Abstract
Background: High-amylose-maize resistant starch type 2 (HAMRS2) is a fermentable dietary fiber known to alter the gut milieu, including the gut microbiota, which may explain the reported effects of resistant starch to ameliorate obesity-associated metabolic dysfunction.
Objective: Our working hypothesis was that HAMRS2-induced microbiome changes alter gut-derived signals (i.e., xenometabolites) reaching the liver via the portal circulation, in turn altering liver metabolism by regulating gene expression and other pathways.
Methods: We used a multi-omics systems biology approach to characterize HAMRS2-driven shifts to the cecal microbiome, liver metabolome, and transcriptome, identifying correlates between microbial changes and liver metabolites under obesogenic conditions that, to our knowledge, have not previously been recognized. Five-week-old male C57BL/6J mice were fed an energy-dense 45% lard-based-fat diet for 10 wk supplemented with either 20% HAMRS2 by weight (n = 14) or rapidly digestible starch (control diet; n = 15).
Results: Despite no differences in food intake, body weight, glucose tolerance, fasting plasma insulin, or liver triglycerides, the HAMRS2 mice showed a 15–58% reduction in all measured liver amino acids, except for Gln, compared with control mice. These metabolites were equivalent in the plasma of HAMRS2 mice compared with controls, and transcripts encoding key amino acid transporters were not different in the small intestine or liver, suggesting that HAMRS2 effects were not simply due to lower hepatocyte exposure to systemic amino acids. Instead, alterations in gut microbial metabolism could have affected host nitrogen and amino acid homeostasis: HAMRS2 mice showed a 62% increase (P < 0.0001) in 48-h fecal output and a 41% increase (P < 0.0001) in fecal nitrogen compared with control mice. Beyond amino acid metabolism, liver transcriptomics revealed pathways related to lipid and xenobiotic metabolism; and pathways related to cell proliferation, differentiation, and growth were affected by HAMRS2 feeding.
Conclusion: Together, these differences indicate that HAMRS2 dramatically alters hepatic metabolism and gene expression concurrent with shifts in specific gut bacteria in C57BL/6J mice.
Keywords: dietary fiber, gut microbiota, metabolomics, transcriptomics, liver
Introduction
A large body of literature shows that certain dietary fibers act as fermentable substrates that alter the gut microbiome and often improve host metabolic phenotype, such as improving insulin sensitivity (1–3). The mechanisms, gut-derived signals, target tissues, and pathways involved in these responses have not been fully elucidated. The best-known gut-derived metabolite signals that affect host metabolism are SCFAs (4); however, there are many other gut-derived metabolites, termed “xenometabolites,” that also likely affect host physiology (5). We reasoned that the liver is a primary target of signals derived from the gut in response to change in the microbiome. This is because xenometabolites, for instance, exit the gut and enter the portal circulation, thus bathing the liver and potentially regulating hepatic metabolism and function to adapt to changing nutrition or gut health.
To investigate the relation between shifts in the gut microbiome and hepatic metabolism, we used the fermentable carbohydrate high-amylose-maize resistant starch type 2 (HAMRS2)15. HAMRS2 is derived from corn that has been naturally selected to contain a higher amylose-to-amylopectin ratio. The linear amylose molecules form granules that partially resist digestion by mammalian enzymes in the small intestine. The remaining ∼60% of undigested HAMRS2 passes into the large intestine where it can be fermented by microbes (6). HAMRS2 alters the gut microbiota (7), increases gut satiety hormones (8), and modulates intestinal gene expression (9). In the liver, HAMRS2 supplementation has been shown to increase glycolysis, cholesterol output, and lipid oxidation and to decrease lipogenesis (10, 11). However, it is not clear whether these differences relate to shifts in specific gut microbes. In the current study, we used a multi-omics approach to obtain a systems-level, comprehensive overview of microbe and host responses to HAMRS2. This enabled the identification of candidate microbes and metabolites that could help explain resistant starch–associated differences in liver function.
Methods
Diet-induced obesity mice and diets.
Four-week-old male C57BL/6J mice (Jackson Laboratory) were individually housed under standard temperature (20–22°C) and light-dark cycle (12 h:12 h) conditions in a specific-pathogen-free facility. Mice were fed Teklad Rodent Diet 2918 (Envigo) for a 1-wk acclimation period, then randomly assigned (n = 15/group) to purified isocaloric experimental diets containing 45% kcal from fat (Teklad Diet TD.08511) (12) and supplemented with rapidly digestible corn starch (control) or 20% HAMRS2 (Hi-Maize 260; Ingredion) (13) by weight of the diet for 10 wk (Supplemental Table 1). One mouse from the HAMRS2 group was excluded from analysis due to abnormally low body weight gain and failure to thrive. The control group was composed of the same mice as reported in a comparison to enzyme-treated wheat bran feeding (14). Mice were given ad libitum access to food and water. Body weight and food intake were recorded every 2–3 d. All animal protocols were approved by the University of California at Davis Institutional Animal Care and Use Committee according to Animal Welfare Act guidelines.
Assays and analyses.
Detailed descriptions of molecular methods, biochemical assays, and microbiome analysis are described in the complementary article that describes enzyme-treated wheat bran effects in mice (14) and are also provided under Materials and Methods in the Supplemental Materials.
Statistical analysis.
Statistical analyses were performed by using GraphPad Prism (version 5.04 for Windows) and the open-source statistical software R (version 3.1.2) (15). Data are presented as means ± SEMs in text. An α level was determined at 0.05 for all statistical tests unless otherwise specified. The significance of microbial percentage abundance and metabolomics data was assessed by using the Mann-Whitney U test, and Benjamini-Hochberg false discovery rate (FDR)–corrected P values ≤0.05 were considered significant (16). Metabolomics data used in multivariate analysis were first assessed for univariate outliers by using Grubb’s test for outliers at α = 0.01. Outliers meeting those criteria were removed and then imputed via the k-nearest neighbors algorithm (17). In total, 103 outliers were removed, which accounted for only 0.4% of the entire metabolomics data. Partial least-squares-discriminate analysis (PLS-DA) from the “pls” package was used to determine variables that discriminate HAMRS2-fed mice from controls. PLS-DA model accuracy was assessed with a cross-validation scheme in which the data were randomly partitioned into training and test data sets encompassing two-thirds and one-third of all animals, respectively. Model fitting and feature selection were determined solely with data from the training set. Training data were scaled and centered to unit variance before model development, whereas test data were scaled and centered by using the means and SDs from the training data. Metabolites of interest were assessed in PLS-DA models with a variable importance in projection (VIP) calculation of ≥1. VIP is a weighted measure of the contribution of each metabolite to discriminate the classification groups, and calculations of ≥1 have been argued to be an adequate threshold to determine discriminant variables in PLS-DA models (18, 19). Furthermore, we used bootstrapping (20) to determine a distribution of VIP scores, and then tested whether the bootstrapped VIP distribution was significantly ≥1 with the use of an independent 1-tailed t test at α = 0.01. Metabolites meeting these criteria were chosen for inclusion in final PLS-DA model development. Model performance was assessed on the basis of the model’s ability to accurately predict the classification of the test set mice (i.e., mice that were held out of model development and feature selection). Final models that used 1 latent variable were able to predict the classification of test mice with 70% accuracy on the basis of plasma metabolites and with 100% accuracy on the basis of liver metabolites. Principal components score plots of plasma and liver metabolites can be found in Supplemental Figure 1. Principal components analysis is unsupervised modeling (i.e., model is not provided with treatment group assignments), whereas PLS-DA is supervised modeling (i.e., model is provided with treatment group assignments). Spearman’s correlation matrices feature the following data: fecal and cecal data and jejunal amino acid transporters identified by Student’s t test as significant at P ≤ 0.05, hepatic genes identified as significantly different from CuffDiff analysis after FDR correction and named by WebGestalt pathway analysis, plasma and liver metabolites, and cecal microbes. Correlations between any 2 variables were made by using individual mouse data (i.e., if variable A was measured in a subset of 10 mice and variable B was measured in the entire treatment group, only mice used in variable A would be used to determine the correlation between variables A and B).
Results
Adiposity, liver TGs, or plasma biochemical variables
Ten weeks of HAMRS2 supplementation on the background of an obesogenic 45%-fat diet did not alter body weight, adiposity, or feed efficiency compared with the control diet (Table 1, Supplemental Figure 2A). There was a significant diet × time interaction on cumulative energy intake (P < 0.0001), with intakes modestly reduced in the HAMRS2-fed mice compared with the control mice starting at day 49 of the ∼70-d feeding intervention (P < 0.0015) (Supplemental Figure 2B). There were no differences in terminal postabsorptive plasma measurements (glucose, insulin, nonesterified FAs, TGs), or liver TGs (Table 1) compared with control mice. No difference in oral-glucose tolerance (glucose AUC) was observed after 8 wk of HAMRS2 feeding. HAMRS2-fed mice did show significantly higher concentrations of hepatic reactive oxygen species.
TABLE 1.
Body weight, adiposity, and plasma biochemical variables in male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk1
| Variable | Control | HAMRS2 | P2 |
| Terminal body weight, g | 34.5 ± 0.69 | 34.1 ± 0.92 | 0.76 |
| Adiposity index3 | 3.15 ± 0.17 | 3.06 ± 0.24 | 0.76 |
| Feed efficiency,4 mg body weight gained/total cumulative kcal consumed | 22.70 ± 0.72 | 22.70 ± 1.29 | 0.99 |
| OGTT (AUC), mg/dL × min | 15,700 ± 1200 | 17,200 ± 1100 | 0.38 |
| Plasma glucose, mg/dL | 138 ± 4.9 | 150 ± 9.5 | 0.07 |
| Plasma insulin, ng/mL | 0.60 ± 0.06 | 0.74 ± 0.12 | 0.27 |
| Plasma nonesterified FAs, mM | 0.24 ± 0.01 | 0.24 ± 0.01 | 0.69 |
| Plasma TGs, mg/dL | 52.6 ± 2.45 | 46.8 ± 2.95 | 0.15 |
| Liver weight, g | 1.19 ± 0.03 | 1.23 ± 0.02 | 0.29 |
| Liver, % of body weight | 3.44 ± 0.06 | 3.61 ± 0.05 | 0.05 |
| Liver TGs, mg TGs/g liver | 46.7 ± 2.95 | 41.5 ± 3.52 | 0.27 |
| Liver reactive oxygen species, mmol DCF ⋅ mg protein−1 ⋅ min−1 | 3.44 ± 0.21 | 4.09 ± 0.17 | 0.03 |
Values are means ± SEMs; n = 15 in the control group, n = 14 in the HAMRS2 group. DCF, dichlorofluorescein; HAMRS2, high-amylose-maize resistant starch type 2; OGTT, oral-glucose-tolerance test.
Derived by using a 2-tailed Student’s t test. P ≤ 0.05 was considered significant.
Adiposity index is the sum of epididymal, retroperitoneal, and subcutaneous fat pads in grams. Plasma and tissue samples were collected from mice in the postabsorptive state after ∼4–8 h food deprivation in the morning.
To convert kcal to kJ, multiply by 4.184.
Resistant starch supplementation significantly alters gut environment
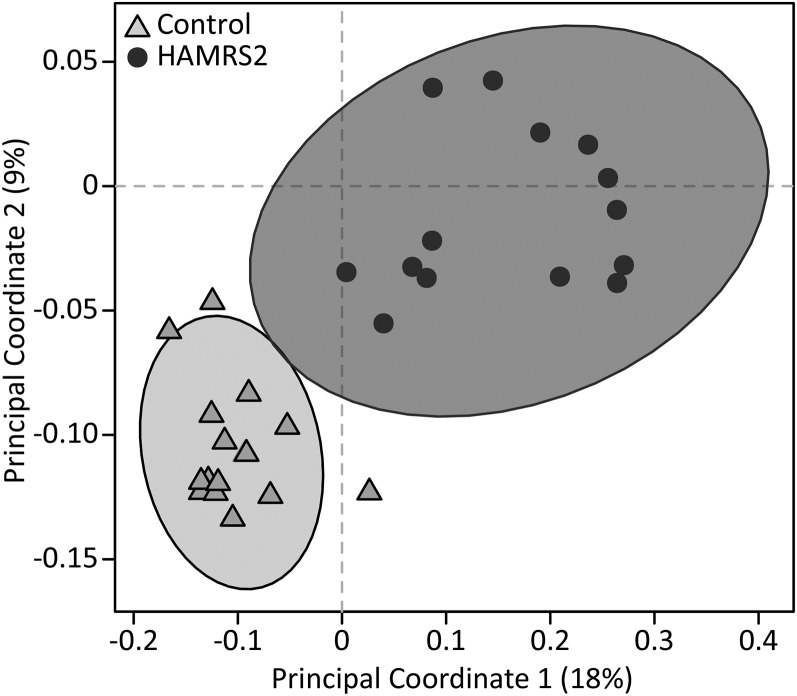
Mice supplemented with HAMRS2 showed significantly increased cecal tissue and cecal content weights as well as increased 48-h fecal output compared with control mice (Table 2), which is consistent with an altered microbiome. HAMRS2-fed mice showed a significant, albeit modest, decrease in cecal pH, despite no difference in measured total or individual cecal SCFAs (acetic, propionic, butyric, isobutyric, valeric, isovaloric, and isocaproic acids). The HAMRS2 group showed significantly altered cecal microbiota (Figure 1), with significantly fewer observed species than in the control group (control: 473 ± 10; HAMRS2: 318 ± 13; P < 0.0001). At the phylum level, the HAMRS2 group showed significantly greater abundances of Bacteroidetes, Tenericutes, and Verrucomicrobia; significantly reduced abundances of Firmicutes and Proteobacteria; and no difference in Actinobacteria (Table 3). The bacteria described below were the main contributors to differences at the phylum level, selected on the basis of representing ≥0.05% abundance in ≥1 treatment group, and were significant after FDR correction. The greater proportion of Bacteroidetes in the HAMRS2 group was driven predominantly by the family Rikenellaceae. The greater proportion of Tenericutes in the HAMRS2 group was driven by the order RF39. The genus Akkermansia accounted for the greater abundance of Verrucomicrobia in the HAMRS-fed mice. The reduced proportion of Firmicutes in the HAMRS2 group was due to reduced abundances of the family Lachnospiraceae and the genus Ruminococcus; there was, however, a greater proportion of the Firmicutes families Ruminococcaceae and Lactobacillaceae in the HAMRS2 group. The reduced proportion of Proteobacteria in the HAMRS2 group was driven by reductions in the Alphaproteobacteria family Caulobacteraceae (this difference was not significant after FDR correction).
TABLE 2.
Fecal output, fecal nitrogen, and cecal characteristics of male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk1
| Variable | Control | HAMRS2 | P2 |
| 48-h Fecal output,3 mg | 517 ± 23.0 | 840 ± 33.5 | <0.0001 |
| Mean 24-h fecal nitrogen,3 mg | 5.74 ± 0.20 | 8.10 ± 0.43 | <0.0001 |
| Mean 24-h dietary nitrogen,3 mg | 88.4 ± 2.36 | 87.6 ± 1.85 | 0.80 |
| Cecal tissue,4 mg | 54.4 ± 2.3 | 83.2 ± 5.8 | <0.0001 |
| Cecal contents, mg | 177 ± 6.3 | 271 ± 12.0 | <0.0001 |
| Cecal pH | 7.9 ± 0.1 | 7.8 ± 0.1 | 0.03 |
| Total cecal SCFAs,5 μmol | 6.15 ± 1.21 | 5.94 ± 0.83 | 0.89 |
Values are means ± SEMs; n = 15 in the control group and n = 14 in the HAMRS2 group unless otherwise indicated. HAMRS2, high-amylose-maize resistant starch type 2.
Derived by using a 2-tailed Student’s t test. P ≤ 0.05 was considered significant.
n = 10/group. Contents were removed from the cecum and tissue weight was recorded.
Contents were removed from the cecum and weight was recorded.
Total SCFAs were quantified by summing concentrations (in millimoles per gram) of acetic, propionic, butyric, isobutyric, valeric, isovaleric, and isocaproic acids and multiplying by total cecal contents (in grams) to obtain total SCFA production in the entire cecal contents.
FIGURE 1.
Unweighted UniFrac Beta-Diversity principal coordinates analysis plot shows the separation between treatment groups on the basis of the cecal microbiota of male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk. Axes represent percentages of the variance that can be accounted for on the basis of cecal microbiota profile. Ellipses represent 95% CIs on the basis of Hotelling’s T2 statistic, and each symbol represents a mouse. Control: n = 15/group; HAMRS2: n = 14/group. HAMRS2, high-amylose-maize resistant starch type 2.
TABLE 3.
Percentage abundances of cecal bacteria phyla and significantly altered taxa (ranked from highest to lowest by percentage difference) in male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk1
| Control | HAMRS2 | Percentage difference (HAM RS2 relative to control)2 | P3 | |
| Phylum | ||||
| Tenericutes | 0.08 ± 0.01 | 0.31 ± 0.04 | 310 | 0.0002 |
| Actinobacteria | 1.19 ± 0.40 | 2.56 ± 0.80 | 115 | 0.06 |
| Bacteroidetes | 30.8 ± 1.86 | 54.2 ± 3.65 | 76 | 0.0001 |
| Verrucomicrobia | 4.22 ± 0.54 | 6.62 ± 0.87 | 57 | 0.0198 |
| Proteobacteria | 0.05 ± 0.02 | 0.03 ± 0.02 | −35 | 0.0198 |
| Firmicutes | 63.6 ± 1.66 | 36.2 ± 3.07 | −43 | <0.0001 |
| Taxon4 | ||||
| p__Bacteroidetes; f__Rikenellaceae | 4.75 ± 0.43 | 29.4 ± 4.15 | 520 | <0.0001 |
| p__Tenericutes; o__RF39 | 0.07 ± 0.01 | 0.33 ± 0.05 | 373 | 0.0003 |
| p__Firmicutes; f__Ruminococcaceae | 3.59 ± 0.62 | 11.00 ± 1.43 | 206 | 0.0003 |
| p__Firmicutes; f__Lactobacillaceae | 1.26 ± 0.53 | 2.94 ± 0.52 | 134 | 0.0020 |
| p__Verrucomicrobia; g__Akkermansia | 4.22 ± 0.54 | 6.45 ± 0.83 | 53 | 0.0348 |
| p__Firmicutes; g__Streptococcus | 0.12 ± 0.01 | 0.09 ± 0.02 | −23 | 0.0192 |
| p__Firmicutes; f__Lachnospiraceae; g__Ruminococcus | 0.53 ± 0.06 | 0.27 ± 0.04 | −48 | 0.0125 |
| p__Firmicutes; g__Oscillospira | 2.44 ± 0.19 | 1.00 ± 0.16 | −59 | 0.0010 |
| p__Firmicutes; f__Ruminococcaceae; g_Ruminococcus | 12.6 ± 0.85 | 4.61 ± 0.93 | −63 | 0.0001 |
| p__Firmicutes; f__Lachnospiraceae | 25.2 ± 1.27 | 7.73 ± 1.40 | −69 | <0.0001 |
| p__Firmicutes; c__Clostridia | 4.40 ± 0.47 | 0.93 ± 0.13 | −79 | 0.0001 |
| p__Firmicutes; o__Coriobacteriales | 3.35 ± 0.45 | 0.56 ± 0.19 | −83 | 0.0003 |
| p__Firmicutes; f__Peptostreptococcaceae | 0.60 ± 0.17 | 0.08 ± 0.05 | −86 | 0.0418 |
Values are means ± SEMs; n = 15 in the control group, n = 14 in the HAMRS2 group. c_, class; f_, family; g_, genus; HAMRS2, high-amylose-maize resistant starch type 2; o_, order; p_, phylum.
Percentage difference = [(HAMRS2 − control)/control] × 100.
Group comparisons were assessed by Mann-Whitney U tests. P values were adjusted for false discovery rate correction. Significance was set at an adjusted P value ≤0.05.
Reported taxa had a minimum of 0.05% mean abundance in each group and an adjusted P value ≤ 0.05. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification).
Resistant starch supplementation significantly alters the plasma and liver metabolomes
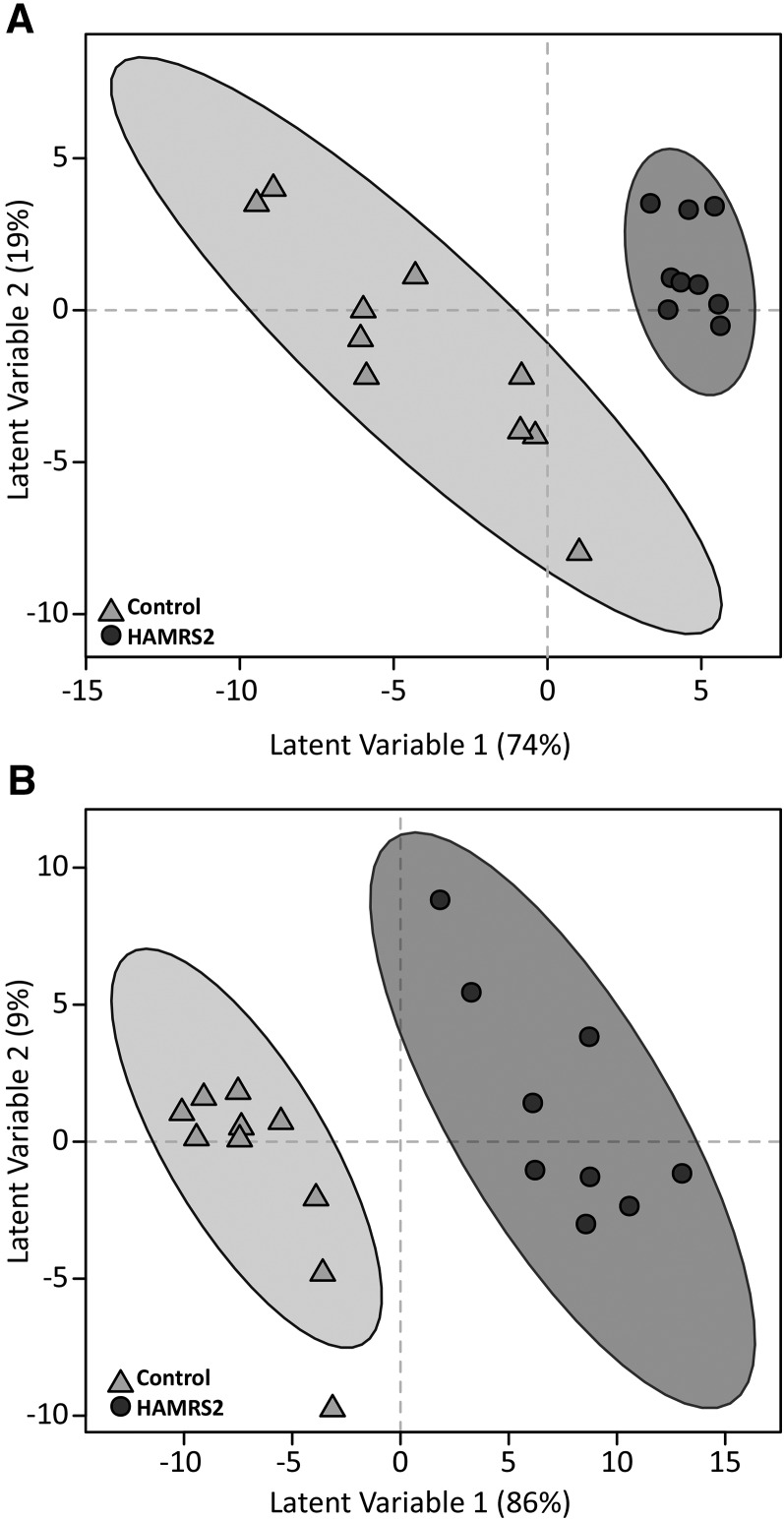
Plasma.
A total of 386 plasma metabolites were detected by using the GC–time-of-flight–MS analytical platform. Of these, 133 metabolites were annotated in the metabolite database, and the remaining metabolites were nonannotated and labeled with a numerical BinBase ID (Supplemental Table 2). A total of 86 metabolites had a mean bootstrapped VIP distribution ≥1 in the PLS-DA model, indicating that they contribute to discrimination of the groups (Figure 2A); of these, 28 metabolites were annotated (Table 4; for brevity only annotated metabolites are shown). The metabolites listed were identified by multivariate modeling as important discriminators between treatment groups, yet only one plasma metabolite, 1,5-anhydroglucitol, was identified as significant by univariate analysis after FDR correction. The HAMRS2 mice had greater abundances of the sugar alcohols sorbitol and 2-deoxyerythritiol and a reduced abundance of 1,5-anhydroglucitol. Glutamine was greater in the plasma of HAMRS2-fed mice than in controls, whereas aspartic acid was reduced. There was a modest reduction in urea of the HAMRS2 mice compared with controls. Greater abundances of the FAs pelargonic acid (C9) and capric acid (C10) were observed in the plasma of the HAMRS2 group, with reduced concentrations of palmitoleic acid (16:1n–7). There was a greater abundance of the ketone body 3-hydroxybutanoic acid (β-hydroxybutyrate) and reduced abundances of fumaric and pyruvic acids.
FIGURE 2.
PLS-DA score plots that show discrimination of male mice fed a 45%-fat diet with or without HAMRS2 supplementation. PLS-DA models were fit with plasma (A) and liver (B) metabolites. Each symbol represents a mouse; ellipses represent 95% CIs on the basis of Hotelling’s T2 statistic. Annotated metabolites contributing to these plots are shown in Tables 4 and 5; nonannotated metabolites are shown in Supplemental Tables 2 and 3. PLS-DA model development used 10 mice in the control group and 9 mice in the HAMRS2 group; therefore, score plots represent results from the PLS-DA model. PLS-DA model validation was performed by using the remaining 5 mice/group. HAMRS2, high-amylose-maize resistant starch type 2; PLS-DA, partial least-squares-discriminant analysis.
TABLE 4.
Postabsorptive plasma metabolite abundances (ranked highest to lowest by percentage difference) in male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk1
|
P4 |
||||||
| Metabolite2 | Control | HAMRS2 | Percentage difference (HAMRS2 relative to control)3 | MWU | MWU-FDR | VIP5 |
| Carbohydrates | ||||||
| Sorbitol | 1160 ± 123 | 1450 ± 113 | 25 | 0.0140 | 0.20 | 1.23 |
| 2-Deoxyerythritol | 5210 ± 316 | 6490 ± 526 | 25 | 0.09 | 0.40 | 1.15 |
| Fructose | 6140 ± 817 | 7580 ± 548 | 24 | 0.09 | 0.40 | 1.26 |
| Ribose | 76 ± 8 | 93 ± 8 | 23 | 0.19 | 0.56 | 1.18 |
| 1,5-Anhydroglucitol | 13,100 ± 690 | 9710 ± 264 | −26 | <0.0001 | <0.0001 | 1.62 |
| Nitrogenous | ||||||
| Glutamine | 40,400 ± 2600 | 50,600 ± 2000 | 25 | 0.0010 | 0.08 | 1.43 |
| N-methylalanine | 6580 ± 478 | 8070 ± 245 | 23 | 0.0120 | 0.20 | 1.54 |
| Urea | 427,000 ± 94,600 | 467,000 ± 48,500 | 9 | 0.98 | 1.00 | 1.39 |
| Lactamide | 407 ± 35 | 361 ± 22 | −11 | 0.33 | 0.67 | 1.11 |
| Aspartic acid | 1700 ± 155 | 1480 ± 78 | −13 | 0.25 | 0.62 | 1.10 |
| Nicotinamide | 570 ± 35 | 458 ± 13 | −20 | 0.0140 | 0.20 | 1.59 |
| Lipids | ||||||
| β-Sitosterol | 292 ± 52 | 403 ± 49 | 38 | 0.13 | 0.48 | 1.63 |
| Pelargonic acid | 5240 ± 540 | 7080 ± 246 | 35 | 0.0040 | 0.17 | 1.47 |
| Capric acid | 1060 ± 105 | 1330 ± 48 | 25 | 0.06 | 0.35 | 1.24 |
| Palmitoleic acid | 1540 ± 111 | 1280 ± 98 | −17 | 0.2010 | 0.58 | 1.17 |
| Octadecanol | 246 ± 21 | 183 ± 11 | −26 | 0.0150 | 0.20 | 1.19 |
| Other | ||||||
| Malonic acid | 2650 ± 289 | 3660 ± 437 | 38 | 0.10 | 0.44 | 1.16 |
| 3-Hydroxybutanoic acid | 13,380 ± 1113 | 18,200 ± 2826 | 36 | 0.11 | 0.45 | 1.42 |
| Benzoic acid | 5020 ± 484 | 6460 ± 257 | 29 | 0.0230 | 0.22 | 1.33 |
| Glyceric acid | 2050 ± 139 | 2380 ± 78 | 16 | 0.06 | 0.35 | 1.17 |
| 2-Hydroxybutanoic acid | 9040 ± 769 | 10470 ± 825 | 16 | 0.22 | 0.59 | 1.64 |
| Threonic acid | 2850 ± 206 | 3300 ± 82 | 16 | 0.0450 | 0.31 | 1.36 |
| 2-Ketoisocaproic acid | 2540 ± 175 | 2920 ± 159 | 15 | 0.0460 | 0.31 | 1.64 |
| 2-Deoxyisotetronic acid | 3470 ± 259 | 3980 ± 139 | 15 | 0.06 | 0.35 | 1.36 |
| Isothreonic acid | 607 ± 33 | 676 ± 29 | 12 | 0.16 | 0.51 | 1.25 |
| 2,3-Dihydroxybutanoic acid | 247 ± 13 | 262 ± 11 | 6 | 0.33 | 0.67 | 1.14 |
| Fumaric acid | 3471 ± 182 | 3247 ± 260 | −7 | 0.31 | 0.65 | 1.10 |
| Pyruvic acid | 16,900 ± 1294 | 12,900 ± 915 | −23 | 0.0290 | 0.25 | 1.76 |
Values are means ± SEMs; n = 15 in the control group, n = 14 in the HAMRS2 group. Only annotated metabolites with mean bootstrapped VIP measurements ≥1 are presented. Nonannotated metabolites are not shown for the sake of brevity but are provided in Supplemental Table 2. FDR, false discovery rate; HAMRS2, high-amylose-maize resistant starch type 2; MWU, Mann-Whitney U; VIP, variable importance in projection.
Metabolite abundances are reported in quantifier ion peak heights in the 0.5-μL extract derived from 15 μL plasma.
Percentage difference = [(HAMRS2 − control)/control] × 100.
Group comparisons were assessed by Mann-Whitney U tests. P values were adjusted for false discovery rate correction. Significance was set at an adjusted P value ≤ 0.05.
VIP was calculated from bootstrapped partial least-squares-discriminant analysis models derived from training data (n = 10 mice/group).
Liver.
A total of 454 liver metabolites were detected, 162 of which were annotated (Supplemental Table 3). The abundances of 131 metabolites were identified by PLS-DA to significantly contribute to separation between treatment groups (Figure 2B); 73 of these metabolites were annotated (Table 5; for brevity only annotated metabolites are shown). Unlike in the plasma, the majority of the metabolites identified as important discriminators by multivariate modeling in the liver were also significant by univariate analysis. Several sugars were reduced in the livers of HAMRS2-fed mice, including the sugar alcohols 1,5-anhydroglucitol, myo-inositol, ribitol, and xylitol. The HAMRS2 breakdown products, maltotriose, maltose, and glucose, were also decreased in HAMRS2-fed mice. There was an almost universal reduction in amino acids and other nitrogenous metabolites in the HAMRS2 group, including creatinine, ornithine, urea, and purine-related metabolites. Many lipid metabolites were also reduced in the HAMRS2 group, including palmitic acid, palmitoleic acid, oleic acid, arachidonic acid, and linoleic acid. Other metabolite differences include reductions in malic and fumaric acids.
TABLE 5.
Postabsorptive liver metabolite abundances (ranked highest to lowest by percentage difference) in male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk1
|
P4 |
||||||
| Metabolite2 | Control | HAMRS2 | Percentage difference (HAMRS2 relative to control)3 | MWU | MWU-FDR | VIP5 |
| Carbohydrates | ||||||
| Myo-inositol | 25,600 ± 1180 | 19,000 ± 794 | −26 | <0.0001 | <0.0001 | 1.64 |
| Galacturonic acid | 15,000 ± 794 | 10,900 ± 692 | −28 | <0.0001 | <0.0001 | 1.51 |
| Hexuronic acid | 14,800 ± 774 | 10,700 ± 678 | −28 | <0.0001 | <0.0001 | 1.55 |
| Flucose | 692,000 ± 29,800 | 463,000 ± 25,900 | −33 | <0.0001 | <0.0001 | 1.68 |
| Maltotriose | 124,000 ± 9290 | 67,500 ± 5310 | −46 | <0.0001 | <0.0001 | 1.20 |
| Galactonic acid | 586 ± 50 | 296 ± 49 | −49 | <0.0001 | <0.0001 | 1.47 |
| lactobionic acid | 25,900 ± 6520 | 12,600 ± 5320 | −51 | 0.0030 | 0.0130 | 1.23 |
| Lellobiose | 2200 ± 325 | 1020 ± 139 | −54 | 0.0010 | 0.0060 | 1.47 |
| Cellobiotol | 104,000 ± 5700 | 47,200 ± 5340 | −55 | <0.0001 | <0.0001 | 1.66 |
| 3,6-Anhydrogalactose | 429 ± 47 | 194 ± 21 | −55 | <0.0001 | <0.0001 | 1.43 |
| Xylitol | 2440 ± 245 | 1100 ± 156 | −55 | <0.0001 | <0.0001 | 1.52 |
| Maltose | 404,000 ± 22,100 | 174,000 ± 22,500 | −57 | <0.0001 | <0.0001 | 1.66 |
| Arabinose | 595 ± 73 | 240 ± 14 | −60 | <0.0001 | <0.0001 | 1.63 |
| Ribose | 5250 ± 844 | 1590 ± 388 | −70 | <0.0001 | <0.0001 | 1.31 |
| Fructose | 34,400 ± 5310 | 8760 ± 3040 | −75 | 0.0030 | 0.0130 | 1.04 |
| Ribitol | 1490 ± 431 | 346 ± 122 | −77 | <0.0001 | <0.0001 | 1.22 |
| Amino acids and derivatives | ||||||
| Alanine | 669,000 ± 41,000 | 536,000 ± 17,600 | −20 | 0.0010 | 0.0060 | 1.38 |
| Cysteine | 1410 ± 189 | 958 ± 90 | −32 | 0.0700 | 0.1830 | 1.10 |
| Tryptophan | 7070 ± 244 | 4730 ± 303 | −33 | <0.0001 | <0.0001 | 1.66 |
| β-Alanine | 3080 ± 204 | 2040 ± 125 | −34 | <0.0001 | <0.0001 | 1.51 |
| Glycine | 207,000 ± 15,200 | 135,000 ± 5850 | −35 | <0.0001 | <0.0001 | 1.52 |
| Isoleucine | 17,000 ± 1180 | 11,000 ± 558 | −35 | <0.0001 | <0.0001 | 1.47 |
| Valine | 32,200 ± 2030 | 20,400 ± 1080 | −36 | <0.0001 | <0.0001 | 1.46 |
| Proline | 18,800 ± 1400 | 11,600 ± 688 | −38 | <0.0001 | <0.0001 | 1.64 |
| Taurine | 275,000 ± 38,200 | 161,000 ± 18,800 | −41 | 0.0290 | 0.0930 | 1.08 |
| Phenylalanine | 6320 ± 644 | 3600 ± 339 | −43 | 0.0030 | 0.0130 | 1.38 |
| Tyrosine | 29300 ± 2330 | 16,600 ± 1850 | −43 | <0.0001 | <0.0001 | 1.30 |
| Leucine | 39700 ± 3070 | 21,600 ± 1720 | −45 | <0.0001 | <0.0001 | 1.65 |
| β-Glutamic acid | 329 ± 45 | 179 ± 22 | −46 | 0.0040 | 0.0170 | 1.33 |
| Threonine | 13,700 ± 1060 | 7300 ± 645 | −47 | <0.0001 | <0.0001 | 1.62 |
| Serine | 22,900 ± 2180 | 11,500 ± 1280 | −50 | <0.0001 | <0.0001 | 1.52 |
| Asparagine | 4860 ± 470 | 2170 ± 159 | −55 | <0.0001 | <0.0001 | 1.72 |
| Methionine | 3530 ± 431 | 1470 ± 274 | −58 | <0.0001 | <0.0001 | 1.40 |
| Other nitrogenous | ||||||
| Spermidine | 488 ± 39 | 391 ± 61 | −20 | 0.0370 | 0.1150 | 1.32 |
| N-methylalanine | 5000 ± 248 | 3990 ± 283 | −20 | 0.0120 | 0.0450 | 1.10 |
| Inosine 5′-monophosphate | 5670 ± 573 | 4390 ± 353 | −23 | 0.1580 | 0.3280 | 1.06 |
| Urea | 63,200 ± 4570 | 47,800 ± 2470 | −24 | 0.0030 | 0.0130 | 1.20 |
| γ-Glutamyl-valine | 186 ± 19 | 127 ± 9 | −32 | 0.0030 | 0.0130 | 1.39 |
| Xanthosine | 905 ± 74 | 604 ± 68 | −33 | 0.0030 | 0.0130 | 1.15 |
| N-acetylmannosamine | 615 ± 28 | 389 ± 17 | −37 | <0.0001 | <0.0001 | 1.70 |
| Creatinine | 5260 ± 360 | 3300 ± 161 | −37 | <0.0001 | <0.0001 | 1.72 |
| Glutamyl-threonine | 1310 ± 65 | 790 ± 48 | −40 | <0.0001 | <0.0001 | 1.72 |
| Allantoic acid | 2540 ± 314 | 1520 ± 208 | −40 | 0.0180 | 0.0640 | 1.45 |
| Nicotinamide | 24,600 ± 1670 | 13,900 ± 522 | −44 | <0.0001 | <0.0001 | 1.65 |
| Pantothenic acid | 1100 ± 102 | 598 ± 72 | −46 | 0.0010 | 0.0060 | 1.58 |
| Ornithine | 13,000 ± 1180 | 6780 ± 650 | −48 | <0.0001 | <0.0001 | 1.62 |
| Inosine | 32,200 ± 2630 | 14,300 ± 1480 | −56 | <0.0001 | <0.0001 | 1.59 |
| Flavin adenine dinucleotide | 420 ± 60 | 185 ± 16 | −56 | 0.0010 | 0.0060 | 1.41 |
| Xanthine | 9110 ± 799 | 3990 ± 577 | −56 | <0.0001 | <0.0001 | 1.54 |
| Uridine | 2010 ± 334 | 727 ± 119 | −64 | <0.0001 | <0.0001 | 1.60 |
| Uracil | 5060 ± 666 | 1560 ± 222 | −69 | <0.0001 | <0.0001 | 1.62 |
| Hypoxanthine | 23,600 ± 3269 | 6640 ± 1720 | −72 | <0.0001 | <0.0001 | 1.35 |
| Lipids | ||||||
| Caprylic acid | 1020 ± 53 | 837 ± 69 | −18 | 0.0850 | 0.2120 | 1.34 |
| Myristic acid | 2610 ± 156 | 2020 ± 84 | −22 | 0.0040 | 0.0170 | 1.39 |
| Isoheptadecanoic acid | 4640 ± 217 | 3400 ± 170 | −27 | 0.0010 | 0.0060 | 1.32 |
| Icosenoic acid | 2120 ± 192 | 1510 ± 110 | −29 | 0.0100 | 0.0390 | 1.29 |
| 1-Monostearin | 623 ± 53 | 385 ± 40 | −38 | 0.0010 | 0.0060 | 1.28 |
| Palmitic acid | 67,500 ± 4720 | 41,200 ± 3000 | −39 | <0.0001 | <0.0001 | 1.37 |
| Linoleic acid | 4510 ± 692 | 1340 ± 405 | −70 | <0.0001 | <0.0001 | 1.22 |
| Arachidonic acid | 17,100 ± 2540 | 4900 ± 1430 | −71 | <0.0001 | <0.0001 | 1.26 |
| Palmitoleic acid | 12,000 ± 2520 | 3360 ± 832 | −72 | 0.0010 | 0.0060 | 1.23 |
| Oleic acid | 18,800 ± 3430 | 4950 ± 1350 | −74 | <0.0001 | <0.0001 | 1.30 |
| Other | ||||||
| 2-Hydroxyglutaric acid | 2060 ± 130 | 1500 ± 97 | −27 | 0.0010 | 0.0060 | 1.32 |
| Glucuronic acid | 5640 ± 554 | 3970 ± 273 | −30 | 0.0410 | 0.1220 | 1.14 |
| Idonic acid | 1310 ± 70 | 903 ± 43 | −31 | <0.0001 | <0.0001 | 1.67 |
| Shikimic acid | 559 ± 79 | 382 ± 40 | −32 | 0.0200 | 0.0700 | 1.13 |
| Phosphoric acid | 94,100 ± 5860 | 55,900 ± 3400 | −41 | <0.0001 | <0.0001 | 1.65 |
| Gluconic acid | 521 ± 59 | 286 ± 48 | −45 | <0.0001 | <0.0001 | 1.17 |
| 2-Oxogluconic acid | 316 ± 47 | 165 ± 14 | −48 | 0.0010 | 0.0060 | 1.20 |
| Fumaric acid | 3860 ± 514 | 1910 ± 289 | −51 | 0.0030 | 0.0130 | 1.07 |
| Malic acid | 4640 ± 855 | 1630 ± 348 | −65 | 0.0020 | 0.0100 | 1.14 |
| Ribonic acid | 1820 ± 310 | 600 ± 144 | −67 | 0.0020 | 0.0100 | 1.16 |
| Glycerol | 46,000 ± 7160 | 13,400 ± 2070 | −71 | <0.0001 | <0.0001 | 1.50 |
Values are means ± SEMs; n = 15 in the control group, n = 14 in the HAMRS2 group. Only annotated metabolites with mean bootstrapped VIP measurements ≥1 are presented. Nonannotated metabolites are not shown for the sake of brevity but are provided in Supplemental Table 3. FDR, false discovery rate; HAMRS2, high-amylose-maize resistant starch type 2; MWU, Mann-Whitney U; VIP, variable importance in projection.
Metabolite abundances are reported in quantifier ion peak heights in the 0.5-μL extract derived from 4 mg liver.
Percentage difference = [(HAMRS2 − control)/control] × 100.
Group comparisons were assessed by Mann-Whitney U tests. P values were adjusted for false discovery rate correction. Significance was set at an adjusted P value ≤ 0.05.
VIP was calculated from bootstrapped partial least-squares-discriminant analysis models derived from training data (n = 10 mice/group).
Resistant starch alters nitrogen pools
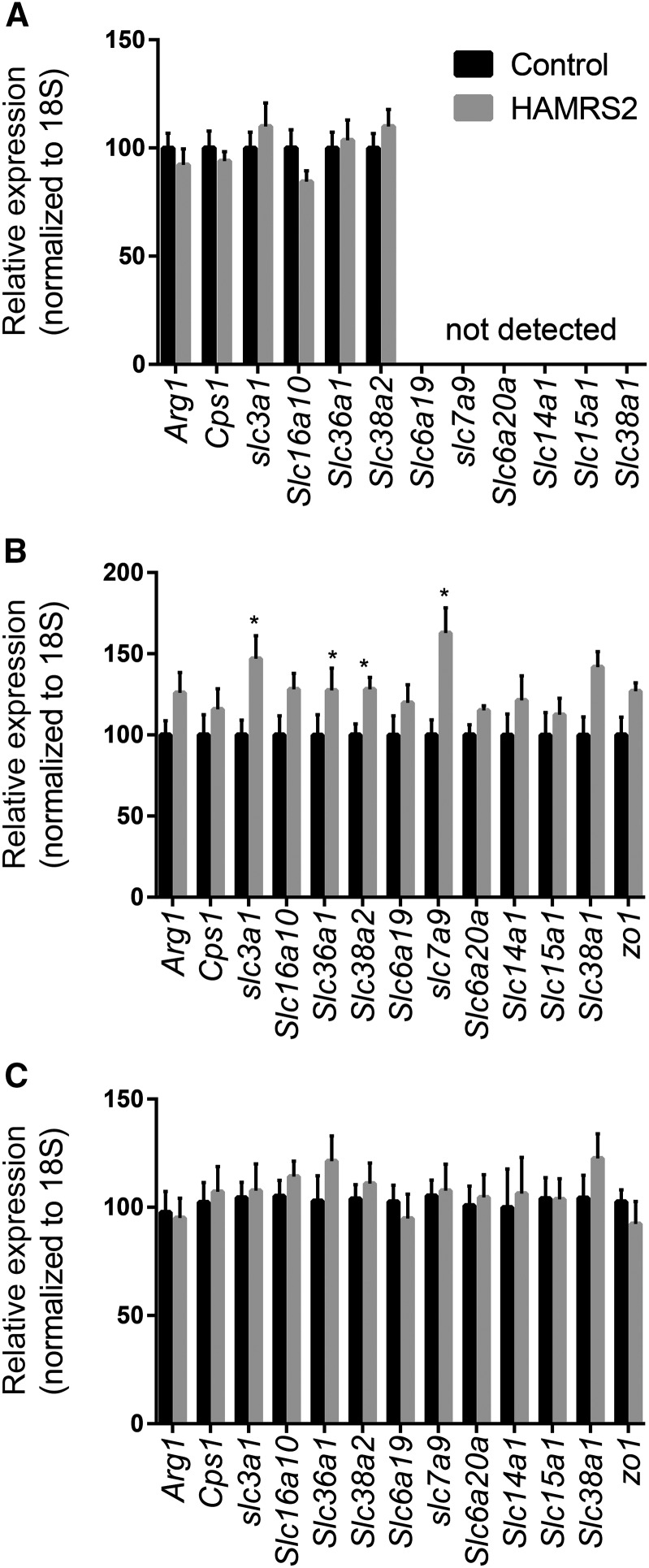
Because of the striking reduction in hepatic amino acids, we considered potential mechanisms that could affect liver amino acid metabolism. This occurred despite being fed equal amounts of a diet formulated to be isonitrogenous compared with the control diet. Decreases in liver amino acids were not reflected in the peripheral blood plasma. Altogether, this suggests that hepatic amino acid status is due to differences occurring in the liver and/or gut—i.e., reduced amino acid transporters or altered enzymology that affect liver exposure. HAMRS2-fed mice excreted significantly more nitrogen during a 48-h fecal collection than did control mice (Table 2), which could contribute to lower hepatic nitrogen metabolite exposure. We also performed qPCR for a representative number of amino acid transporters, and urea cycle transcripts were evaluated in the liver, jejunum, and ileum (Figure 3A–C). Transcript abundances for 4 amino acid transporters were elevated in the jejunum of HAMRS-fed mice, and these transcripts were either not expressed or not changed in the liver, providing evidence that the decreased liver amino acids may not be due to reductions in splanchnic amino acid transport.
FIGURE 3.
Gene expression of urea and amino acid transporters and urea cycle enzymes in the liver (A), jejunum (B), and ileum (C) of male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk. Control: n = 15/group; HAMRS2: n = 14/group. *P ≤ 0.05. Data are from qPCR analysis and expressed relative to the mean value in the control group. Arg1, arginase 1; Cps1, carbamoyl-phosphate synthetase I; HAMRS2, high-amylose-maize resistant starch type 2; Slc, solute carrier; zo1, zona occluden 1.
Resistant starch significantly alters hepatic metabolic gene expression
To further explore the biochemical pathways and mechanisms underlying the unique HAMRS2-related metabolite patterns in the liver, global liver transcriptomics analyses were conducted. Sixty-three protein-coding genes in the liver were considered significantly differentially expressed between treatment groups after FDR correction and an additional 312 genes had an unadjusted P ≤ 0.05 (Supplemental Table 4). Pathway analysis of genes that maintained significance after FDR correction revealed that 9 Kyoto Encyclopedia of Genes and Genomes pathways were affected by HAMRS2 treatment (Table 6). Pathway analysis was also conducted including genes with unadjusted P ≤ 0.05 and is shown in Supplemental Table 5. Fourteen genes were validated by qPCR (Figure 3A, Supplemental Table 6). Several cytochrome P450 enzymes involved in drug/xenobiotic and lipid metabolism were increased in HAMRS2-fed mice. HAMRS2 feeding also affected pathways related to Jak-STAT, TGF-β, and Wnt signaling. No clear patterns associated with amino acid metabolism were apparent.
TABLE 6.
Hepatic gene expression pathways (genes ranked highest to lowest by percentage difference) affected in male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk1
| Mean, FPKMs |
||||
| Pathway2 | Definition | Control | HAMRS2 | Percentage difference (HAMRS2 relative to control)3 |
| Metabolism of xenobiotics by cytochrome P450 (KEGG pathway: 00980) (C = 77; O = 3; E = 0.08; R = 35.78; rawP = 8.50 × 10−5; adjP = 0.0003) | ||||
| Cyp2b9 | Cytochrome P450, family 2, subfamily b, polypeptide 9 | 3.2 | 9.6 | 204 |
| Cyp2c37 | Cytochrome P450, family 2, subfamily c, polypeptide 37 | 66.0 | 93.9 | 42 |
| Cyp2c54 | Cytochrome P450, family 2, subfamily c, polypeptide 54 | 103 | 133 | 29 |
| Jak-STAT signaling pathway (KEGG pathway: 04630) (C = 153; O = 5; E = 0.17; R = 30.01; rawP = 7.52 × 10−7; adjP = 1.35 × 10−5) | ||||
| Spry4 | Sprouty homolog 4 (Drosophila) | 2.2 | 2.9 | 32 |
| Il6ra | IL-6 receptor, α | 9.7 | 7.4 | −24 |
| Crebbp | CREB binding protein | 3.9 | 2.9 | −27 |
| Cish | Cytokine inducible SH2-containing protein | 16.8 | 9.9 | −41 |
| Myc | Myelocytomatosis oncogene | 4.4 | 2.2 | −50 |
| TGF-β signaling pathway (KEGG pathway: 04350) (C = 85; O = 4; E = 0.09; R = 43.22; rawP = 2.42 × 10−6; adjP = 1.37 × 10−5) | ||||
| Rock1 | Rho-associated coiled-coil containing protein kinase 1 | 12.5 | 9.5 | −24 |
| Crebbp | CREB binding protein | 3.9 | 2.9 | −27 |
| Inhba | Inhibin β-A | 2.4 | 1.5 | −39 |
| Myc | Myelocytomatosis oncogene | 4.4 | 2.2 | −50 |
| Wnt signaling pathway (KEGG pathway: 04310) (C = 154; O = 3; E = 0.17; R = 17.89; rawP = 0.0007; adjP = 0.0014) | ||||
| Rock1 | Rho-associated coiled-coil containing protein kinase 1 | 12.5 | 9.5 | −24 |
| Crebbp | CREB binding protein | 3.9 | 2.9 | −27 |
| Myc | Myelocytomatosis oncogene | 4.4 | 2.2 | −50 |
| Vascular smooth muscle contraction (KEGG pathway: 04270) (C = 123; O = 3; E = 0.13; R = 22.40; rawP = 0.0003; adjP = 0.0007) | ||||
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | 189 | 245 | 30 |
| Rock1 | Rho-associated coiled-coil containing protein kinase 1 | 12.5 | 9.5 | −24 |
| Ppp1r12a | Protein phosphatase 1, regulatory (inhibitor) subunit 12A | 5.7 | 4.1 | −27 |
| Arachidonic acid metabolism (KEGG pathway: 00590) (C = 90; O = 4; E = 0.10; R = 40.82; rawP = 3.04 × 10−6; adjP = 1.37 × 10−5) | ||||
| Cyp2b9 | Cytochrome P450, family 2, subfamily b, polypeptide 9 | 3.2 | 9.6 | 204 |
| Cyp2c37 | Cytochrome P450, family 2. subfamily c, polypeptide 37 | 66.0 | 93.9 | 42 |
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | 189 | 245 | 30 |
| Cyp2c54 | Cytochrome P450, family 2, subfamily c, polypeptide 54 | 103 | 133 | 29 |
| Retinol metabolism (KEGG pathway: 00830) (C = 77; O = 4; E = 0.08; R = 47.71; rawP = 1.63 × 10−6; adjP = 1.37 × 10−5) | ||||
| Cyp2b9 | Cytochrome P450, family 2, subfamily b, polypeptide 9 | 3.2 | 9.6 | 204 |
| Cyp2c37 | Cytochrome P450, family 2. subfamily c, polypeptide 37 | 66.0 | 93.9 | 42 |
| Cyp4a10 | Cytochrome P450, family 4, subfamily a, polypeptide 10 | 189 | 245 | 30 |
| Cyp2c54 | Cytochrome P450, family 2, subfamily c, polypeptide 54 | 103 | 133 | 29 |
| Biosynthesis of unsaturated FAs (KEGG pathway: 01040) (C = 25; O = 2; E = 0.03; R = 73.47; rawP = 0.0003; adjP = 0.0007) | ||||
| Acot3 | Acyl-CoA thioesterase 3 | 9.0 | 13.2 | 47 |
| Elovl6 | ELOVL family member 6, elongation of long-chain FAs (yeast) | 15.5 | 12.1 | −22 |
| Linoleic acid metabolism (KEGG pathway: 00591) (C = 46; O = 2; E = 0.05; R = 39.93; rawP = 0.0012; adjP = 0.0022) | ||||
| Cyp2c37 | Cytochrome P450, family 2. subfamily c, polypeptide 37 | 66.0 | 93.9 | 42 |
| Cyp2c54 | Cytochrome P450, family 2, subfamily c, polypeptide 54 | 103 | 133 | 29 |
Pathways are derived from liver transcriptomics data. Parameters—organism: mus musculus; ID type: gene_symbol; reference set: mmusculus_genome; statistic: hypergeometric; significance level: Top10; multiple test correction: Benjamini-Hochberg; minimum number of genes for a category: 2. Genes included have an adjusted P value ≤ 0.05. adjP, P value adjusted by the multiple test adjustment; FPKM, fragment per kilobase of transcript per million; HAMRS2, high-amylose-maize resistant starch type 2; KEGG, Kyoto Encyclopedia of Genes and Genomes; rawP, P value from hypergeometric test.
The C, O, E, and R in parentheses indicate the number of reference genes in the category, number of genes in the gene set and also in the category, the expected number in the category, and the ratio of enrichment, respectively.
Percentage difference = [(HAMRS2 − control)/control] × 100.
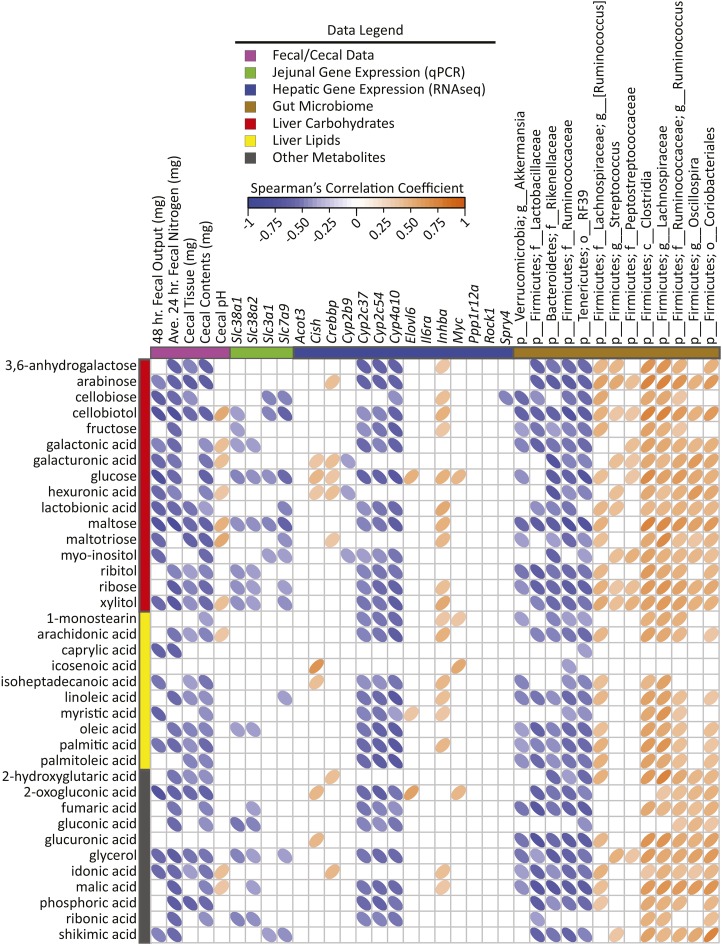
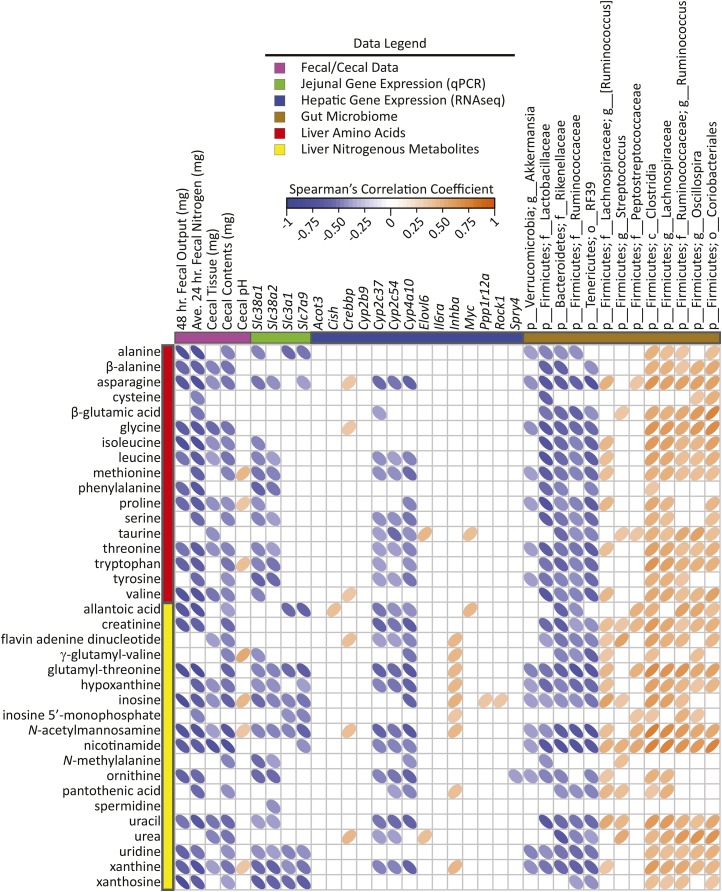
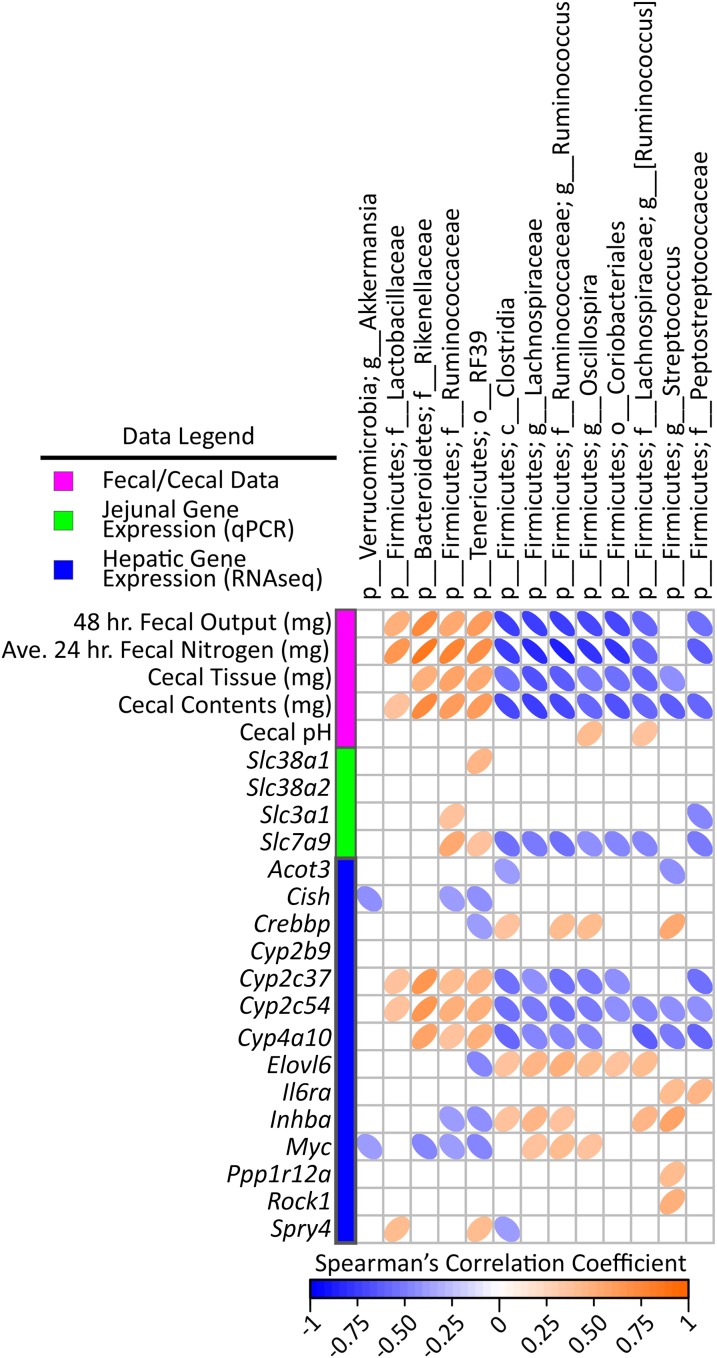
To identify new potential connections between liver metabolism, gene regulation, and gut microbes, cross-correlation plots of hepatic gene transcripts compared with PLS-DA–selected liver metabolites and differentially abundant gut microbes are shown in Figures 4 and 5. Distinct correlation patterns were present between gut microbes, liver metabolites, and gene expression data (Figures 4 and 5). Fecal output, fecal nitrogen, cecal tissue, and cecal content weight showed negative correlations with liver metabolites from all classes (i.e., carbohydrates, lipids, and nitrogenous metabolites). Negative correlations also existed between hepatic cytochrome P450 enzyme expression and liver metabolites from all classes. Jejunal expression of the amino acid transporters Slc38a1 and Slc38a2 showed negative correlations with many nitrogenous liver metabolites as well as several carbohydrates, including glucose.
FIGURE 4.
Spearman’s correlation matrix of fecal/cecal data, hepatic gene expression, and cecal bacteria abundances compared with liver carbohydrates, lipids, and miscellaneous metabolites selected in PLS-DA models of male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). Metabolites were selected on the basis of having a mean bootstrapped VIP ≥1. The direction of ellipses represents positive or negative correlations and the width of the ellipse represents the strength of correlation (narrow ellipse = stronger correlation). Acot3, acyl-CoA thioesterase 3; Ave., average; c_, class; Cish, cytokine inducible SH2-containing protein; Crebbp, CREB binding protein; Cyp, cytochrome P450; Elovl6, ELOVL family member 6, elongation of long-chain FAs (yeast); f_, family; g_, genus; HAMRS2, high-amylose-maize resistant starch type 2; Il6ra, IL-6 receptor α Inhba, inhibin β-A; Myc, myelocytomatosis oncogene; o_, order; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; Ppp1r12a, protein phosphatase 1, regulatory (inhibitor) subunit 12A; Rock1, rho-associated coiled-coil containing protein kinase 1; Slc, solute carrier; Spry4, sprouty homolog 4 (Drosophila); VIP, variable importance in projection.
FIGURE 5.
Spearman’s correlation matrix of cecal/fecal data, hepatic gene expression, and gut bacterial abundances compared with liver nitrogenous metabolites selected in PLS-DA models of male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). Metabolites were selected on the basis of having a mean bootstrapped VIP ≥1. The direction of ellipses represents positive or negative correlations and the width of the ellipse represents the strength of correlation (narrow ellipse = stronger correlation). Acot3, acyl-CoA thioesterase 3; Ave., average; c_, class; Cish, cytokine inducible SH2-containing protein; Crebbp, CREB binding protein; Cyp, cytochrome P450; Elovl6, ELOVL family member 6, elongation of long-chain FAs (yeast); f_, family; g_, genus; HAMRS2, high-amylose-maize resistant starch type 2; Il6ra, IL-6 receptor α Inhba, inhibin β-A; Myc, myelocytomatosis oncogene; o_, order; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; Ppp1r12a, protein phosphatase 1, regulatory (inhibitor) subunit 12A; Rock1, rho-associated coiled-coil containing protein kinase 1; Slc, solute carrier; Spry4, sprouty homolog 4 (Drosophila); VIP, variable importance in projection.
Potential connections between specific cecal bacteria, hepatic gene expression, and plasma and liver metabolites
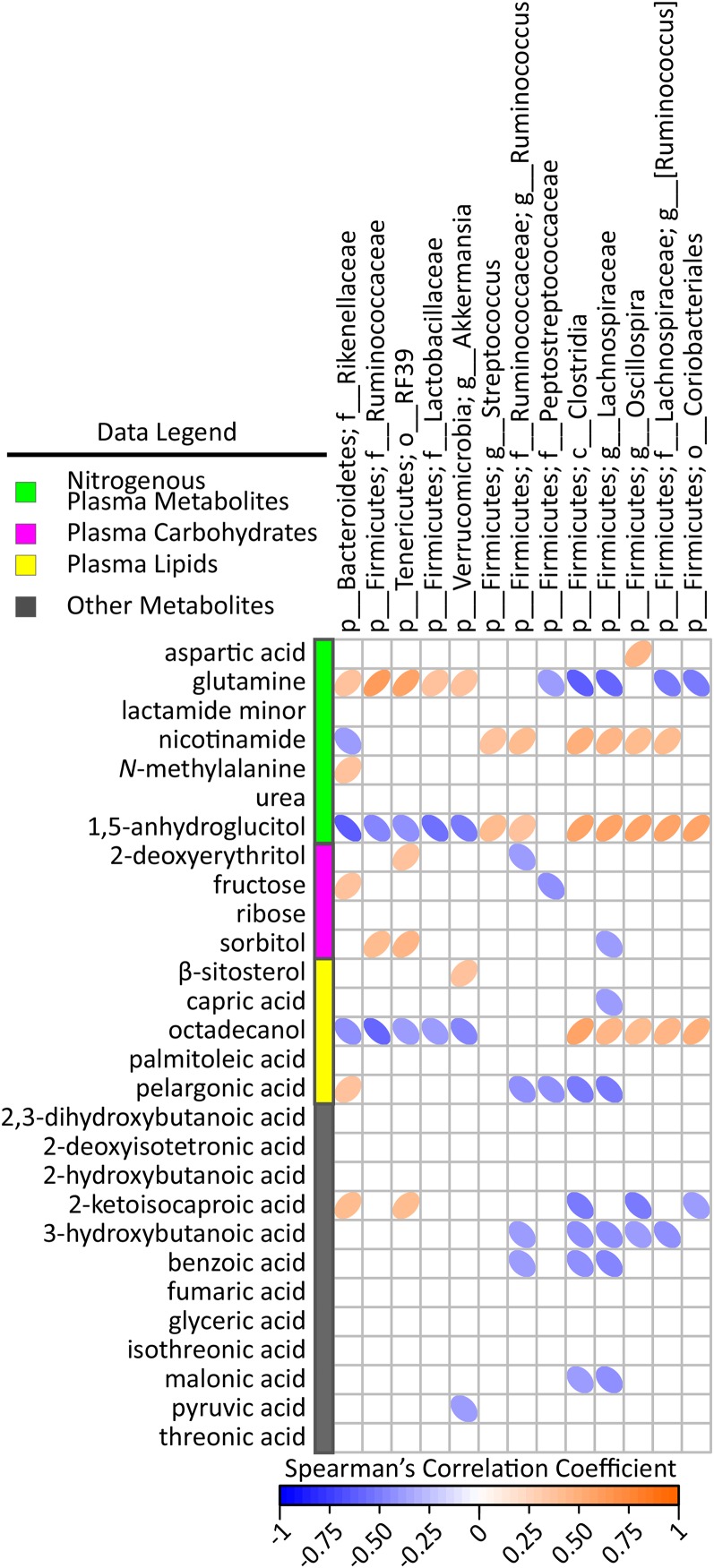
To further identify candidate microbes and metabolite messengers that associate with hepatic molecular physiology, we examined cross-correlation plots among specific cecal bacteria, gut-relevant meta-data, and hepatic variables. With respect to gene transcripts (from the pathways described above) (Figure 6), several correlations existed among fecal output, fecal nitrogen, cecal tissue and cecal content weight and hepatic expression of cytochrome P450 enzymes compared with cecal bacteria abundances. There were a few, selective correlations among plasma metabolites and cecal bacteria abundances (i.e., glutamine, 1,5-anhydroglucitol, and octadecanol) (Figure 7).
FIGURE 6.
Spearman’s correlation matrix of cecal bacteria compared with fecal/cecal data and hepatic gene expression in male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk. Bacteria included had a minimum of 0.05% mean abundance in each group and an adjusted Mann-Whitney U P value ≤ 0.05. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). The direction of ellipses represents positive or negative correlations and the width of the ellipse represents the strength of correlation (narrow ellipse = stronger correlation). Acot3, acyl-CoA thioesterase 3; Ave., average; c_, class; Cish, cytokine inducible SH2-containing protein; Crebbp, CREB binding protein; Cyp, cytochrome P450; Elovl6, ELOVL family member 6, elongation of long-chain FAs (yeast); f_, family; g_, genus; HAMRS2, high-amylose-maize resistant starch type 2; Il6ra, IL-6 receptor α Inhba, inhibin β-A; Myc, myelocytomatosis oncogene; o_, order; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; Ppp1r12a, protein phosphatase 1, regulatory (inhibitor) subunit 12A; Rock1, rho-associated coiled-coil containing protein kinase 1; Slc, solute carrier; Spry4, sprouty homolog 4 (Drosophila).
FIGURE 7.
Spearman’s correlation matrix of cecal bacteria compared with plasma carbohydrates, lipids, nitrogenous, and miscellaneous metabolites selected in PLS-DA models of male mice fed a 45%-fat diet with or without HAMRS2 supplementation for 10 wk. Bacteria included had a minimum of 0.05% mean abundance in each group and an adjusted Mann-Whitney U P value ≤ 0.05. Bacteria are listed to the lowest level of classification (i.e., if the last taxon assignment is f_, “family” is the lowest level of classification). Metabolites were selected on the basis of having a mean bootstrapped VIP ≥1. The direction of ellipses represents positive or negative correlations and the width of the ellipse represents the strength of correlation (narrow ellipse = stronger correlation). c_, class; f_, family; g_, genus; HAMRS2, high-amylose-maize resistant starch type 2; o_, order; p_, phylum; PLS-DA, partial least-squares-discriminant analysis; VIP, variable importance in projection.
Discussion
To our knowledge, this study provides the first comprehensive assessment of HAMRS2-induced differences in hepatic metabolism and relates these to shifts in the cecal microbiota. HAMRS2-induced differences in hepatic metabolism are of interest because the liver is the first organ to be bathed in gut-derived endogenous metabolites and xenometabolites via the portal vein. Because of this intimate connection, the liver is likely to be affected by interventions that alter the gut milieu, such as resistant starch or fiber supplementation. Several studies have shown HAMRS2 supplementation alters the gut microbiota in humans (7, 21) and in animal models (22, 23) and can lead to differences in gut gene expression profiles in rodent (24) and porcine (9, 25) models. There is a paucity of studies that determine how these differences in the gut milieu affect liver metabolism (10, 11). To address this knowledge gap, we used a multi-omics systems approach to characterize the blood and liver metabolome, liver transcriptome, and cecal microbiota after HAMRS2 feeding. We observed several significant associations between hepatic metabolite profiles, gene expression patterns, and specific cecal bacterial populations, providing candidate pathways and microbes that may be involved in effects of resistant starch on host physiology.
A particular strength of the experiment is that HAMRS2 elicited differences in hepatic metabolism without a difference in body weight, adiposity, oral glucose tolerance, or liver TG accumulation—thereby eliminating all of these as potential confounding factors for the observed differences in liver metabolism and cecal bacteria populations. HAMRS2-induced changes in body weight and composition have been inconsistent, with some studies observing a decrease in body weight and/or adiposity (26–28) whereas other studies reported no difference (29, 30). These disparate outcomes in response to HAMRS2 may be due to differences in animal models, dose of HAMRS2, and/or duration of the intervention. One potential explanation for no difference in body weight observed in the present study may be impaired fermentation due to the high amount of fat in the diet. High-fat diets have been shown to impair carbohydrate fermentation (31). Although we did not observe a difference in total or individual SCFAs between the HAMRS2-supplemented mice and controls, there was evidence for very active microbiome fermentation (see below). Altogether, we conclude that the differences in liver metabolism observed herein in response to HAMRS2 were due to SCFA- and body weight–independent events.
Despite no difference in cecal SCFAs, there was a small but significant decrease in the cecal pH of HAMRS2-fed mice, which may be due to other fermentation products such as lactate or succinate (32). In addition to decreased cecal pH, HAMRS2-fed mice also showed increased cecal tissue and cecal content weights. Together, these differences indicate increased microbial fermentation (31). Differences in the cecal microbiota profile were observed at the phylum and lower taxonomic levels. The HAMRS2-fed mice showed significantly greater proportions of Bacteroidetes and reduced levels of Firmicutes compared with controls. Studies in humans and animal models have found increases in the Bacteroidetes-to-Firmicutes ratio with high-fiber feeding (7, 33, 34). This phylum-level shift has been associated with positive health outcomes (35, 36); however, some studies have found the opposite ratio to be associated with improved health outcomes (37, 38). The greatest shift in the cecal microbiota was due to an increase in the Bacteroidetes family Rikenellaceae in the HAMRS2-fed mice. This family was found to be significantly reduced in mice fed a fiber-deficient diet compared with mice fed a standard diet (39). The majority of Firmicutes were decreased in the HAMRS2 mice, with the exception of a greater abundance in 2 families, Ruminococcaceae and Lactobacillaceae. Although the specific bacteria that drove the increase in Ruminococcaceae are unknown, there is 1 species within this family shown to consistently increase with resistant starch feeding, Ruminococcus bromii (7, 40). R. bromii is considered a keystone species, meaning that it is responsible for initial carbohydrate degradation, freeing smaller degradation products for other bacteria to utilize, thereby promoting cross-feeding (41, 42). This particular species possesses several amylose-degrading enzymes arranged in what has been termed an “amylosome,” allowing it flourish on substrates such as HAMRS2 (43). Increases in Lactobacillaceae abundance have been observed with high-fiber feeding (44), and members from this family are regarded as beneficial bacteria (45). The main contributor to the reduced overall abundance of Firmicutes in the HAMRS2 group was Lachnospiraceae. This drastic reduction has also been observed in the cecal contents and feces of rats fed HAMRS2 (46). Several of these cecal bacteria shifts showed significant correlations with hepatic gene expression, blood and liver metabolites, as well as cecal and fecal characteristics, suggesting that factors affected by their activities regulate host physiology. These potential associations are described in more detail below.
The most striking difference in metabolites was the almost universal decrease in liver amino acids and nitrogenous metabolites in the HAMRS-fed mice. This difference was not reflected in the plasma, indicating that this was not due to lower concentrations of circulating amino acids in peripheral blood; this may reflect liver-specific effects rather than systemic effects. It is possible that the liver amino acid phenotype was due to alterations in hepatic and/or gut epithelium amino acid transport and/or metabolism. However, gene expression for a variety of representative amino acid transporters remained unchanged or even higher in response to HAMRS2 in the ileum and jejunum. Furthermore, there were no changes in the gene expression of liver amino acid transporters detected by RNAseq or validation qPCR (Supplemental Figure 3). HAMRS2-fed mice did show increased fecal output with greater amounts of nitrogen in the feces. Alterations in nitrogen pools, such as increased fecal nitrogen and decreased blood concentrations of nitrogenous metabolites (i.e., urea, ammonia, indoles), have been observed in humans (47, 48) and animal models (46, 49, 50) with fiber supplementation. Dietary fibers have been used as a treatment for chronic kidney disease and reduced hepatic amino acids may play a role in reducing the nitrogen burden on the kidneys (51).
Other studies have shown increases in factors related to protein synthesis in the gut of HAMRS2-fed animals (24); HAMRS2-fed mice from this study did show an increase in cecal tissue weight. Fiber-induced increases in microbial and host gut tissue growth may serve to sequester nitrogen in the gut, leading to increased nitrogen excretion in the feces and reduced blood concentrations of metabolites such as urea (52). We observed a modest reduction in plasma urea of HAMRS2-fed mice, and PLS-DA modeling identified plasma urea as an important discriminator between the HAMRS2 and control groups. Urea was significantly reduced in the liver of HAMRS2-fed mice by both univariate and multivariate analysis, and notably, this was accompanied by no significant differences in liver urea cycle enzyme gene expression. It is therefore most likely that urea flux and nitrogen balance—through processes associated with microbial ecology—affected net hepatic amino acid pool size in response to HAMRS2. Indeed, bacteria that increased in the HAMRS2 group (Rikenellaceae, Lactobacillaceae, and the Firmicutes family Ruminococcaceae) showed negative correlations with liver nitrogenous metabolites, and HAMRS2 promotes the growth of bacteria that require ammonia as their primary nitrogen source; this ammonia is largely supplied by endogenous urea (46). Whole-genome sequencing of human fecal samples revealed that fiber supplementation enriched bacterial amino acid metabolism pathways (34), and this was concurrent with a decrease in several amino acid degradation products (i.e., ammonia, indole, and phenol-containing compounds) (53). Several negative correlations also existed among jejunal expression of the ubiquitously expressed sodium-coupled neutral amino acid transporters Scl38a1 and Slc38a2 (54) and nitrogenous liver metabolites. In the liver, Slc38a2 has been shown to play an important role in ammonia metabolism and urea synthesis (55), but the physiologic significance of our observations remains to be evaluated.
Another apparently novel observation was that cytochrome P450 enzymes, related to drug/xenobiotic metabolism and lipid metabolism, were among the hepatic genes most affected by HAMRS2 feeding. Cytochrome P450 (Cyp) family 2, subfamily b, polypeptide 9 (Cyp2b9) showed the greatest difference in hepatic gene expression, with a >200% increase in gene expression in the HAMRS2-fed mice compared with controls. Cyp2b enzymes metabolize a variety of endogenous and exogenous compounds and their expression is influenced by several factors, including strain, sex, age, chemical exposure, and nutrient status (56, 57). In addition to Cyp2b9, 3 lipid-metabolizing cytochrome P450 enzymes were also increased by HAMRS2 feeding: Cyp2c37 [metabolizes arachidonic acid to 12-hydroxyeicosatetraenoic acid (58)], Cyp2c54 [metabolizes arachidonic acid to epoxyeicosatrienoic acids and linoleic acid to epoxyoctadecenoic acids (59)], and Cyp4a10 [metabolizes arachidonic acid to 20-hydroxyeicosatetranoic acid (60)]. Many of these arachidonic acid–derived lipids act as signaling molecules and possess anti-inflammatory activity (61, 62). Studies in murine models have shown hepatic expression of Cyp2b9 (63) and Cyp4a10 (64) to play a role in intestinal inflammation. Notably, all 3 arachidonic acid–metabolizing genes showed negative correlations in the liver with hepatic abundance of arachidonic acid. Similar differences in Cyp enzyme expression were also found when comparing control mice from this study with mice fed enzyme-treated wheat bran (14). To our knowledge, the connection between dietary fiber and Cyp2 or Cyp4 hepatic enzyme expression has not been previously reported. These genes also displayed several significant correlations with specific cecal bacteria (i.e., a positive correlation with Lactobacillaceae and a negative correlation with Clostridia), indicating that hepatic cytochrome gene expression may be influenced by these microbes. The bacteria-derived molecular regulators of Cyp enzyme gene expression in the liver (and perhaps other tissues including gut) and the subsequent impact of these events on immune function remain to be elucidated.
With regard to limitations of the current study, translating results from this and similar rodent studies to the human condition is potentially limited because rodents engage in coprophagy. It is not clear how coprophagy affects the microbiota and metabolite profile, especially in relation to nitrogen cycling. Another limitation particular to this study was the inability to assess amino acid transport and flux of other metabolites across gut epithelia ex vivo via Ussing chambers. Ussing chambers may have provided more insight into potential HAMRS2-associated amino acid, ammonia, and urea transport as a functional readout to complement intestinal gene expression results. The study was not designed a priori to test specific metabolite intestinal fluxes but instead to gain an understanding of potential pathways influenced by resistant starch feeding. Overall, the results herein are hypothesis-generating and allow for more focused experiments to understand the mechanisms underlying the novel amino acid, P450, and other liver phenotypes that were observed in response to resistant starch feeding in mice.
In summary, HAMRS2 fostered the growth of a select number of cecal bacterial taxa, and these microbiota shifts were concurrent with alterations in liver metabolite profiles and hepatic gene expression without affecting SCFA composition. A notable finding was that HAMRS2-supplemented mice showed increased concentrations of fecal nitrogen and decreased abundances of several nitrogenous liver metabolites, including amino acids. HAMRS2 and other fibers have been shown to alter nitrogen cycling by decreasing circulating concentrations of nitrogen and increasing fecal nitrogen. Fiber supplementation has proven useful in ameliorating symptoms of chronic kidney disease (51, 65), a disease characterized by an altered gut microbiota (66) and elevated concentrations of circulating nitrogenous products (67). In addition to shifting nitrogen pools, HAMRS2 also upregulated a number of cytochrome P450 enzymes involved in arachidonic metabolism and downregulated genes involved in regulating cell growth, differentiation, and apoptosis. The unbiased “omics” approach herein highlights the potential role of xenometabolites or other microbiome-derived signals in driving the effects of dietary resistant starch on the metabolism and function of the liver.
Acknowledgments
We thank Michael Blackburn and Kikumi Ono-Moore (Arkansas Children's Nutrition Center) for assistance with plasma TG and nonesterified FA measurements as well as Pieter Oort (Western Human Nutrition Research Center) for technical assistance. DAK, RJM, and SHA designed the research and had primary responsibility for the final content; DAK, EBK, MLG, TND, RJM, and SHA conducted the research; MLM, MJK, and KEBK provided the essential materials; DAK, BDP, RJM, and SHA analyzed the data; DAK wrote the manuscript; and DAK, BDP, MLM, KEBK, RJM, and SHA edited the manuscript. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: Cyp, cytochrome P450; FDR, false discovery rate; HAMRS2, high-amylose-maize resistant starch type 2; PLS-DA, partial least-squares-discriminant analysis; VIP, variable importance in projection.
References
- 1.Cani PD, Delzenne NM. The role of the gut microbiota in energy metabolism and metabolic disease. Curr Pharm Des 2009;15:1546–58. [DOI] [PubMed] [Google Scholar]
- 2.Cani PD, Lecourt E, Dewulf EM, Sohet FM, Pachikian BD, Naslain D, De Backer F, Neyrinck AM, Delzenne NM. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. Am J Clin Nutr 2009;90:1236–43. [DOI] [PubMed] [Google Scholar]
- 3.Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN. Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr 2005;82:559–67. [DOI] [PubMed] [Google Scholar]
- 4.Natarajan N, Pluznick JL. From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. Am J Physiol Cell Physiol 2014;307:C979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang LS, Davies SS. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome Med 2016;8:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annison G, Topping DL. Nutritional role of resistant starch: chemical structure vs physiological function. Annu Rev Nutr 1994;14:297–320. [DOI] [PubMed] [Google Scholar]
- 7.Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 2010;5:e15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Martin RJ, Tulley RT, Raggio AM, McCutcheon KL, Shen L, Danna SC, Tripathy S, Hegsted M, Keenan MJ. Dietary resistant starch upregulates total GLP-1 and PYY in a sustained day-long manner through fermentation in rodents. Am J Physiol Endocrinol Metab 2008;295:E1160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haenen D, Zhang J, da Silva CS, Bosch G, van der Meer IM, van Arkel J, van den Borne JJ, Gutiérrez OP, Smidt H, Kemp B. A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine. J Nutr 2013;143:274–83. [DOI] [PubMed] [Google Scholar]
- 10.Polakof S, Díaz-Rubio ME, Dardevet D, Martin J-F, Pujos-Guillot E, Scalbert A, Sebedio J-L, Mazur A, Comte B. Resistant starch intake partly restores metabolic and inflammatory alterations in the liver of high-fat-diet-fed rats. J Nutr Biochem 2013;24:1920–30. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Zhang Y, Shi R, Zhou Z, Wang F, Strappe P. A novel gene network analysis in liver tissues of diabetic rats in response to resistant starch treatment. Springerplus 2015;4:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas AP, Dunn TN, Drayton JB, Oort PJ, Adams SH. A high calcium diet containing nonfat dry milk reduces weight gain and associated adipose tissue inflammation in diet-induced obese mice when compared to high calcium alone. Nutr Metab 2012;9:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tulley RT, Appel MJ, Enos TG, Hegsted M, McCutcheon KL, Zhou J, Raggio AM, Jeffcoat R, Birkett A, Martin RJ, et al. . Comparative methodologies for measuring metabolizable energy of various types of resistant high amylose corn starch. J Agric Food Chem 2009;57:8474–9. [DOI] [PubMed] [Google Scholar]
- 14.Kieffer DA, Piccolo BD, Marco ML, Kim EB, Goodson ML, Keenan MJ, Dunn TN, Knudsen KEB, Adams SH, Martin RJ. Obese mice fed a diet supplemented with enzyme-treated wheat bran display marked shifts in the liver metabolome concurrent with altered gut bacteria. J Nutr 2016;146:2445–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2014. [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B 1995;57:289–300. [Google Scholar]
- 17.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics 2001;17:520–5. [DOI] [PubMed] [Google Scholar]
- 18.Mehmood T, Liland KH, Snipen L, Sæbø S. A review of variable selection methods in partial least squares regression. Chemom Intell Lab Syst 2012;118:62–9. [Google Scholar]
- 19.Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 2001;58:109–30. [Google Scholar]
- 20. Canty A, Ripley B. Boot: Bootstrap R (S-Plus) functions. R package version 2.0.1 [Internet]. 2015 [cited 2015 Sep 15]. Available from: https://cran.r-project.org/package=boot.
- 21.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, et al. . Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 2011;5(2):220–30. [cited 2015 Jul 25]. Available from: http://www.nature.com/ismej/journal/v5/n2/suppinfo/ismej2010118s1.html. [DOI] [PMC free article] [PubMed]
- 22.Wang X, Brown I, Khaled D, Mahoney M, Evans A, Conway P. Manipulation of colonic bacteria and volatile fatty acid production by dietary high amylose maize (amylomaize) starch granules. J Appl Microbiol 2002;93:390–7. [DOI] [PubMed] [Google Scholar]
- 23.Tachon S, Zhou J, Keenan M, Martin R, Marco M. The intestinal microbiota in aged mice is modulated by dietary resistant starch and correlated with improvements in host responses. FEMS Microbiol Ecol 2013;83:299–309. [DOI] [PubMed] [Google Scholar]
- 24.Keenan MJ, Martin RJ, Raggio AM, McCutcheon KL, Brown IL, Birkett A, Newman SS, Skaf J, Hegsted M, Tulley RT. High-amylose resistant starch increases hormones and improves structure and function of the gastrointestinal tract: a microarray study. J Nutrigenet Nutrigenomics 2012;5:26–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haenen D, Souza da Silva C, Zhang J, Koopmans SJ, Bosch G, Vervoort J, Gerrits WJJ, Kemp B, Smidt H, Müller M, et al. . Resistant starch induces catabolic but suppresses immune and cell division pathways and changes the microbiome in the proximal colon of male pigs. J Nutr 2013;143:1889–98. [DOI] [PubMed] [Google Scholar]
- 26.Keenan MJ, Janes M, Robert J, Martin RJ, Raggio AM, McCutcheon KL, Pelkman C, Tulley R, Goita MF, Durham HA. Resistant starch from high amylose maize (HAM‐RS2) reduces body fat and increases gut bacteria in ovariectomized (OVX) rats. Obesity (Silver Spring) 2013;21:981–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keenan MJ, Zhou J, McCutcheon KL, Raggio AM, Bateman HG, Todd E, Jones CK, Tulley RT, Melton S, Martin RJ, et al. . Effects of resistant starch, a non-digestible fermentable fiber, on reducing body fat. Obesity (Silver Spring) 2006;14:1523–34. [DOI] [PubMed] [Google Scholar]
- 28.Toden S, Bird AR, Topping DL, Conlon MA. High red meat diets induce greater numbers of colonic DNA double-strand breaks than white meat in rats: attenuation by high-amylose maize starch. Carcinogenesis 2007;28:2355–62. [DOI] [PubMed] [Google Scholar]
- 29.Topping DL, Gooden JM, Brown IL, Biebrick DA, McGrath L, Trimble RP, Choct M, Illman RJ. A high amylose (amylomaize) starch raises proximal large bowel starch and increases colon length in pigs. J Nutr 1997;127:615–22. [DOI] [PubMed] [Google Scholar]
- 30.Kleessen B, Stoof G, Proll J, Schmiedl D, Noack J, Blaut M. Feeding resistant starch affects fecal and cecal microflora and short-chain fatty acids in rats. J Anim Sci 1997;75:2453–62. [DOI] [PubMed] [Google Scholar]
- 31.Charrier JA, Martin RJ, McCutcheon KL, Raggio AM, Goldsmith F, Goita MF, Senevirathne RN, Brown IL, Pelkman C, Zhou J. High fat diet partially attenuates fermentation responses in rats fed resistant starch from high‐amylose maize. Obesity (Silver Spring) 2013;21:2350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc 2003;62:67–72. [DOI] [PubMed] [Google Scholar]
- 33.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. . Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014;20:159–66. [DOI] [PubMed] [Google Scholar]
- 34.Holscher HD, Caporaso JG, Hooda S, Brulc JM, Fahey GC, Swanson KS. Fiber supplementation influences phylogenetic structure and functional capacity of the human intestinal microbiome: follow-up of a randomized controlled trial. Am J Clin Nutr 2015;101:55–64. [DOI] [PubMed] [Google Scholar]
- 35.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–31. [DOI] [PubMed] [Google Scholar]
- 36.Jeffery IB, O’Toole PW, Ohman L, Claesson MJ, Deane J, Quigley EM, Simren M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012;61:997–1006. [DOI] [PubMed] [Google Scholar]
- 37.Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sorensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 2010;5:e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, Queipo-Ortuño MI. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med 2013;11:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, et al. . Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 2013;144:967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Birt DF, Boylston T, Hendrich S, Jane J-L, Hollis J, Li L, McClelland J, Moore S, Phillips GJ, Rowling M, et al. . Resistant starch: promise for improving human health. Adv Nutr 2013;4:587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012;3:289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 2012;6:1535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, Louis P, Henrissat B, Juge N, Koropatkin NM, et al. . Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. MBio 2015;6:e01058–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maathuis A, Hoffman A, Evans A, Sanders L, Venema K. The effect of the undigested fraction of maize products on the activity and composition of the microbiota determined in a dynamic in vitro model of the human proximal large intestine. J Am Coll Nutr 2009;28:657–66. [DOI] [PubMed] [Google Scholar]
- 45.Gill H, Prasad J. Probiotics, immunomodulation, and health benefits. In: Bioactive components of milk. New York: Springer; 2008. p. 423–54. [DOI] [PubMed] [Google Scholar]
- 46.Kalmokoff M, Zwicker B, O’Hara M, Matias F, Green J, Shastri P, Green-Johnson J, Brooks SPJ. Temporal change in the gut community of rats fed high amylose cornstarch is driven by endogenous urea rather than strictly on carbohydrate availability. J Appl Microbiol 2013;114:1516–28. [DOI] [PubMed] [Google Scholar]
- 47.Birkett A, Muir J, Phillips J, Jones G, O’Dea K. Resistant starch lowers fecal concentrations of ammonia and phenols in humans. Am J Clin Nutr 1996;63:766–72. [DOI] [PubMed] [Google Scholar]
- 48.Bliss DZ, Stein TP, Schleifer CR, Settle RG. Supplementation with gum arabic fiber increases fecal nitrogen excretion and lowers serum urea nitrogen concentration in chronic renal failure patients consuming a low-protein diet. Am J Clin Nutr 1996;63:392–8. [DOI] [PubMed] [Google Scholar]
- 49.Younes H, Demigné C, Behr S, Rémésy C. Resistant starch exerts a lowering effect on plasma urea by enhancing urea N transfer into the large intestine. Nutr Res 1995;15:1199–210. [Google Scholar]
- 50.Kieffer DA, Piccolo BD, Vaziri ND, Liu S, Lau WL, Khazaeli M, Nazertehrani S, Moore ME, Marco ML, Martin RJ, et al. . Resistant starch alters gut microbiome and metabolomic profiles concurrent with amelioration of chronic kidney disease in rats. Am J Physiol Renal Physiol 2016;310:F857–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaziri ND, Liu SM, Lau WL, Khazaeli M, Nazertehrani S, Farzaneh SH, Kieffer DA, Adams SH, Martin RJ. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS One 2014;9:e114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Younes H, Alphonse J-C, Behr SR, Demigné C, Rémésy C. Role of fermentable carbohydrate supplements with a low-protein diet in the course of chronic renal failure: experimental bases. Am J Kidney Dis 1999;33:633–46. [DOI] [PubMed] [Google Scholar]
- 53.Boler BM, Rossoni Serao MC, Bauer LL, Staeger MA, Boileau TW, Swanson KS, Fahey GC. Digestive physiological outcomes related to polydextrose and soluble maize fibre consumption by healthy adult men. Br J Nutr 2011;106:1864–71. [DOI] [PubMed] [Google Scholar]
- 54.Mackenzie B, Erickson JD. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflugers Arch 2004;447:784–95. [DOI] [PubMed] [Google Scholar]
- 55.Brosnan ME, Brosnan JT. Hepatic glutamate metabolism: a tale of 2 hepatocytes. Am J Clin Nutr 2009;90(Suppl):857S–61S. [DOI] [PubMed] [Google Scholar]
- 56.Glue P, Clement RP. Cytochrome P450 enzymes and drug metabolism—basic concepts and methods of assessment. Cell Mol Neurobiol 1999;19:309–23. [DOI] [PubMed] [Google Scholar]
- 57.Nemoto N, Sakurai J. Glucocorticoid and sex hormones as activating or modulating factors for expression of Cyp2b-9 and Cyp2b-10 in the mouse liver and hepatocytes. Arch Biochem Biophys 1995;319:286–92. [DOI] [PubMed] [Google Scholar]
- 58.Luo G, Zeldin DC, Blaisdell JA, Hodgson E, Goldstein JA. Cloning and expression of murine CYP2Cs and their ability to metabolize arachidonic acid. Arch Biochem Biophys 1998;357:45–57. [DOI] [PubMed] [Google Scholar]
- 59.Wang H, Zhao Y, Bradbury JA, Graves JP, Foley J, Blaisdell JA, Goldstein JA, Zeldin DC. Cloning, expression, and characterization of three new mouse cytochrome p450 enzymes and partial characterization of their fatty acid oxidation activities. Mol Pharmacol 2004;65:1148–58. [DOI] [PubMed] [Google Scholar]
- 60.Powell PK, Wolf I, Jin R, Lasker JM. Metabolism of arachidonic acid to 20-hydroxy-5,8,11, 14-eicosatetraenoic acid by P450 enzymes in human liver: involvement of CYP4F2 and CYP4A11. J Pharmacol Exp Ther 1998;285:1327–36. [PubMed] [Google Scholar]
- 61.Campbell WB. New role for epoxyeicosatrienoic acids as anti-inflammatory mediators. Trends Pharmacol Sci 2000;21:125–7. [DOI] [PubMed] [Google Scholar]
- 62.Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer 2006;6:947–60. [DOI] [PubMed] [Google Scholar]
- 63.Chaluvadi MR, Nyagode BA, Kinloch RD, Morgan ET. TLR4-dependent and -independent regulation of hepatic cytochrome P450 in mice with chemically induced inflammatory bowel disease. Biochem Pharmacol 2009;77:464–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nyagode BA, Williams IR, Morgan ET. Altered inflammatory responses to Citrobacter rodentium infection, but not bacterial lipopolysaccharide, in mice lacking the Cyp4a10 or Cyp4a14 genes. Inflammation 2014;37:893–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sirich TL, Plummer NS, Gardner CD, Hostetter TH, Meyer TW. Effect of increasing dietary fiber on plasma levels of colon-derived solutes in hemodialysis patients. Clin J Am Soc Nephrol 2014;9:1603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, Ni Z, Nguyen T-H, Andersen GL. Chronic kidney disease alters intestinal microbial flora. Kidney Int 2013;83:308–15. [DOI] [PubMed] [Google Scholar]
- 67.Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, Argiles A. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol 2012;23:1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]