Abstract
Odorant-binding proteins (OBPs) and chemosensory proteins (CSPs) are endowed with several different functions besides being carriers for pheromones and odorants. Based on a previous report of a CSP acting as surfactant in the proboscis of the moth Helicoverpa armigera, we revealed the presence of orthologue proteins in two other moths Plutella xylostella and Chilo suppressalis, as well as two butterflies Papilio machaon and Pieris rapae, using immunodetection and proteomic analysis. The unusual conservation of these proteins across large phylogenetic distances indicated a common specific function for these CSPs. This fact prompted us to search for other functions of these proteins and discovered that CSPs are abundantly expressed in the eyes of H. armigera and possibly involved as carriers for carotenoids and visual pigments. This hypothesis is supported by ligand-binding experiments and docking simulations with retinol and β-carotene. This last orange pigment, occurring in many fruits and vegetables, is an antioxidant and the precursor of visual pigments. We propose that structurally related CSPs solubilise nutritionally important carotenoids in the proboscis, while they act as carriers of both β-carotene and its derived products 3-hydroxyretinol and 3-hydroxyretinal in the eye. The use of soluble olfactory proteins, such as CSPs, as carriers for visual pigments in insects, here reported for the first time, parallels the function of retinol-binding protein in vertebrates, a lipocalin structurally related to vertebrate odorant-binding proteins.
Keywords: Odorant-binding protein, Chemosensory protein, Proboscis, Lepidoptera, Proteomics, Vision.
Introduction
Odorant-binding proteins (OBPs) and chemosensory proteins (CSPs) are two families of small soluble proteins with affinity for several different organic compounds 1-2. OBPs, and to some extent also CSPs, have been mostly associated to insect olfaction and are believed to be involved in carrying hydrophobic odorants and pheromones from the external environment to the membrane of chemosensory neurons across the aqueous lymph of chemosensilla 3-5.
However, in recent years' experimental evidence has been accumulating on functions of both OBPs and CSPs unrelated to chemosensing and even to chemical communication. It is not surprising that proteins of both classes, while performing a role of carriers for semiochemicals from the environment to sensory structures, might well be involved in the action of delivering the same chemical compounds from glands to the external environment. Several examples of such double role are reported in the literature in different species of insects 6-12. This situation parallels what had been known for a long time for OBPs of vertebrates, which, besides being active in the nasal mucosa, are also expressed in several glands and excreted in biological fluids, such as urine and saliva, complexed with species specific pheromones 13-15.
More intriguing is the expression of OBPs and CSPs in organs not dealing with chemical communication. In fact, these proteins have been shown to be involved in development 16-17, immune protection 18-19 and dietary function 20. Interestingly, the first member of the CSP family, before such proteins were discovered in insect antennae, was reported as a regenerating factor in the legs of cockroaches 16. Another CSP is required for a correct development of the embryo in the honey bee 1-7, while OBPs with unknown functions have been reported in the egg shell of the mosquito Aedes aegypti 21. Some CSPs might also act in a sort of immune protection against insecticides, as their genes have been reported to be upregulated in the gut of some insect species by such chemicals 18-19. Another role, beneficial to the diet, has been reported for an OBP of the house fly Phormia regina. This protein, abundantly present in the oral disk, might solubilise important fatty acids 20.
All these functions, although unrelated to chemical communication, may still be linked to the binding capacity of both OBPs and CSPs for all sorts of hydrophobic chemicals. This is certainly the case when these proteins act as sequestering agents for noxious compounds or as nutrient solubilisers, but it is also likely that in development they might play the role of carriers for specific hormones.
Different is the situation where OBPs and CSPs are used not for their binding properties but for other characteristics. So far only one of such examples has been reported in the literature, a CSP extremely abundant in the proboscis of two Helicoverpa species, that has been suggested to facilitate sucking through a surfactant action 22.
In this work, we started to investigate the presence and the role of CSPs in the proboscis of other Lepidopteran species and ended up discovering that the same or similar CSPs are expressed in the eyes, where they can reasonably act as carriers for visual pigments. This finding mirrors the use of lipocalins structurally similar to vertebrates OBPs for transport of retinol across the blood stream in vertebrates.
Materials and Methods
Insects
Cotton bollworm Helicoverpa armigera, striped rice stem borer Chilo suppressalis, and diamondback moth, Plutella xylostella, were reared at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences and at the Institute of Zoology, Chinese Academy of Sciences, Beijing, China. Butterflies Papilio machaon and Pieris rapae were collected in the campus of China Agricultural University, Beijing. Proboscises and eyes were collected from 3-day-old moths and from live butterflies, and immediately used or frozen in liquid nitrogen.
Reagents
All enzymes, unless otherwise stated, were from Thermo Scientific. Oligonucleotides were custom synthesised and plasmids were sequenced at Sheng Gong, Beijing, China. Urea was purchased from Euroclone and trypsin from Promega (Sequencing Grade Modified Trypsin). The hand-made desalting/purification STAGE column were prepared using three C18 Empore Extraction Disks (3M). The benzoates used in binding studies were synthesised as previously reported 23. All other chemical reagents and ligands, unless stated otherwise, were purchased from Sigma-Aldrich and of reagent grade.
In gel digestion and protein identification
Protein extracts from proboscises (equivalent of 3 individuals for H. armigera, 2 for P. machaon, 100 for P. xylostella and 100 for C. suppressalis) and from eyes (equivalent of 1 individual for H. armigera) were separated on a 14% SDS-PAGE gel.
Bands from gels were cut as reported in Figures 2 and 4. Slices were transferred to 1.5 mL microcentrifuge tubes and washed three times for 10 min in acetonitrile, then in 0.1 M aqueous ammonium bicarbonate. The slices were then incubated for 30 min in the dark with 55 mM iodoacetamide in 0.1 M ammonium bicarbonate and washed successively in acetonitrile and in 0.1 M ammonium bicarbonate. Digestion was performed overnight at 37°C using 1ng/µL of modified trypsin (Promega, Madison, WI, USA) in 10 mM ammonium bicarbonate. The reaction was blocked by the addition of 10 % TFA and the supernatant was recovered for HPLC-MS analyses.
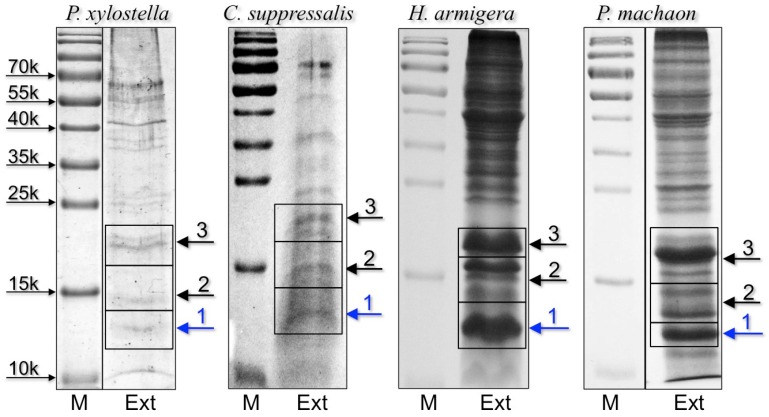
Figure 2.
SDS-PAGE separation of crude extracts from proboscis of the four-species subjected to proteomic analysis. Bands were excised from the gel, proteins were extracted and processed as reported in the Material and Methods section, then analysed by LC/MS-MS. OBPs and CSPs were detected only in band 1 of each gel and the results are summarised in Table 1. Supplementary files S2-S5 report all the proteins identified in the three bands for each species.
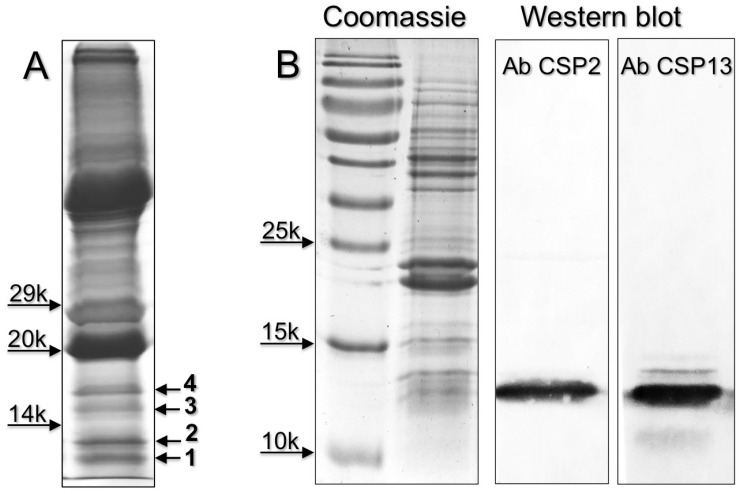
Figure 4.
(A) SDS-PAGE separation of a crude extracts from the eyes of H. armigera (loaded the equivalent of 2 eyes). Four bands, indicated with numbers 1-4, were excised and subjected to proteomic analysis, as reported in the Material and Methods section. OBPs and CSPs were detected only in band 1 and 2 and listed in Table 2. Supplementary file S6 reports all the proteins identified in the four bands. (B) Detection of HarmCSP2 and HarmCSP13 in the eyes of H. armigera (loaded the equivalent of 1 eye per lane). Primary antiserum and HRP-linked secondary antiserum were used at 1:1000 dilution.
Each peptide mixture was submitted to nanoLC-nanoESI-MS/MS analysis on an Ultimate 3000 HPLC (Dionex, San Donato Milanese, Milano, Italy) coupled to a LTQ-Orbitrap mass spectrometer (Thermo Fisher, Bremen, Germany). Samples were injected directly into a homemade nano column packed with Aeris Peptide XB-C18 phase (75 μm i.d. × 15 cm, 3.6 μm, 100Å, Phenomenex, Torrance, CA, USA) and eluted with a flow rate of 300 nL/min. The elution mobile phases compositions were: aqueous 0.1% formic acid/acetonitrile 97/3 (phase A) and 20/80 (phase B). The elution programme was: time 0: 2% B; 40 minutes: 2% B; 68 minutes: 15% B; 168 minutes: 25% B; 228 minutes: 35% B; 273 minutes: 50% B; 274 minutes: 90%B; 288 minutes: 90% B; 289 minutes: 2% B; 309 minutes: 2% B. Mass spectra were acquired in positive ion mode, setting the spray voltage at 1.8 kV, the capillary voltage and temperature respectively at 45 V and 200 °C, and the tube lens at 130 V. Data were acquired in data dependent mode with dynamic exclusion enabled (repeat count 2, repeat duration 15 seconds, exclusion duration 30 seconds). Survey MS scans were recorded in the Orbitrap analyzer in the mass range 300-2000 m/z at a 15,000 nominal resolution at m/z = 400; then up to five most intense ions in each full MS scan were fragmented (isolation width: 3 m/z, normalized collision energy: 30) and analyzed in the IT analyzer. Monocharged ions did not trigger MS/MS experiments.
The acquired data were searched with Mascot 2.4 search engine (Matrix Science Ltd., London, UK) against a non-redundant NCBI protein database, and selecting 'Other Methazoa' (containg all Methazoa except for Chordata, Drosophila melanogaster, Caenorhabditis elegans) as taxonomy parameter.
Searches were performed allowing: (i) up to three missed cleavage sites, (ii) 10 ppm of tolerance for the monoisotopic precursor ion and 0.8 mass unit for monoisotopic fragment ions, (iii) carbamidomethylation of cysteine and oxidation of methionine as variable modification. Peptide significance threshold was set at 0.05 and only peptides with scores higher than identity or extensive homology were considered.
RNA extraction and cDNA synthesis
Total RNA was extracted from antennae and other tissues using Trizol Reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. The first-strand cDNA was synthesized from 2 μg of total RNA using an oligo-dT primer and the Revert Aid First Strand cDNA Synthesis Kit (Fermentas, Glen Burnie, MD) and following the manufacturer's protocol. The product was either used directly for PCR amplification or stored at -70°C.
Bacterial expression and purification of proteins
All four proteins were expressed in E. coli BL21 cells at 37°C. For HarmCSP2 (acc no. HQ874659) and HarmCSP13 (acc no. AF368375), PCR products bearing at their ends restriction enzyme sequences were ligated into pET30 vector, after digestion with the appropriate enzymes. For the expression of HarmCSP4 (acc no. HQ874664) and HarmCSP11 (acc no. JX305306) the two bases CG were added at the 5'-end for blunt ligation into MscI site of pET22 vector. The primers used for amplification of the sequences and the enzymes sites included are listed in Supplementary Table S1. As a result, HarmCSP2 presented a His-tag segment at its N-terminus, while the other three CSPs only contained a single additional residue at their N-terminus with respect to their mature protein sequences.
Cells were harvested by centrifugation (15 min at 3,500 rpm) and suspended in 50 mM Tris buffer, 0.5 M NaCl, 1 mM PMSF. For HarmCSP2 and HarmCSP13, cells were broken by sonication. After centrifugation (1 h at 12,000 rpm), HarmCSP2 was mostly present in the pellet and was recovered by solubilisation in 8 M Urea and 1 mM DTT. The protein was purified by affinity chromatography on Ni Sepharose columns (GE Healthcare Biosciences, Uppsala, Sweden) and renatured by extensive dialysis against Tris buffer. HarmCSP13, instead, was mostly present in the supernatant and was purified by two chromatographic steps on anion exchange resin DE-52 (Whatman).
HarmCSP4 and HarmCSP11, instead, were expressed in the periplasmic space and extracted by osmotic shock. The bacterial pellet from 1 L of culture was suspended in 60 mL of 20% sucrose, 1 mM EDTA in 50 mM Tris, pH 7.4 and stirred for 10 min. After centrifugation at 10,000 rpm for 10 min, the pellet was resuspended in 40 mL of cold 5 mM calcium chloride in 50 mM Tris, pH 7.4 and stirred for 10 min in ice. After a second centrifugation in the same conditions, the recombinant protein was obtained in the supernatant. The proteins were purified by anion-exchange chromatography on DE-52 (Whatman) followed by a second step on Mono-Q (GE-Healthcare), along with standard protocols previously adopted for other CSPs and OBPs 24-25.
Preparation of antisera and immunodetection
Polyclonal antisera were custom made in rabbits at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China, by intracutaneous injection of H. armigera purified recombinant proteins.
For Western blot immunodetection, proteins and extracts were separated on 14% SDS-PAGE gels and transferred to Nitrocellulose membranes using a semi-dry system, at the constant potential of 30 V for 20 minutes. Membranes were treated with polyclonal antisera at the dilution of 1:1000 (or 1:500 for extracts of species different from the H. armigera) for 2 hours at room temperature in 0.02% Tween Tris buffer containing 5% of skimmed dry milk. After 3x5 min washings, membranes were incubated in 1:1000 HRP-linked secondary antiserum in the same buffer for one hour at room temperature. Development was accomplished by treatment with 0.05% 4-chloronaphthol and hydrogen peroxide.
For “drink blot” experiments, moths and butterflies were allowed to drink from Nitrocellulose membranes soaked in sugar solutions. The membrane then was treated as described for Western blot experiments. Alternatively, the HRP-linked second antiserum was replaced with FITC-linked second antiserum and, after washing, membranes were directly observed under a fluorescence microscope. In control experiments, primary antiserum was replaced by pre-immune serum at the same concentration.
Fluorescence measurements
Emission fluorescence spectra were recorded on a Horiba scientific Fluoromax-4 spectrofluorometer at room temperature in a right-angle configuration, with a 1 cm light path quartz cuvette and 5 nm slits for both excitation and emission. Proteins were dissolved in 50 mM Tris-HCl buffer, pH 7.4, while ligands were added as 1 mM methanol solutions.
Ligand-binding experiments
The affinity of the fluorescent reporter N-phenyl-1-naphthylamine (1-NPN) to each protein was measured by titrating a 2 μM solution of the protein with aliquots of 1 mM ligand in methanol to final concentrations of 2-12 μM. The probe was excited at 337 nm and emission spectra were recorded between 380 and 450 nm. The affinities of other ligands were measured in competitive binding assays, where a solution of the protein and 1-NPN, both at the concentration of 2 μM, was titrated with 1 mM methanol solutions of each competitor to final concentrations of 2-16 μM. Due to the poor solubility of β-carotene in methanol, a 0.2 mM solution was used for this ligand. Dissociation constants for 1-NPN and the stoichiometry of binding were obtained processing the data with Prism software.
Molecular modeling and docking
Three-dimensional models of HarmCSP2, HarmCSP4, HarmCSP11 and HarmCSP13 were generated using the online tool SWISS MODEL 26-28 and the structure of Mamestra brassicae A6 complexed with three molecules of 12-Br-dodecanol 29 (PDB id: 1N8V) as template. Docking was performed by the on-line tool SWISS DOCK using default parameters 30. Models were visualised with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311) 31.
Results
Conserved CSPs in the proboscis of Lepidoptera
Following a previous report on a CSP with surfactant activity in the proboscis of Helicoverpa moths 22, we decided to investigate on the presence of similar proteins in the proboscis of two other moths, Plutella xylostella and Chilo suppressalis, both currently reared in the laboratory of Institute of Plant Protection, and two easily available butterflies Papilio machaon, whose genome has been recently sequenced 32, and Pieris rapae. We applied the same technique of “drink-blot”, where a moth is allowed to drink from a nitrocellulose membrane soaked in a sucrose solution. The membrane is then developed as in colorimetric detection of ordinary Western-blot experiments to reveal protein traces left by the insect on touching the surface with its proboscis.
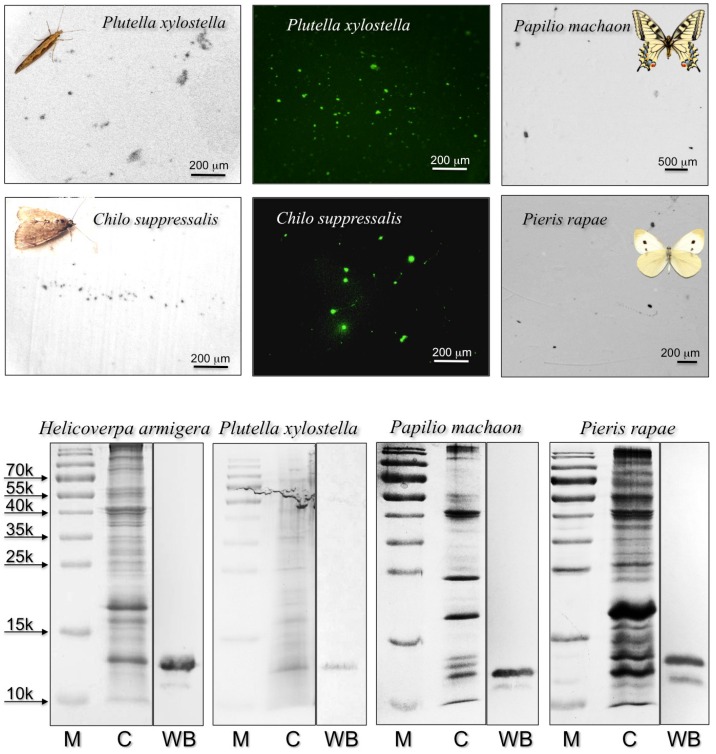
When using a polyclonal antiserum against H. armigera CSP4, we detected a large number of tiny spots, indicating that proteins similar enough to HarmCSP4 to cross-react with the antiserum were abundant in the proboscis of all four species (Figure 1). In parallel control experiments, the primary antiserum was replaced by pre-immune serum (Supplementary Figure S3). In Western blot experiments, the same antiserum stained bands of the expected size in crude proboscis extracts after electrophoretic separation in denaturing conditions (Figure 1). The quality and the specificity of the two newly prepared antisera (against HarmCSP2 and HarmCSP13) were checked in Western blot experiments, using the corresponding recombinant proteins (Supplementary Figure S4).
Figure 1.
Upper: “drink-blot” experiments performed with two species of moths and two species of butterflies. The insects were allowed to drink from a sugar solution absorbed by pieces of nitrocellulose membrane. The membranes were treated, as in Western blot experiments, with the antiserum against HarmCSP4 as the first antibody, then with HRP-linked or FITC-linked second antiserum. Parallel experiments where the first antiserum was replaced with pre-immune serum did not show clear spots when treated with FITC-linked second antiserum (Supplementary Figure S3). Lower: SDS-PAGE and Western blot experiments on crude extracts from the proboscis of the species indicated. The antiserum against HarmCSP4 reacts with a band at the expected molecular mass of around 13 kDa. M: protein ladder; C: crude extract stained with Coomassie; WB: Western blot reaction.
Proteomic analysis of soluble proteins in the proboscis of Lepidoptera
To identify the orthologue CSPs detected in immunostaining experiments and to gather more detailed information on the protein composition of proboscises, we performed a proteomic analysis on extracts from the proboscis of all species, except for P. rapae, for which we could not find in the databases any sequence information regarding olfactory proteins. We selected three bands based on low molecular weigth from electrophoretic gels of crude proboscis extracts to be analysed by LC/MS-MS (Figure 2).
The OBPs and CSPs identified in this experiment (all from band 1 of each species) are listed in Table 1, while the complete proteomic data are reported in Supplementary files S2-S5. The raw data obtained by the MS and MS/MS analysis were searched against a NCBI non-redundant protein database containing all Methazoa except for Chordata, Drosophila melanogaster and Caenorhabditis elegans.
Table 1.
Chemosensory and Odorant-binding proteins identified in slices 1 of SDS-PAGE gels (Figure 2) from proboscises extracts. Together with the description reported in the database, the sequence coverage and the number of peptides found in our analysis are also indicated.
| Species | NCBI acc. | Description | Seq. cov. % | MW | Pept |
|---|---|---|---|---|---|
| Papilio machaon | XP_014365771 gi|943967648 | ejaculatory bulb-specific protein 3-like [P. machaon] | 34.7 | 14167 | 6 |
| Papilio machaon | XP_013137230 gi|909563971 | ejaculatory bulb-specific protein 3-like [P. polytes] | 8.6 | 14805 | 1 |
| Chilo suppressalis | AHC05675 gi|564969555 | chemosensory protein [C.suppressalis] | 24.6 | 14540 | 6 |
| Chilo suppressalis | AHC05678 gi|564969624 | chemosensory protein [C. suppressalis] | 10.2 | 12630 | 1 |
| Chilo suppressalis | AHC05672 gi|564969460 | chemosensory protein [C. suppressalis] | 9.8 | 13332 | 1 |
| Helicoverpa armigera | AEX07269 gi|365919044 | CSP4 [H. armigera] | 25.8 | 14657 | 9 |
| Helicoverpa armigera | AFR92095 gi|405117278 | chemosensory protein 11 [H. armigera] | 32 | 14529 | 5 |
| Helicoverpa armigera | AEB54587 gi|328879860 | OBP6 [H. armigera] | 17.7 | 15897 | 2 |
| Helicoverpa armigera | AEX07265 gi|365919036 | CSP2 [H. armigera] | 18.3 | 13662 | 3 |
| Helicoverpa armigera | AEB54580 gi|328879846 | OBP1 [H. armigera] | 8.8 | 15960 | 1 |
| Plutella xylostella | XP_011568454 gi|768412729 | ejaculatory bulb-specific protein 3-like [P. xylostella] | 15.6 | 14768 | 1 |
| Plutella xylostella | NP_001292453 gi|770075633 | general odorant-binding protein 56d-like precursor [P. xylostella] | 18.8 | 15186 | 2 |
We detected the presence of two OBPs in H. armigera (HarmOBP1, acc. no. AEB54580, and HarmOBP6, acc. no. AEB54587) and one in P. xylostella (PxylOBP, acc. no. XP_011568454), besides one or more CSPs in each of the four species. HarmOBP1 and HarmOBP6 are about 60% identical to each other, but very different from PxylOBP. Best similarity for these two proteins is found in the database with an OBP expressed in the brain of the moth Galleria mellonella, named “sericotropin” and reported to stimulate RNA synthesis in silk glands (acc. no. AAA85090). It is very unlikely that these proteins could perform such function in the proboscis of H. armigera or P. xylostella. Instead, they might act as carriers for some yet unknown nutrients. We cannot also exclude a role of these OBPs in chemosensing, based on the observation that chemosensilla have been described in the proboscis of Lepidoptera 33-35.
The eight CSPs found in the proboscis of the four species represent a more homogeneous group, with five of them sharing around 70% of identical amino acids and most likely responsible for the observed cross-reactivity in drink-blot and Western blot experiments. All the eight CSPs are aligned in Figure S1. Several other sequences, identified in our analysis (Tables S2-S5), include proteins of muscle, general enzymes, cuticular proteins and lipid carrier proteins, some of which might be worth of further attention as potential carriers of hydrophobic nutrients. The high similarity of CSPs in phylogenetically distant species 36 prompted us to search for specific roles of these proteins beyond the previously observed surfactant effect 22.
Proteomic analysis of soluble proteins in the eyes of H. armigera
Based on our previous observation that the antiserum against CSP4 showed significant cross-reactivity only with the head extract, besides the proboscis 22, we searched for this protein in the pump, a cavity directly connected to the proboscis 34, and in the eyes. In Western blot experiments we were able to stain proteins of the same apparent masses in proboscis and eyes, but not in the pump (Figure 3).
Figure 3.
Detection of electrophoretic bands reacting with the antiserum against HarmCSP4 in crude extract of proboscis (P), pump (p) and eyes (E) of H. armigera. When a moth was allowed to drink from a solution of a fluorescent compound (A), strong fluorescence was observed in a cavity connected to the proboscis and situated just behind the eye, a pump for pushing a liquid through the proboscis (B) Cross-reactivity is observed in the proboscis sample, due to high concentrations of HarmCSP4 (Tables 1 and S2), and in the eyes, where HarmOBP11 (75% identical with HarmCSP4) has been detected in proteomic analysis (Tables 2 and S6). (C) Coomassie stained gel. (D) Western blot with both first antiserum against HarmCSP4 and HRP-linked secondary antiserum diluted 1:1000.
Next we performed a proteomic investigation on the eyes of H. armigera, and analysed low molecular weight bands excised from SDS-PAGE gel of crude eyes extracts (Figure 4). The OBPs and CSPs identified are listed in Table 2, while all the proteins detected in the four bands are reported in Supplementary file S6. We were able to identify three OBPs and three CSPs, all in bands 1 and 2. HarmOBP1, also detected in the proboscis, and HarmOBP5 (acc. no. AEB54581), only found in eyes, are more than 60% identical between each other and with HarmOBP6, another OBP of the proboscis. HarmOBP17 (acc. no. AFI57166), on the other hand, is 75% identical with the OBP identified in the proboscis of P. xylostella, both belonging to the sub-class of C-minus OBPs with only 4 conserved cysteines 37.
Table 2.
Chemosensory and Odorant-binding proteins identified in slices of SDS-PAGE gels (Figure 4) from eyes extracts of H. armigera. Together with the description reported in the database, the sequence coverage and the number of peptides found in our analysis are also indicated.
| Slice | NCBI acc. | Description | Seq. cov. % | MW | Pept |
|---|---|---|---|---|---|
| 1 | AEX07265 gi|365919036 | CSP2 [H. armigera] | 35 | 13662 | 4 |
| 1 | gi|14091480 | CSP13 [H. armigera] | 28.3 | 14580 | 4 |
| 1 | AEB54580 gi|328879846 | OBP1 [H. armigera] | 8.8 | 15960 | 1 |
| 1 | AFR92095 gi|405117278 | CSP11 [H. armigera] | 10.9 | 14529 | 1 |
| 1 | AFI57166 gi|385275511 | OBP17 [H. armigera] | 8.9 | 15237 | 1 |
| 2 | AFR92095 gi|405117278 | CSP11 [H. armigera] | 21.9 | 14529 | 2 |
| 2 | AEB54581 gi|328879848 | OBP5 [H. armigera] | 19.7 | 15967 | 4 |
| 2 | AEX07265 gi|365919036 | CSP2 [H. armigera] | 33.3 | 13662 | 4 |
Among the CSPs, HarmCSP2, also detected in proboscis, is very abundant in eyes, as suggested by the high emPAI 38 (Exponentially Modified Protein Abundance Index) value (Table S6). Also, highly represented is HarmCSP13 and, to a lower extent, HarmCSP11. This last protein was also found in the proboscis and is 75% identical with HarmCSP4, the major protein of the proboscis, thus explaining the cross-reaction in Western blot experiments with the eyes extract.
These results reveal some connection between proboscis and eyes, as the two organs express ligand carrier proteins (OBPs and CSPs) either identical or highly similar. We decided to further investigate the function of the CSPs, which are also well conserved across Lepidoptera, on the hypothesis that they might act as carriers for visual pigments (3-hydroxyretinol and 3-hydroxyretinal) in the eyes and for their precursors (β-carotene and other carotenoids) important for the diet 39-40 in the proboscis.
Therefore, first we produced recombinant HarmCSP2, HarmCSP13 and HarmCSP11 and confirmed in Western blot experiments (Figure 4) that the first two proteins are expressed in the eyes of H. armigera, while the presence HarmCSP11 had been detected by the antiserum against HarmCSP4 (Figure 3). Next, we used the three recombinant proteins, together with the previously reported HarmCSP4 22 in ligand binding experiments.
Ligand-binding experiments and docking simulations
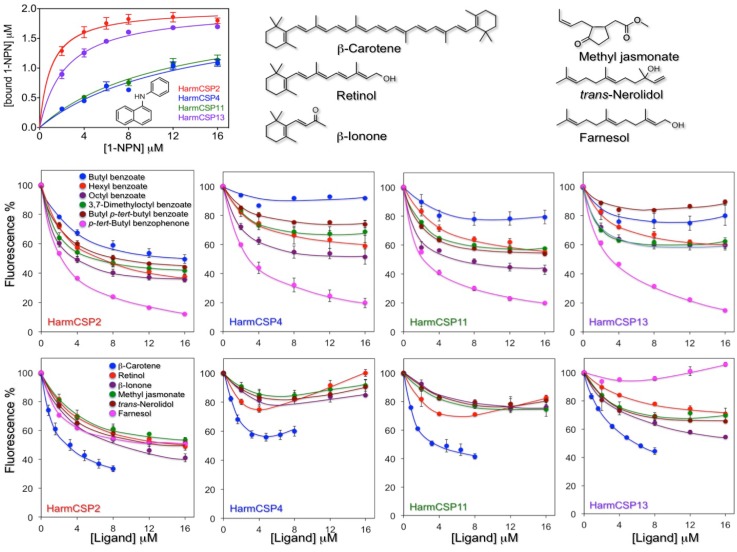
Both HarmCSP4 and HarmCSP11 bind the fluorescent probe 1-NPN (N-phenyl-1-naphthylamine) rather weakly, with dissociation constants of 8.2 and 12.1 μM respectively. By contrast, HarmCSP2 and HarmCSP13 show much better affinity to 1-NPN with dissociation constants of 1.02 and 2.34 μM, respectively (Figure 5).
Figure 5.
Binding properties of the four CSPs identified in the proboscis and eyes of H. armigera. Upper left panel: binding to 1-NPN (structure reported in the panel). HarmCSP4 and HarmCSP11 showed moderate affinity to 1-NPN with dissociation constants of 8.2 (SEM 4.0) and 12.1 (SEM 2.9) μM, respectively. HarmCSP2 and HarmCSP13, instead, bind strongly the fluorescent probe with dissociation constants of 1.02 (SEM 0.23) and 2.34 (SEM 0.25) μM, respectively. Competitive binding experiments (panels of the second and third row) were performed with two series of ligands. The first includes a number of benzoates and an aromatic ketone to evaluate the size requirement for a good fitting and reveals that all four proteins show preference for large ligands. The second group comprises some naturally occurring terpenes and terpenoids. Among these, β-carotene binds well to the four CSPs, followed by retinol. In both series of experiments HarmCSP2 showed a broader specificity with respect to the other three proteins. The structures of the ligands in the second series are shown in the upper region of the figure.
To evaluate the size requirements for a good fitting, we then performed competitive binding with a series of benzoates, also including an aromatic ketone. Among these compounds, octyl benzoate and p-tert-butylbenzophenone proved to be the best ligands for all four proteins, suggesting the presence of relatively large cavities.
The second group of chemicals includes β-carotene, retinol and structurally related terpenoids β-ionone, trans-nerolidol and farnesol, as well the common plant volatile methyl jasmonate. Within this group, the best ligand for all our four proteins was β-carotene, with retinol also showing appreciable affinity. The other chemicals did not exhibit significant binding except for HarmCSP2, which instead showed good affinity to all the tested ligands, proving to be the least specific of the four CSPs (Figure 5). In our experiments, we decided to use retinol instead of 3-hydroxyretinol, the pigment adopted by insects, due to the difficulty in obtaining a sample of this compound, combined with its poor stability. We also did not include retinal (or its 3-hydroxy derivative) because we found that the absorbance of this chemical has a very strong quenching effect on the fluorescence of 1-NPN.
We observe that some competitive binding curves, such as that of β-carotene with HarmCSP4 and those of retinol with HarmCSP4 and HarmCSP11, exhibited a peculiar effect, first decreasing at low concentrations and then increasing at concentrations above 5-6 μM. Such behaviour has been observed several times in the literature and has been explained with formation of micelles of the ligand, that could bind in their cavity molecules of the fluorescent probe, thus producing an increase in fluorescence 41-42. However, this explanation hardly adapts to β-carotene, which, being a hydrocarbon, cannot form micelles. In this case, perhaps, we could speculate that a molecule of 1-NPN could enter the binding cavity together with one of β-carotene. This idea is based on the fact that the CSP of Mamestra brassicae, similar in structure to our proteins, has been reported to be exceptionally flexible and able to swell, swallowing in the process three molecules of 12-Br-dodecanol 29 (PDB id: 1N8V).
When we modeled the four HarmCSPs (Figure S2), in fact, the programme selected as the best template the M. brassicae CSP (MbraCSP) in its complexed form with three molecules of ligand (amino acid identities between MbraCSP and HarmCSPs #2, 4, 11, 13 were 38, 54, 51, 89%, respectively). Using these models of the four CSPs we then performed docking simulations with β-carotene, 3-hydroxyretinol and 3-hydroxyretinal (the visual pigments used by insect instead of retinol and retinal used by vertebrates 43) and found that all three molecules can fit well inside the binding pockets.
Discussion
The aim of this work was to understand why CSPs abundantly found in the proboscis of Lepidoptera were conserved among phylogenetically distant species, including moths and butterflies. We tentatively answer this question suggesting that such proteins may help solubilising important hydrophobic nutrients, in particular β-carotene, a precursor of visual pigments (retinol and retinal) and growth and development factors (retinol and retinoic acid). This hypothesis is supported by the fact that same or similar proteins are expressed in the eyes, and by the observation that the four CSPs we have identified in proboscis and/or eyes of H. armigera, all bind β-carotene and, to some extent, retinol (Figure 5).
We believe that the most interesting aspect of our research is having found that proteins commonly regarded as carriers for odorants and pheromones, such as OBPs and CSPs, are likely involved in the visual process in insects, besides being carrier for diet carotenoids. This link between chemoreception and vision parallels a similar phenomenon in vertebrates, where retinol-binding proteins are members of the large family of lipocalins, that include vertebrate OBPs 44-45. To our knowledge, this is the first report of soluble olfactory proteins in the eyes of insects and confirms once again that during evolution successful proteins belonging to the same family can become adapted to various physiologically unrelated functions. A retinoid-binding protein of 273 amino acids PINTA (acc. no. AAO62632) had been reported in Drosophila 46-47 and a retinol-binding protein of 235 amino acids has been identified in the butterfly Papilio xuthus 48-49 (acc. no. AB070628). Both these proteins are very different in size and sequences from OBPs or CSPs. Certainly, the complex chain of events leading from β-carotene to visual pigments requires several binding proteins to transport the hydrophobic molecules generated during the enzymatic cascade across cells and intercellular fluids.
In particular, CSPs might be particularly suitable to bind and transport the large molecules of β-carotene and possibly other carotenoids, as compared to OBPs, thanks to their relatively high flexibility. In fact, the two disulfide bridges present in CSPs do not put any constrain on the protein folding, as instead is the case with the three interlocked bridges of insect OBPs. The flexibility of CSPs and their capacity of increasing the volume of the binding pocket to accommodate large ligands was elegantly demonstrated with the X-ray structure of the M. brassicae CSP complexed with three molecules of 12-Br-dodecanol 29.
β-Carotene is an important nutrient for insects and can be obtained from pollen, which contains between 10 and 200 ppm of this compound, besides several other carotenoids 50-51. It is known that the major source of nutrition of adult Lepidoptera is nectar, which contains mainly sugar, together with small amounts of salts, amino acids and other minor compounds 52. There are only few examples of Lepidoptera that in their adult stage rely on pollen as important food source. Butterflies of the genus Heliconius uptake pollen grains with their proboscis and with the help of saliva extruded from the proboscis, assisted by mechanical action, manage to break the pollen and extract amino acids and other nutrients from its grains 53-55. Unique is the case of Gelechiidae moths, primitive Lepidoptera that are endowed with a specific enzyme to dissolve sporopollenin, main constituent of the pollen wall 56.
Apart from these special cases, very little information is available in the literature on the role of pollen in the diet of adult Lepidoptera, although it is generally assumed that the majority of species can feed on lipids on the surface of pollen grain as well as on nutrients that become available through the germination pores. In such context, the abundant CSPs of the proboscis could help solubilising hydrophobic compounds, including β-carotene and other carotenoids and extracting them from the pollen grains. Such hypothesis, however, still needs to be experimentally verified.
Supplementary Material
Figures S1-S4, Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31230062 to G. Wang and 31471777 to C.-Z. Wang).
References
- 1.Vogt RG, Riddiford LM. Pheromone binding and inactivation by moth antennae. Nature. 1981;293:161–163. doi: 10.1038/293161a0. [DOI] [PubMed] [Google Scholar]
- 2.Angeli S, Ceron F, Scaloni A, Monti M, Monteforti G, Minnocci A. et al. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur J Biochem. 1999;262:745–754. doi: 10.1046/j.1432-1327.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- 3.Vogt RG. Blomquist GJ, Vogt RG. Eds, Insect Pheromone Biochemistry and Molecular Biology. Elsevier Academic Press, London; 2003. Biochemical diversity of odor detection: OBPs, ODEs and SNMPs; pp. 391–446. [Google Scholar]
- 4.Pelosi P, Zhou JJ, Ban LP, Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63:1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 6.Jacquin-Joly E, Vogt RG, François MC, Nagnan-Le Meillour P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem Senses. 2001;26:833–844. doi: 10.1093/chemse/26.7.833. [DOI] [PubMed] [Google Scholar]
- 7.Li S, Picimbon JF, Ji SD, Kan YC, Qiao CL, Zhou JJ. et al. Multiple functions of an odorant-binding protein in the mosquito Aedes aegypti. Biochem Biophys Res Comm. 2008;372:464–468. doi: 10.1016/j.bbrc.2008.05.064. [DOI] [PubMed] [Google Scholar]
- 8.Iovinella I, Dani FR, Niccolini A, Sagona S, Michelucci E, Gazzano A. et al. Differential Expression Of Odorant-Binding Proteins In The Mandibular Glands Of The Honey Bee According To Caste And Age. J Proteome Res. 2011;10:3439–3449. doi: 10.1021/pr2000754. [DOI] [PubMed] [Google Scholar]
- 9.Dani FR, Michelucci E, Francese S, Mastrobuoni G, Cappellozza S, La Marca G. et al. Odorant-binding proteins and Chemosensory proteins in pheromone detection and release in the silkmoth Bombyx mori. Chem Senses. 2011;36:335–344. doi: 10.1093/chemse/bjq137. [DOI] [PubMed] [Google Scholar]
- 10.Sun YL, Huang LQ, Pelosi P, Wang CZ. Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species. PLoS One. 2012;7:e30040. doi: 10.1371/journal.pone.0030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou XH, Ban LP, Iovinella I, Zhao LJ, Gao Q, Felicioli A. et al. Diversity, abundance and sex-specific expression of chemosensory proteins in the reproductive organs of the locust Locusta migratoria manilensis. Biol Chem. 2013;394:43–54. doi: 10.1515/hsz-2012-0114. [DOI] [PubMed] [Google Scholar]
- 12.Gu SH, Wu KM, Guo YY, Pickett JA, Field LM, Zhou JJ. et al. Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics. 2013;14:636. doi: 10.1186/1471-2164-14-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavaggioni A, Mucignat-Caretta C. Major urinary proteins, r2u-globulins and aphrodisin. Biochim Biophys Acta. 2000;1482:218–228. doi: 10.1016/s0167-4838(00)00149-7. [DOI] [PubMed] [Google Scholar]
- 14.Marchese S, Pes D, Scaloni A, Carbone V, Pelosi P. Lipocalins of boar salivary glands binding odours and pheromones. Eur J Biochem. 1988;252:563–568. doi: 10.1046/j.1432-1327.1998.2520563.x. [DOI] [PubMed] [Google Scholar]
- 15.Pelosi P, Iovinella I, Felicioli A, Dani FR. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 2014;5:320. doi: 10.3389/fphys.2014.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitabayashi AN, Arai T, Kubo T, Natori S. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana American cockroach. Insect Biochem Mol Biol. 1998;28:785–790. doi: 10.1016/s0965-1748(98)00058-7. [DOI] [PubMed] [Google Scholar]
- 17.Maleszka J, Forêt S, Saint R, Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee Apis mellifera. Dev Genes Evol. 2007;217:189–196. doi: 10.1007/s00427-006-0127-y. [DOI] [PubMed] [Google Scholar]
- 18.Liu GX, Xuan N, Chu D, Xie HY, Fan ZX, Bi YP. et al. Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch Insect Biochem Physiol. 2014;85:137–151. doi: 10.1002/arch.21148. [DOI] [PubMed] [Google Scholar]
- 19.Xuan N, Guo X, Xie HY, Lou QN, Lu XB, Liu GX. et al. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2015;22:203–219. doi: 10.1111/1744-7917.12116. [DOI] [PubMed] [Google Scholar]
- 20.Ishida Y, Ishibashi J, Leal WS. Fatty acid solubilizer from the oral disk of the blowfly. PLoS One. 2013;8:e51779. doi: 10.1371/journal.pone.0051779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marinotti O, Ngo T, Kojin BB, Chou SP, Nguyen B, Juhn J. et al. Integrated proteomic and transcriptomic analysis of the Aedes aegypti eggshell. BMC Dev Biol. 2014;14:15. doi: 10.1186/1471-213X-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu YL, Guo H, Huang L-Q, Pelosi P, Wang C-Z. Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J Exp Biol. 2014;217:1821–1826. doi: 10.1242/jeb.102020. [DOI] [PubMed] [Google Scholar]
- 23.Qiao H, Tuccori E, He X, Gazzano A, Field L, Zhou JJ. et al. Discrimination of alarm pheromone (E)-beta-farnesene by aphid odorant-binding proteins. Insect Biochem Mol Biol. 2009;39:414–419. doi: 10.1016/j.ibmb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Ban LP, Scaloni A, D'Ambrosio C, Zhang L, Yan YH, Pelosi P. Biochemical characterisation and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell Mol Life Sci. 2003;60:390–400. doi: 10.1007/s000180300032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvello M, Guerra N, Brandazza A, D'Ambrosio C, Scaloni A, Dani FR. et al. Soluble proteins of chemical communication in the social wasp Polistes dominulus. Cell Mol Life Sci. 2003;60:1933–1943. doi: 10.1007/s00018-003-3186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peitsch M. Protein modeling by E-mail. Bio/Technology. 1995;13:658–660. [Google Scholar]
- 27.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modeling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 28.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Research. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campanacci V, Lartigue A, Hallberg BM, Jones TA, Giudici-Orticoni MT, Tegoni M. et al. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc Natl Acad Sci USA. 2003;29:5069–5074. doi: 10.1073/pnas.0836654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC. et al. UCSF Chimera, a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Li X, Fan D, Zhang W, Liu G, Zhang L, Zhao L. et al. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat Commun. 2015;6:8212. doi: 10.1038/ncomms9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue TA, Asaoka K, Seta K, Imaeda D, Ozaki M. Sugar receptor response of the food-canal taste sensilla in a nectar-feeding swallowtail butterfly, Papilio xuthus. Naturwissenschaften. 2009;96:355–363. doi: 10.1007/s00114-008-0483-8. [DOI] [PubMed] [Google Scholar]
- 34.Krenn HW. Feeding mechanisms of adult Lepidoptera: structure, function, and evolution of the mouthparts. Annu Rev Entomol. 2010;55:307–327. doi: 10.1146/annurev-ento-112408-085338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haverkamp A, Bing J, Badeke E, Hansson BS, Knaden M. Innate olfactory preferences for flowers matching proboscis length ensure optimal energy gain in a hawkmoth. Nat Commun. 2016;7:11644. doi: 10.1038/ncomms11644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Regier JC, Mitter C, Zwick A, Bazinet AL, Cummings MP, Kawahara AY. et al. A large-scale, higher-level, molecular phylogenetic study of the insect order Lepidoptera (moths and butterflies) PLoS One. 2013;8:e58568. doi: 10.1371/journal.pone.0058568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou JJ, Huang W, Zhang GA, Pickett JA, Field LM. “Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene. 2004;327:117–129. doi: 10.1016/j.gene.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 38.IshihamaY Oda Y, Tabata T Sato T, Nagasu T Rappsilber J. et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Zheng SJ, Snoeren TA, Hogewoning SW, van Loon JJ, Dicke M. Disruption of plant carotenoid biosynthesis through virus-induced gene silencing affects oviposition behaviour of the butterfly Pieris rapae. New Phytol. 2010;186:733–745. doi: 10.1111/j.1469-8137.2010.03213.x. [DOI] [PubMed] [Google Scholar]
- 40.Almbro M, Dowling DK, Simmons LW. Effects of vitamin E and beta-carotene on sperm competitiveness. Ecol Lett. 2011;14:891–895. doi: 10.1111/j.1461-0248.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- 41.Sun YF, De Biasio F, Qiao HL, Iovinella I, Yang SX, Ling Y. et al. Two odorant-binding proteins mediate the behavioural response of aphids to the alarm pheromone (E)-ß-farnesene and structural analogues. PLoS One. 2012;7:e32759. doi: 10.1371/journal.pone.0032759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leal GM, Leal WS. Binding of a fluorescence reporter and a ligand to an odorant-binding protein of the yellow fever mosquito, Aedes aegypti. F1000 Res. 2014;3:305. doi: 10.12688/f1000research.5879.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babino D, Golczak M, Kiser PD, Wyss A, Palczewski K, von Lintig J. The Biochemical Basis of Vitamin A3 Production in Arthropod Vision. ACS Chem Biol. 2016;11:1049–1057. doi: 10.1021/acschembio.5b00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monaco HL. The transthyretin-retinol-binding protein complex. Biochim Biophys Acta. 2000;1482:65–72. doi: 10.1016/s0167-4838(00)00140-0. [DOI] [PubMed] [Google Scholar]
- 45.Newcomer ME, Ong DE. Plasma retinol binding protein: structure and function of the prototypic lipocalin. Biochim Biophys Acta. 2000;1482:57–64. doi: 10.1016/s0167-4838(00)00150-3. [DOI] [PubMed] [Google Scholar]
- 46.Wang T, Montell C. Rhodopsin formation in Drosophila is dependent on the PINTA retinoid-binding protein. J Neurosci. 2005;25:5187–5194. doi: 10.1523/JNEUROSCI.0995-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T, Jiao Y, Montell C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J Cell Biol. 2007;177:305–316. doi: 10.1083/jcb.200610081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakakuwa M, Arikawa K, Ozaki K. A novel retinol-binding protein in the retina of the swallowtail butterfly, Papilio xuthus. Eur J Biochem. 2003;270:2436–2445. doi: 10.1046/j.1432-1033.2003.03614.x. [DOI] [PubMed] [Google Scholar]
- 49.Wakakuwa M, Ozaki K, Arikawa K. Immunohistochemical localization of Papilio RBP in the eye of butterflies. J Exp Biol. 2004;207:1479–1486. doi: 10.1242/jeb.00913. [DOI] [PubMed] [Google Scholar]
- 50.Schulte F, Maeder J, Kroh LW, Panne U, Kneipp J. Characterization of Pollen Carotenoids with in situ and High-Performance Thin-Layer Chromatography Supported Resonant Raman Spectroscopy. Anal Chem. 2009;81:8426–8433. doi: 10.1021/ac901389p. [DOI] [PubMed] [Google Scholar]
- 51.Denisow B, Denisow-Pietrzyk M. Biological and therapeutic properties of bee pollen. A review. J Sci Food Agric. 2016;96:4303–4309. doi: 10.1002/jsfa.7729. [DOI] [PubMed] [Google Scholar]
- 52.Nepi M, Stpiczyńska M. The complexity of nectar: secretion and resorption dynamically regulate nectar features. Naturwissenschaften. 2008;95:177–184. doi: 10.1007/s00114-007-0307-2. [DOI] [PubMed] [Google Scholar]
- 53.Gilbert LE. Pollen feeding and reproductive biology of Heliconius butterflies. Proc Natl Acad Sci USA. 1972;69:1403–1407. doi: 10.1073/pnas.69.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eberhard SH, Hrassnigg N, Crailsheim K, Krenn HW. Evidence of protease in the saliva of the butterfly Heliconius melpomene (L.) (Nymphalidae, Lepidoptera) J Insect Physiol. 2007;53:126–131. doi: 10.1016/j.jinsphys.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harpel D, Cullen DA, Ott SR, Jiggins CD, Walters JR. Pollen feeding proteomics: Salivary proteins of the passion flower butterfly, Heliconius melpomene. Insect Biochem Mol Biol. 2015;63:7–13. doi: 10.1016/j.ibmb.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 56.Luo S, Li Y, Chen S, Zhang D, Renner SS. Gelechiidae moths are capable of chemically dissolving the pollen of their host plants: first documented sporopollenin breakdown by an animal. PLoS One. 2011;6:e19219. doi: 10.1371/journal.pone.0019219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S4, Table S1.
Table S2.
Table S3.
Table S4.
Table S5.
Table S6.