Abstract
Background: Enzyme-treated wheat bran (ETWB) contains a fermentable dietary fiber previously shown to decrease liver triglycerides (TGs) and modify the gut microbiome in mice. It is not clear which mechanisms explain how ETWB feeding affects hepatic metabolism, but factors (i.e., xenometabolites) associated with specific microbes may be involved.
Objective: The objective of this study was to characterize ETWB-driven shifts in the cecal microbiome and to identify correlates between microbial changes and diet-related differences in liver metabolism in diet-induced obese mice that typically display steatosis.
Methods: Five-week-old male C57BL/6J mice fed a 45%-lard–based fat diet supplemented with ETWB (20% wt:wt) or rapidly digestible starch (control) (n = 15/group) for 10 wk were characterized by using a multi-omics approach. Multivariate statistical analysis was used to identify variables that were strong discriminators between the ETWB and control groups.
Results: Body weight and liver TGs were decreased by ETWB feeding (by 10% and 25%, respectively; P < 0.001), and an index of liver reactive oxygen species was increased (by 29%; P < 0.01). The cecal microbiome showed an increase in Bacteroidetes (by 42%; P < 0.05) and a decrease in Firmicutes (by 16%; P < 0.05). Metabolites that were strong discriminators between the ETWB and control groups included decreased liver antioxidants (glutathione and α-tocopherol); decreased liver carbohydrate metabolites, including glucose; lower hepatic arachidonic acid; and increased liver and plasma β-hydroxybutyrate. Liver transcriptomics revealed key metabolic pathways affected by ETWB, especially those related to lipid metabolism and some fed- or fasting-regulated genes.
Conclusions: Together, these changes indicate that dietary fibers such as ETWB regulate hepatic metabolism concurrently with specific gut bacteria community shifts in C57BL/6J mice. It is proposed that these changes may elicit gut-derived signals that reach the liver via enterohepatic circulation, ultimately affecting host liver metabolism in a manner that mimics, in part, the fasting state.
Keywords: dietary fiber, gut microbiota, metabolomics, transcriptomics, xenobiotic
Introduction
The world is facing unprecedented rates of overweight, obesity, and comorbidities such as type 2 diabetes and nonalcoholic fatty liver disease (NAFLD)15 (1–4). Therefore, the need for affordable, accessible, and safe approaches to body-weight control and improved metabolic health are in high demand. Changes in dietary patterns, such as increased dietary fiber intake, have been shown to be effective in body-weight reduction and improved metabolic function, at least in some contexts (5–8). Despite the proposed benefits of increased dietary fiber intake, most adults consume less than half the recommended amount (9) and the 2010 Dietary Guidelines for Americans named dietary fiber as a nutrient of concern (10). One way in which dietary fibers improve metabolic health is by altering the gut microbiota (11, 12). The human gut microbiota is characterized by trillions of microbes that possess far more protein-coding genes than the human genome (13, 14). Unlike the human genome, which is largely fixed, the gut microbiome is plastic and can be greatly altered depending on the type of diet the host is consuming (15). There is mounting evidence that the gut microbiota participate in extensive cross-talk with the host via production of xenometabolites (e.g., SCFAs, modified amino acids, and other factors) as well as influence host gene expression (16). Importantly, the production of xenometabolites is highly influenced by host diet (17, 18). It is likely that specific xenometabolites and other microbe-derived factors serve as signals to host tissues, including the liver.
Several studies have shown that shifts in the gut microbiota can result in differences in circulating blood metabolites and alterations in liver metabolism (19, 20). Because the liver is in close proximity to the gut, receives portal blood rich in gut-derived xenometabolites and other factors, and plays a major role in regulating metabolism, an understanding of how changes in the gut microbiota affect liver physiology and biochemistry is of great interest. Therefore, we set out to characterize how the consumption of the dietary fiber enzyme-treated wheat bran (ETWB) alters the gut microbiota and liver metabolome. ETWB is made by treating wheat bran with heat and enzymes, resulting in a mixture of arabinoxylan oligosaccharides (AXOSs), high–molecular weight arabinoxylans, cellulose, and lignin (21). The treated wheat bran is more prone to bacterial degradation than is wheat bran in its native form (18, 19). Human and animal studies have shown that the consumption of ETWB can alter the gut microbiota, reduce body fat, and improve glucose and insulin homeostasis (21). However, there is a paucity of information with regard to what effects ETWB has on the liver, a major organ in terms of whole-body metabolic homeostasis. Therefore, in addition to testing the whole-body effects of ETWB on typically measured metabolic outcomes in a diet-induced obesity (DIO) mouse model, a first-ever, to our knowledge, comprehensive landscape of liver metabolism in response to ETWB was characterized through the application of transcriptomics and metabolomics tools. We hypothesized that improvements in whole-body metabolic health indexes would be associated with alterations in pathways related to liver lipid metabolism (i.e., reduced steatosis) and glucose homeostasis. Correlation and multivariate statistical analyses were used to identify candidate microbes and metabolites that drive ETWB-associated differences in liver metabolism.
Methods
DIO mice and diets.
Four-week-old male C57BL/6J mice (Jackson Laboratory) were individually housed under standard temperature (20–22°C) and light-dark cycle (12 h:12 h) conditions in a specific pathogen–free facility. Mice were fed Teklad Rodent Diet 2918 (Envigo) with an energy content of 24%, 58%, and 18% of protein, carbohydrate, and fat, respectively, for a 1-wk acclimation period, then randomly assigned (n = 15/group) to purified experimental diets containing 45% kcal from fat (Teklad; TD.08511) (22) and supplemented with rapidly digestible corn starch (control) or ETWB for 10 wk (Supplemental Table 1). The wheat bran was treated with xylanases and cellulases to increase the content of AXOSs (DuPont Industrial Biosciences Danisco A/S). Thus, the ETWB contained a mix of AXOSs and high-molecular-weight dietary fiber polysaccharides predominantly as arabinoxylan and cellulose. Mice were given ad libitum access to food and water. Body weight and food intake were recorded every 2–3 d. All animal protocols were approved by the University of California at Davis (UC Davis) Institutional Animal Care and Use Committee according to Animal Welfare Act guidelines.
Oral-glucose-tolerance test.
At study week 8 an oral-glucose-tolerance test was performed on a subset of randomly selected mice (n = 10/group). After 14 h of overnight food deprivation mice were orally administered a sterile solution of 25% glucose in water (1 g glucose/kg body weight). Blood glucose was measured by using a OneTouch Ultra 2 Blood Glucose Meter (LifeScan) at 0 (baseline), 15, 30, 60, and 120 min after gavage.
Tissue harvest.
At week 10, mice were briefly feed-deprived (between 4 and 8 h, starting at 0400) before being anesthetized via isoflurane inhalation (3% in oxygen), and blood was collected by cardiac exsanguination by using EDTA-treated syringes. Mice did not survive this procedure. Blood was centrifuged at 10,000 × g for 2 min at room temperature. Plasma was collected and flash-frozen in liquid nitrogen. Epididymal fat pads, retroperitoneal fat pads, femoral subcutaneous fat pads, liver, and cecum were excised, weighed, and flash-frozen in liquid nitrogen. All tissues were stored at −70°C. Total fat pad weights were used as an index of adiposity.
Plasma assays.
Plasma glucose was assessed by using the Glucose Enzymatic Assay Kit (Sigma). Insulin was assessed by using the low-range Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chemistry). Nonesterified FAs (NEFAs) were assessed with the use of the NEFA-HR (2) microtiter procedure (Wako Diagnostics). TGs were assessed by using the L-type M Triglyceride Microtiter procedure (Wako Diagnostics). All assays were performed according to manufacturers’ instructions.
Liver TGs.
Liver lipid extraction was performed by using a modified Folch method (23). Briefly, liver tissue (∼100 mg) was homogenized in 1 mL of a 2:1 (vol:vol) chloroform:methanol solution. The organic phase was evaporated by using a GeneVac EZ-2, then reconstituted in 1 mL isopropanol. TG content of lipid extracts was measured by using an enzymatic assay kit (TR0100; Sigma) according to manufacturer’s instructions.
Reactive oxygen species.
Reactive oxygen species (ROS) formation was estimated as described in previous reports (24). Briefly, frozen liver tissue (∼50 mg) was sonicated for 20 s in ice-cold PBS. Aliquots of 0.5 mL were incubated with 0.5 mL of 5-μM 2′,7′-dichlorofluorescein-diacetate at 37°C for 60 min. Dichlorofluorescein fluorescence, the fluorescent product of dichlorofluorescein diacetate, was recorded at the end of the incubation period at an excitation wavelength of 488 nm and an emission wavelength of 525 nm in a Perkin-Elmer LS55 luminescence spectrometer. A standard curve was constructed by using increasing concentrations of dichlorofluorescein. Results were expressed as nanomoles of dichlorofluorescein formed per milligram of protein per minute. Protein concentrations were measured according to the method described by Lowry et al. (25).
Cecal pH and SCFAs.
Contents were removed from the cecum and flash-frozen in liquid nitrogen and stored at −70°C. Cecal contents were thawed on ice and homogenized in distilled water (0.5 g wet sample to 5 mL water). A combination electrode was used to measure pH. Samples were then acidified with 200 μL of a 25% (wt:wt) solution of metaphosphoric acid that contained 2 g 2-ethyl-butyric acid/L that served as an internal standard for SCFA content. Solids in the homogenized samples were separated by centrifugation at 8000 g for 10 min. Samples were filtered through a Millipore filter (MILX HA 33 mm, 0.45-μm MCE STRL; Fisher SLHA 033SS). SCFAs in the effluent were analyzed by GLC, similar to Barry et al. (26). Briefly, the column used was an Alltech Econo-cap EC-1000, 100% polyethylene glycol acid modified with dimensions of 15 m × 0.53 mm with a film thickness of 1.20 μm. Settings for temperature control were as follows: 115°C for 0.1 min, temperature was increased at a rate of 10°C/min up to 150°C and held for 0.1 min, then this temperature was increased at 11°C/min up to 170°C and held for 2 min. The injector temperature was 250°C.
Cecal microbiota.
Total cecal DNA was extracted by mechanical (0.1 mm zirconia/silica beads; BioSpec) and enzymatic lysis (lysozyme; Fischer) followed by DNA purification with the use of the QIAamp DNA stool mini kit (Qiagen). Five nanograms of DNA was used to amplify the V4 region of the 16S ribosomal RNA (rRNA) gene with 32 PCR cycles at 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s by using barcoded forward 515 (GTGCCAGCMGCCGCGGTAA) and reverse 806 (GGACTACHVGGGTWTCTAAT) (27, 28). Sequencing of pooled 250-bp paired-end amplicons was performed by using Illumina MiSeq at the UC Davis Genome Center. Raw Illumina FASTQ files were demultiplexed and quality-filtered with QIIME (Quantitative Insights into Microbial Ecology) software [version 1.6.0; split_libraries_fastq.py, parameter settings: –r 3 –p 0.75 –q 3 –n 0 (29)]. High-quality paired reads were assembled to obtain a single sequence with the use of a short-read assembler, FLASH (30). Assembled reads were used for operational taxonomic unit (OTU) picking. OTUs sharing ≥97% nucleotide identity were identified by using a closed-reference OTU picking process according to a 16S rRNA sequence database [Greengenes version 12_10 (31)] and further analyzed in QIIME.
Metabolomics.
The details of sample handling, metabolite detection, and analysis have been previously described (32). Briefly, untargeted metabolomics was performed at the UC Davis West Coast Metabolomics Center on plasma and liver samples by using GC–time-of-flight–MS to detect primary metabolites (i.e., purines, amino acids, sugars, etc.). For plasma, 15 μL was added to 1 mL ice-chilled extraction solution (acetonitrile:isopropanol:water, 3:3:2) and mixed on a vortex for 10 s. This same procedure was conducted by using an ∼4 mg sample of frozen liver tissue. Samples were centrifuged for 2 min at 14,000 × g (Eppendorf 5415D), and 500 μL supernatant was evaporated (Labconco Centrivap) to complete dryness. For derivitization, 10 μL methoxyamine hydrochloride (Aldrich) was added to dried samples and left on a shaker for 90 min at 30°C, and then 91 μL of 100:1 N-methyl-N-(trimethylsilyl)-trifluoroacetamide (Aldrich):FA methyl ester mixture was added. Samples were left on the shaker for 30 min at 37°C. Samples (0.5 μL) were separated by using an Agilent 6890 gas chromatograph equipped with a 30 m × 0.25 mm i.d., Rtx5Sil-MS column with 0.25 μm 5% diphenyl film and a 10-m integrated guard column (Restek). Chromatography was performed with a constant flow of 1 mL/min while increasing the oven temperature from 50°C to 330°C over a 22-min total run time. Mass spectra were acquired on a Leco Pegasus IV time-of-flight mass spectrometer with a 280°C transfer line, a 250°C ion source, and −70 eV electron ionization impact. The acquisition rate was 17 spectra s−1 with a scan mass range of 85–500 Da. Result files were exported to servers and processed by the Fiehn laboratory’s metabolomics database, known as BinBase (33). Database entries in BinBase were matched against the Fiehn mass spectral library of 1200 authentic metabolite spectra by using retention index and mass spectrum information or the National Institute of Standards and Technology commercial library. Identified metabolites were reported if present in ≥50% of the samples (as defined in the SetupX database) (34). Each metabolite was normalized by the sum of identified metabolite quantifier ion peak heights present in each individual sample. These relative abundances were used for subsequent statistical analysis.
RNA sequencing (RNAseq).
Total RNA was extracted from ∼30 mg liver tissue (subset of n = 13/group randomly selected from n = 15/group for RNAseq and qPCR validation) by using the Qiagen RNEasy Mini RNA Purification Kit according to the manufacturer’s instructions (Qiagen). RNA abundance was quantified by using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies). mRNA was isolated by using the NEBNext Poly(A) Magnetic Isolation Module (New England BioLabs). First- and second-strand synthesis was performed with the use of the NEBNext Ultra Directional RNA Library Prep Kit. Adaptor ligation for Illumina was performed by using the NEBNext Multiplex Oligos for Illumina. Purification of cDNA, ligation, and PCR reactions was performed by using AMPure XP beads (Agencourt BioSciences). RNA and DNA library quality was assessed by using the Caliper GX at the UC Davis Genome Center. Library sequencing was performed on an Illumina Hiseq 2500 at University of California, Berkeley. Library quality was assessed by using the FastQC algorithm (version 0.11.3). Sequences were aligned to the mouse genome (GRC38/m10) by using Tophat (version 2.0.14) and the Bowtie 2 aligner (version 2.2.5). Differential gene expression was determined with the use of the Cuffdiff algorithm that is part of the Cufflinks analysis suite (version 2.2.1). Pathway analysis was performed by using WebGestalt (35, 36). Genes were selected for pathway analysis input if the gene had a CuffDiff unadjusted P value ≤0.05 and both treatment groups had a mean of ≥1 fragment per kilobase of transcript per million. Analysis parameters were as follows: Organism: Mus musculus, Id Type: gene_symbol, Reference Set: mmusculus_genome, Statistic: Hypergeometric, Significance Level: Top10, Multiple Test Correction: Benjamini-Hochberg, Minimum number of genes for a category: 2.
Validation qPCR.
RNA was isolated as described in the section above. Methods used were similar to that previously described (22). Briefly, the Superscript III First-Strand Synthesis System (Invitrogen) was used to synthesize cDNA from total RNA according to the manufacturer’s instructions. Taqman primers and 6-carboxyfluorescein-minor groove binder labeled probes (Supplemental Table 2) were assayed in triplicate for each sample on a ViiA7 instrument (Applied Biosystems). Reactions were carried out in a 384-well format. Each well contained the following: 8 ng air-dried cDNA, 1× Master Mix (ABI Universal Master Mix), and 1× specific primer-probe mix. Cycle conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s or 60°C for 1 min. Relative gene expression was determined by using the Δ-ΔCt method as previously described (37). Eukaryotic 18S rRNA (Life Technologies) was used as the endogenous reference gene. A comparison of gene expression determined by RNAseq and qPCR can be found in Supplemental Figure 1.
Statistical analysis.
Statistical analyses were performed by using GraphPad Prism (version 5.04 for Windows) and the open-source statistical software R (version 3.1.2) (38). The significance of microbial percentage abundance and metabolomics data was assessed by using the Mann-Whitney U test, and Benjamini-Hochberg false discovery rate (FDR)–corrected P values ≤0.05 were considered significant (39). Multivariate analyses of metabolomics data were performed by using partial least-squares-discriminate analysis (PLS-DA) from the R package “pls” (40). Metabolomics data were log-transformed, and each metabolite was assessed for univariate outliers by using Grubb’s test with the R package “outliers” (41). Outliers were removed if determined to be significant at α = 0.01. In total, 157 outliers were removed, which accounted for only 0.6% of the entire metabolomics data. Removed outliers were imputed by using k-nearest neighbors from the Bioconductor “impute” package (42). PLS-DA model accuracy was assessed with a cross-validation scheme in which the data were randomly partitioned into training and test data sets encompassing two-thirds and one-third of mice, respectively. Model fitting and feature selection were determined solely with data from the training set. Training data were scaled and centered to unit variance before model development, whereas test data were scaled and centered by using the means and SDs from the training data. Metabolites of interest were assessed in PLS-DA models with variable importance in projection (VIP) scores, a weighted measure of the contribution of each metabolite to discriminate the classification groups. A VIP score ≥1 has been argued to be an adequate threshold to determine discriminant variables in PLS-DA models (43, 44). Furthermore, we used bootstrapping (45) to determine a distribution of VIP scores, and then tested whether the bootstrapped VIP distribution was significantly ≥1 by using an independent 1-tailed t test. Metabolites meeting these criteria were chosen for inclusion in final PLS-DA model development. Model performance was assessed on the basis of the model’s ability to accurately predict the classification of the test set mice (e.g., control compared with ETWB) by using data from the test set. Final models that used 2 components were able to predict the classification of test mice with 70% accuracy on the basis of plasma metabolites and 80% accuracy on the basis of liver metabolites. Principal components analysis score plots of plasma and liver metabolites can be found in Supplemental Figure 2. Principal components analysis is unsupervised modeling (i.e., model is not provided with treatment group assignments), whereas PLS-DA is supervised modeling (i.e., model is provided with treatment group assignments). Spearman’s correlation matrices feature the following data: adiposity, cecal weight, liver TGs, and liver ROS identified by Student’s t test as significant at P ≤ 0.05; hepatic genes identified as significantly different from CuffDiff analysis after FDR correction and named by WebGestalt pathway analysis; plasma and liver metabolites; and cecal microbes.
Results
Adiposity and liver TGs
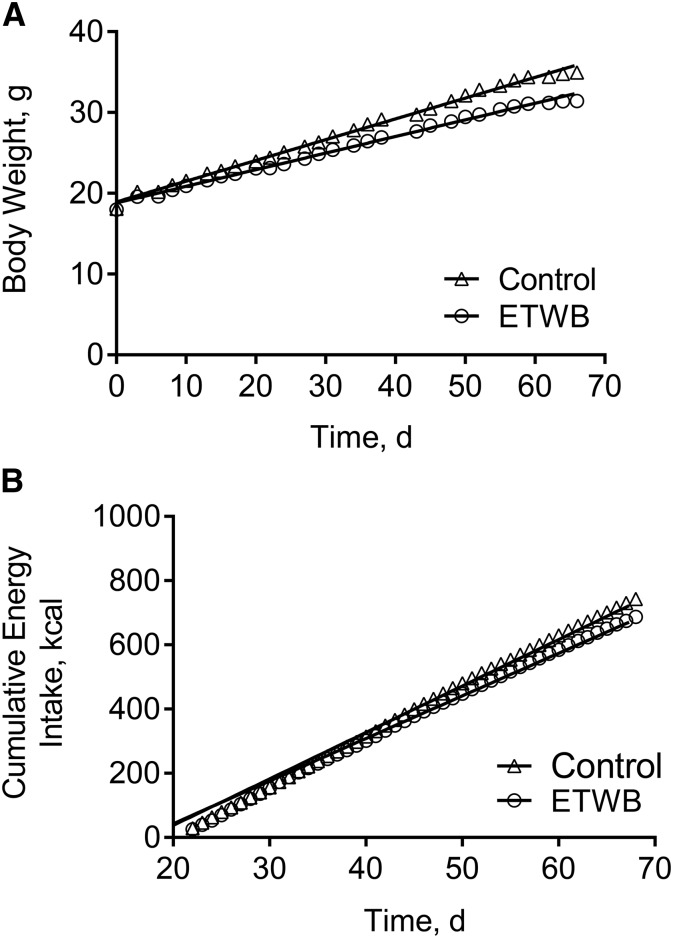
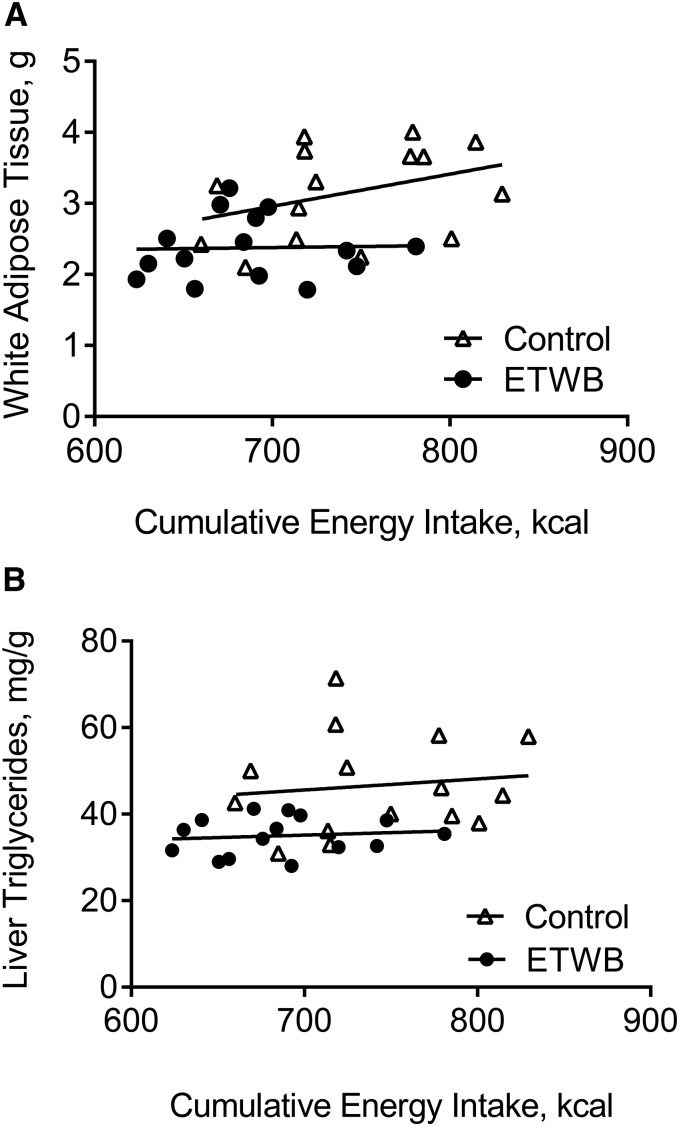
Mice supplemented with ETWB showed significantly reduced body weight, adiposity index (summed fat pad weights), and feed efficiency compared with control mice (Figure 1A, Table 1). There was a significant diet × time interaction on cumulative energy intake (P < 0.0002), with mean cumulative energy intake reduced in the ETWB-fed mice compared with control mice starting at day 60 of the ∼70-d feeding intervention (P < 0.05) (Figure 1B). There was no difference in AUCs with an oral-glucose-tolerance test (15,701 ± 1206 and 15,601 ± 694 mg/dL × min for control and ETWB mice, respectively) performed at week 8 of the study and no difference in terminal postabsorptive plasma glucose, insulin, TGs, or NEFAs (Table 1). Liver TGs were significantly reduced and liver ROS were significantly increased in ETWB mice (Table 1). No significant correlations between energy intake and adiposity index or energy intake and liver TGs in the control or ETWB groups were observed, but interestingly, the ETWB group maintained lower adiposity and liver TGs at any given cumulative energy intake (Figure 2A, B), consistent with reduced feed efficiency.
FIGURE 1.
Body weight (A) and cumulative energy intake (B) in male mice (n = 15/group) fed a 45%-fat diet with or without a 20% (by wt) ETWB supplement for 10 wk. To convert kcal to kJ, multiply by 4.184. ETWB, enzyme-treated wheat bran.
TABLE 1.
Body weight, energy efficiency, and plasma and liver characteristics of male mice fed a 45%-fat diet with or without an ETWB supplement for 10 wk1
| Variable | Control | ETWB | P2 |
| Terminal body weight, g | 34.5 ± 0.69 | 31.1 ± 0.39 | 0.0003 |
| Adiposity index3 | 3.15 ± 0.17 | 2.37 ± 0.11 | 0.0007 |
| Cumulative energy intake,4 kcal/10 wk | 743 ± 14 | 687 ± 12 | 0.0044 |
| Cumulative energy intake, kcal/g terminal body weight | 21.9 ± 0.4 | 22.5 ± 0.5 | 0.32 |
| Feed efficiency,4 mg body weight gained/total kcal consumed | 22.7 ± 0.7 | 20.1 ± 0.7 | 0.0133 |
| Plasma glucose, mg/dL | 138 ± 4.9 | 154 ± 10 | 0.09 |
| Plasma insulin, ng/mL | 0.60 ± 0.06 | 0.62 ± 0.06 | 0.79 |
| Plasma NEFAs, mM | 0.24 ± 0.01 | 0.24 ± 0.01 | 0.65 |
| Plasma TGs, mg/dL | 52.6 ± 2.5 | 46.5 ± 3.6 | 0.18 |
| Liver weight, g | 1.19 ± 0.03 | 1.18 ± 0.01 | 0.74 |
| Liver, % of body weight | 3.44 ± 0.06 | 3.80 ± 0.06 | 0.0003 |
| Liver TGs, mg/g | 46.7 ± 3.0 | 35.0 ± 1.1 | 0.0009 |
| Liver ROS, mmol DCF · mg protein−1 · min−1 | 3.44 ± 0.21 | 4.58 ± 0.29 | 0.0035 |
Values are means ± SEMs, n = 15/group. DCF, dichlorofluorescein; ETWB, enzyme-treated wheat bran; NEFA, nonesterified fatty acid; ROS, reactive oxygen species.
Derived by using 2-tailed Student’s t test. P < 0.05 was considered significant.
Adiposity index is the sum of epididymal, retroperitoneal, and subcutaneous fat pads in grams. Samples were collected from mice in the postabsorptive state after ∼4–8 h food deprivation in the morning.
To convert kcal to kJ, multiply by 4.184.
FIGURE 2.
Correlations between cumulative energy intake and total white adipose tissue weight (A) and liver TG concentrations (B) in male mice (n = 15/group) fed a 45%-fat diet with or without an ETWB supplement (20% by wt) for 10 wk. To convert kcal to kJ, multiply by 4.184. ETWB, enzyme-treated wheat bran.
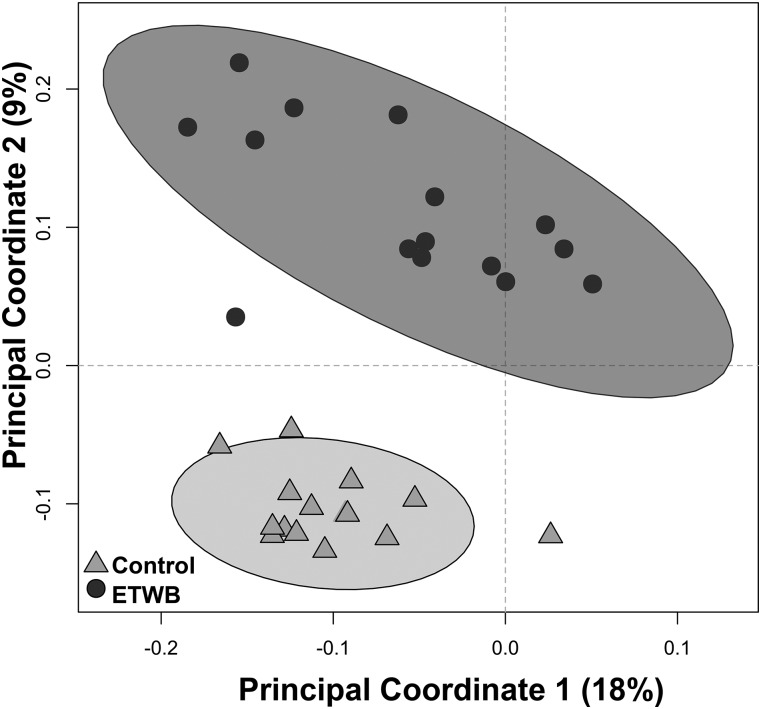
ETWB supplementation significantly alters gut environment
Cecal tissue, cecal contents, and 48-h fecal output were significantly increased in ETWB mice compared with the control group (Table 2). No differences in cecal pH or cecal SCFAs (acetic, propionic, butyric, isobutyric, valeric, isovaloric, and isocaproic acids) were observed. Despite no difference in the number of cecal bacteria taxa present between groups (data not shown), there were significant shifts in cecal bacteria at both the phylum and lower taxonomic levels (Figure 3, Table 3). Compared with control mice, the ETWB mice had significantly greater proportions of Bacteroidetes and Tenericutes and significantly lower proportions of Firmicutes, Proteobacteria, and Verrucomicrobia and no difference in Actinobacteria. The bacteria described below were the main contributors to differences at the phylum level and survived FDR correction. The families S24-7 and Rikenellaceae contributed to the greater percentage abundances in the Bacteroidetes phylum in the ETWB group. The order RF39 accounted for the higher proportions of Tenericutes in the ETWB-fed mice. Firmicutes from the genus Turicibacter and the class Clostridia were reduced in the cecum of the ETWB mice. The family Enterobacteraceae accounted for the reduced percentage abundance in the phylum Proteobacteria in the ETWB mice. Proportions of the phylum Verrucomicrobia were reduced in the ETWB mice due to reductions in the genus Akkermansia.
TABLE 2.
Fecal and cecal characteristics of male mice fed a 45%-fat diet with or without an ETWB supplement for 10 wk1
| Variable | Control | ETWB | P2 |
| Fecal output,3 mg/48 h | 517 ± 23 | 688 ± 14 | <0.0001 |
| Cecal tissue,4 mg | 54.4 ± 2.3 | 76.4 ± 3.3 | <0.0001 |
| Cecal contents, mg | 177 ± 6 | 247 ± 10 | <0.0001 |
| Cecal pH | 7.9 ± 0.1 | 7.8 ± 0.1 | 0.24 |
| Total cecal SCFAs,5 μmol | 6.2 ± 1.2 | 5.1 ± 0.7 | 0.44 |
Values are means ± SEMs, n = 15/group unless otherwise noted. ETWB, enzyme-treated wheat bran.
Derived by using 2-tailed Student’s t test. P ≤ 0.05 was considered significant.
n = 10/group.
Contents removed from the cecum and tissue weight were recorded.
Total SCFAs quantified by summing concentrations (mmol/g) of acetic, propionic, butyric, isobutyric, valeric, isovaleric, and isocaproic acids
FIGURE 3.
Unweighted UniFrac Beta-Diversity Principal Coordinates Analysis plot shows the separation between treatment groups on the basis of the cecal microbiota of male mice (n = 15/group) fed a 45%-fat diet with or without an ETWB supplement (20% by wt). Axes represent percentages of the variance that can be accounted for on the basis of cecal microbiota profile. Ellipses represent 95% CIs on the basis of Hotelling’s T2 statistic. Each symbol represents 1 mouse. ETWB, enzyme-treated wheat bran.
TABLE 3.
Percent abundance of cecal bacteria phyla and significantly altered taxa in male mice fed a 45%-fat diet with or without an ETWB supplement for 10 wk1
| Control | ETWB | Percentage difference (ETWB relative to control)2 | P3 | |
| Phylum | ||||
| Tenericutes | 0.08 ± 0.01 | 0.20 ± 0.04 | 150 | 0.0042 |
| Bacteroidetes | 30.8 ± 1.9 | 43.8 ± 2.7 | 42 | 0.0014 |
| Firmicutes | 63.6 ± 1.7 | 53.5 ± 3.0 | −16 | 0.0079 |
| Actinobacteria | 1.19 ± 0.40 | 0.99 ± 0.28 | −17 | 1.00 |
| Verrucomicrobia | 4.22 ± 0.54 | 1.49 ± 0.40 | −65 | 0.0003 |
| Proteobacteria | 0.05 ± 0.02 | 0.01 ± 0.01 | −80 | 0.0015 |
| Taxon4 | ||||
| p__Firmicutes; g__Dorea | 0.06 ± 0.01 | 0.30 ± 0.07 | 416 | 0.0008 |
| p__Tenericutes; o__RF39 | 0.07 ± 0.01 | 0.19 ± 0.04 | 175 | 0.0056 |
| p__Firmicutes; g__Adlercreutzia | 0.09 ± 0.01 | 0.18 ± 0.02 | 99 | 0.0115 |
| p__Bacteroidetes; f__Rikenellaceae | 4.75 ± 0.43 | 9.41 ± 0.88 | 98 | 0.0010 |
| p__Firmicutes; f__Ruminococcaceae | 3.59 ± 0.62 | 5.81 ± 0.49 | 62 | 0.0057 |
| p__Bacteroidetes; f__S24-7 | 26.1 ± 2.0 | 34.4 ± 2.3 | 32 | 0.0392 |
| p__Verrucomicrobia; g__Akkermansia | 4.22 ± 0.54 | 1.49 ± 0.40 | −65 | 0.0007 |
| p__Firmicutes; c__Clostridia | 4.40 ± 0.47 | 1.50 ± 0.18 | −66 | <0.0001 |
| p__Firmicutes; g__Streptococcus | 0.12 ± 0.10 | 0.04 ± 0.36 | −72 | <0.0001 |
| p__Firmicutes; g__Turicibacter | 5.91 ± 0.88 | 1.08 ± 0.27 | −82 | 0.0010 |
| p__Firmicutes; o__Clostridiales | 0.95 ± 0.18 | 0.08 ± 0.10 | −91 | <0.0001 |
| p__Firmicutes; f__Peptostreptococcaceae | 0.60 ± 0.17 | 0.01 ± 0.01 | −98 | 0.0384 |
Values are means ± SEMs, n = 15/group. c_, class; ETWB, enzyme-treated wheat bran; f_, family; g_, genus; o_, order; p_, phylum.
Percentage difference = [(ETWB − control)/control] × 100.
Group comparisons were assessed by Mann-Whitney U tests. P values were adjusted for false discovery rate correction. Significance was set at an adjusted P value ≤0.05.
Reported taxa included had a minimum of 0.05% mean abundance in each group and an adjusted P value ≤0.05. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification).
Plasma and liver metabolome
Plasma.
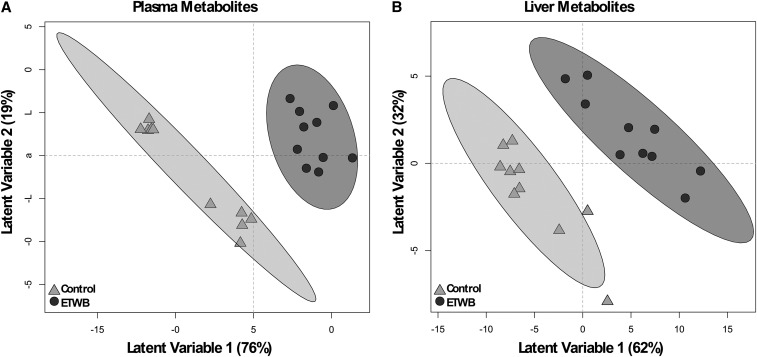
A total of 386 plasma metabolites were detected by using the GC–time-of-flight–MS analytical platform. Of these, 133 metabolites were annotated in the metabolite database; the remaining metabolites were nonannotated and labeled with a numerical BinBase ID (Supplemental Table 3). A total of 78 metabolites had a mean bootstrapped VIP distribution of ≥1 in the PLS-DA model and thus contributed to discrimination of the diet groups; of these, 34 metabolites were annotated (Figure 4A, Table 4; for brevity only annotated metabolites are shown). The plasma metabolite with the highest VIP, 2-deoxyerythritol, also had the greatest percentage difference of all carbohydrate metabolites in plasma. In addition, the sugar alcohols ribitol and xylitol were higher in ETWB-fed mouse plasma, whereas 1,5-anhydroglucitol was reduced in the ETWB group. The plasma lipid metabolite with the greatest percentage difference was β-sitosterol in the ETWB-fed mice. Other plasma lipids and lipid derivatives that were higher in the ETWB group include pelargonic acid (C9); capric acid (C10); 1-monostearin, linoleic acid (C18:2n–6); and the ketone body β-hydroxybutyrate (3-hydroxybutanoic acid). Adenosine-5-phosphate had the greatest percentage difference of any nitrogenous metabolite in the plasma in the ETWB-fed mice compared with controls. Several other nitrogenous metabolites were reduced in the ETWB mouse plasma including the following: 2-hydroxyglutaric acid, 5-methoxytryptamine, creatinine, and taurine. Other metabolite differences in the ETWB-fed mice included higher abundances of hydrocinnamic acid (3-phenylpropanoic acid) and 2-ketoisocaproic acid as well as differences in the organic acids lactic acid and pyruvic acid.
FIGURE 4.
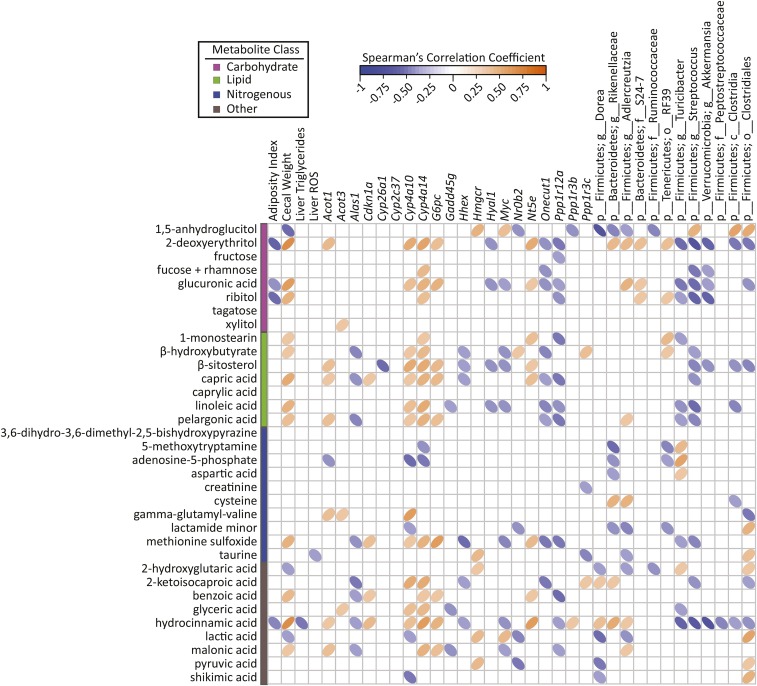
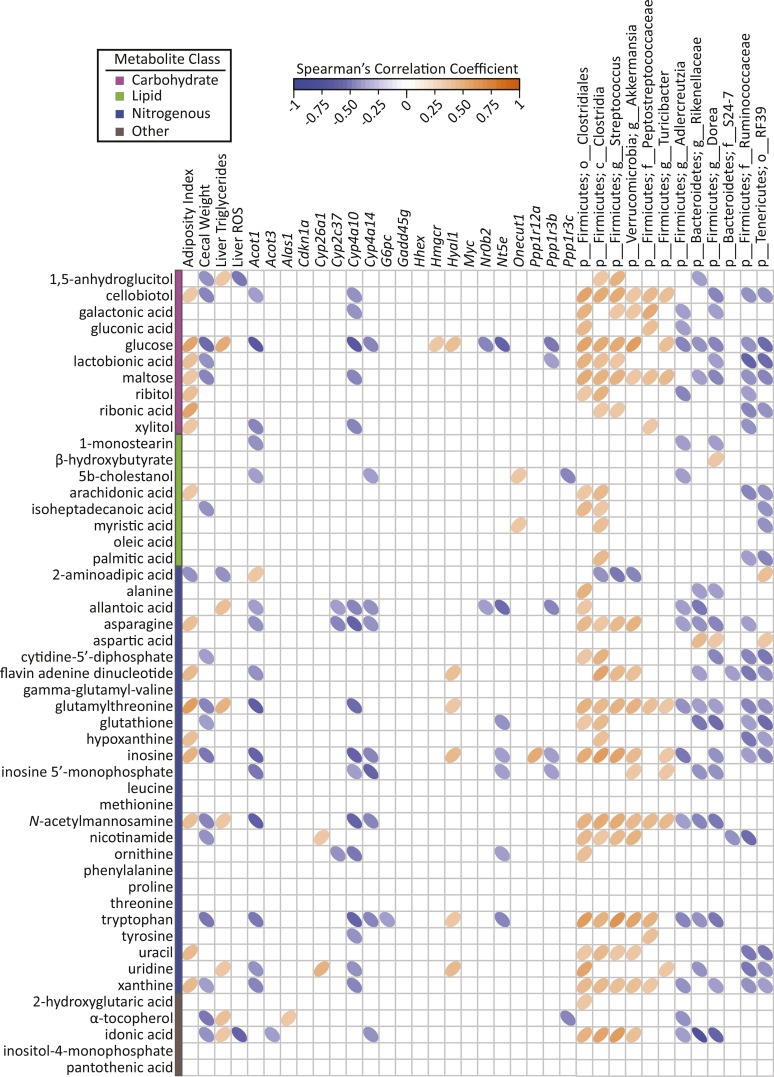
PLS-DA score plots based on abundances of plasma (A) and liver (B) metabolites of male mice fed a 45%-fat diet with or without an ETWB supplement. Ellipses represent 95% CIs on the basis of Hotelling’s T2 statistic; each symbol represents 1 mouse. Annotated metabolites contributing to these plots can be found in Tables 4 and 5; nonannotated metabolites can be found in Supplemental Table 2. Metabolomics analyses were performed on samples from 15 mice/group: a model was developed with the use of 10 mice/group and model validation was performed by using 5 mice/group (Supplemental Figure 1). ETWB, enzyme-treated wheat bran; PLS-DA, partial least-squares-discriminant analysis.
TABLE 4.
Postabsorptive plasma metabolite abundances (ranked highest to lowest by percentage difference) in male mice fed a 45%-fat diet with or without an ETWB supplement for 10 wk1
|
P4 |
||||||
| Metabolite2 | Control | ETWB | Percentage difference (ETW B relative to control)3 | MWU | MWU-FDR | VIP5 |
| Carbohydrates | ||||||
| 2-Deoxyerythritol | 5270 ± 321 | 7910 ± 444 | 50 | <0.0001 | <0.0001 | 2.12 |
| Ribitol | 657 ± 34 | 835 ± 57 | 27 | 0.0240 | 0.35 | 1.28 |
| Glucuronic acid | 863 ± 36 | 1060 ± 50 | 23 | 0.0030 | 0.23 | 1.69 |
| Tagatose | 33 ± 3 | 40 ± 5 | 19 | 0.58 | 0.92 | 1.42 |
| Fucose + rhamnose | 2910 ± 115 | 3170 ± 109 | 9 | 0.09 | 0.65 | 1.10 |
| Fructose | 6200 ± 818 | 6690 ± 773 | 8 | 0.62 | 0.92 | 1.29 |
| Xylitol | 284 ± 15 | 302 ± 15 | 7 | 0.33 | 0.84 | 1.08 |
| 1,5-Anhydroglucitol | 13,200 ± 690 | 9800 ± 343 | −26 | <0.0001 | <0.0001 | 1.73 |
| Lipids | ||||||
| β-Sitosterol | 294 ± 52 | 593 ± 80 | 101 | 0.0040 | 0.25 | 1.32 |
| Pelargonic acid | 5280 ± 539 | 7910 ± 963 | 50 | 0.0290 | 0.35 | 1.45 |
| Linoleic acid | 365 ± 38 | 537 ± 40 | 47 | 0.0070 | 0.25 | 1.40 |
| β-Hydroxybutyric acid (3-hydroxybutanoic acid) | 13,400 ± 1110 | 19,600 ± 1950 | 46 | 0.0150 | 0.34 | 2.02 |
| 1-Monostearin | 100 ± 11 | 134 ± 17 | 34 | 0.25 | 0.65 | 1.48 |
| Capric acid | 1070 ± 105 | 1422 ± 84 | 33 | 0.0210 | 0.35 | 1.51 |
| Caprylic acid | 4419 ± 201 | 4350 ± 256 | −2 | 0.62 | 0.92 | 1.33 |
| Nitrogenous | ||||||
| γ-Glutamyl-valine | 88 ± 9 | 137 ± 20 | 56 | 0.10 | 0.66 | 1.52 |
| Cysteine | 1680 ± 126 | 2170 ± 176 | 29 | 0.07 | 0.61 | 1.31 |
| Methionine sulfoxide | 895 ± 54 | 1060 ± 58 | 19 | 0.02 | 0.35 | 1.07 |
| Aspartic acid | 1710 ± 156 | 1530 ± 123 | −10 | 0.37 | 0.84 | 1.06 |
| 2-Hydroxyglutaric acid | 1120 ± 48 | 986 ± 48 | −12 | 0.0290 | 0.35 | 1.06 |
| 3,6-Dihydro-3,6-dimethyl-2,5-bishydroxypyrazine | 564 ± 57 | 493 ± 45 | −13 | 0.44 | 0.86 | 1.18 |
| Creatinine | 15,500 ± 1180 | 13,300 ± 1410 | −14 | 0.33 | 0.84 | 1.25 |
| Lactamide minor | 407 ± 35 | 322 ± 19 | −21 | 0.0340 | 0.38 | 1.47 |
| Taurine | 13,900 ± 1480 | 10,100 ± 1390 | −27 | 0.0410 | 0.42 | 1.16 |
| 5-Methoxytryptamine | 2840 ± 739 | 1610 ± 559 | −43 | 0.14 | 0.70 | 1.07 |
| Adenosine-5-phosphate | 791 ± 272 | 189 ± 55 | −76 | 0.12 | 0.70 | 1.16 |
| Other | ||||||
| Hydrocinnamic acid (3-phenylproanoic acid) | 199 ± 25 | 403 ± 59 | 102 | 0.0010 | 0.10 | 1.87 |
| 2-Ketoisocaproic acid | 2540 ± 174 | 3160 ± 254 | 24 | 0.0330 | 0.38 | 1.67 |
| Malonic acid | 2660 ± 289 | 3260 ± 275 | 22 | 0.10 | 0.65 | 1.05 |
| Benzoic acid | 5060 ± 483 | 6140 ± 291 | 21 | 0.07 | 0.61 | 1.23 |
| Glyceric acid | 2060 ± 140 | 2390 ± 142 | 16 | 0.12 | 0.69 | 1.31 |
| Pyruvic acid | 16,900 ± 1290 | 13,000 ± 1230 | −23 | 0.07 | 0.59 | 1.58 |
| Lactic acid | 355,000 ± 22500 | 270,000 ± 18400 | −24 | 0.0070 | 0.25 | 1.90 |
| Shikimic acid | 3470 ± 675 | 2090 ± 387 | −40 | 0.11 | 0.67 | 1.24 |
Values are means ± SEMs, n = 15/group. Only annotated metabolites that had mean bootstrapped VIP measurements ≥1 are presented. Nonannotated metabolites are not shown for the sake of brevity but are provided in Supplemental Table 3. ETWB, enzyme-treated wheat bran; FDR, false discovery rate; MWU, Mann-Whitney U test; VIP, variable importance in projection.
Metabolite abundances reported in quantifier ion peak height in the 0.5-μL extract derived from 15 μL plasma.
Percentage difference = [(ETWB − control)/control] × 100.
Group comparisons were assessed by MWUs. P values were adjusted for FDR correction. Significance was set at an adjusted P value ≤0.05.
VIP was calculated from bootstrapped partial-least-squares discriminant analysis models derived from training data (n = 10 mice/group).
Liver.
A total of 454 liver metabolites were detected, 162 of which were annotated (Supplemental Table 4). The abundances of 110 metabolites were identified by PLS-DA to significantly contribute to separation between treatment groups; 49 of these metabolites were annotated (Figure 4B, Table 5; for brevity only annotated metabolites are shown). As in plasma, several sugars and sugar alcohols were reduced in the livers of ETWB-fed mice including the following: glucose, maltose, 1,5-anhydroglucitol, ribitol, and xylitol. Many lipid liver metabolites were also reduced in the ETWB group, including 1-monostearin, arachidonic acid, isoheptadecanoic acid, myristic acid (C14), oleic acid, and palmitic acid. Just as in plasma, the liver abundance of the ketone body β-hydroxybutyrate was higher in ETWB-fed mice. The following amino acids were reduced in the ETWB group: alanine, asparagine, leucine, methionine, phenylalanine, proline, threonine, tryptophan, and tyrosine. Aspartic acid was higher in the ETWB group. The following purine- and pyrimidine-related metabolites were reduced in the ETWB group: cytidine-5′-diphosphate, uridine, allantoic acid, inosine, inosine-5′-monophosphate, xanthine, and hypoxanthine. The antioxidants α-tocopherol and glutathione were reduced in ETWB livers.
TABLE 5.
Postabsorptive liver metabolite abundances (ranked highest to lowest by percentage difference) in male mice fed a 45%-fat diet with or without an ETWB supplement for 10 wk1
|
P4 |
||||||
| Metabolite2 | Control | ETWB | Percentage difference (ETWB relative to control)3 | MWU | MWU-FDR | VIP5 |
| Carbohydrates | ||||||
| Gluconic acid | 521 ± 59 | 400 ± 67 | −23 | 0.07 | 0.35 | 1.35 |
| 1,5-Anhydroglucitol | 2350 ± 199 | 1744 ± 95 | −26 | 0.0130 | 0.19 | 1.11 |
| Glucose | 692,000 ± 29,800 | 512,000 ± 30,800 | −26 | 0.0000 | <0.0001 | 1.72 |
| Galactonic acid | 586 ± 50 | 421 ± 63 | −28 | 0.0210 | 0.22 | 1.73 |
| Xylitol | 2440 ± 245 | 1680 ± 183 | −31 | 0.0410 | 0.30 | 1.24 |
| Cellobiotol | 104,000 ± 5690 | 66,000 ± 8290 | −37 | 0.0010 | 0.0450 | 1.73 |
| Maltose | 404,000 ± 22,100 | 254,000 ± 33,500 | −37 | 0.0020 | 0.06 | 1.72 |
| Ribonic acid | 1820 ± 310 | 1000 ± 227 | −45 | 0.0610 | 0.35 | 1.19 |
| Lactobionic acid | 25,900 ± 6520 | 12,400 ± 4710 | −52 | 0.01 | 0.13 | 1.52 |
| Ribitol | 1490 ± 431 | 708 ± 232 | −53 | 0.06 | 0.34 | 1.20 |
| Lipids | ||||||
| β-Hydroxybutyric acid (3-hydroxybutanoic acid) | 1250 ± 107 | 1560 ± 1410 | 25 | 0.1150 | 0.43 | 1.45 |
| 5b-Cholestanol | 505 ± 39 | 428 ± 41 | −15 | 0.2130 | 0.56 | 1.11 |
| Myristic acid | 2610 ± 156 | 2200 ± 125 | −16 | 0.0670 | 0.35 | 1.07 |
| Isoheptadecanoic acid | 4640 ± 217 | 3880 ± 316 | −16 | 0.0620 | 0.35 | 1.29 |
| 1-Monostearin | 623 ± 53 | 493 ± 37 | −21 | 0.0740 | 0.35 | 1.38 |
| Palmitic acid | 67,500 ± 4715 | 52,800 ± 6540 | −22 | 0.0610 | 0.35 | 1.11 |
| Oleic acid | 18,800 ± 3430 | 12,700 ± 3340 | −33 | 0.2020 | 0.55 | 1.04 |
| Arachidonic acid | 17,100 ± 2540 | 9600 ± 2046 | −44 | 0.0330 | 0.27 | 1.17 |
| Nitrogenous | ||||||
| Aspartic acid | 6650 ± 1490 | 9430 ± 1250 | 42 | 0.07 | 0.35 | 1.40 |
| 2-Aminoadipic acid | 464 ± 56 | 646 ± 57 | 39 | 0.0160 | 0.20 | 1.51 |
| γ-Glutamyl-valine | 186 ± 19 | 169 ± 17 | −9 | 0.44 | 0.70 | 1.08 |
| Alanine | 715,000 ± 21,500 | 629,000 ± 23,300 | −12 | 0.0100 | 0.17 | 1.51 |
| Threonine | 13,700 ± 1060 | 11,800 ± 857 | −13 | 0.20 | 0.55 | 1.07 |
| Phenylalanine | 6320 ± 644 | 5450 ± 465 | −14 | 0.35 | 0.65 | 1.09 |
| Leucine | 39,700 ± 3070 | 33,300 ± 2420 | −16 | 0.15 | 0.46 | 1.15 |
| Proline | 18,700 ± 1400 | 15,700 ± 1070 | −16 | 0.12 | 0.43 | 1.17 |
| Tyrosine | 29,300 ± 2330 | 23,500 ± 2100 | −20 | 0.09 | 0.39 | 1.04 |
| Ornithine | 12,300 ± 1180 | 10,200 ± 1100 | −21 | 0.11 | 0.42 | 1.38 |
| Tryptophan | 7070 ± 244 | 5500 ± 242 | −22 | <0.0001 | <0.0001 | 1.91 |
| Methionine | 3530 ± 431 | 2730 ± 407 | −23 | 0.20 | 0.55 | 1.13 |
| Inosine 5′-monophosphate | 5670 ± 573 | 4160 ± 474 | −27 | 0.0450 | 0.31 | 1.13 |
| N-acetylmannosamine | 617 ± 28 | 449 ± 31 | −27 | 0.0010 | 0.05 | 1.67 |
| Glutamylthreonine | 1300 ± 65 | 937 ± 70 | −28 | 0.0020 | 0.06 | 1.65 |
| Nicotinamide | 24,600 ± 1665 | 17,400 ± 1170 | −29 | 0.0020 | 0.06 | 1.14 |
| Allantoic acid | 2540 ± 314 | 1690 ± 221 | −33 | 0.0450 | 0.31 | 1.12 |
| Xanthine | 9110 ± 799 | 5750 ± 703 | −37 | 0.0070 | 0.14 | 1.49 |
| Asparagine | 4860 ± 470 | 3050 ± 291 | −37 | 0.0030 | 0.08 | 1.67 |
| Inosine | 32,200 ± 2630 | 19,700 ± 2060 | −39 | <0.0001 | <0.0001 | 1.43 |
| Hypoxanthine | 23,600 ± 3270 | 13,900 ± 2650 | −41 | 0.0290 | 0.26 | 1.13 |
| Uracil | 5060 ± 666 | 2920 ± 404 | −42 | 0.0190 | 0.22 | 1.29 |
| Flavin adenine dinucleotide | 420 ± 60 | 239 ± 32 | −43 | 0.0190 | 0.22 | 1.26 |
| Cytidine-5′-diphosphate | 6350 ± 949 | 3590 ± 787 | −43 | 0.0190 | 0.22 | 1.17 |
| Uridine | 2010 ± 334 | 1100 ± 139 | −45 | 0.0080 | 0.15 | 1.63 |
| Glutathione | 2200 ± 716 | 545 ± 107 | −75 | 0.0150 | 0.20 | 1.33 |
| Other | ||||||
| Inositol-4-monophosphate | 462 ± 24 | 424 ± 34 | −8 | 0.27 | 0.61 | 1.20 |
| 2-Hydroxyglutaric acid | 2060 ± 130 | 1730 ± 165 | −16 | 0.07 | 0.35 | 1.16 |
| Pantothenic acid | 1100 ± 102 | 918 ± 124 | −17 | 0.20 | 0.55 | 1.34 |
| Idonic acid | 1310 ± 70 | 961 ± 45 | −27 | <0.0001 | <0.0001 | 1.86 |
| α-Tocopherol | 1770 ± 197 | 1180 ± 100 | −33 | 0.0370 | 0.29 | 1.38 |
Values are means ± SEMs, n = 15/group. Only annotated metabolites that had mean bootstrapped VIP measurements ≥1 are presented. Nonannotated metabolites are not shown for the sake of brevity but are provided in Supplemental Table 4. ETWB, enzyme-treated wheat bran; FDR, false discovery rate; MWU, Mann-Whitney U test; VIP, variable importance in projection.
Metabolite abundances reported in quantifier ion peak height in the 0.5-μL extract derived from 4 mg liver tissue.
Percentage difference = [(ETWB − control)/control] × 100.
Group comparisons were assessed by MWUs. P values were adjusted for FDR correction. Significance was set at an adjusted P value ≤0.05.
VIP was calculated from bootstrapped partial least-squares-discriminant analysis models derived from training data (n = 10 mice/group).
Alterations in hepatic metabolic gene expression
A transcriptomics study was conducted to determine if gene expression patterns associated with specific biochemical pathways might explain ETWB-related metabolite shifts observed in the liver. Fifty-eight protein-coding genes in the liver were considered significantly differentially expressed between treatment groups after FDR correction and an additional 392 genes had an unadjusted P value ≤0.05 (Supplemental Table 5). Pathway analysis of genes that maintained significance after FDR correction revealed that 10 Kyoto Encyclopedia of Genes and Genomes pathways were affected by ETWB treatment (Table 6). Genes involved in lipid metabolism were generally increased in the following pathways: arachidonic acid metabolism, biosynthesis of unsaturated FAs, and retinol metabolism. Insulin signaling and cell cycle pathways were also affected by ETWB feeding. Pathway analysis was also conducted including genes with unadjusted P values ≤0.05, which is shown in Supplemental Table 6. Eight genes were validated by qPCR (Supplemental Figure 1). Cross-correlation plots of hepatic gene transcripts identified as significantly different from CuffDiff analysis after FDR correction and named by WebGestalt pathway analysis and PLS-DA selected plasma and liver metabolites are shown in Figures 5 and 6, respectively.
TABLE 6.
Hepatic gene expression pathways (genes ranked highest to lowest by percentage difference) affected in male mice fed a 45%-fat diet with or without an ETWB supplement for 10 wk1
| Mean, FPKMs |
||||
| Pathway2 | Definition | Control | ETWB | Percentage difference (ETWB relative to control)3 |
| Metabolic pathways (KEGG pathway 1100) (C = 1184; O = 8; E = 1.15; R = 6.98; rawP = 1.79 × 10−5; adjP = 0.0002) | ||||
| Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | 284 | 410 | 44 |
| Nt5e | 5′ Nucleotidase, ecto | 1.6 | 2.2 | 43 |
| Cyp2c37 | Cytochrome P450, family 2, subfamily c, polypeptide 37 | 66.0 | 86.7 | 31 |
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | 189 | 247 | 31 |
| G6pc | Glucose-6-phosphatase, catalytic | 55.1 | 72.0 | 31 |
| Hmgcr | 3-Hydroxy-3-methylglutaryl-CoA reductase | 19.3 | 14.4 | −25 |
| Hyal1 | Hyaluronoglucosaminidase 1 | 13.4 | 9.6 | −28 |
| Alas1 | Aminolevulinic acid synthase 1 | 190 | 134 | −30 |
| Arachidonic acid metabolism (KEGG pathway 590) (C = 90; O = 3; E = 0.09; R = 34.44; rawP = 9.50 × 10−5; adjP = 0.0006) | ||||
| Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | 284 | 410 | 44 |
| Cyp2c37 | Cytochrome P450, family 2, subfamily c, polypeptide 37 | 66.0 | 86.7 | 31 |
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | 189 | 247 | 31 |
| Biosynthesis of unsaturated FAs (KEGG pathway 1040) (C = 25; O = 2; E = 0.02; R = 82.66; rawP = 0.0003; adjP = 0.0008) | ||||
| Acot3 | Acyl-CoA thioesterase 3 | 9.0 | 12.2 | 35 |
| Acot1 | Acyl-CoA thioesterase 1 | 19.6 | 26.4 | 34 |
| Bile secretion (KEGG pathway 4976) (C = 71; O = 2; E = 0.08; R = 26.29; rawP = 0.0027;adjP = 0.0049) | ||||
| Nr0b2 | Nuclear receptor subfamily 0, group B, member 2 | 31.8 | 45.9 | 44 |
| Hmgcr | 3-Hydroxy-3-methylglutaryl-CoA reductase | 19.3 | 14.4 | −25 |
| Retinol metabolism (KEGG pathway 830) (C = 77; O = 4; E = 0.07; R = 53.67; rawP = 1.01× 10−6; adjP = 2.02 × 10−5) | ||||
| Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | 284 | 410 | 44 |
| Cyp2c37 | Cytochrome P450, family 2, subfamily c, polypeptide 37 | 66.0 | 86.7 | 31 |
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | 189 | 247 | 31 |
| Cyp26a1 | Cytochrome P450, family 26, subfamily a, polypeptide 1 | 3.7 | 2.2 | −40 |
| Insulin signaling pathway (KEGG pathway 4910) (C = 137; O = 3; E = 0.15; R = 20.44; rawP = 0.0004; adjP = 0.0010) | ||||
| Ppp1r3b | Protein phosphatase 1, regulatory (inhibitor) subunit 3B | 24.7 | 33.1 | 34 |
| Ppp1r3c | Protein phosphatase 1, regulatory (inhibitor) subunit 3C | 16.9 | 22.3 | 32 |
| G6pc | Glucose-6-phosphatase, catalytic | 55.1 | 72.0 | 31 |
| Maturity Onset Diabetes of the Young (KEGG pathway 4950) (C = 26; O = 2; E = 0.03; R = 79.48; rawP = 0.0003; adjP = 0.0008) | ||||
| Hhex | Hematopoietically expressed homeobox | 66.0 | 42.6 | −36 |
| Onecut1 | One cut domain, family member 1 | 10.1 | 6.3 | −38 |
| Vascular smooth muscle contraction (KEGG pathway 4270) (C = 123; O = 3; E = 0.12; R = 25.20; rawP = 0.0002; adjP = 0.0008) | ||||
| Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | 284 | 410 | 44 |
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | 189 | 247 | 31 |
| Ppp1r12a | Protein phosphatase 1, regulatory (inhibitor) subunit 12A | 5.7 | 3.9 | −30 |
| Cell cycle (KEGG pathway 4110) (C = 127; O = 3; E = 0.12; R = 24.41; rawP = 0.0003; adjP = 0.0008) | ||||
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (P21) | 1.6 | 2.5 | 59 |
| Gadd45g | Growth arrest and DNA-damage-inducible 45γ | 11.2 | 7.9 | −29 |
| Myc | Myelocytomatosis oncogene | 4.4 | 2.0 | −55 |
| Bladder cancer (KEGG pathway 5219) (C = 43; O = 2; E = 0.04; R = 48.06; rawP = 0.0008; adjP = 0.0018) | ||||
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (P21) | 1.6 | 2.5 | 59 |
| Myc | Myelocytomatosis oncogene | 4.4 | 2.0 | −55 |
Pathways are derived from liver transcriptomics data. Parameters: Organism: Mus musculus, Id Type: gene_symbol, Reference Set: mmusculus_genome, Statistic: Hypergeometric, Significance Level: Top10, Multiple Test Correction: Benjamini-Hochberg, Minimum number of genes for a category: 2. Genes included have an adjusted P value ≤0.05. adjP, P value adjusted by the multiple test adjustment; ETWB, enzyme-treated wheat bran; FPKM, fragment per kilobase of transcript per million; KEGG, Kyoto Encyclopedia of Genes and Genomes; rawP, P value from hypergeometric test.
The C, O, E, and R in parentheses indicate the number of reference genes in the category, number of genes in the gene set and also in the category, the expected number in the category, and the ratio of enrichment, respectively.
Percentage difference = [(ETWB − control)/control] × 100.
FIGURE 5.
Spearman’s correlation matrix of metadata, hepatic gene expression, and cecal bacteria compared with PLS-DA–selected plasma metabolites in male mice (n = 15/group) fed a 45%-fat diet with or without an ETWB supplement. Bacteria included had a minimum of 0.05% mean abundance in each group and an adjusted Mann-Whitney U P value ≤0.05. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). The direction of ellipses represents positive or negative correlations and the width of ellipses represents the strength of correlation (narrow ellipse = stronger correlation). Acot, acyl-CoA thioesterase; Alas1, aminolevulinic acid synthase 1; c_, class; Cdkn1a, cyclin-dependent kinase inhibitor 1A ; Cyp, cytochrome P450; ETWB, enzyme-treated wheat bran; f_, family; g_, genus; Gadd45g, growth arrest and DNA-damage-inducible 45γ G6pc, glucose-6-phosphatase, catalytic; Hhex, hematopoietically expressed homeobox; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; Hyal1, hyaluronoglucosaminidase 1; Myc, myelocytomatosis oncogene; Nr0b2, nuclear receptor subfamily 0, group B, member 2; Nt5e, 5′ nucleotidase, ecto; o_, order; Onecut1, one cut domain, family member 1; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; Ppp1r, protein phosphatase 1, regulatory (inhibitor); ROS, reactive oxygen species.
FIGURE 6.
Spearman’s correlation matrix of metadata, hepatic gene expression, and cecal bacteria compared with PLS-DA–selected liver metabolites in male mice (n = 15/group) fed a 45%-fat diet with or without an ETWB supplement. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). The direction of ellipses represents positive or negative correlations and the width of ellipses represents the strength of correlation (narrow ellipse = stronger correlation). Acot, acyl-CoA thioesterase; Alas1, aminolevulinic acid synthase 1; c_, class; Cdkn1a, cyclin-dependent kinase inhibitor 1A ; Cyp, cytochrome P450; ETWB, enzyme-treated wheat bran; f_, family; g_, genus; Gadd45g, growth arrest and DNA-damage-inducible 45γ G6pc, glucose-6-phosphatase, catalytic; Hhex, hematopoietically expressed homeobox; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; Hyal1, hyaluronoglucosaminidase 1; Myc, myelocytomatosis oncogene; Nr0b2, nuclear receptor subfamily 0, group B, member 2; Nt5e, 5′ nucleotidase, ecto; o_, order; Onecut1, one cut domain, family member 1; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; Ppp1r, protein phosphatase 1, regulatory (inhibitor); ROS, reactive oxygen species.
Potential connections between specific cecal bacteria and host metabolic health indexes, hepatic gene expression, and plasma and liver metabolites
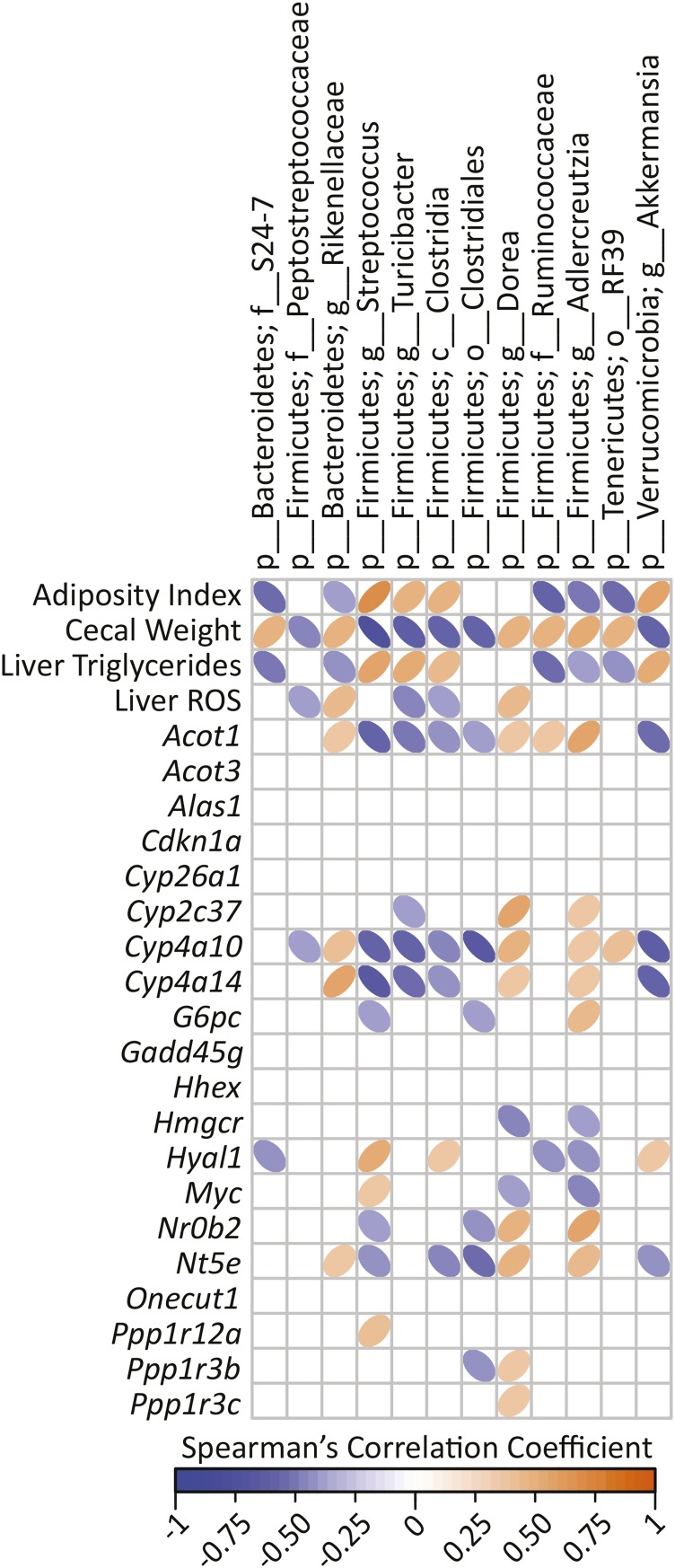
A goal of this experiment was to identify candidate gut bacteria subpopulations that drive whole-body and liver metabolic phenotypes. A cross-correlation plot revealed significant correlations among specific cecal bacteria and adiposity, cecal weight, liver TGs, and liver ROS as well as hepatic gene transcripts (Figure 7). Five bacterial taxonomic groups (Bacteroidetes families S24-7 and Rikellenaceae and the following Firmicutes: order RF39, family Ruminococcaceae, and genus Adlercreutzia) had negative correlations with adiposity index and liver TGs and a positive correlation with cecal weight. These same 5 taxa had significantly greater percentage abundances in the ETWB mice. Conversely, 4 bacterial taxa had the opposite relation (Firmicutes genera Streptococcus, Turicibacter, the class Clostridia, and the genus Akkermansia from the phylum Verrucomicrobia); these taxa were significantly reduced in the ETWB mice. Furthermore, these same 4 taxa had negative correlations with liver cytochrome P450 gene transcripts. To further consider potential contributions of gut bacterial shifts to modifying the host metabolome, we performed correlations among cecal bacteria compared with PLS-DA–selected plasma and liver metabolites, which are presented in Figures 5 and 6, respectively.
FIGURE 7.
Spearman’s correlation matrix of cecal bacteria compared with metadata and hepatic gene expression in male mice (n = 15/group) fed a 45%-fat diet with or without an ETWB supplement. Bacteria included had a minimum of 0.05% mean abundance in each group and an adjusted Mann-Whitney U P value ≤0.05. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). The direction of ellipses represents positive or negative correlations and the width of ellipses represents the strength of correlation (narrow ellipse = stronger correlation). Acot, acyl-CoA thioesterase; Alas1, aminolevulinic acid synthase 1; c_, class; Cdkn1a, cyclin-dependent kinase inhibitor 1A; Cyp, cytochrome P450; ETWB, enzyme-treated wheat bran; f_, family; g_, genus; Gadd45g, growth arrest and DNA-damage-inducible 45γ G6pc, glucose-6-phosphatase, catalytic; Hhex, hematopoietically expressed homeobox; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; Hyal1, hyaluronoglucosaminidase 1; Myc, myelocytomatosis oncogene; Nr0b2, nuclear receptor subfamily 0, group B, member 2; Nt5e, 5′ nucleotidase, ecto; o_, order; Onecut1, one cut domain, family member 1; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; Ppp1r, protein phosphatase 1, regulatory (inhibitor); ROS, reactive oxygen species.
Discussion
This study provides, for the first time to our knowledge, a comprehensive picture of the changes in hepatic metabolic and molecular physiology outcomes associated with microbiota shifts in response to a diet rich in fiber; in this case, ETWB. These observations provide candidate pathways that may be relevant to the mechanisms underlying the effects of dietary fiber on whole-body metabolism. Animal models and humans supplemented with AXOSs, the primary component of ETWB, at amounts between 5% and 10% (wt:wt) of rodent diets or 6–15 g/d in humans, exhibited a broad range of health improvements including the following: decreased body weight and liver TGs, increased gut satiety hormones, improved glucose homeostasis and immunomodulatory activities, and increased abundances of putatively beneficial gut bacteria (46–50). Consistent with this, mice supplemented with ETWB in the current study gained less weight over time and had decreased liver TGs. These decreases may partially be accounted for by decreased energy intake and/or energy utilization; however, there are additional mechanisms involved because there was little to no correlation between cumulative energy intake and adiposity index or liver TGs. Future studies are warranted to test the impact of ETWB on energetics and energy balance in the context of equivalent body weight.
One possibility is that ETWB leads to increased fecal energy loss (51). Fecal energy was not measured, because the intent of this study was to identify shifts in liver metabolism and cecal bacteria induced by ETWB feeding; however, fecal output was significantly increased in the ETWB-fed mice consistent with higher energy loss via this route. Nevertheless, mice remained in positive energy balance and gained weight during the study. The reduced concentrations of liver TGs observed in the ETWB-fed mice concurrent with increased liver ROS [which increase with enhanced fat oxidation (52)] may indicate increased hepatic mitochondrial β-oxidation. This idea is supported by significantly increased plasma and liver concentrations of the ketone body β-hydroxybutyrate in the ETWB group. Reduced concentrations of the antioxidants α-tocopherol and glutathione in the liver of ETWB-fed mice may be due to increased hepatic ROS concentrations. Overall, these differences suggest that ETWB supplementation led to more robust liver fat combustion, similar to a more “fasted” metabolic state, despite mice being in positive energy balance.
Similarly, several gene expression differences suggest that ETWB shifts liver metabolism toward a fasted phenotype (i.e., increased hepatic β-oxidation and/or reductions in TG synthesis), despite mice being in positive energy balance and gaining weight. Transcriptomics data revealed a reduction in the expression of FA synthase (Fasn); a decrease in Ces3b, which participates in VLDL assembly (53); and an increase in Cd36, which is involved in FA uptake (54) (Supplemental Table 5). In addition, acyl-CoA thioesterase 1 and 3 (Acot1 and Acot3, respectively) were significantly increased in the ETWB group and Acot4 was also increased. Acot proteins belong to a class of enzymes that cleave thioester bonds of acyl-CoAs, leading to the release of CoA and NEFAs, thus regulating lipid metabolism (55). The expression of several Acot genes are upregulated during feed deprivation in mice, including Acot1, Acot3, and Acot4 (56). In addition, the expression of the cytochrome p450 enzymes, Cyp4a10 and Cyp4a14, were significantly increased by ETWB feeding. These enzymes have been shown to increase hepatic lipid oxidation in mice with diet-induced steatohepatitis that lack the main hepatic cytochrome lipid-metabolizing enzyme Cyp2e1 (57). In addition, mice fed a high-fat diet supplemented with brown rice extract (58) or quercetin (59) displayed increased hepatic gene expression of Cyp4a10 along with decreased serum and liver TGs and increased levels of genes involved in hepatic lipid metabolism. Hepatic gene expression levels of Cyp4a10 and Cyp4a14 in this study correlated with a variety of liver metabolites (Figure 6), including a strong negative correlation with glucose. Interestingly, neither Cyp4a10 nor Cyp4a14 correlated with liver or plasma concentrations of the ketone body β-hydroxybutyrate; a positive correlation between these variables was thought to have existed on the basis of lipid oxidation increasing ketone body concentrations (60). However, liver ROS did have a significant positive correlation with Cyp4a14 (Spearman correlation r = 0.42, P = 0.0351) and a positive but nonsignificant relation with Cyp4a10 (Spearman correlation r = 0.39, P = 0.06). Increases in Cyp4a10 expression have been observed in the small intestine of mice after being feed-deprived for 24 h (61). Metabolism of hepatic lipids via the cytochrome p450 pathway is of interest not only because it may act to decrease liver lipid accumulation but because it can result in the production of anti-inflammatory arachidonic acid–derived metabolites, such as epoxyeicosanoids.
In addition to alterations in hepatic FA metabolism, there were also marked differences in glucose metabolism in response to ETWB feeding. Glucose concentration was reduced in the livers of ETWB-fed mice, despite plasma glucose concentrations remaining unchanged. Possibly associated with this observation was a significant increase in glucose-6-phosphatase mRNA in the livers of ETWB-fed mice. This enzyme, which is typically more active in the fasted state, participates in gluconeogenesis and glycogenolysis and therefore plays an important role in glucose homeostasis through liver glucose output (62). ETWB-fed mice also showed increased expression of the rate-limiting enzyme for gluconeogenesis, phosphoenolpyruvate carboxykinase 1 (Pck1), which is upregulated during fasting (63). However, not all gene expression differences were consistent with a fasted phenotype. The glycogen-targeting regulatory subunits of protein phosphatases, Ppp1r3b and Ppp1r3c, were increased in the ETWB group; these transcripts have been previously reported to increase in the liver during feeding and to decrease during feed deprivation (64).
The mechanisms by which dietary fiber affects whole-body and hepatic physiology remain to be fully elucidated but are likely driven in part by cecal bacterial populations, which, in turn, could alter specific microbe- and host-derived signaling factors. As expected, ETWB supplementation significantly altered the cecal microbiota. Increases in the Bacteroidetes-to-Firmicutes ratio, as observed in the ETWB-fed mice, have been reported upon feeding high-fiber diets (65) and after weight loss (66). An increase in Bacteroidetes may partially be due to the increased repertoire of carbohydrate-degrading enzymes in the Bacteroidetes phylum (67, 68). The increase in Bacteroidetes was driven by increases in the families Rikenellaceae and S24-7, which have been reported to increase after high-fiber feeding (69, 70). Decreased abundance of the mucin-degrading bacteria (71) Akkermansia in the ETWB group may be of interest because it has been reported to decrease with obesity (72) and appears to have a differential growth response depending on fiber source. Akkermansia was reported to increase in aged mice fed resistant starch (73); however, another study found this bacteria to be highest in fiber-free–fed rats compared with rats supplemented with pectin or guar gum (74). A thinning of the host mucin layer concurrent with an increase in Akkermansia has been observed when feeding low-carbohydrate diets to mice (75), and this bacteria may therefore play a role in gut barrier function.
Because the liver is the first organ to be bathed in portal blood derived from the gut, we reason that these gut microbe–associated signals will have a profound impact on hepatic (patho)physiology. The most well-established example of gut-derived regulatory factors is the often-observed increase in SCFAs in response to dietary fiber intake. SCFAs serve as fuel or receptor ligands that have been implicated in influencing many processes including the following: gastrointestinal tract growth (76), gut-derived satiety hormones (77), and immune system modulation (78) [also reviewed in (79, 80)]. However, SCFAs are not the sole regulators of host physiology: changes in the gut microbiome lead to alterations in many microbe-derived metabolites, often called “xenobiotics” but more accurately termed “xenometabolites” (81). Indeed, in the current study, there were marked ETWB-related changes in host physiology and liver metabolism that were concurrent with gut microbiome shifts, with no difference in total cecal SCFA abundance, indicating that factors independent of SCFAs were involved. Potential explanations for the lack of change in SCFAs include the following: impairment of microbial fermentation due to the presence of high fat in the diet (82) and increased uptake and utilization by colonic epithelial cells and/or bacteria. Beyond SCFAs, we identified a variety of candidate xenometabolites that are altered by changes in the gut bacteria and that reach the systemic circulation. For example, plasma concentrations of the microbial phenol degradation product 3-phenylpropanoic acid (83, 84) positively correlated with Rikenellaceae and negatively correlated with several other cecal bacteria (Figure 5). Microbial degradation of ferulic acid, a purportedly beneficial phenol found in high abundances in wheat bran, has been shown to increase concentrations of 3-phenylpropanoic acid in vitro (85). Bioaccessibility of ferulic acid by the host is limited and microbial degradation is needed to free this compound in amounts sufficient to increase circulating concentrations (86). Rikenellaceae has not previously been reported to play a role in (poly)phenol degradation (87).
Many other significant correlations among cecal bacteria, metabolites, and host phenotype (cecal tissue weight, liver TGs, hepatic genes, and plasma and liver metabolites) were found. However, it remains to be established in future studies which, if any, of the metabolites identified in this report have bioactivities that—directly or indirectly—affect the observed ETWB-associated changes in adiposity, hepatic TG content, lipid and glucose metabolism, and gene regulation. With that said, 5 bacterial taxa that increased with ETWB feeding had negative correlations with adiposity index and liver TGs and a positive correlation with cecal weight (Bacteroidetes families S24-7 and Rikellenaceae and the following Firmicutes order RF39, family Ruminococcaceae, and genus Adlercreutzia). Interestingly, some of these bacterial taxa also correlated with purine metabolites. Ruminococcaceae and RF39 negatively correlated with inosine and hypoxanthine, whereas Clostridia, which had a positive correlation with adiposity and liver TGs, also had a positive correlation with inosine and hypoxanthine. Factors produced by these taxa may affect hepatic and whole-body physiology and therefore warrant further study.
ETWB feeding elicited changes in hepatic gene expression related to cell cycle as identified by pathway analysis. The ETWB group showed increased expression of genes reported to be related to cell cycle, such as Cdkn1a and Gdf2, and a decrease in Gadd45g, Plk3, and Myc. The functional ramifications of these changes are not clear; however, it is perhaps relevant that dietary fiber intake has been associated with decreased incidence of cancer, particularly colon cancer (88–90). In addition to cell cycle, genes involving immune function were also altered by ETWB feeding, such as decreases in Cish and Ikbke. Interestingly, the consumption of arabinoxylans has been shown to alter innate and adaptive immune function in human and animal models (50). Such observations provide a foundation for hypothesis testing to determine if ETWB and fiber feeding influences hepatic cell cycle and inflammation outcomes.
In conclusion, we have shown that mice that consumed a 45%-fat diet supplemented with 20% ETWB by weight of the diet for 10 wk showed decreased adiposity, lower liver TGs, increased liver ROS, marked differences in plasma and liver metabolites, and changes in hepatic gene expression, including those related to lipid metabolism. It is acknowledged that interpretations related to ETWB effects on the microbiome and metabolome in the current DIO model are confounded by lower weight and adiposity in mice fed this fiber. Thus, future experiments could focus on weight-independent effects of ETWB (i.e., testing parameters at an earlier treatment time point or performing dose-response experiments). Our results help form the foundation for this. If the results in DIO mice are shown in humans, it would support the investigation of ETWB as a potential dietary means to help mitigate or prevent NAFLD and related metabolic disorders. Concurrent with the obesity epidemic, there is an alarming increase in NAFLD in both the adult and pediatric populations and diet is known to greatly affect obesity and related comorbidities (91). Future research should investigate the cellular mechanisms by which specific microbes and xenometabolites associated with ETWB feeding (identified herein) regulate gut, liver, and whole-body systems.
Acknowledgments
We thank Michael Blackburn and Kikumi Ono-Moore (Arkansas Children's Nutrition Center) for assistance with plasma TG and NEFA measurements as well as Pieter Oort (Western Human Nutrition Research Center) for technical assistance. DAK, SHA, and RJM designed the research and had primary responsibility for the final content; DAK, EBK, MLG, TND, SHA, and RJM conducted the research; MLM, MJK, and KEBK provided essential materials; DAK, BDP, SHA, and RJM analyzed the data; DAK wrote the manuscript; and DAK, BDP, MLM, KEBK, SHA, and RJM edited the manuscript. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AXOS, arabinoxylan oligosaccharide; DIO, diet-induced obesity; ETWB, enzyme-treated wheat bran; Fasn, fatty acid synthase; FDR, false discovery rate; NAFLD, nonalcoholic liver disease; NEFA, nonesterified fatty acid; OTU, operational taxonomic unit; Pck1, phosphoenolpyruvate carboxykinase 1; PLS-DA, partial least-squares-discriminant analysis; ROS, reactive oxygen species; RNAseq, RNA sequencing; rRNA, ribosomal RNA; UC Davis, University of California at Davis; VIP, variable importance in projection.
References
- 1.Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obesity 2008;32:1431–7. [DOI] [PubMed] [Google Scholar]
- 2.Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther 2011;34:274–85. [DOI] [PubMed] [Google Scholar]
- 3.Welsh JA, Karpen S, Vos MB. Increasing prevalence of nonalcoholic fatty liver disease among United States adolescents, 1988–1994 to 2007–2010. J Pediatr 2013;162:496–500, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 2004;27:1047–53. [DOI] [PubMed] [Google Scholar]
- 5.Pal S, Khossousi A, Binns C, Dhaliwal S, Ellis V. The effect of a fibre supplement compared to a healthy diet on body composition, lipids, glucose, insulin and other metabolic syndrome risk factors in overweight and obese individuals. Br J Nutr 2011;105:90–100. [DOI] [PubMed] [Google Scholar]
- 6.Babiker R, Merghani TH, Elmusharaf K, Badi RM, Lang F, Saeed AM. Effects of gum arabic ingestion on body mass index and body fat percentage in healthy adult females: two-arm randomized, placebo controlled, double-blind trial. Nutr J 2012;11:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson MD, Wright JW, Loizon E, Debard C, Vidal H, Shojaee-Moradie F, Russell-Jones D, Umpleby AM. Insulin-sensitizing effects on muscle and adipose tissue after dietary fiber intake in men and women with metabolic syndrome. J Clin Endocrinol Metab 2012;97:3326–32. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Cao Y, Wang C, Sun B. Wheat bran xylooligosaccharides improve blood lipid metabolism and antioxidant status in rats fed a high-fat diet. Carbohydr Polym 2011;86:1192–7. [Google Scholar]
- 9.King DE, Mainous AG III, Lambourne CA. Trends in dietary fiber intake in the United States, 1999–2008. J Acad Nutr Diet 2012;112:642–8. [DOI] [PubMed] [Google Scholar]
- 10.USDA. Dietary guidelines for Americans 2010. 7th ed Washington (DC): USDA; 2010. [Google Scholar]
- 11.Delzenne NM, Neyrinck AM, Cani PD. Gut microbiota and metabolic disorders: how prebiotic can work? Br J Nutr 2013;109(Suppl 2):S81–5. [DOI] [PubMed] [Google Scholar]
- 12.Graf D, Di Cagno R, Fåk F, Flint HJ, Nyman M, Saarela M, Watzl B. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis 2015;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464(7285):59–65. [cited 2014 Aug 24]. Available from: http://www.nature.com/nature/journal/v464/n7285/suppinfo/nature08821_S1.html. [DOI] [PMC free article] [PubMed]
- 15.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004;101:15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keenan MJ, Zhou J, McCutcheon KL, Raggio AM, Bateman HG, Todd E, Jones CK, Tulley RT, Melton S, Martin RJ, et al. Effects of resistant starch, a non-digestible fermentable fiber, on reducing body fat. Obesity (Silver Spring) 2006;14:1523–34. [DOI] [PubMed] [Google Scholar]
- 18.Jensen MT, Cox RP, Jensen BB. Microbial production of skatole in the hind gut of pigs given different diets and its relation to skatole deposition in backfat. Anim Sci 1995;61:293–304. [Google Scholar]
- 19.Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, Martin P, Philippe C, Walker F, Bado A, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 2013;62:1787–94. [DOI] [PubMed] [Google Scholar]
- 20.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 2009;106:3698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingerslev AK, Theil PK, Hedemann MS, Laerke HN, Bach Knudsen KE. Resistant starch and arabinoxylan augment SCFA absorption, but affect postprandial glucose and insulin responses differently. Br J Nutr 2014;111:1564–76. [DOI] [PubMed] [Google Scholar]
- 22.Thomas AP, Dunn TN, Oort PJ, Grino M, Adams SH. Inflammatory phenotyping identifies CD11d as a gene markedly induced in white adipose tissue in obese rodents and women. J Nutr 2011;141:1172–80. [DOI] [PubMed] [Google Scholar]
- 23.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497–509. [PubMed] [Google Scholar]
- 24.Ali SF, LeBel CP, Bondy SC. Reactive oxygen species formation as a biomarker of methylmercury and trimethyltin neurotoxicity. Neurotoxicology 1992;13:637–48. [PubMed] [Google Scholar]
- 25.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265–75. [PubMed] [Google Scholar]
- 26.Barry KA, Wojcicki BJ, Bauer LL, Middelbos IS, Vester Boler BM, Swanson KS, Fahey GC. Adaptation of healthy adult cats to select dietary fibers in vivo affects gas and short-chain fatty acid production from fiber fermentation in vitro. J Anim Sci 2011;89:3163–9. [DOI] [PubMed] [Google Scholar]
- 27.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 2011;108(Suppl 1):4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin X, Yan Y, Kim EB, Lee B, Marco ML. Short communication: effect of milk and milk containing Lactobacillus casei on the intestinal microbiota of mice. J Dairy Sci 2014;97:2049–55. [DOI] [PubMed] [Google Scholar]
- 29.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 2013;10:57–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011;27:2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiehn O, Kind T. Metabolite profiling in blood plasma. Methods Mol Biol 2007;358:3–17. [DOI] [PubMed] [Google Scholar]
- 33.Fiehn O, Wohlgemuth G, Scholz M. Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. In: Ludäscher B, Raschid L, editors. Data integration in the life sciences. Berlin, Heidelberg (Germany): Springer; 2005. p. 224–39.
- 34.Scholz M, Fiehn O. SetupX—a public study design database for metabolomic projects. Stanford (CA): Shriram Center for BioE & ChemE. Pac Symp Biocomput 2007:169–80. [PubMed] [Google Scholar]
- 35.Zhang B, Kirov S, Snoddy J.. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33(Web Server issue):W741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Duncan D, Shi Z, Zhang B.. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013;41(Web Server issue):W77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oort PJ, Warden CH, Baumann TK, Knotts TA, Adams SH. Characterization of Tusc5, an adipocyte gene co-expressed in peripheral neurons. Mol Cell Endocrinol 2007;276:24–35. [DOI] [PubMed] [Google Scholar]
- 38.R Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2014. [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300. [Google Scholar]
- 40. Mevik B, Wehrens, R., Liland, KH. Pls: partial least squares and principal component regression. J Stat Software; 2013.
- 41.Komsta L. Outliers: tests for outliers. R package version 0.14. [Internet]; 2011 [cited 2014 Aug 8]. Available from: http://CRAN.R-project.org/package=outliers.
- 42. Wong J. Imputation: imputation. R package version 2.0.1 [Internet]. [cited 2014 Aug 8]. Available from: http://CRAN.R-project.org/package=imputation.
- 43.Mehmood T, Liland KH, Snipen L, Sæbø S. A review of variable selection methods in partial least squares regression. Chemom Intell Lab Syst 2012;118:62–9. [Google Scholar]
- 44.Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 2001;58:109–30. [Google Scholar]
- 45.Canty A, Ripley B. Boot: Bootstrap R (S-Plus) functions. R package version 2.0.1. [Internet]; 2015 [cited 2015 Sep 15]. Available from: https://cran.r-project.org/package=boot.
- 46.Neyrinck AM, Van Hee VF, Piront N, De Backer F, Toussaint O, Cani PD, Delzenne NM. Wheat-derived arabinoxylan oligosaccharides with prebiotic effect increase satietogenic gut peptides and reduce metabolic endotoxemia in diet-induced obese mice. Nutr Diabetes 2012;2:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neyrinck AM, Possemiers S, Druart C, Van de Wiele T, De Backer F, Cani PD, Larondelle Y, Delzenne NM. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS One 2011;6:e20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu ZX, Walker KZ, Muir JG, O’Dea K. Arabinoxylan fibre improves metabolic control in people with Type II diabetes. Eur J Clin Nutr 2004;58:621–8. [DOI] [PubMed] [Google Scholar]
- 49.Broekaert WF, Courtin CM, Verbeke K, Van de Wiele T, Verstraete W, Delcour JA. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit Rev Food Sci Nutr 2011;51:178–94. [DOI] [PubMed] [Google Scholar]
- 50.Zhang S, Li W, Smith CJ, Musa H. Cereal-derived arabinoxylans as biological response modifiers: extraction, molecular features, and immune-stimulating properties. Crit Rev Food Sci Nutr 2015;55:1035–52. [DOI] [PubMed] [Google Scholar]
- 51.Kristensen M, Jensen MG, Aarestrup J, Petersen K, Søndergaard L, Mikkelsen MS, Astrup A. Flaxseed dietary fibers lower cholesterol and increase fecal fat excretion, but magnitude of effect depend on food type. Nutr Metab (Lond) 2012;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reddy JK, Mannaerts GP. Peroxisomal lipid metabolism. Annu Rev Nutr 1994;14:343–70. [DOI] [PubMed] [Google Scholar]
- 53.Lian J, Wei E, Wang SP, Quiroga AD, Li L, Di Pardo A, van der Veen J, Sipione S, Mitchell GA, Lehner R. Liver specific inactivation of carboxylesterase 3/triacylglycerol hydrolase decreases blood lipids without causing severe steatosis in mice. Hepatology 2012;56:2154–62. [DOI] [PubMed] [Google Scholar]
- 54.Xu S, Jay A, Brunaldi K, Huang N, Hamilton JA. CD36 enhances fatty acid uptake by increasing the rate of intracellular esterification but not transport across the plasma membrane. Biochemistry 2013;52:7254–61. [DOI] [PubMed] [Google Scholar]
- 55.Hunt MC, Alexson SEH. The role acyl-CoA thioesterases play in mediating intracellular lipid metabolism. Prog Lipid Res 2002;41:99–130. [DOI] [PubMed] [Google Scholar]
- 56.Ellis JM, Bowman CE, Wolfgang MJ. Metabolic and tissue-specific regulation of acyl-CoA metabolism. PLoS One 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leclercq IA, Farrell GC, Field J, Bell DR, Gonzalez FJ, Robertson GR. CYP2E1 and CYP4A as microsomal catalysts of lipid peroxides in murine nonalcoholic steatohepatitis. J Clin Invest 2000;105:1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang H-H, Park M-Y, Kim H-W, Lee Y-M, Hwang K-A, Park J-H, Park D-S, Kwon O. Black rice (Oryza sativa L.) extract attenuates hepatic steatosis in C57BL/6 J mice fed a high-fat diet via fatty acid oxidation. Nutr Metab (Lond) 2012;9:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoek-van den Hil EF, Keijer J, Bunschoten A, Vervoort JJ, Stankova B, Bekkenkamp M, Herreman L, Venema D, Hollman PC, Tvrzicka E, et al. Quercetin induces hepatic lipid omega-oxidation and lowers serum lipid levels in mice. PLoS One 2013;8:e51588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cahill GF. Fuel metabolism in starvation. Annu Rev Nutr 2006;26:1–22. [DOI] [PubMed] [Google Scholar]
- 61.van den Bosch HM, Bunger M, de Groot P, van der Meijde J, Hooiveld G, Muller M. Gene expression of transporters and phase I/II metabolic enzymes in murine small intestine during fasting. BMC Genomics 2007;8:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nordlie RC, Foster JD, Lange AJ. Regulation of glucose production by the liver. Annu Rev Nutr 1999;19:379–406. [DOI] [PubMed] [Google Scholar]
- 63.She P, Shiota M, Shelton KD, Chalkley R, Postic C, Magnuson MA. Phosphoenolpyruvate carboxykinase is necessary for the integration of hepatic energy metabolism. Mol Cell Biol 2000;20:6508–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo X, Zhang Y, Ruan X, Jiang X, Zhu L, Wang X, Ding Q, Liu W, Pan Y, Wang Z, et al. Fasting-induced protein phosphatase 1 regulatory subunit contributes to postprandial blood glucose homeostasis via regulation of hepatic glycogenesis. Diabetes 2011;60:1435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444(7122):1022–3. [cited 2014 Sep 18]. Available from: http://www.nature.com/nature/journal/v444/n7122/suppinfo/4441022a_S1.html. [DOI] [PubMed]
- 67.Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Micro 2013;11(7):497–504. [cited 2014 Sep 18]. Available from: http://www.nature.com/nrmicro/journal/v11/n7/abs/nrmicro3050.html#supplementary-information. [DOI] [PubMed]
- 68.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 2012;10:323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee SM, Han HW, Yim SY. Beneficial effects of soy milk and fiber on high cholesterol diet-induced alteration of gut microbiota and inflammatory gene expression in rats. Food Funct 2015;6:492–500. [DOI] [PubMed] [Google Scholar]
- 70.Zhong Y, Nyman M, Fak F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol Nutr Food Res 2015;59:2066–76. [DOI] [PubMed] [Google Scholar]
- 71.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 2004;54:1469–76. [DOI] [PubMed] [Google Scholar]
- 72.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA 2013;110:9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tachon S, Zhou J, Keenan M, Martin R, Marco ML. The intestinal microbiota in aged mice is modulated by dietary resistant starch and correlated with improvements in host responses. FEMS Microbiol Ecol 2013;83:299–309. [DOI] [PubMed] [Google Scholar]
- 74.Jakobsdottir G, Xu J, Molin G, Ahrné S, Nyman M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS One 2013;8:e80476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Earle K A, Billings G, Sigal M, Lichtman J S, Hansson G C, Elias J E, Amieva M R, Huang K C, Sonnenburg J L. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 2015;18:478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakata T. Stimulatory effect of short-chain fatty acids on epithelial cell proliferation in the rat intestine: a possible explanation for trophic effects of fermentable fibre, gut microbes and luminal trophic factors. Br J Nutr 1987;58:95–103. [DOI] [PubMed] [Google Scholar]
- 77.Zhou J, Martin RJ, Tulley RT, Raggio AM, McCutcheon KL, Shen L, Danna SC, Tripathy S, Hegsted M, Keenan MJ. Dietary resistant starch upregulates total GLP-1 and PYY in a sustained day-long manner through fermentation in rodents. Am J Physiol Endocrinol Metab 2008;295:E1160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013;504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. In: Frederick WA, editor. Adv Immunol 2014;121:91–119. [DOI] [PubMed]
- 80.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 2015;11:577–91. [DOI] [PubMed] [Google Scholar]
- 81.Campbell C, Grapov D, Fiehn O, Chandler CJ, Burnett DJ, Souza EC, Casazza GA, Gustafson MB, Keim NL, Newman JW. Improved metabolic health alters host metabolism in parallel with changes in systemic xeno-metabolites of gut origin. PLoS One 2014;9:e84260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charrier JA, Martin RJ, McCutcheon KL, Raggio AM, Goldsmith F, Goita M, Senevirathne RN, Brown IL, Pelkman C, Zhou J, et al. High fat diet partially attenuates fermentation responses in rats fed resistant starch from high-amylose maize. Obesity (Silver Spring) 2013;21:2350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Selma MV, Espín JC, Tomás-Barberán FA. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem 2009;57:6485–501. [DOI] [PubMed] [Google Scholar]
- 84.Ou K, Sarnoski P, Schneider KR, Song K, Khoo C, Gu L. Microbial catabolism of procyanidins by human gut microbiota. Mol Nutr Food Res 2014;58:2196–205. [DOI] [PubMed] [Google Scholar]
- 85.Anson NM, Selinheimo E, Havenaar R, Aura AM, Mattila I, Lehtinen P, Bast A, Poutanen K, Haenen GR. Bioprocessing of wheat bran improves in vitro bioaccessibility and colonic metabolism of phenolic compounds. J Agric Food Chem 2009;57:6148–55. [DOI] [PubMed] [Google Scholar]
- 86.Zhao Z, Egashira Y, Sanada H. Digestion and absorption of ferulic acid sugar esters in rat gastrointestinal tract. J Agric Food Chem 2003;51:5534–9. [DOI] [PubMed] [Google Scholar]
- 87.Duda-Chodak A, Tarko T, Satora P, Sroka P. Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review. Eur J Nutr 2015;54:325–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bradbury KE, Appleby PN, Key TJ. Fruit, vegetable, and fiber intake in relation to cancer risk: findings from the European Prospective Investigation into Cancer and Nutrition (EPIC). Am J Clin Nutr 2014;100(Suppl 1):394S–8S. [DOI] [PubMed] [Google Scholar]
- 89.Aune D, Chan DSM, Lau R, Vieira R, Greenwood DC, Kampman E, Norat T. Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. BMJ 2011;343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Z, Xu G, Ma M, Yang J, Liu X. Dietary fiber intake reduces risk for gastric cancer: a meta-analysis. Gastroenterology 2013;145:113–20, e3. [DOI] [PubMed] [Google Scholar]
- 91.Schwimmer JB, Deutsch R, Kahen T, Lavine JE, Stanley C, Behling C. Prevalence of fatty liver in children and adolescents. Pediatrics 2006;118:1388–93. [DOI] [PubMed] [Google Scholar]