillin (100 μg mL−1) at 30 °C with shaking at 150 rpm. Overnight cultures of E. coli MT102 (pJBA132) were diluted 10 times in AB minimal medium88. A volume of 200 μL of diluted E. coli MT102 (pJBA132) was added to each well in the microtiter plate that contained extract, or oxo-hexanoyl homoserine lactone (as a control). Plates were incubated for 5 h at 30 °C at 100 rpm, and GFP fluorescence (485 nm excitation, 535 nm emission) was determined using a CLARIOstar plate reader (BMG Labtech, Ortenberg, Germany) as previously described37. Relative fluorescence units were determined by subtracting the background fluorescence (media blank), and fluorescent intensities were normalized to mg of protein present in whole cell extracts. Protein concentration was determined by the bicinchoninic acid method (see Microscopy and DNase I and Proteinase K treatments). Slides for microscopy were prepared using biomass from the wells of the microtitre plates immediately following fluorescence determinations.
Proteomics
Proteomics was performed based on previously described methods38,89. Cells were grown in medium A (with ammonium) or medium B (without ammonium) that contained peptone (0.025% w/v) and yeast extract (0.005% w/v). After cells reached mid-logarithmic phase (4 d), half of the culture volume was removed and cells harvested to obtain a whole cell pellet and an extracellular fraction (supernatant). The remaining half of the culture was grown until 14 d, and cells were harvested and processed as for log phase cells. Quantitative proteomics was performed using 8-plex iTRAQ labelling90 according to manufacturer’s instructions (Sciex, Framingham, MA, USA) using a specific labelling design (Table S1). In brief, a total of four biological replicates were prepared for each growth condition. The growth conditions were: medium A, log phase (day 4); medium A, stationary phase (day 14); medium B, log phase (day 4); medium B, stationary phase (day 14). For each 8-plex iTRAQ labelling run, two of the biological replicates was used from each growth condition. The approach was used for proteins from whole cell extracts and the extracellular fraction, and two labeling experiments were performed for each fraction. This resulted in a total of four 8-plex iTRAQ labelling experiments and a total of 32 protein samples analyzed. Samples were analyzed using a TripleTOF 5600+ hybrid tandem mass spectrometer (ABSciex, Foster City, USA), and data acquired in information-dependent acquisition mode with Analyst TF 1.7 software (AB Sciex, Foster City, USA). Each 8-plex iTRAQ labelling experiment was run twice to provide two technical replicates. By running technical replicates of each 8-plex iTRAQ labelling experiment, a total of four datasets were generated for whole cell extracts and extracellular fractions of each growth condition. Each of the four datasets for the specific growth condition and fraction were combined and searched with ProteinPilot software 4.5 (AB Sciex, Foster City, USA) against the local Hrr. lacusprofundi ACAM34 protein FASTA database to identify proteins. A minimum unused score of 1.3 was accepted for protein identification, representing 95% confidence in correct sequence identification.
For quantitative analysis of relative abundance level changes, data were considered statistically significant when p was less than 0.05 and the error factor was less than 2. The weighted average mean and standard deviation of differential abundance between iTRAQ reporter ion ratios were calculated91. In addition, an average weighted abundance ratio of 1.5-fold or more was used as the cutoff for differential abundance for assessing proteins involved in biofilm formation. Typically the iTRAQ proteomics literature cites a minimum fold change of 1.292,93. In this study, we adopted a more conservative approach in selecting a fold change of 1.5, in addition to a minimum p value of 0.05. Pearson’s correlation analysis between biological replicates, technical replicates and labelling experiment replicates was performed using SPSS 22.0 software (Fig. S6). By comparing differential abundance between log phase and stationary phase in medium B, and differential abundance between stationary phase cultures from medium A and medium B, the core set of proteins common to both comparisons represented proteins specific to biofilm development under both the growth phase and growth medium conditions that were tested. In some cases, proteins with 1.2–1.5 fold change were considered if they were from functional categories represented by the core set of proteins (Table 2). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository94 with the dataset identifier PXD004202. Additional methodological details are provided in Supplementary Information.
All proteins of relevance were manually functionally annotated based on a previously described approach95. InterPro platform (http://www.ebi.ac.uk/interpro/) was used to predict protein domains or signal peptides, and features diagnostic of protein function. Transmembrane domains were predicted by TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and annotation as a membrane protein required a positive result from TMHMM. Proteins with N-terminal signal peptides and/or a single C-terminal transmembrane helix and/or homology to experimentally characterized cell surface proteins (e.g. S-layer proteins) were annotated as cell surface/envelope proteins according to a previously described approach5.
Additional Information
How to cite this article: Liao, Y. et al. Morphological and proteomic analysis of biofilms from the Antarctic archaeon, Halorubrum lacusprofundi. Sci. Rep. 6, 37454; doi: 10.1038/srep37454 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Australian Research Council [DP150100244]. Y.L. and J.Y. were funded by the China Scholarship Council (File No. 201206910027 and 201206230085, respectively). Mass spectrometry results were obtained at the Bioanalytical Mass Spectrometry Facility within the Analytical Centre of the University of New South Wales. This work was undertaken using infrastructure provided by NSW Government co-investment in the National Collaborative Research Infrastructure Scheme. Subsidized access to this facility is gratefully acknowledged. We thank the PRIDE team and ProteomeXchange for efficiently processing and hosting the mass spectrometry data. We also acknowledge Dr. Simon Hager’s assistance from the Electron Microscopy Unit of the University of New South Wales.
Footnotes
The authors declare no competing financial interests.
Author Contributions R.C. conceived the research. Y.L. performed all the experiments except B.P.B. designed the quorum sensing assay, J.C. performed fluorescence-based microplate assays for quorum sensing detection and J.Y. performed SEM. A.P. and M.J.W. assisted with the mass spectrometry for proteomics. Y.L., R.C. and T.J.W. designed experiments performed by Y.L. and analyzed the data and interpreted the findings. R.C., Y.L. and T.J.W. wrote the manuscript. All authors vetted the manuscript and viewed the final version.
References
- DeMaere M. Z. et al. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc. Natl. Acad. Sci. USA 110, 16939–16944 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P. J. Primary productivity of a hypersaline Antarctic lake. Mar. Freshw. Res. 29, 717–724 (1978). [Google Scholar]
- Ferris J. M. & Burton H. R. The annual cycle of heat content and mechanical stability of hypersaline Deep Lake, Vestfold Hills, Antarctica. Hydrobiologia 165, 115–128 (1988). [Google Scholar]
- Barker R. J. Physical and chemical parameters of Deep Lake, Vestfold Hills, Antarctica. 5 (Australian Government Publishing Service, 1981). [Google Scholar]
- Tschitschko B. et al. Antarctic archaea–virus interactions: metaproteome-led analysis of invasion, evasion and adaptation. ISME J. 1, 1–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen J. S., Burmølle M., Hansen L. H. & Sørensen S. J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65, 183–195 (2012). [DOI] [PubMed] [Google Scholar]
- Reid I. N. et al. Terrestrial models for extraterrestrial life: methanogens and halophiles at Martian temperatures. Int. J. Astrobiol. 5, 89–97 (2006). [Google Scholar]
- Fröls S., Dyall-Smith M. & Pfeifer F. Biofilm formation by haloarchaea. Environ. Microbiol. 14, 3159–3174 (2012). [DOI] [PubMed] [Google Scholar]
- Williams T. J. et al. Microbial ecology of an Antarctic hypersaline lake: genomic assessment of ecophysiology among dominant haloarchaea. ISME J. 8, 1645–1658 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschitschko B. et al. Ecophysiological distinctions of haloarchaea from a hypersaline Antarctic lake as determined by metaproteomics. Appl. Environ. Microbiol. 82, 3165–3173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.-C. & Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- Elasri M. O. & Miller R. V. Study of the response of a biofilm bacterial community to UV radiation. Appl. Environ. Microbiol. 65, 2025–2031 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaka K., Crawford R. J., Nazarenko E. L. & Ivanova E. P. Bacteria extracellular polysaccharides. Adv. Exp. Med. Biol. 715, 213–226 (2011). [DOI] [PubMed] [Google Scholar]
- Rudolph C., Wanner G. & Huber R. Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl. Environ. Microbiol. 67, 2336–2344 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger R., Moissl C., Amann T., Rudolph C. & Huber R. New insights into the lifestyle of the cold-loving SM1 euryarchaeon: natural growth as a monospecies biofilm in the subsurface. Appl. Environ. Microbiol. 72, 192–199 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph C., Moissl C., Henneberger R. & Huber R. Ecology and microbial structures of archaeal/bacterial strings-of-pearls communities and archaeal relatives thriving in cold sulfidic springs. FEMS Microbiol. Ecol. 50, 1–11 (2004). [DOI] [PubMed] [Google Scholar]
- Moissl C., Rachel R., Briegel A., Engelhardt H. & Huber R. The unique structure of archaeal ‘hami’, highly complex cell appendages with nano-grappling hooks. Mol. Microbiol. 56, 361–370 (2005). [DOI] [PubMed] [Google Scholar]
- Fröls S. Archaeal biofilms: widespread and complex. Biochem. Soc. Trans. 41, 393–398 (2013). [DOI] [PubMed] [Google Scholar]
- Davies D. G. et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 280, 295–298 (1998). [DOI] [PubMed] [Google Scholar]
- Parsek M. R. & Greenberg E. P. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13, 27–33 (2005). [DOI] [PubMed] [Google Scholar]
- Waters C. M. & Bassler B. L. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346 (2005). [DOI] [PubMed] [Google Scholar]
- Paggi R. A., Martone C. B., Fuqua C. & De Castro R. E. Detection of quorum sensing signals in the haloalkaliphilic archaeon Natronococcus occultus. FEMS Microbiol. Lett. 221, 49–52 (2003). [DOI] [PubMed] [Google Scholar]
- Tommonaro G., Abbamondi G. R., Iodice C., Tait K. & De Rosa S. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb. Ecol. 63, 490–495 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang G. et al. Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. ISME J. 6, 1336–1344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C., Potrykus J., Wexler M., Bond P. L. & Dopson M. Biofilm development in the extremely acidophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Extremophiles 14, 485–491 (2010). [DOI] [PubMed] [Google Scholar]
- Koerdt A. et al. Macromolecular fingerprinting of Sulfolobus species in biofilm: A transcriptomic and proteomic approach combined with spectroscopic analysis. J. Proteome Res. 10, 4105–4119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli R. Cold-adapted archaea. Nat. Rev. Microbiol. 4, 331–343 (2006). [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R. & Lappin-Scott H. M. Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 (1995). [DOI] [PubMed] [Google Scholar]
- Franzmann P. D. et al. Halobacterium lacusprofundi sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst. Appl. Microbiol. 11, 20–27 (1988). [Google Scholar]
- Chimileski S., Franklin M. J. & Papke R. T. Biofilms formed by the archaeon Haloferax volcanii exhibit cellular differentiation and social motility, and facilitate horizontal gene transfer. BMC Biol. 12, 65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch C. B., Tolker-nielsen T., Ragas P. C. & Mattick J. S. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002). [DOI] [PubMed] [Google Scholar]
- Tetz G. V., Artemenko N. K. & Tetz V. V. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 53, 1204–1209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. L., Hanman K., Reuter M., Betts R. P. & van Vliet A. H. M. Campylobacter jejuni biofilms contain extracellular DNA and are sensitive to DNase I treatment. Front. Microbiol. 6, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. K. & Rao T. S. Dispersal of Bap-mediated Staphylococcus aureus biofilm by proteinase K. J. Antibiot. (Tokyo). 266, 55–60 (2013). [DOI] [PubMed] [Google Scholar]
- Ðapa T. et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195, 545–555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen U. T. & Burrows L. L. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int. J. Food Microbiol. 187, 26–32 (2014). [DOI] [PubMed] [Google Scholar]
- Charlesworth J., Kimyon O., Manefield M. & Burns B. P. Detection and characterization of N-acyl-l-homoserine lactones using GFP-based biosensors in conjunction with thin-layer chromatography. J. Microbiol. Methods 118, 164–167 (2015). [DOI] [PubMed] [Google Scholar]
- Williams T. J. et al. Defining the response of a microorganism to temperatures that span its complete growth temperature range (− 2 C to 28 C) using multiplex quantitative proteomics. Environ. Microbiol. 13, 2186–2203 (2011). [DOI] [PubMed] [Google Scholar]
- Brown M. R. W., Allison D. G. & Gilbert P. Resistance of bacterial biofilms to antibiotics a growth-rate related effect? J. Antimicrob. Chemother. 22, 777–780 (1988). [DOI] [PubMed] [Google Scholar]
- Wentland E. J., Stewart P. S., Huang C. & McFeters G. A. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12, 316–321 (1996). [DOI] [PubMed] [Google Scholar]
- Yeom J., Shin J. H., Yang J. Y., Kim J. & Hwang G.-S. 1H NMR-based metabolite profiling of planktonic and biofilm cells in Acinetobacter baumannii 1656-2. PLoS One 8, e57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. Facing extremes: archaeal surface-layer (glyco) proteins. Microbiology 149, 3347–3351 (2003). [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., Jones G. M., Kandiba L., Nair D. B. & Eichler J. S-layer glycoproteins and flagellins: reporters of archaeal posttranslational modifications. Archaea 2010, 612948 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S. V. & Meyer B. H. The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426 (2011). [DOI] [PubMed] [Google Scholar]
- Kaminski L. & Eichler J. Haloferax volcanii N-glycosylation: delineating the pathway of dTDP-rhamnose biosynthesis. PLoS One 9, e97441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier M., Vacheron M. J., Guinand M. & Michel G. Purification and partial characterization of the extracellular gamma-D-glutamyl-(L)meso-diaminopimelate endopeptidase I, from Bacillus sphaericus NCTC 9602. Eur. J. Biochem. 148, 539–543 (1985). [DOI] [PubMed] [Google Scholar]
- Hourdou M. L. et al. Characterization of the sporulation-related gamma-D-glutamyl-(L)meso-diaminopimelic-acid-hydrolysing peptidase I of Bacillus sphaericus NCTC 9602 as a member of the metallo(zinc) carboxypeptidase A family. Modular design of the protein. Biochem. J. 292, 563–570 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß B. M. et al. Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch. Microbiol. 192, 715–728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugabi R. et al. The role of activated acetate intermediates in the control of Escherichia coli biofilm amounts. Webmedcentral 3, pii: 3577 (2012). [PMC free article] [PubMed] [Google Scholar]
- White A. P. et al. A global metabolic shift is linked to Salmonella multicellular development. PLoS One 5, e11814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara M., Yamazaki T., Oshima T. & Koga Y. sn-Glycerol-1-phosphate-forming activities in Archaea: separation of archaeal phospholipid biosynthesis and glycerol catabolism by glycerophosphate enantiomers. J. Bacteriol. 181, 1330–1333 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M. E. et al. Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: Carbon and energy flow contribute to the distinct biofilm growth state. BMC Genomics 13, 138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. et al. The gene yggE functions in restoring physiological defects of Escherichia coli cultivated under oxidative stress conditions. Appl. Environ. Microbiol. 71, 2762–2765 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuña L. G., Calderón I. L., Elías A. O., Castro M. E. & Vásquez C. C. Expression of the yggE gene protects Escherichia coli from potassium tellurite-generated oxidative stress. Arch. Microbiol. 191, 473–476 (2009). [DOI] [PubMed] [Google Scholar]
- Park S. & Imlay J. A. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185, 1942–1950 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamuri P., Mehta N., Burk A. & Maier R. J. Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and fur. J. Bacteriol. 188, 5325–5330 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysz M. A. et al. Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70, 6098–6112 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier F., Laine B. & Sautiere P. Characterization of the chromosomal protein MC1 from the thermophilic archaebacterium Methanosarcina sp. CHTI 55 and its effect on the thermal stability of DNA. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 951, 149–156 (1988). [DOI] [PubMed] [Google Scholar]
- Seaver L. C. & Imlay J. A. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183, 7173–7181 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Camper A. K., Ehrlich G. D., Costerton J. W. & Davies D. G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmokoff M. et al. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188, 4312–4320 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T. K. et al. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics 10, 3130–3141 (2010). [DOI] [PubMed] [Google Scholar]
- Seneviratne C. J., Wang Y., Jin L., Abiko Y. & Samaranayake L. P. Candida albicans biofilm formation is associated with increased anti-oxidative capacities. Proteomics 8, 2936–2947 (2008). [DOI] [PubMed] [Google Scholar]
- Hahn J., Inamine G., Kozlov Y. & Dubnau D. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol. Microbiol. 10, 99–111 (1993). [DOI] [PubMed] [Google Scholar]
- Facius D. & Meyer T. F. Novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol. Microbiol. 10, 699–712 (1993). [DOI] [PubMed] [Google Scholar]
- Chimileski S., Dolas K., Naor A., Gophna U. & Papke R. T. Extracellular DNA metabolism in Haloferax volcanii. Front. Microbiol. 5, 57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans B., Anantharaman V., Aravind L. & Koonin E. V. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3, 1625–1650 (2004). [DOI] [PubMed] [Google Scholar]
- Dominski Z. Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 42, 67–93 (2007). [DOI] [PubMed] [Google Scholar]
- Alzbutas G., Kaniusaite M. & Lagunavicius A. Enhancement of DNaseI salt tolerance by mimicking the domain structure of DNase from an extremely halotolerant bacterium Thioalkalivibrio sp. K90mix. PLoS One 11, e0150404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilain S., Pretorius J. M., Theron J. & Brözel V. S. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl. Environ. Microbiol. 75, 2861–2868 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M. K. et al. Gene expression profile of early in vitro biofilms of Streptococcus pneumoniae. Microbiol. Immunol. 56, 621–629 (2012). [DOI] [PubMed] [Google Scholar]
- De Vriendt K. et al. Proteomics of Shewanella oneidensis MR-1 biofilm reveals differentially expressed proteins, including AggA and RibB. Proteomics 5, 1308–1316 (2005). [DOI] [PubMed] [Google Scholar]
- Rice K. C. et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 104, 8113–8118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. et al. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J. Bacteriol. 196, 2355–2366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okshevsky M. & Meyer R. L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 41, 341–352 (2015). [DOI] [PubMed] [Google Scholar]
- Atkinson S., Cámara M. & Williams P. N-Acylhomoserine lactones, quorum sensing and biofilm development in Gram-negative bacteria. In, The Biofilm Mode of Life: Mechanisms and Adaptations (eds Kjelleberg S. & Givskov M.) 95–122 (Wymondham, UK: Horizon Bioscience, 2007). [Google Scholar]
- Oosthuizen M. C. et al. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68, 2770–2780 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M. E. & O’toole G. A. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P. Pseudomonas: Genomics and Molecular Biology 1–244 (Horizon Scientific Press, 2008). [Google Scholar]
- Liao Y. et al. Developing a genetic manipulation system for the Antarctic archaeon, Halorubrum lacusprofundi: investigating acetamidase gene function. Sci. Rep. 6, 34639 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns D. & Dyall-Smith M. 22 Cultivation of haloarchaea. Methods Microbiol. 35, 535–552 (2006). [Google Scholar]
- Ye J., Nielsen S., Joseph S. & Thomas T. High-resolution and specific detection of bacteria on complex surfaces using nanoparticle probes and electron microscopy. PLoS One 10, e0126404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith M. The Halohandbook: Protocols for Halobacterial Genetics. http://www.haloarchaea.com/resources/handbook (2009).
- Stepanović S. et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis 115, 891–899 (2007). [DOI] [PubMed] [Google Scholar]
- Adetunji V. O. & Isola T. O. Crystal violet binding assay for assessment of biofilm formation by Listeria monocytogenes and Listeria spp. on wood, steel and glass surfaces. Glob. Vet. 6, 6–10 (2011). [Google Scholar]
- Shaw P. D. et al. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94, 6036–6041 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J. B. O. et al. GFP-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67, 575–585 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J. & Maaløe O. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23, 99–112 (1967). [Google Scholar]
- Williams T. J. et al. Global proteomic analysis of the insoluble, soluble, and supernatant fractions of the psychrophilic archaeon Methanococcoides burtonii part I: the effect of growth temperature. J. Proteome Res. 9, 640–652 (2009). [DOI] [PubMed] [Google Scholar]
- Phanstiel D., Unwin R., McAlister G. C. & Coon J. J. Peptide quantification using 8-plex isobaric tags and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 15, 1693–1698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. et al. Proteomic analysis of rabbit tear fluid: Defensin levels after an experimental corneal wound are correlated to wound closure. Proteomics 7, 3194–3206 (2007). [DOI] [PubMed] [Google Scholar]
- Lim Y. A. et al. Abeta and human amylin share a common toxicity pathway via mitochondrial dysfunction. Proteomics 10, 1621–1633 (2010). [DOI] [PubMed] [Google Scholar]
- Wang H. J., Chen S. F. & Lo W. Y. Identification of cofilin-1 induces G0/G1 arrest and autophagy in angiotensin-(1-7)-treated human aortic endothelial cells from iTRAQ quantitative proteomics. Sci. Rep. 6, 35372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A. et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. A. et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME J. 3, 1012–1035 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.