Abstract
Different plant cultivars of the same genus and species can exhibit vastly different genetic transformation efficiencies. However, the genetic factors underlying these differences in transformation rate remain largely unknown. In barley, ‘Golden Promise’ is one of a few cultivars reliable for Agrobacterium-mediated transformation. By contrast, cultivar ‘Haruna Nijo’ is recalcitrant to genetic transformation. We identified genomic regions of barley important for successful transformation with Agrobacterium, utilizing the ‘Haruna Nijo’ × ‘Golden Promise’ F2 generation and genotyping by 124 genome-wide SNP markers. We observed significant segregation distortions of these markers from the expected 1:2:1 ratio toward the ‘Golden Promise’-type in regions of chromosomes 2H and 3H, indicating that the alleles of ‘Golden Promise’ in these regions might contribute to transformation efficiency. The same regions, which we termed Transformation Amenability (TFA) regions, were also conserved in transgenic F2 plants generated from a ‘Morex’ × ‘Golden Promise’ cross. The genomic regions identified herein likely include necessary factors for Agrobacterium-mediated transformation in barley. The potential to introduce these loci into any haplotype of barley opens the door to increasing the efficiency of transformation for target alleles into any haplotype of barley by the TFA-based methods proposed in this report.
Cultivated barley (Hordeum vulgare) is diploid cereal crop with a well-characterized genome of 5.1 Gbp1. Barley ranks fourth among cereals in production worldwide2 and is used for food, brewing and animal feed. Due to its importance as a crop, numerous agronomical and industrial traits of barley have been genetically analyzed. Genetic transformation is an essential tool in genetics; incorporating DNA into an organism’s genome allows examination of endogenous genes as well as introduction of novel traits. Unfortunately, genetic analysis in barley remains limited by the technical challenges involved in transformation, in stark contrast to the ease of Agrobacterium-mediated transformation in other plant species such as Arabidopsis thaliana and rice (Oryza sativa). The most commonly employed method of Agrobacterium-mediated transformation in barley was developed by Tingay et al.3, using immature embryos of ‘Golden Promise’ as an explant. Other than this technique, pollen culture of ‘Igri’ is one of a few alternative efficient methods for barley transformation4. This situation poses a great challenge for complementation analysis in barley, because analysis of traits of interest is restricted to those alleles non-functional in ‘Golden Promise’ or ‘Igri’. Analysis of functional alleles generally requires development of a genetic substitution line of the target allele from the donor into ‘Golden Promise’ via multiple generations of back-crossing. Recently Yeo et al.5 developed a new barley line ‘Golden SusPtrit’ (SG062N) that combines the transformation ability of ‘Golden Promise’ with the high susceptibility to rust disease inherited from ‘SusPtrit’, to develop a line of barley amenable for functional studies of non-host and partial resistance to fungal rust. The enhanced rate of Agrobacterium-mediated transformation in this recombined line suggested the existence of genetic factors responsible for the transformation efficiency of ‘Golden Promise’. That study also compared the genomic differences between a transformation-susceptible line (SG062N) and an insusceptible line (SG133N) in the same double haploid (DH) mapping population, to predict the genomic regions responsible for Agrobacterium-mediated transformation in ‘Golden Promise’5.
Four quantitative trait loci (QTLs) responsible for regeneration of green shoots from calli in barely were previously identified in a ‘Steptoe’ × ‘Morex’ cross by Mano et al.6, then Bregitzer & Campbell7 identified eight QTLs using same population. In addition Mano & Komatsuda8 identified three QTLs for shoot regeneration and two QTLs for callus growth in other population, ‘Azumamugi’ × ‘Kanto Nakate Gold’, although no genetic factors for transformation ability itself were specifically investigated. In Brassica oleracea, Cogan et al.9 identified three QTLs involved in Agrobacterium rhizogenes-mediated root transformation using mature seed. The genes responsible have not yet been identified, although two of three loci are speculated to be paralogs of each other originated by genome duplication around these regions10. In Solanum tuberosum, El-Kharbotly et al.11 found a locus involved in transformation efficiency linked with the R1 gene (resistance to Phytophthora infestans). In the case of rice, Nishimura et al.12 isolated a gene encoding ferredoxin-nitrate reductase (NiR) involved in a major QTL, promoter of shoot regeneration (PSR). In that case, the efficiency of regeneration in ‘Koshihikari’ was increased by introducing the intact genomic region or over-expressed cDNA of the NiR gene from ‘Kasalath’ in the callus derived from mature seed. Interestingly, Tyagi et al.13 reported that a QTL on chromosome 6H in barley, responsible for green shoot regeneration, was located close to the HvNiR locus (AK371794).
The efficiency of Agrobacterium-mediated transformation is heavily dependent on environmental and technical factors as well as the type and genetic makeup of the plant material under investigation. Technical factors include culture conditions, medium composition, type of Agrobacterium strain, and the specific binary vector and selection marker employed (reviewed by Cheng et al.14). The type of tissue used for transformation also plays an important role, as well as genetic factors that dictate interactions with Agrobacterium and subsequent development, including genes responsible for integration of T-DNA elements, cell division under selective conditions, and regeneration from callus14. Nam et al.15 reported that the efficiency of T-DNA integration into host genome depended on Arabidopsis ecotypes.
Recently, genome editing technologies, such as Trans Activator-Like Effector Nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated proteins 9 (CRISPR/Cas9) have been developed in plants. However, the utility of these technologies in barley is limited by the fact that transformation is necessary as the first step to introduce those nucleases in most plant species, as reported by Budhagatapalli et al.16 and Lawrenson et al.17
In this study, we mapped the genetic factors responsible for Agrobacterium-mediated transformation in ‘Golden Promise’ to develop a method that may be used for transformation, and thus, analysis of any barley gene. Instead of using genetic populations with fixed genotypes such as recombinant inbred lines (RILs) or double haploid (DH) lines, which requires phenotyping a large number of individuals under constant conditions, we used ‘Haruna Nijo’ (HN) × ‘Golden Promise’ (GP) derived F2 explants generated from individual immature embryos (Figure S1). We used the recombinant progeny of a cross GP × HN as a mapping population to identify the genetic factors that promote Agrobacterium-mediated transformation and demonstrated these genetic factors in an alternate cross between ‘Morex’ × GP. We propose that these genetic markers can be used to identify easily transformable progeny plants of the recalcitrant barley genotypes after crossing with GP, thus enabling gene/allele testing in the desirable background.
Results
Generation of transgenic HN × GP plants
We usually achieve approximately 10% efficiency of transformation using immature embryo of GP in our method. By the same experimental method, we didn’t obtain any transgenic plants from 261 immature embryo of HN. In addition, we tried to use immature embryos of BC3F8 recombinant chromosome substitute lines derived from a cross between GP and HN, with three rounds of backcrossing to HN as an explant for transformation, but did not obtain any transgenic plants from 4,661 immature embryos of these lines. Here, the F2 immature embryos derived from a cross between HN and GP (HN × GP) were inoculated with Agrobacterium tumefaciens strain AGL1 carrying a binary vector, pIG121-Hm, bearing genes for GUS (ß-glucuronidase) and HPT (Hygromycin PhosphoTransferase)18. From 3,013 immature embryos, 293 hygromycin-resistant calli were observed after selection on induction/selection medium. Green shoots were regenerated from 64 of those calli on regeneration medium. A total of 60 independent plants produced healthy roots on hygromycin-selective rooting medium, and were transferred to pots and grown in a controlled environment. DNA was isolated from leaves of the 60 candidate plants and used for subsequent genotyping and PCR analysis, which showed that all 60 candidate plants were PCR-positive for both GUS and HPT transgenes (Figure S2A,B). These data demonstrating incorporation of the transgenes suggested that all 60 candidates carried the genetic factors from GP that allowed Agrobacterium-mediated transformation.
SNP marker analysis of transgenic HN × GP plants
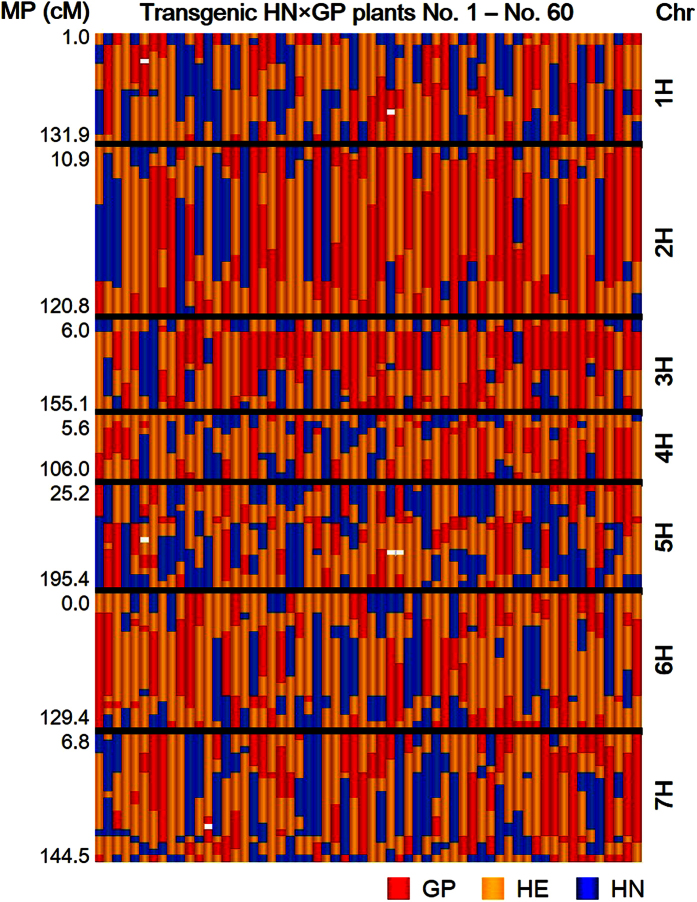
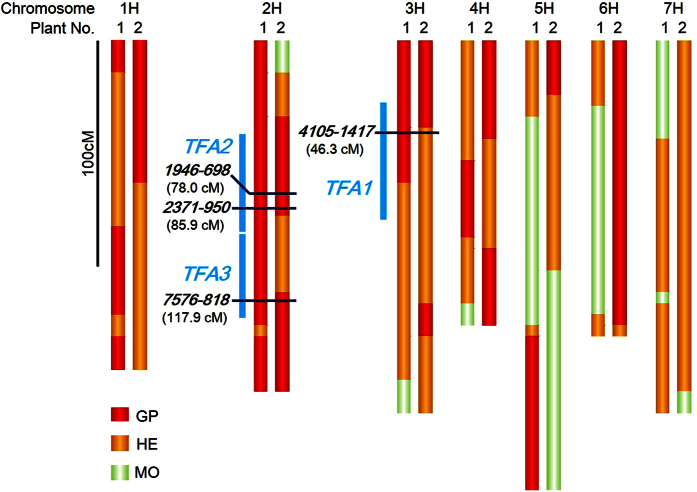
The genotypes of the 60 transgenic HN × GP plants were determined using a 124-marker subset of the 384-SNP Illumina GoldenGate® platform from the barley oligonucleotide pooled assay 1 (BOPA1)19. The genetic positions of all markers in this assay are available in a consensus genetic map19. The genotyping data for the transgenic HN × GP plants is presented in Table S1 and the genotype based on marker order in each chromosome is presented in Fig. 1. Each transgenic HN × GP plant had a unique genotype, indicating that these 60 plants resulted from independent transformation events.
Figure 1. Graphical overview of genotypes in transgenic HN × GP plants.
Genotyping of transgenic HN × GP plants from 1–60 (from left to right) was performed using the 384-SNP platform Illumina GoldenGate® assay. One hundred and twenty-four SNP markers are shown according to marker position (MP) in each chromosome (Chr) based on the consensus genetic distance of barley 1,19. Alleles from Haruna Nijo (HN), heterozygous regions (HE), and alleles from Golden Promise (GP) are shown as blue, yellow and red, respectively. The number on left show the end MP (cM) in each Chr.
Allele frequency and analysis of segregation distortion
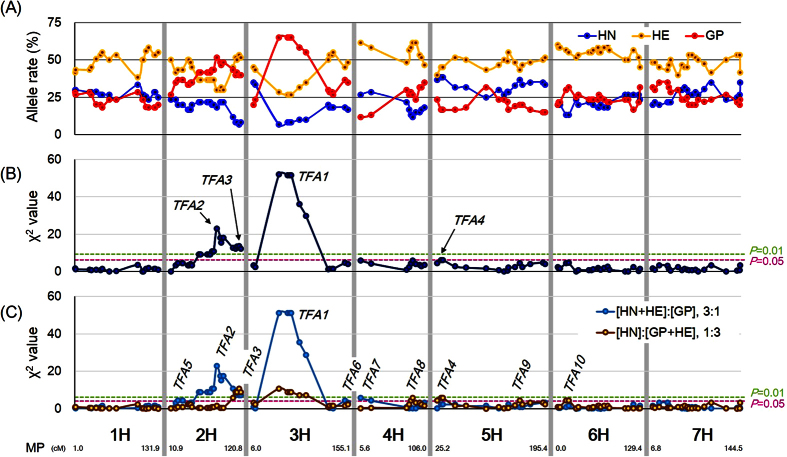
The allele frequency was calculated based on the genotyping data and mapped to the corresponding chromosomes (Fig. 2A). The Mendelian law of independent assortment predicts that the alleles homozygous for GP, heterozygous (HE), and homozygous for HN should segregate in a 1:2:1 ratio. The GP allele displayed a frequency of over 40% in the regions between markers 2580-1456 (position: 55.0 cM) and 3256-1196 (120.8 cM) in chromosome 2H and between 4105-1417 (46.3 cM) and 8020-87 (88.8 cM) in 3H (Fig. 2A and Table S1).
Figure 2. Allele and segregation analysis of SNP markers in transgenic HN × GP plants.
Allele segregation (A) and Chi-square (χ2) values (B,C) were calculated by genotyping 124 SNP markers in transgenic HN × GP plants. SNP markers are shown from short arm (left) to long arm (right) in each chromosome according to the order and genetic distance of markers in genome information of barley1,19. The number (cM) on bottom shows the end marker position (MP) in each chromosome. (A) Blue, yellow, and red line plots show the percentage of ‘Haruna Nijo’ (HN), heterozygous (HE) and Golden Promise (GP) alleles for each SNP marker. (B) Dark blue line plots show χ2 values calculated using 1:2:1 as the expected segregation ratio. Lime green and magenta dotted lines indicate statistical significance at the 1% and 5% levels, respectively (df = 2). (C) Light blue and brown line plots show χ2 values calculated using 3:1 as the expected segregation ratios for [HN + HE]:[GP] and [GP + HE]:[HN], respectively. Lime green and magenta dotted lines indicate statistical significance at the 1% and 5% levels, respectively (df = 1). TFAs (TFA1-10) indicate the loci showing segregation distortion.
Chi-square (χ2) test was used to identity segregation distortions caused by selection for transformation and regeneration (Fig. 2B,C). These χ2 values for the expected Mendelian ratio (HN:HE:GP = 1:2:1) were plotted with the marker positions on the barley genome (Fig. 2B). The maximum χ2 value was 52.1, with marker 4105-1417 on chromosome 3H, while the surrounding region between 4105-1417 (46.3 cM) and 8020-87 (88.8 cM) showed statistically significant segregation distortion (df = 2, p < 0.01). We named this region as TFA1 (Transformation Amenability 1). Another region with significant segregation distortion was observed on chromosome 2H between 2580-1456 (55.0 cM) and 3256-1196 (120.8 cM) (df = 2, p < 0.01). This region included two peaks with markers 6117-1507 (82.8 cM, χ2 = 22.9) and 7576-818 (117.9 cM, χ2 = 13.6), and these loci were named TFA2 and TFA3, respectively. An increased frequency of GP alleles at these regions suggested that a factor for transformation efficiency in GP was located there. Interestingly, an increased frequency of the HN allele was observed in a region of 5H between 8377-1022 (30.99 cM) and 4684-775 (34.25 cM), and was named as TFA4. The χ2 values of loci with significant segregation distortion are summarized in Table 1.
Table 1. Summary of detected TFAs.
| Locus | Chr No. | Interval (cM) | Peak position (cM) | Peak Marker | Allele frequency HN:HE:GP | χ2 value | ||
|---|---|---|---|---|---|---|---|---|
| 1:2:1a | 3:1b ([HN + HE]:GP) | 3:1b ([GP + HE]:HN) | ||||||
| TFA1 | 3H | 46.31–88.82 | 46.3 | 4105-1417 | 4:17:39 | 52.10** | 51.20** | 10.76** |
| TFA2 | 2H | 54.95–90.10 | 82.6 | 6117-1507 | 11:18:31 | 22.93** | 22.76** | 1.42 |
| TFA3 | 2H | 90.10–120.80 | 117.9 | 7576-818 | 4:32:24 | 13.60** | 7.20** | 10.76** |
| TFA4 | 5H | 25.23–34.25 | 31.0 | 8377-1022 | 23:27:10 | 6.23* | 2.22 | 5.69* |
| 34.3 | 4684-775 | |||||||
| TFA5 | 2H | 21.61–29.15 | 21.6 | 1865-396 | 12:26:22 | 4.4 | 4.36* | 0.8 |
| 28.4 | 7747-1056 | |||||||
| 29.2 | 7032-201 | |||||||
| TFA6 | 3H | — | 150.4 | 7818-967 | 11:27:22 | 4.63 | 4.36* | 1.42 |
| TFA7 | 4H | 5.55–21.61 | 5.6 | ABC14522-1-8-350 | 16:37:07 | 5.97 | 5.69* | 0.09 |
| TFA8 | 4H | 84.30–87.49 | 87.5 | 4564-604 | 7:37:16 | 5.97 | 0.09 | 5.69* |
| TFA9 | 5H | — | 155.1 | ConsensusGBS0451-1 | 22:26:12 | 4.4 | 0.8 | 4.36* |
| TFA10 | 6H | 12.54–16.97 | 17.0 | 1769-545 | 8:33:19 | 4.63 | 1.42 | 4.36* |
a,bAsterisks indicate statistical significance by Chi-square test (adf = 2, bdf = 1, **p < 0.05, **p < 0.01).
To examine the direction of dominance of the identified loci, the dominance segregation ratio (3:1) was used for χ2 tests (Fig. 2C). Using the assumption that the GP allele was functionally recessive ([HN + HE]:[GP] = 3:1), the regions or markers showing significant distortion were observed between markers 1865-396 (21.6 cM) and 7032-201 (29.2 cM) in 2H, at 7818-967 (150.4 cM) in 3H and between ABC14522-1-8-350 (5.5 cM) and 2055-947 (21.6 cM) in 4H, in addition to TFA1, TFA2 and TFA3. These additional loci were named as TFA5, TFA6 and TFA7, respectively (light blue line in Fig. 2C and Table 1). Meanwhile, using the assumption that the GP allele was dominant ([GP + HE]:[HN] = 3:1), loci between 4986-1214 (84.3 cM) and 4564-604 (87.5 cM) in 4H, at ConsensusGBS0451-1 (155.1 cM) in 5H and between ConsensusGBS0346-1 (12.5 cM) and 1769-545 (17.0 cM) in 6H were identified, alongside TFA1, TFA3 and TFA4. These new loci were named as TFA8, TFA9 and TFA10, respectively (brown line in Fig. 2C and Table 1). Interestingly, the χ2 value for TFA2 was not significant under the GP-dominant condition, although it showed significant segregation distortion under [HN + HE]:[GP] = 3:1.
Cloning and genotyping of the HvNiR gene
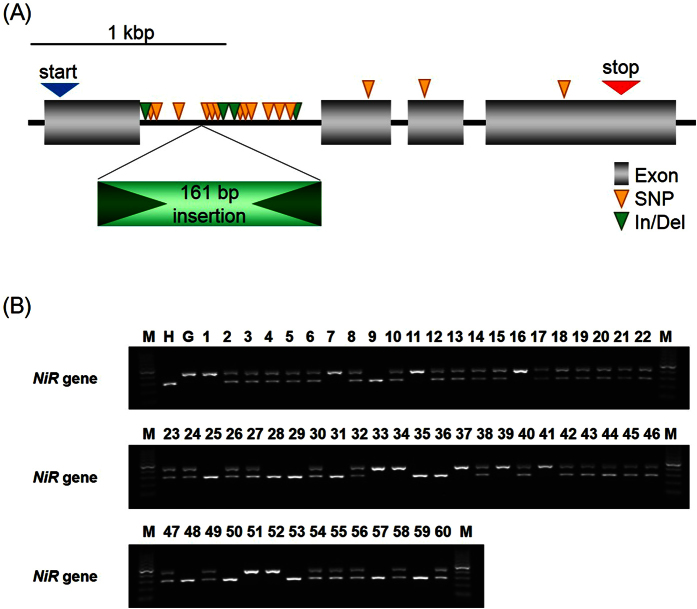
To examine whether the barley ortholog of rice NiR played a role in transformation efficiency, we isolated the NiR ortholog from HN and GP for comparison between these two cultivars. The putative HvNiR gene, including predicted start and stop codons, was 2,965 bp in HN and 3,097 bp in GP. Analysis of the HvNiR genes in HN and GP revealed 4 exons and 3 introns, and a comparison of the DNA sequence between HN and GP yielded 14 SNPs in introns and 3 synonymous SNPs in exons (Fig. 3A). In addition, the HN allele had a 7-bp and a 23-bp insertion in the first intron, while the GP allele had a 161-bp insertion homologous to a retrotransposon-like sequence in the first intron (Fig. 3A). Despite these differences, the predicted amino acid sequences for HvNiR in HN and GP were identical.
Figure 3. Genic structure and allele of the HvNiR gene in transgenic HN × GP plants.
(A) The structure of HvNiR genic region including introns, start and stop codons. Yellow and green wedges show SNPs and Insertions/Deletions, respectively, between ‘Haruna Nijo’ and ‘Golden Promise’. A 161 bp insertion of retrotransposon-like sequence was present in the first intron of HvNiR in ‘Golden Promise’. (B) Genotyping of transgenic HN × GP plants was performed using HvNiR-specific primers. The expected sizes of PCR fragments were 280 bp and 417 bp in alleles of ‘Haruna Nijo’ (H) and ‘Golden Promise’ (G), respectively. M, 100 bp ladder marker; 1–60, individual transgenic HN × GP plants. Original gel image is available from Supplementary Information.
To prove whether the insertions in the first intron of HvNiR might be related to transformation efficiency, we designed a codominant sequence-tagged-site (STS) marker to amplify the region spanning the GP-specific 161-bp insertion for genotyping, and performed an STS analysis using DNA from the 60 transgenic HN × GP plants. Analysis of the alleles revealed a segregation ratio of 12:37:11 (HN:HE:GP), which fits a monofactorial Mendelian ratio (Fig. 3B, χ2 = 3.3 at 1:2:1 ratio) in the absence of segregation distortion.
Genome-wide locus-locus interactions
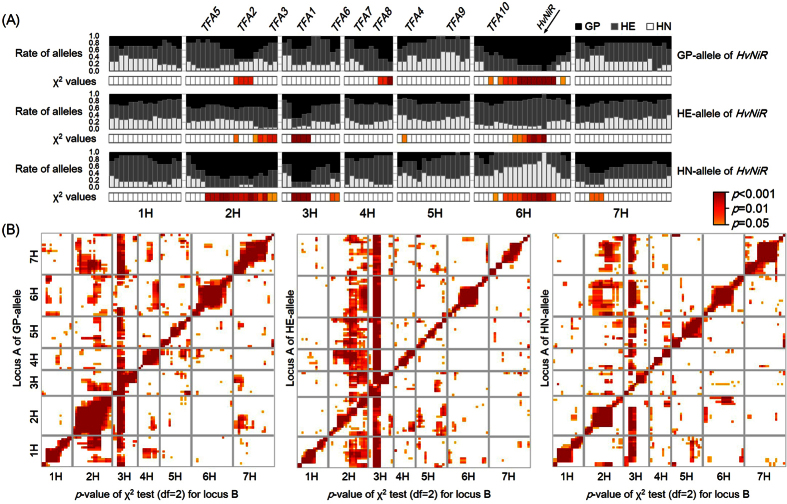
Normally, alleles corresponding to opposing phenotype data (e.g., resistant vs. sensitive to a given stress) are used for locus-locus interaction analysis. Since we used transgenic plants to map transformation efficiency, i.e. most loci have transformation-amenable alleles, there is selection bias in our mapping populations, and the standard software does not serve to detect locus-locus interactions. To analyze locus-locus interaction in the transgenic HN × GP plants, we first grouped genotypes according to the allele type of a single marker, we then calculated the segregation ratio and χ2 value of other markers relative to the first marker. For instance, using the HvNiR locus, the HN × GP plants were placed into groups of 11 plants for GP, 37 plants for HE and 12 plants for HN based on the alleles at HvNiR (top, middle and bottom panels in Fig. 4A, respectively), we then compared the ratio (band chart) and p-values of χ2 test (heat map, df = 2) of alleles in each of the other loci. Because HvNiR was located on the long arm of chromosome 6H, the frequency of alleles of this region matched the frequency of HvNiR. On chromosome 2H, distortion favoring the GP allele was observed around TFA2 in GP- and HN-type HvNiR groups but not in the HE-type group (Fig. 4A). Also around TFA3, GP and HE alleles were dominant in HN- and HE-type HvNiR groups, but not in the GP-type group. Interestingly, there was no segregation distortion around TFA1 among the GP-type HvNiR plants. Additional segregation distortion was also observed on long arm of chromosome 4H in GP-type HvNiR plants (Fig. 4A).
Figure 4. Analysis of locus-locus interactions in transgenic HN × GP plants.
(A) Percentages of ‘Golden Promise’ (GP, black)-, heterozygous (HE, dark gray)- and ‘Haruna Nijo’ (HN, light gray) alleles were calculated among transgenic HN × GP plants having GP (top), HE (middle) and HN (bottom) alleles of HvNiR and shown as band charts. One hundred and ten SNP markers and the HvNiR gene marker are shown from short arm (left) to long arm (right) of the chromosomes. TFAs (TFA1-10) indicate the loci showing segregation distortion in Fig. 2. The heat maps based on Chi-square (χ2) tests of the markers (df = 2) are shown under each band chart. (B) Heat maps were made based on p-values of χ2 tests (df = 2) representing significance of segregation distortions in transgenic HN × GP plants. The markers (locus A) were grouped into GP (left panel), HE (middle panel) and HN (right panel) alleles and ordered on vertical axis from short arm (bottom) to long arm (top) in each chromosome. Then, χ2 tests (df = 2) were performed for all other markers (locus B) ordered on horizontal axis from short arm (left) to long arm (right) in each chromosome; the p-values representing degree of significance are indicated by color grade.
Using above method, χ2 tests for all of the non-redundant markers (111 in total) were performed in the transgenic HN × GP plants. First, we counted the number of the allele combinations between one locus (locus A) and another (locus B) (e.g. GP-HN for locus A-B), then calculated χ2 values (df = 2) for segregation of locus B among each type of allele on locus A. Fig. 4B shows the heat maps of p-values representing significant segregation distortion of locus B on the horizontal axis against locus A on the vertical axis, which are separated into the cases of GP (left panel), HE (middle panel) and HN alleles (right panel). A segregation distortion was observed around TFA1 as locus B against almost all of the markers, except for the region in chromosome 6H including HvNiR for which the HN × GP plants had the GP allele (left panel in Fig. 4B). The plants having the GP allele in this region of 6H showed segregation distortion in the long arm of 4H as well as at TFA2 (left panel in Fig. 4B). Apart from these loci, the major segregation distortions were observed at 1H-4H, 2H-7H, 3H-7H, 4H-5H, 7H-2H and 7H-4H with GP-allele markers as locus A (left panel in Fig. 4B). With HN-allele markers as locus A in HN × GP plants, segregation distortions were observed at 1H-2H, 1H-5H, 2H-6H, 3H-2H, 6H-2H and 7H-2H in addition to ones with TFA1 (right panel in Fig. 4B). Particularly the plants with the HN alleles in 6H and 7H as locus A showed widespread segregation distortion in 2H as locus B.
Recursive test for genetic factors of transformation amenability by an alternative cross
To demonstrate the TFAs, particularly TFA1, TFA2 and TFA3, as important genetic factors for transformation in barley, we performed Agrobacterium-mediated transformation on F2 immature embryos derived from a cross between ‘Morex’ (MO) and ‘Golden Promise’ (MO × GP). Two independent transgenic MO × GP plants from 1,722 immature embryos were obtained and genotyped using the GoldenGate® platform. The genotypes of these transgenic MO × GP plants and the locations of TFA1, TFA2 and TFA3 are illustrated in Fig. 5. Most of chromosomes 1H, 2H, 3H and 4H corresponded to the homozygous GP or heterozygous genotype in both plants; however, large regions in chromosomes 5H, 6H and 7H corresponded to the MO genotype in one or both plants. At TFA1, one marker 4105-1417 detected both GP and heterozygous alleles in the MO × GP transgenic plants. Additionally, the markers 1946-698 (78.0 cM) and 2371-950 (85.9 cM), on TFA2, and marker 7576-818 (117.9 cM) in TFA3, all detected the GP allele. These results confirm that GP alleles of TFA1, TFA2 and TFA3 are effective for transformation in an alternative haplotype of barley.
Figure 5. Graphical genotypes in transgenic MO × GP plants.
Genotyping of two transgenic MO × GP plants, No. 1 and No. 2, was performed by 384-SNP platform Illumina GoldenGate® assay. One hundred and seventy-eight SNPs markers were positioned from short arm (above) to long arm (bottom) of chromosomes according to the order of marker position in the consensus genetic distance of barley 1,19. Alleles of Morex (MO), hetero (HE) and Golden Promise (GP) were shown as green, yellow and red, respectively. Light blue bars and markers with position (cM) indicate the regions and nearest marker of TFL1, TFL2 and TFL3, respectively.
Discussion
The success of genetic transformation in plants depends on various factors. The immature embryo of ‘Golden Promise’ (GP) is the most reliable material reported for Agrobacterium-mediated transformation in barley20. Previously, we attempted to use BC3F8 recombinant chromosome substitution lines for transformation assays to identify possible genetic factors regulating Agrobacterium-mediated transformation in barley. However, we did not obtain a single transgenic plant from 4,661 immature embryos, likely due to insufficient factors in the genetic material favoring transformation. In this study, to increase the chance of generating a population of transformants for further study, we employed a different approach, utilizing the F2 generation of immature embryos derived from a cross between ‘Haruna Nijo’ (HN) and GP as the material for Agrobacterium-mediated transformation. With this new approach, we successfully obtained 60 transgenic barley plants (HN × GP) from 3,013 F2 individuals. Utilizing these 60 transgenic plants for genome-wide analysis, we identified regions of the genome that display significant segregation distortion of alleles from the expected 1:2:1 ratio. We found major regions of segregation distortion in chromosomes 2H and 3H, which we named TFA1, TFA2 and TFA3, where the GP allele was highly over represented in the transformants (Fig. 2B). The relatively higher rate of transformation in heterozygote individuals also suggests that some the possible factors regulating transformation efficiency exhibit dominance (Fig. 2A). Importantly, the GP alleles of these TFAs were conserved in two transgenic plants derived from an alternative cross, MO × GP (Fig. 5). This report is the first identifying the loci underlying transformation amenability in barley by measuring segregation distortion of alleles in the F2 population. We also confirmed that organelle genomes likely do not play a role in transformation amenability, since all HN × GP and MO × GP transgenic plants originated from a cross using GP as the pollen parent. To confirm that the distortion was not caused by a reproductive barrier between HN and GP, we performed a SNP analysis using the F10 generation of RILs derived from the same cross (Table S2). We found that three genomic regions, on 2H, 3H and 5H, in the RILs were distorted; however, these regions were in different positions from TFAs and were caused by overaccumulation of the HN allele, rather than the GP allele (Table S2). The distortions observed in the RILs could be due local adaptation of the HN genotype to the field in Kurashiki, Japan during generation advancement of the population. Overall, however, these findings indicate that the distortions that led to the identification of the TFAs were not caused by abnormal segregation between GP and HN genotypes due to a reproductive barrier.
Several genetic factors that affected the efficiency of Agrobacterium-mediated transformation have been documented, for example, factors responsible for interaction with Agrobacterium, growth rate of calli, as well as regeneration ability14,20. Rikiishi et al.21 reported that barley cultivar ‘Lenins’ has higher regeneration ability compared to some other cultivars, but that trait alone is not enough for efficient transformation. Yeo et al.5 suggested that a region close to TFA2 on chromosome 2H, of the line SG062N, may affect transformation efficiency. The locations of TFA6 and TFA8 also correspond with regions previously reported by Yeo et al.5 Mano et al.6, Mano & Komatsuda8 and Bregitzer & Campbell7, reported that QTLs promoting green shoot regeneration from callus in barley were observed on chromosomes 1H, 2H (Qsr1), 3H (Qsr2), 5H (Qsr4), 6H (Qsr3) and 7H. Because TFA2 was mapped close to Qsr1, this locus could be related to regeneration as well as increased transformation efficiency. TFA3 was mapped adjacent to Qcg1, and the function of TFA3 might be involved in propagation ability of calli under hygromycin selection. We found that TFA1 was mapped between Qsr2 and Qcg2. Thus, we speculate that TFA1 may also be involved in regeneration or growth efficiency of calli. In dicot plants, Nam et al.15 mentioned that the efficiency of integration of T-DNA into Arabidopsis genome was dependent on its genotype/ecotype and controlled by a single major locus. Also Sparrow et al.22,23 reported that the genetic analysis of Agrobacterium susceptibility using a doubled haploid mapping population of Brassica oleracea. They found the genetic segregation of efficiency of GUS expression implying the integration of T-DNA, and the related QTL for crown gall formation. While the exact function of TFAs in barley is still unknown, it is possible that some of them play similar roles in responding to Agrobacterium including integration of T-DNA into barley genome, cell propagation and tissue regeneration. Interestingly, TFA2 and TFA3 showed different patterns of segregation distortion based on 3:1 segregation ratios (Table 1, Fig. 3C). We speculate that GP allele function may be recessive at TFA2 but dominant at TFA3 for transformation amenability. Dominance of the HN allele was observed at TFA4, TFA7 and TFA9. This result indicated that there could be negative factors for transformation in GP at these loci, or possibly positive factors in HN, in which regeneration levels are moderate24,25. TFA4 was mapped near the locus ari-e.GP, which is a marker of the semi-dwarf phenotype due gibberellin insensitivity26,27. Therefore, this locus might control the growth and development of callus tissue as well as plant biomass. Number of transgenic plants in all combinations of alleles at TFA4, TFA7 and TFA9 and Chi-square values among genotypes were shown in Figure S3 and Table S3, respectively. Every single locus segregation in TFA4, TFA7 and TFA9 showed significant difference at least one of the co-dominant or dominant segregations (Fig. 2 and Table S3). The allelic combination of TFA4 and TFA9 also showed statistical significance, indicating that the combination of HN-alleles of TFA4 and TFA9 might have positive epistatic effect for transformation amenability. The result encourage us that the substitution of HN allele of TFA4 and TFA9, from GP allele may develop more efficient transformation amenable lines. As well, the regeneration level of MO was severely low in our condition25. If we perform further analysis using MO × GP, we may find another factors which Morex has as negative for regeneration. This might be reason why we obtained only two transgenic plants from 1722 immature embryos of MO × GP. A gene encoding ferredoxin-nitrate reductase (NiR) contributes to increased regeneration and transformation efficiency in rice12. Tyagi et al.13 reported that an e-QTL in barley for green shoot regeneration was located on the marker HVSMEI0013e16r2, near the NiR gene on chromosome 6H. To study the effect of NiR on Agrobacterium-mediated transformation in barley, we isolated the barley orthologue of NiR (HvNiR) in GP, which contained 3 introns (Fig. 3A). The HN HvNiR allele differed from the GP allele by 3 synonymous SNPs in exons, and 12 SNPs and 5 insertion/deletions in the first intron. We found that the segregation of alleles on HvNiR followed the 1:2:1 Mendelian ratio in transgenic HN × GP plants, as would be expected if the alleles were independently assorted (Fig. 3B). We further analyzed locus-locus interaction between HvNiR and TFAs (Fig. 4A,B) and found that the plants with the GP allele at HvNiR showed segregation distortion in TFA2 but not in TFA1 and TFA3. In the case of the HN allele at HvNiR, plants showed segregation distortion on TFA1 as well as the region including TFA2 and TFA3. We speculate that HvNiR may complement the function of TFA1, TFA2 and TFA3, although HvNiR itself did not show a significant χ2 value for distortion (Fig. 4A,B). These results suggest that HvNiR may contribute to transformation efficiency in barley, as well as in rice.
Currently, if researchers want to carry functional analyses of genes for which GP does not carry a suitable haplotype, they generally develop substitution barley lines backcrossed with GP for transformation. For instance, Deng et al.28 made BC3F2 barley lines carrying a VRN1-HA (VRN1, vernalization 1; HA, haemagglutinin epitope tag) transgene for analysis to discover the target genes of VRN1, which encodes a MADS box transcription factor. To do this, the authors first had to generate BC2F1 plants of winter-type GP introgressed with vernalization-requiring genome sets by backcrossing twice before crossing with the transgenic GP. Such studies require a substantial amount of time to develop the plants necessary to conduct further analysis.
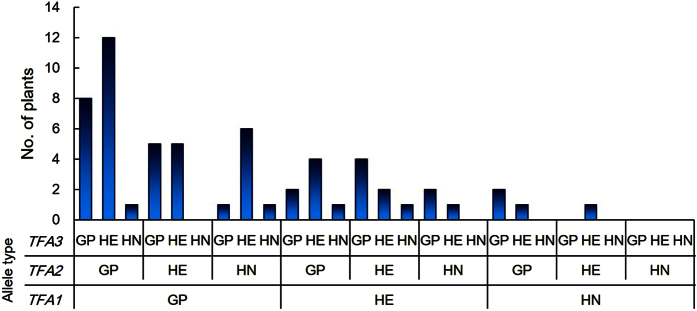
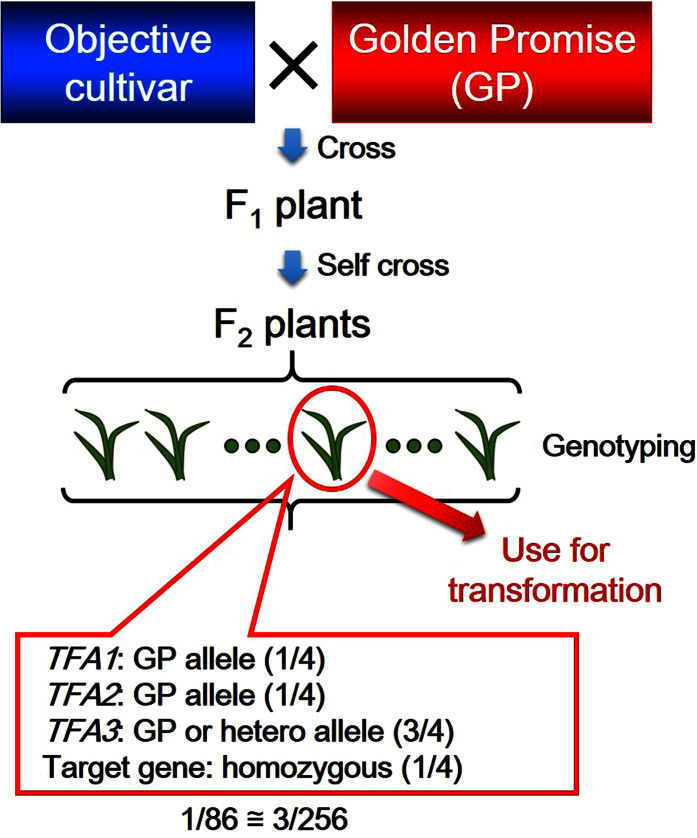
The identification and characterization of the TFAs should enable the scientific community to conduct complementation experiments relatively rapidly in any haplotype of barley, which has thus far been limited to the alleles of transformation-efficient cultivars such as ‘Golden Promise’ and ‘Igri’. We have demonstrated the ability to conduct transformation from any haplotype using the F2 generation derived from a cross between the haplotype of interest and GP (Fig. 5). Figure 6 shows the number of transgenic HN × GP plants with various allele combinations in TFA1, TFA2 and TFA3. Among 60 transgenic HN × GP plants, 20 plants had only the GP alleles in TFA1 and TFA2, and the GP allele or heterozygosity in TFA3. While our study did not identify the specific genes necessary for barley transformation, we have demonstrated that these specific allele combinations at the identified TFA regions increase the rate of successful transformation. We hereby propose our method as a possible universal complementation system for the barely research community. This TFA-based method involves genotyping F2 plants to collect the necessary alleles of TFAs segregating in the population as well as the target allele to be analyzed (Fig. 7). Importantly, the desired combination should be fairly common, with approximately one out of every 86 F2 plants harboring the correct sets of TFA alleles as well as the target allele. In our estimation, it takes in total approximately 13 months minimum to obtain transgenic T0 plants from the first cross in our system; 1 month to harvest matured F1 seed; 5 months to harvest F2 seeds on the F1 plant; 4 months to perform genotyping and to harvest immature embryo for transformation; 3 months to generate transgenic T0 plants.
Figure 6. Frequency of transgenic HN × GP plants classified by genotype at TFA1, TFA2 and TFA3.
Transgenic HN × GP plants were grouped by the haplotype of TFA1, TFA2 and TFA3 and counted.
Figure 7. The schematic overview of proposed TFA-based universal complementation system in barley.
For complementation tests of target genes, F2 plants would be generated from a cross between the objective cultivar with the target gene and cv. ‘Golden Promise’. Among the F2 plants, genotyping would be carried out to identify plants with the GP allele at TFA1 and TFA2, and GP/heterozygous at TFA3, as well as homozygous for the target gene. One plant fitted to the purpose is predicted to be found among approximately each 86 F2 plants.
In conclusion, in this study, we generated transgenic barley plants from the progeny of a cross between ‘Haruna Nijo’ and ‘Golden Promise’. Genotypic analysis of the HN × GP transgenic plants, revealed 3 significant and 7 suggestive QTLs represented by segregation distortion regions, which we named TFAs for Transformation Amenability. Because these transgenic plants were generated via successful Agrobacterium-mediated transformation events, we believe that these segregation distortions identified regions in the genome containing genetic factors that are necessary for or enhance successful transformation. To demonstrate the role of TFAs in producing transgenic plants, we also showed the presence of the TFAs in transgenic plants generated from a different cross (MO × GP). Inter-locus interactions were observed for several genomic regions, particularly between TFAs and chromosome 6H including the HvNiR gene. We anticipate utilization of these TFAs to enable more efficient analysis of barley genomes, genes, and alleles, not only in ‘Golden Promise’, but in all barley cultivars and hybrid progenies.
Methods
Plant materials
The hybrid F1 barley plants (HN × GP or MO × GP) were derived from ‘Haruna Nijo’ (HN) or ‘Morex’ (MO), respectively, as a seed parent and ‘Golden Promise’ (GP) as a pollen parent. Plants were grown under 16 hours of daylight per 24 hour cycle and 15 °C/13 °C (day/night). The caryopses were harvested for isolation of immature embryos approximately 14 days after pollination.
Agrobacterium-mediated transformation
Transformation in barley was performed following the protocol of Hensel et al.29 The dissected immature embryos were soaked in 100 mg/l acetosyringone solution at 43 °C for 4 minutes before inoculation with Agrobacterium following Hiei et al.30 and Zheng et al.31 Agrobacterium tumefaciens strain AGL1 carrying the binary vector pIG121-Hm18 bearing the ß-glucuronidase (GUS) and hygromycin phosphotransferase (HPT) genes was used for inoculation.
DNA analysis by PCR
Genomic DNA was extracted from the leaves of barley plants using the DNeasy Plant mini kit (QIAGEN, Germany). PCR amplification was performed with the following program: initial denaturation at 95 °C for 2 min, 28 cycles of denaturation at 95 °C for 30 sec, annealing at 55 °C (for the GUS gene) or 60 °C (for HPT) for 30 sec, and extension at 72 °C for 30 sec, with a final extension at 72 °C for 10 min. The 10-μl reaction mixture for PCR included 50 ng genomic DNAs as a template, 1 × GoTaq® Green Master Mix (Promega, USA) and 0.5 μM each specific primer, as follows: gus4 (5′-AACAGTTCCTGATTAACCACAAACC-3′) and gus5 (5′-GCCAGAAGTTCTTTTTCCAGTACC-3′) for the GUS gene, and hph1 (5′-GCTGGGGCGTCGGTTTCCACTATCGG-3′) and hph2 (5′-CGCATAACAGCGGTCATTGACTGGAGC-3′) for the HPT gene.
Cloning the HvNiR gene
To clone the HvNiR gene (Acc. No. LC097010 and LC097011 and for GP and HN, respectively), PCR amplification was performed with following program: initial denaturation at 98 °C for 2 min, 30 cycles of denaturation at 98 °C for 10 sec, primer annealing at 55 °C for 15 sec, and extension at 72 °C for 150 sec, with a final extension at 72 °C for 10 min. The 30-μl reaction mixture for PCR included 50 ng genomic DNA from ‘Haruna Nijo’ or ‘Golden Promise’ as a template, 1 × PrimeSTAR® Max DNA Polymerase (TAKARA, Japan) and 0.5 μM specific primers HvNiR-F1 (5′-AACCACAAGCAGCATCCATG-3′) and HvNiR-R1 (5′-GAGATCATCAGGAGAAGGAG-3′). PCR products were cloned into pCRTM4-TOPO® (Invitrogen, USA) and sequenced according to manufacturer’s protocol using the TOPO® TA Cloning® Kit for Sequencing (Invitrogen, USA).
Genotyping
The 384-SNP platform of Illumina GoldenGate® oligonucleotide pool assays19 was employed for genotyping all transgenic plants generated in this study using genomic DNA extracted with the DNeasy Plant mini kit, according to the manufacturer’s protocol (Illumina, USA).
For genotyping with the HvNiR gene marker, PCR amplification was performed with a modified touchdown PCR program32: initial denaturation at 95 °C for 2 min, 10 cycles of melting at 95 °C for 30 sec, annealing at 65 °C decreased 1 °C/cycle for 20 sec, and extension at 72 °C for 30 sec, followed by 20 cycles of denaturation at 95 °C for 10 sec, annealing at 55 °C for 20 sec and extension at 72 °C for 30 sec, with a final extension at 72 °C for 10 min. The 10-μl reaction mixture for PCR included 50 ng genomic DNA as template, 1 × GoTaq® Green Master Mix and 0.5 μM specific primers HvNiR-GPspeF (5′-CCCGCATGCATATCCCATAG-3′) and HvNiR-GPspeR (5′-CCCGGACTAGTCCAAGATAC-3′). PCR products were analyzed by electrophoresis with 1.5% agarose gels (Wako, Japan) in TBE running buffer.
Detection of distorted segregation
Chi-square (χ2) tests were performed to identify genome regions displaying significant segregation distortion using the expected Mendelian ratio of 1:2:1 (df = 2) for the null hypothesis as well as assumed dominant ratios of 3:1 or 1:3 (df = 1). Microsoft® Excel® was used for all calculations, charts, and heat maps.
Additional Information
How to cite this article: Hisano, H. and Sato, K. Genomic regions responsible for amenability to Agrobacterium-mediated transformation in barley. Sci. Rep. 6, 37505; doi: 10.1038/srep37505 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Mr. Yukoh Hiei, Dr. Toshihiko Komari (Japan Tobacco, Iwata, Japan), Dr. Masanao Sato (IAB, Keio Univ., Tsuruoka, Japan), Dr. Kazuyoshi Takeda and Dr. Daisuke Saisho (IPSR, Okayama Univ., Kurashiki, Japan) for discussing the results of this work. Also we thank Ms. Yuka Motoi and Ms. Hiromi Ando for technical assistance. Barley seeds were provided by the National Bioresource Project of Barley, MEXT of Japan. This work was supported by Japanese Society for the Promotion of Science (JSPS) KAKENHI, Grant Numbers 24880025, 26850004 and 16K18634.
Footnotes
The authors declare no competing financial interests.
Author Contributions H.H. and K.S. designed this study and raised funding. H.H. collected data and drafted the manuscript. Both authors contributed to writing the manuscript and approved the final version.
References
- The International Barley Genome Sequencing Consortium (IBGSC). A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716 (2012). [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO), FAOSTAT, Food and agricultural commodities production., available from: http://faostat3.fao.org/browse/rankings/commodities_by_regions/E (Date of access: 11/Oct/2016) (2013).
- Tingay S. et al. Agrobacterium tumefaciens-mediated barley transformation. The Plant J. 11, 1369–1376 (1997). [Google Scholar]
- Kumlehn J., Serazetdinova L., Hensel G., Becker D. & Loerz H. Genetic transformation of barley (Hordeum vulgare L.) via infection of androgenetic pollen cultures with Agrobacterium tumefaciens. Plant Biotechnol. J. 4, 251–261 (2006). [DOI] [PubMed] [Google Scholar]
- Yeo F. K. S. et al. Golden SusPtrit: a genetically well transformable barley line for studies on the resistance to rust fungi. Theor. Appl. Genet. 127, 325–337 (2014). [DOI] [PubMed] [Google Scholar]
- Mano Y., Takahashi H., Sato K. & Takeda K. Mapping genes for callus growth and shoot regeneration in barley (Hordeum vulgare L.). JPN J. Breed. 46, 137–142 (1996). [Google Scholar]
- Bregitzer P. & Campbell R. D. Genetic markers associated with green and albino plant regeneration from embryogenic barley callus. Crop Sci. 41, 173–179 (2001). [Google Scholar]
- Mano Y. & Komatsuda T. Identification of QTLs controlling tissue-culture traits in barley (Hordeum vulgare L.). Theor. Appl. Genet. 105, 708–715 (2002). [DOI] [PubMed] [Google Scholar]
- Cogan N. et al. Identification of genetic factors controlling the efficiency of Agrobacterium rhizogenes-mediated transformation in Brassica oleracea by QTL analysis. Theor. Appl. Genet. 105, 568–576 (2002). [DOI] [PubMed] [Google Scholar]
- Cogan N. O. I. et al. Identification and characterization of QTL controlling Agrobacterium-mediated transient and stable transformation of Brassica oleracea. Plant Biotechnol. J. 2, 59–69 (2004). [DOI] [PubMed] [Google Scholar]
- El-Kharbotly A., Jacobsen E., Stiekema W. J. & Pereira A. Genetic localisation of transformation competence in diploid potato. Theor. Appl. Genet. 91, 557–562 (1995). [DOI] [PubMed] [Google Scholar]
- Nishimura A. et al. Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proc. Nat. Acad. Sci. USA 102, 11940–11944 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi N., Dahleen L. S. & Bregitzer P. Candidate genes within tissue culture regeneration QTL revisited with a linkage map based on transcript-derived markers. Crop Sci. 50, 1697–1707 (2010). [Google Scholar]
- Cheng M., Lowe B. A., Spencer T. M., Ye X. & Armstrong C. L. Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. In Vitro Cell. Dev. Biol. Plant 40, 31–45 (2004). [Google Scholar]
- Nam J., Matthysse A. G. & Gelvin S. B. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9, 317–333 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhagatapalli N., Rutten T., Gurushidze M., Kumlehn J. & Hensel G. Targeted modification of gene function exploiting homology-directed repair of TALEN-mediated double-strand breaks in barley. G3: Genes Genom. Genet. 5, 1857–1863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson T. et al. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T. & Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994). [DOI] [PubMed] [Google Scholar]
- Close T. J. et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood W. A. Advances and remaining challenges in the transformation of barley and wheat. J. Exp. Bot. 63, 1791–1798 (2012). [DOI] [PubMed] [Google Scholar]
- Rikiishi K., Matsuura T., Maekawa M., Noda K. & Takeda K. Genetic analysis of tissue culture traits in barley cv. ‘Lenins’. Plant Breed. 122, 99–104 (2003). [Google Scholar]
- Sparrow P. A. C., Dale P. J. & Irwin J. A. The use of phenotypic markers to identify Brassica oleracea genotypes for routine high-throughput Agrobacterium-mediated transformation. Plant Cell Rep. 23, 64–70 (2004a). [DOI] [PubMed] [Google Scholar]
- Sparrow P. A. C., Townsend T. M., Arthur A. E., Dale P. J. & Irwin J. A. Genetic analysis of Agrobacterium tumefaciens susceptibility in Brassica oleracea. Theor. Appl. Genet. 108, 644–650 (2004b). [DOI] [PubMed] [Google Scholar]
- Furusho M., Suenaga K. & Nakajima K. Production of haploid barley plants from barley × maize and barley × Italian ryegrass crosses. JPN J. Breed. 41, 175–179 (1991). [Google Scholar]
- Hisano H., Matsuura T., Mori I. C., Yamane M. & Sato K. Endogenous hormone levels affect the regeneration ability of callus derived from different organs in barley. Plant Physiol. Biochem. 99, 66–72 (2016). [DOI] [PubMed] [Google Scholar]
- Forster B. P. Mutation genetics of salt tolerance in barley: An assessment of Golden Promise and other semi-dwarf mutants. Euphytica 120, 317–328 (2002). [Google Scholar]
- Liu H. et al. An evaluation of genotyping by sequencing (GBS) to map the Breviaristatum-e (ari-e) locus in cultivated barley. BMC Genomics 15, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W. et al. Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 6 (2015). [DOI] [PubMed] [Google Scholar]
- Hensel G., Valkov V., Middlefell-Williams J. & Kumlehn J. Efficient generation of transgenic barley: The way forward to modulate plant–microbe interactions. J. Plant Physiol. 165, 71–82 (2008). [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ishida Y., Kasaoka K. & Komari T. Improved frequency of transformation in rice and maize by treatment of immature embryos with centrifugation and heat prior to infection with Agrobacterium tumefaciens. Plant Cell Tiss. Org. Cult. 87, 233–243 (2006). [Google Scholar]
- Zheng L. et al. Isolation and characterization of a barley yellow stripe-like gene, HvYSL5. Plant Cell Physiol. 52, 765–774 (2011). [DOI] [PubMed] [Google Scholar]
- Don R. H., Cox P. T., Wainwright B. J., Baker K. & Mattick J. S. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19, 4008 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.