Abstract
Cigarette smoke (CS) is a major risk of chronic obstructive pulmonary disease (COPD), contributing to airway inflammation. Our previous study revealed that silymarin had an anti-inflammatory effect in CS-exposed mice. In this study, we attempt to further elucidate the molecular mechanisms of silymarin in CS extract (CSE)-induced inflammation using human bronchial epithelial cells. Silymarin significantly suppressed autophagy activation and the activity of ERK/p38 mitogen-activated protein kinase (MAPK) pathway in Beas-2B cells. We also observed that inhibiting the activity of ERK with specific inhibitor U0126 led to reduced autophagic level, while knockdown of autophagic gene Beclin-1 and Atg5 decreased the levels of ERK and p38 phosphorylation. Moreover, silymarin attenuated CSE-induced upregulation of inflammatory cytokines TNF-α, IL-6 and IL-8 which could also be dampened by ERK/p38 MAPK inhibitors and siRNAs for Beclin-1 and Atg5. Finally, we validated decreased levels of both autophagy and inflammatory cytokines (TNF-α and KC) in CS-exposed mice after silymarin treatment. The present research has demonstrated that CSE-induced autophagy in bronchial epithelia, in synergism with ERK MAPK pathway, may initiate and exaggerate airway inflammation. Silymarin could attenuate inflammatory responses through intervening in the crosstalk between autophagy and ERK MAPK pathway, and might be an ideal agent treating inflammatory pulmonary diseases.
Cigarette smoke (CS) contains nearly 5,000 chemicals, most of which are etiological factors in the development of pulmonary diseases, such as chronic obstructive pulmonary disease (COPD). CS exposure has been shown to induce an abnormal inflammatory response in the small airways and alveoli, contributing to airway remodeling and subsequent reduction of the airflow, which is the main characteristic of COPD1. It has been demonstrated that bronchial epithelium acts as a main source of overproduction of various cytokines, chemokines and adhesion molecules that modulate other elements of the airway wall and immune cells against CS2,3. Therefore, mapping the molecular mechanisms for CS-induced alterations in bronchial epithelial cells may offer clues into COPD pathogenesis and treatment.
Macroautophagy (generally referred to as autophagy), a genetically programmed and evolutionarily conserved degradation process4, occurs under various cellular stresses, such as starvation, hypoxia and DNA damage5. How autophagy shapes the inflammatory response has been discussed in various disease models6,7,8,9, and several studies have focused on the functional link between autophagy and inflammation-associated pulmonary pathogenesis, which suggest a critical role of autophagy in inflammation regulation in pulmonary diseases like asthma and acute lung injury (ALI)10,11. On the other hand, emerging investigations have demonstrated that autophagy can be activated by CS in lung cells12,13, acting as a deleterious process during COPD pathogenesis such as the apoptosis of lung epithelial cells13,14. mucin production15 and impaired mucociliary clearance16,17. As abnormal inflammatory response is another well-defined feature of airway epithelium in COPD, we hypothesized that autophagy may also regulate CS-induced inflammation in airway epithelial cells.
Silymarin, a flavonoid compound extracted from the milk thistle (Silybum marianum)18, is suggested as an anti-inflammatory and antioxidant agent19. Our previous study has suggested that pretreatment with silymarin could attenuate CS-induced lung inflammation and oxidative stress in mice, possibly involving ERK/p38 MAPK pathways20, which implied a promising role of silymarin in treating CS-induced airway inflammation. Recently, Liu et al. have reported that silibin, the main component of silymarin, protected UVB-irradiated L929 cells from apoptotic death by repressing the activation of autophagy21. Besides, silibin could inhibit the activation of ERK/p38 MAPK pathways, subsequently decreasing the expression of autophagic genes in an influenza A virus infection model22. These evidences indicate that autophagy pathway might be a potential target of silymarin.
In this study, we attempt to elucidate the biological functions of silymarin in CSE-induced inflammation using human bronchial epithelial cell line (Beas-2B). Particularly, we investigated whether silymarin could attenuate CSE-induced inflammatory response via affecting activation of autophagy. Furthermore, we also explored the potential crosstalk between autophagy and ERK/p38 MAPK pathways.
Material and Methods
Reagents
Silymarin with a purity of 99% was obtained from Sigma (St Louis, MO, USA) and was dissolved in dimethylsulfoxide (DMSO) ( Sigma, St Louis, MO, USA) to make a stock solution. It was diluted with DMEM with the DMSO concentration kept below 0.1% in cell culture, which had no detectable effects on cells. U0126 and SB203580 were purchased from Cell Signaling Technology (Danvers, MA, USA) and Selleck Chemicals (Houston, TX, USA), respectively. Primary antibodies against LC3B, phospho-ERK, phospho-p38, β-actin and horseradish peroxidase-conjugated second antibodies were all pruchased from Cell Signaling Technology. The SuperSignal West Pico Chemiluminescent Substrate for horseradish peroxidase enzyme was obtained from Pierce (Rockford, IL, USA).
Preparation of CSE
CSE was prepared as previously described23. Briefly, mainstream smoke from 15 cigarettes (Jiao Zi, Chengdu Cigarette Factory, Chengdu, China; 1.1 mg nicotine and 11 mg tar per cigarette) was drawn slowly into a 50-ml syringe and bubbled through 50 ml of DMEM pre-warmed in a water bath at 37 °C. The preparation, considered to be 100% CSE, was titrated to pH 7.4 and sterilized with a 0.22 μm filter (Millipore, Bedford, MA, USA), and then stored at −80 °C. Serum-free cell culture medium was used to dilute 100% CSE to the required CSE concentrations.
Cell culture
Beas-2B cells obtained from the American Type Culture Collections (Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin G sodium and 50 μg/ml streptomycin sulfate (Invitrogen, Carlsbad, CA, USA) at 37 °C in 5% CO2 -enriched air. All experiments were performed on logarithmically growing cells. Cell layers were 70–80% confluent at the time of CSE exposure.
Cell viability assay
Cells were seeded at an initial density of 5000 cells per well onto 96-well plates and allowed to attach overnight, before treated with indicated concentrations of CSE or silymarin or in combination (cells were pretreated with silymarin 1 h before CSE exposure). After 24 h, 10 μl reagent of Cell Counting Kit-8 (CCK8, Dojindo Laboratories, Tokyo, Japan) was added to each well and incubated for 2 h. Absorbance values were recorded using a Model 680 microplate reader (Bio-Rad, Hercules, CA, USA) at an absorbance of 450 nm.
Transient transfection of siRNA
Beas-2B cells were cultured in 12-well plate overnight to 40–60% confluence in complete medium without antibiotics. Validated small-interfering RNA (siRNA) oligonucleotides specific to Atg5, Beclin-1 and negative control siRNA were purchased from Sigma (St Louis, MO, USA). Transient transfections were performed using HiPerFect Transfection Reagent (Qiagen,Valencia, CA, USA) according to the manufacturer’s protocols. Briefly, cells were incubated with siRNA-HiPerFect complexes for 16 h. The medium was then removed and replaced with fresh complete medium. After transfection for 24 h, cells were exposed to CSE. The control and CSE groups received the same volume transfection reagents in parallel to avoid stress-induced secretion of cytokines in response to transfection.
Fluoresecence microscope imaging of autophagy after pEGFP-LC3 transfection
The pEGFP-LC3 plasmid expressing the autophagosome marker protein microtubule-associated protein 1 light chain 3 beta (LC3) (Addgene plasmid 21073) was kindly provided by Tamotsu Yoshimori, Department of Genetics, Osaka University Graduate School of Medicine, Japan24. Cells were transfected with the plasmid by use of Attractene (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions and were maintained on coverslips in 24-well plates. After transfection for 24 h, cells were treated with indicated concentrations of CSE or silymarin or in combination (cells were pretreated with silymarin 1 h before CSE exposure). DAPI (Sigma, St Louis, MO, USA) was used to stain the nuclei. The images of cells were acquired using a Nikon fluorescence microscope. Autophagy was detected by monitoring the formation of fluorescent puncta of autophagosomes in pEGFP-LC3 transfected cells.
Treatment of Animals
Animals were treated as previously described20. Briefly, specific pathogen-free male BALB/c mice (6–8 weeks) were randomly divided into four groups (n = 6 per group): control group (C), which received placebo and was not exposed to CS; CS-exposed group (CS), which received placebo and was exposed to CS; CS-exposed low-dose silymarin group (CS+ SiL), which received 25 mg/kg silymarin (i.p., q. d.) and was subsequently exposed to CS; and CS-exposed high- dose silymarin group (CS+SiH), which received 50 mg/kg silymarin (i.p., q. d.) and was subsequently exposed to CS. For cigarette smoke exposure experiment, animals were exposed to mainstream CS at 250 mg total particulate matter (TPM)/m3 for 4 h per day, 6 days per week for 4 weeks using a Baumgartner-Jaeger CSM2082i automated cigarette smoke generator (CH Technologies, Westwood, NJ, USA). A commercially available cigarette was employed (Jiaozi, China Tobacco Chuanyu Industrial Co., Ltd.; 1.1 mg nicotine and 11 mg tar per cigarette). In parallel, C group were exposed to air following the same schedule. After CS exposure for 4 weeks, the mice in all groups were sacrificed with intraperitoneal 3% sodium pentobarbital, followed by exsanguination from the right ventricle to allow tissue sample collection. All experimental protocols involving the mice were in accordance with the Guide for the Care and Use of Laboratory Animals.The study protocol was approved by the Animal Care and Use Committee of West China Hospital.
Collection of Bronchoalveolar Lavage Fluid (BALF)
The right lung was lavaged three times with 0.5 ml of sterile saline, with a recovery rate of 95%. The lavage fluid was centrifuged at 1,000 g for 5 min, and the supernatant was stored at −80 °C for analysis of cytokines using enzyme-linked immunosorbent assay (ELISA).
Western Blot analysis
Cells or lung tissues were lysed in RIPA buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 2 mM sodium fluoride, 2 mM EDTA, 0.1% SDS, and PMSF. Protein concentrations were determined by a BCA protein assay kit (Thermo Fisher Scientific Inc., MA, USA). Total protein (20 μg) was fractionated by 10% SDS polyacrylamide gel electrophoresis and transferred to PVDF membranes. Membranes were blocked for 1 h at room temperature with 5% BSA in TBS-Tween and incubated overnight at 4 °C with the appropriate primary antibodies. After incubation with horseradish peroxidase-conjugated second antibodies, the immune complexes were detected with SuperSignal West Pico chemiluminescent substrate. Band intensities were quantified using computerized image analysis (QuantityOne-software, Bio-Rad, Hercules, CA, USA).
Real-Time PCR
The total RNA was isolated from Beas-2B cells using the Total RNA Kit I (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s protocol. cDNA was synthesized from using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Quantitative real-time RT-PCR analysis was conducted in triplicates by the CFX96 real-time PCR detection system using SsoFast EvaGreen Supermix according to the manufacturer’s specifications (Bio-Rad, Hercules, CA, USA). The primer sequences were as follows: Atg5 forward 5′-TAGTATCATCCCACAGCCAACAG-3′, reverse 5′-AGTAAGACCAGCCCAGTTGCC-3′; Beclin-1 forward 5′-GCTGAGGGATGGAAGGGTCT-3′, reverse 5′-CGCCTGGGCTGTGGTAAGT-3′; β-actin forward 5′-CTGCCAAGTGGGTGGTATAGAG-3′, reverse 5′-GGTAGTCCATAGTGAAGGCGAAC-3′. The efficiency of primers for Atg5, Beclin-1 and β-actin were 102%, 89.5% and 114%, respectively. All data were normalized to β-actin gene expression, and relative expression levels were determined using the 2 −ΔΔCt method.
Enzyme-linked immunosorbent assays (ELISA)
Levels of cytokine release (TNF-α, IL-6 and IL-8 in culture supernatants; TNF-α, IL-6 and KC in BALF) were determined by ELISA kit (ExCell Biology, Shanghai, China) according to the manufacturer’s protocol. The absorbance of 570 nm was measured by a Model 680 microplate reader (Bio-Rad, Hercules, CA, USA).
Statistical analysis
All values were expressed as mean ± SEM. Differences among multiple groups were analyzed by one-way analysis of variance (ANOVA). Significance was defined by a p value < 0.05 (two-tailed). All statistical calculations were carried out in SPSS (version 22.0, SPSS Inc., USA).
Results
Evaluation of cell viability following exposure to CSE and silymarin
Cells were exposed to various concentrations of CSE (0% to 10%) for 24 h, after which cell growth was determined by CCK8 assay. The results showed that CSE inhibited the growth of cells in a dose-dependent manner and exerted cytotoxic effect when the concentration exceeded 6% (Supplementary Fig. S1). Next, we treated Beas-2B cells with 6% CSE or silymarin (10, 20, 30 or 40 μM) or in combination for 24 h. The results revealed that the viability of Beas-2B cells was dose-dependently improved by silymarin treatment (Fig. 1). However, due to the obvious growth inhibitory effect of silymarin at concentrations higher than 30 μM, silymarin at a concentration of 20 μM was selected for further study (Fig. 1).
Figure 1. Effect of silymarin on cell viability in CSE-exposed Beas-2B, as determined by CCK8 assay.
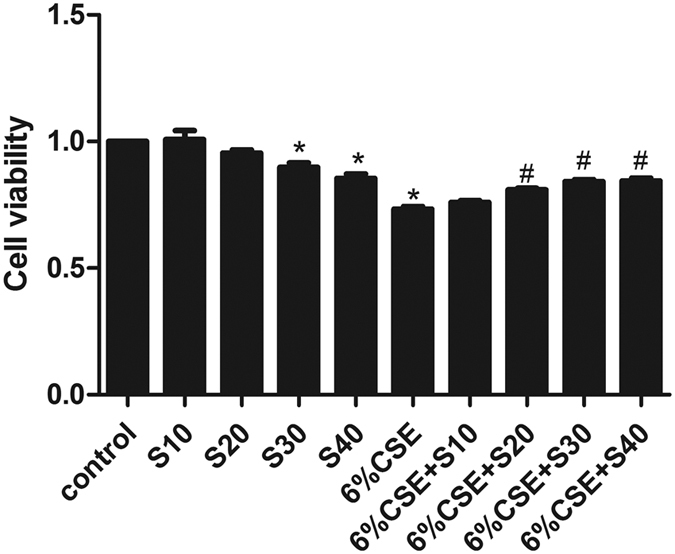
Results are representative of three independent experiments. Values are expressed as mean ± SEM (n = 3). *P < 0.05 with respect to the control group; #P < 0.05 with respect to the CSE-exposed group. S, silymarin.
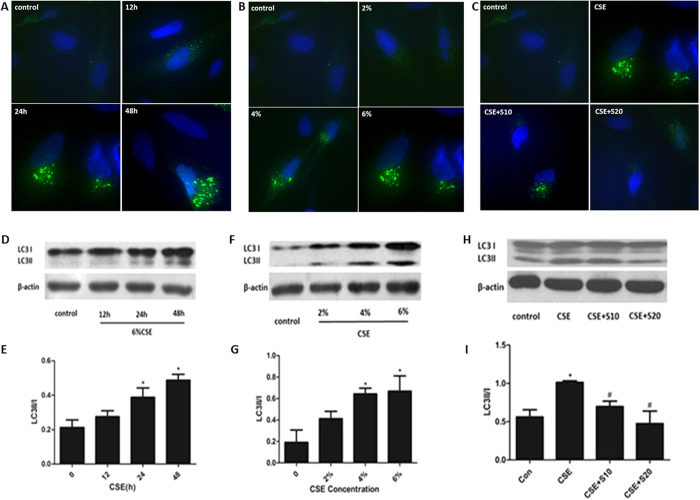
Autophagy was activated by CSE and reversed by silymarin in Beas-2B cells
Autophagy can be activated in cells in response towards various stresses, such as oxidative stress, hypoxia, nutrient deprivation, DNA damage and intracellular pathogens25,26,27. In the present study, CSE activated the autophagic process in time-and dose-dependent manner, as shown by the marked increase of pEGFP-LC3 puncta in Beas-2B cells (Fig. 2A,B) and accelerated conversion of LC3-I to LC3-II in the western-blot analysis (Fig. 2D–G). However, pretreatment with silymarin (10 μM or 20 μM) attenuated this induction of autophagy (Fig. 2C,H,I) without changing the basal level of autophagy (Supplementary Fig. S2).
Figure 2. Autophagy activation in response to CSE and silymarin treatment.
Beas-2B cells were treated with 6%CSE for different time period, indicated concentrations of CSE for 24 h or both 6%CSE and silymarin (10 μM, 20 μM) for 24 h. Formation of pEGFP-LC3 puncta in Beas-2B cells was analyzed by immunofluorescence under fluorescence microscopy (×400) (A–C). Expressions of LC3I and LC3II were measured by Western Blot (D,F,H). Densitometry was performed and the ratio of LC3II/I were calculated (E,G,I). Results are representative of three independent experiments. Values are expressed as mean ± SEM (n = 3). *P < 0.05 with respect to the control group; #P < 0.05 with respect to the CSE-exposed group. S, silymarin.
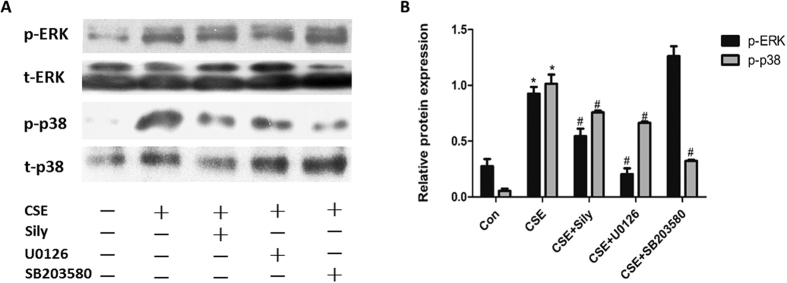
Silymarin suppressed the activity of ERK/p38 MAPK pathway in CSE-treated Beas-2B cells
We next investigated whether silymarin possessed the ability to inhibit ERK/p38 MAPK pathway in CSE-treated Beas-2B cells. The phosphorylation levels of ERK and p38 were measured by Western-blot. As shown in Fig. 3, both ERK and p38 phosphorylation levels were enhanced in the CSE-treated cells, while pretreatment with silymarin effectively prevented the CSE-induced upregulation of p-ERK and p-p38, compared with the inhibitors of ERK/p38 MAPK pathway, U0126 and SB203580. However, silymarin did not change the basal levels of ERK and p38 phosphorylation (Supplementary Fig. S3).
Figure 3. Effect of silymarin on ERK/p38 MAPK pathway in CSE-exposed Beas-2B cells.
Cells were pretreated with silymarin (20 μM), U0126 (10 μM) or SB203580 (10 μM) before exposed to 6%CSE for 24 h. (A) Phosphorylated and total levels of ERK and p38 were measured by Western Blot. (B) Densitometry was performed and the ratio of p-p38/t-p38 and p-ERK/t-ERK were calculated. Results are representative of three independent experiments. Values are expressed as mean ± SEM (n = 3). *P < 0.05 with respect to the control group; #P < 0.05 with respect to the CSE-exposed group.
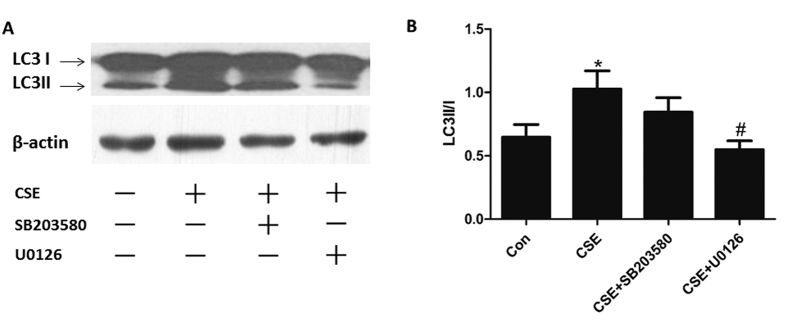
ERK MAPK pathway regulated autophagy activation in CSE-treated Beas-2B cells
Some reports have shown that autophagy could be regulated by ERK and p38 MAPK pathways under different conditions28,29,30. In this study, suppressing the activity of ERK signaling pathway with its specific inhibitor U0126 led to reduced autophagic level, as reflected by the blocking of LC3 conversion in CSE-treated Beas-2B cells. However, p38 MAPK inhibitor SB203580 showed no significant influence on the ratio of LC3II to LC3I in this model (Fig. 4A,B).
Figure 4. ERK MAPK pathway regulated autophagy activation in CSE-treated Beas-2B cells.
Cells were pretreated with U0126 (10 μM) or SB203580 (10 μM) before exposed to 6%CSE for 24 h. (A) Expressions of LC3I and LC3II were measured by Western Blot. (B) Densitometry was performed and the ratio of LC3II/I were calculated. Results are representative of three independent experiments. Values are expressed as mean ± SEM (n = 3). *P < 0.05 with respect to the control group; #P < 0.05 with respect to the CSE-exposed group.
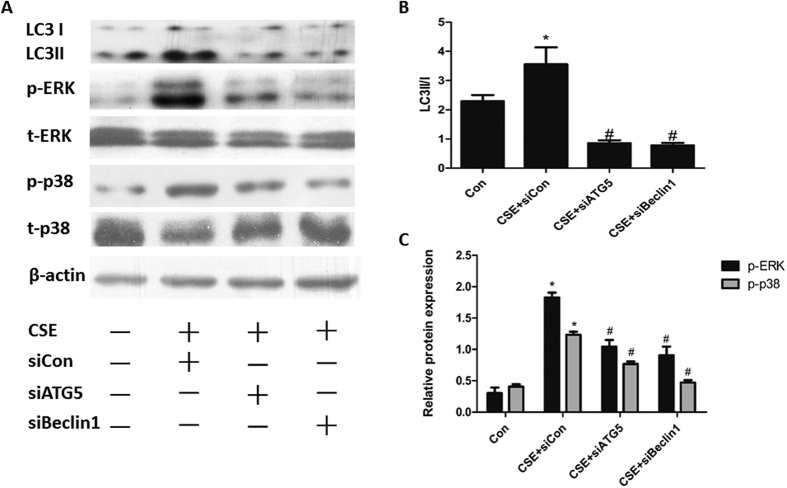
Blocking autophagy suppressed CSE-induced ERK and p38 phosphorylation in Beas-2B cells
To explore the potential interaction between autophagy and ERK/p38 MAPK pathways in Beas-2B cells, knockdown of the autophagic gene Beclin-1 and Atg5 by siRNA were used to repress the induction of autophagic flux. Transfection of Beclin-1 and Atg5 siRNA, efficiently and specifically reduced Beclin-1 and Atg5 mRNA expression, respectively (Supplementary Fig. S4). Further, treatment with Beclin-1 and Atg5 siRNA at 100 nM before 6% CSE stimulation reduced the conversion of LC3 proteins (Fig. 5A,B). By selective knockdown of these autophagy related genes, we observed decreased levels of ERK and p38 phosphorylation (Fig. 5A,C) when CSE presented, suggesting that blocking autophagy might inhibit the ERK and p38 axis.
Figure 5. Blocking autophagy suppressed CSE-induced ERK and p38 phosphorylation in Beas-2B cells.
Cells were transfected with Atg5 siRNA (100 nM), Beclin-1 siRNA (100 nM) or negative control siRNA before exposed to 6%CSE for 24 h. (A) Expressions of LC3I and LC3II, phosphorylated and total levels of ERK and p38 were measured by Western Blot. Densitometry was performed and (B) the ratio of LC3II/I, (C) p-p38/t-p38 and p-ERK/t-ERK were calculated. Results are representative of three independent experiments. Values are expressed as mean ± SEM (n = 3). *P < 0.05 with respect to the control group; #P < 0.05 with respect to the CSE-exposed group.
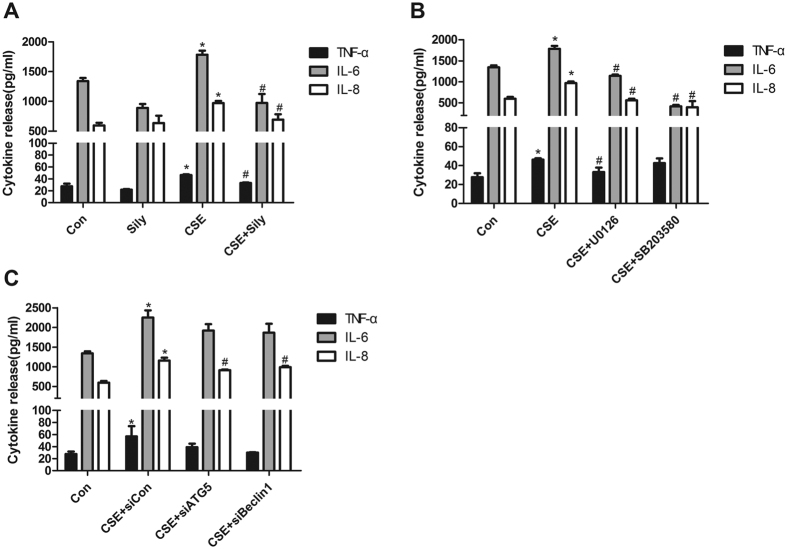
Silymarin attenuated CSE-stimulated inflammatory cytokine release in an autophagy- and ERK/p38 MAPK-dependent manner
ELISA for TNF-α, IL-6 and IL-8 in the culture supernatant was used to determine the effect of silymarin on CSE-induced pro-inflammatory response. As shown in Fig. 6A, silymarin pretreatment significantly reduced the CSE-induced increases of TNF-α, IL-6 and IL-8 release in the supernatants.
Figure 6. Silymarin attenuated CSE-stimulated inflammatory cytokine release in an autophagy- and ERK/p38 MAPK-dependent manner.
(A) Beas-2B cells pretreated with silymarin (20 μM), (B) ERK/p38 MAPK inhibitors U0126(10 μM) and SB203580(10 μM), or (C) negative control/Atg5/Beclin-1 siRNA (100 nM) were exposed to 6%CSE for 24 h. The secretion of TNF-α, IL-6 and IL-8 in supernatants were detected by ELISA. Data were averaged from a duplicate of each sample and from three independent experiments. Values are expressed as mean ± SEM (n = 3). *P < 0.05 with respect to the control group; #P < 0.05 with respect to the CSE-exposed group.
To further investigate whether the anti-inflammatory effects of silymarin were partly mediated by autophagy or ERK/p38 MAPK pathways, selective knockdown of autophagy related genes (Atg5 and Beclin-1) were utilized to inhibit autophagy flux, while selective inhibitors for ERK (U0126) and p38 (SB203580) were also used before CSE treatment. The results indicated that inhibiting ERK/p38 MAPK could decrease the CSE-induced release of IL-6 and IL-8 (Fig. 6B), whereas blocking autophagy significantly decreased the production of proinflammatory cytokine IL-8 (Fig. 6C). Combined with findings of previous parts of this study, these results suggested that ERK/p38 MAPK and autophagy signaling may regulate CSE-induced inflammatory cytokine release, while inhibitory effect of silymarin on these pathways could attenuate such CS-related airway inflammation.
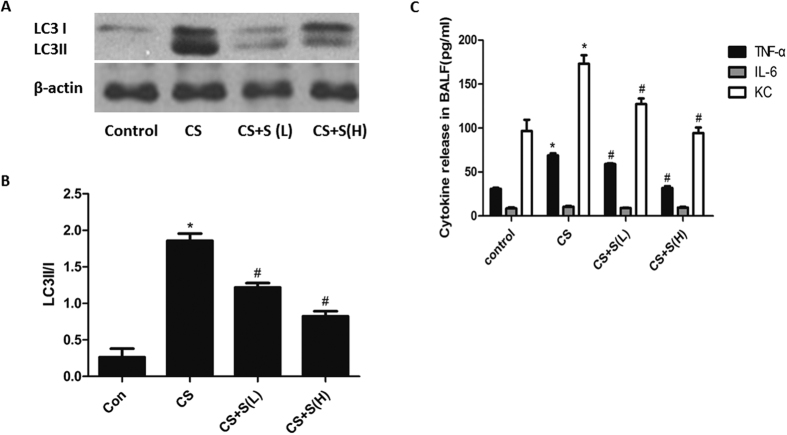
Silymarin inhibited autophagy in a mouse model of airway inflammation
Results from our previous work have implied that silymarin could decrease the activity of ERK/p38 MAPK pathway in CS-treated mice20. Using this model, we further validated whether silymarin influenced activation of autophagy and regulated CS-induced airway inflammation. The results demonstrated that the LC3II/I in lung tissues of mice were increased after CS exposure for 4 weeks, which could be reverted by silymarin treatment in a dose-dependent manner (Fig. 7A,B). Moreover, CS-induced elevation of KC in BALF, along with TNF-α, could be decreased by silymarin pretreatment (Fig. 7C).
Figure 7. Effect of silymarin on autophagy activation and inflammatory cytokine release in mice lungs.
(A) Expressions of LC3I and LC3II were measured by Western Blot. (B) Densitometry was performed and the ratio of LC3II/I were calculated. (C) The secretion of TNF-α, IL-6 and KC in BALF were detected by ELISA. Results are representative of three independent experiments. Values are expressed as mean ± SEM (n = 3). *P < 0.05 with respect to the control group; #P < 0.05 with respect to the CSE-exposed group.
Discussion
Silymarin administration has been used for various inflammatory conditions. The therapeutic potential of silymarin in several pulmonary disease models has recently attracted attention. Silymarin reduced sepsis-induced oxidative damage in lung tissues and inhibited the release of inflammatory mediators31. Silibinin, the main component of silymarin, can also protect against OVA-induced airway inflammation by downregulating pro-inflammatory and Th2 cytokines32. A more recent paper reported that silymarin was able to increase ciliary beat frequency and mucociliary clearance in vivo33. Consistent with our previous study in CS-exposed mice which showed that silymarin could dampen airway inflammatory responses and oxidative stress, the present study further demonstrated its anti-inflammatory effect and the possible underlying mechanisms in CSE-induced Beas-2B cells, which may be more related to human COPD as bronchial epithelial cells play a crutial role in disease pathogenesis.
Autophagy is initiated by induction of several autophagy genes including those that express microtubule-associated protein light chain 3 (LC3), Beclin-1, and other autophagy-related proteins, playing a critical role in the maintenance of intracellular homeostasis under both physiological and pathological conditions34,35,36. Increasing evidence has suggested cross-talk between autophagy and apoptosis in lung epithelial cells subjected to CSE treatment37. Down-regulation of the activated autophagic and apoptotic pathways controls lung cell death and the emphysema development in mice exposed to CS13,14,38. Although autophagy is recently emerging as an important process for the regulation of innate immunity and inflammation, the role of autophagy in the mechanisms of lung inflammation has been controversial with studies that have suggested both protective and injurious aspects. Rescue of dysfunctional autophagy has been shown to play a protective role against hyperinflammatory responses from cystic fibrosis cells39, while autophagy deficiency in alveolar macrophages resulted in greater silica-induced inflammation40. In contrast, inhibition of autophagy in lung macrophages abolished mechanical ventilation-induced nucleotide-binding oligomerization domain-like receptor containing pyrin domain 3 (NLRP3) inflammasome activation and lung inflammatory injury41. In avian influenza A-infected lung epithelial cells, blocking autophagy not only reduced the cell death42,43, but also decreased the production of proinflammatory cytokines43. Besides, genetic blockage of autophagy could markedly reduce PM2.5-induced airway inflammation and mucus hyperproduction44. Whether autophagy is protective or detrimental in lung inflammation seems to depend on the extent of its activation, specific stimuli/circumstances, and specific cell type. Consistent with the latter studies, our data implied that autophagy was deleterious in CSE-induced inflammation in bronchial epithelia, as autophagy inhibition resulted in attenuated proinflammatory cytokine release. These findings were further supported by a recent study showing that inhibition of autophagy by 14, 15-epoxyeicosatrienoic acid (EET) suppressed CS-induced inflammation in Beas-2B cells45. The anti-inflammatory effect of inhibiting autophagy may be exerted by the subsequent accumulation of p62 and ubiquitinated proteins46, which then promotes translocation of Nrf2 into the nucleus47 and regulates antioxidant defenses in inflammatory lung diseases. Thus, targeting autophagy signaling (i.e. silymarin administration or 14, 15-EET) in airway epithelial cells might be effective for CS-induced airway inflammation and exacerbation of various chronic lung diseases.
MAPKs, including p38, ERK and c-Jun N-terminal kinase (JNK), are members of a ubiquitous protein serine/threonine kinase family responsible for signal transduction in eukaryotic organisms48,49,50. It is well established that MAPK activation is implicated in the production of many inflammatory mediators present in the COPD lung or CSE-induced airway epithelial cells51,52,53. Some reports have demonstrated that MAPK pathway could regulate the level of autophagy. Addition of the MEK/ERK inhibitor U0126, blocked the dissociation of Bcl-2–Beclin1 in the setting of enhanced autophagy, suggesting that ERK1/2-mediated phosphorylation of Bcl-2 regulated starvation-induced autophagy30. ERK1/2 can be also suppressed by amino acids to inhibit autophagy29. Moreover, p38 MAPK inhibitor was shown to directly abrogate palmitate-induced conversion of LC3I to LC3II28. Our data revealed that suppression of ERK phosphorylation reduced autophagy in CSE-treated Beas-2B cells, implying a positively regulating role of ERK MAPK in CSE-induced autophagy. Meanwhile, we found that inhibition of autophagy led to a decreased level of ERK phosphorylation, suggesting that CSE-induced inflammatory response in Beas-2B cells might be induced and strengthened by a positive feedback loop of ERK MAPK pathway and autophagy activation. Although the complex interactions exist between ERK MAPK and autophagy, it is speculated that ERK-mTOR signaling may play a role in regulating autophagy. Most of autophagy induction conditions such as low cellular energy levels have been shown to inhibit mTOR activity, suggesting a tight, inverse coupling of autophagy induction and mTOR activation54. Wang et al. reported that transiently or moderately activated MEK/ERK inhibited mTOR activity, so as to cause moderately enhanced Beclin-1 and cytoprotective autophagy, while sustained MEK/ERK activation caused cytodestructive autophagy55. However, reports concerning the direct effect of autophagy on the activation of ERK are limited. One recent study by Martinez-Lopez et al. showed that deleting Atg7 or Atg5 or blocking LC3 lipidation or Atg5-Atg12 conjugation decreased ERK phosphorylation56, indicating that the cellular availability of autophagic structures could determine the degree of ERK phosphorylation. In their study, it was speculated that LC3-II-positive membranes and Atg5-Atg12-positive preautophagosomes might serve as cellular signaling platforms which would facilitate ERK phosphorylation by promoting coordination of the Raf-MEK-ERK cascade. Therefore, interfering this loop at the same time might achieve a better prevention against CS-induced airway inflammation.
In summary, the present research has demonstrated that CSE-induced autophagy activation in bronchial epithelia, acting in synergism with ERK MAPK pathway, may initiate and exaggerate airway inflammation. Silymarin could attenuate inflammatory responses, at least in part, via suppressing ERK/p38 MAPK pathway activity and autophagy. Thus, therapeutic improvements in CS-associated airway inflammation might be facilitated through intervening in the crosstalk between autophagy and ERK MAPK pathway, and silymarin is likely to be an ideal agent for use in inflammatory pulmonary diseases.
Additional Information
How to cite this article: Li, D. et al. Silymarin attenuates cigarette smoke extract-induced inflammation via simultaneous inhibition of autophagy and ERK/p38 MAPK pathway in human bronchial epithelial cells. Sci. Rep. 6, 37751; doi: 10.1038/srep37751 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grants 31171103, 81230001, and 81470236 to Fuqiang Wen) and National Key Technology Research and Development Program of the Ministry of Science and Technology of China during the “12th Five-Year Plan” (2012BAI05B02, 2014BAI08B04). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions D.D.L.: performed experiments, analyzed data and contributed to the writing of the manuscript. J.H.: performed experiments, analyzed data and revised the manuscript. T.W.: performed experiments, analyzed data and revised the manuscript. X.Z.: performed experiments and analyzed data. L.L.: performed experiments. H.W.: performed experiments. Y.Q.W.: analyzed data. D.X.: contributed reagents. F.Q.W.: designed the study, analyzed data and took the lead in writing the manuscript.
References
- Chung K. F. & Adcock I. M. Multifaceted mechanisms in COPD: inflammation, immunity, and tissue repair and destruction. The European respiratory journal 31, 1334–1356, doi: 10.1183/09031936.00018908 (2008). [DOI] [PubMed] [Google Scholar]
- Thorley A. J. & Tetley T. D. Pulmonary epithelium, cigarette smoke, and chronic obstructive pulmonary disease. International journal of chronic obstructive pulmonary disease 2, 409–428 (2007). [PMC free article] [PubMed] [Google Scholar]
- de Vries M. et al. Pim1 kinase protects airway epithelial cells from cigarette smoke-induced damage and airway inflammation. American journal of physiology. Lung cellular and molecular physiology 307, L240–251, doi: 10.1152/ajplung.00156.2013 (2014). [DOI] [PubMed] [Google Scholar]
- Xie Z. & Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nature cell biology 9, 1102–1109, doi: 10.1038/ncb1007-1102 (2007). [DOI] [PubMed] [Google Scholar]
- Wang K. Autophagy and apoptosis in liver injury. Cell cycle 14, 1631–1642, doi: 10.1080/15384101.2015.1038685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456, 264–268, doi: 10.1038/nature07383 (2008). [DOI] [PubMed] [Google Scholar]
- Chang C. P., Su Y. C., Hu C. W. & Lei H. Y. TLR2-dependent selective autophagy regulates NF-kappaB lysosomal degradation in hepatoma-derived M2 macrophage differentiation. Cell death and differentiation 20, 515–523, doi: 10.1038/cdd.2012.146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K. et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology 12, 222–230, doi: 10.1038/ni.1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razani B. et al. Autophagy links inflammasomes to atherosclerotic progression. Cell metabolism 15, 534–544, doi: 10.1016/j.cmet.2012.02.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. P. et al. Participation of autophagy in acute lung injury induced by seawater. Experimental lung research 39, 441–452, doi: 10.3109/01902148.2013.845626 (2013). [DOI] [PubMed] [Google Scholar]
- Pham D. L. et al. Association of autophagy related gene polymorphisms with neutrophilic airway inflammation in adult asthma. The Korean journal of internal medicine 31, 375–385, doi: 10.3904/kjim.2014.390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. W. et al. Cigarette smoke-induced autophagy is regulated by SIRT1-PARP-1-dependent mechanism: implication in pathogenesis of COPD. Archives of biochemistry and biophysics 500, 203–209, doi: 10.1016/j.abb.2010.05.013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. H. et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PloS one 3, e3316, doi: 10.1371/journal.pone.0003316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. H. et al. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proceedings of the National Academy of Sciences of the United States of America 107, 18880–18885, doi: 10.1073/pnas.1005574107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij N., Chandramani P., Westphal C. V., Hole R. & Bodas M. Cigarette smoke induced autophagy-impairment accelerates lung aging, COPD-emphysema exacerbations and pathogenesis. American journal of physiology. Cell physiology, ajpcell 00110 02016, doi: 10.1152/ajpcell.00110.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan S. M., Lam H. C., Ryter S. W. & Choi A. M. “Ciliophagy”: The consumption of cilia components by autophagy. Autophagy 10, 532–534, doi: 10.4161/auto.27641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H. C. et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest 123, 5212–5230, doi: 10.1172/JCI69636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z. et al. Silymarin protects against acute ethanol-induced hepatotoxicity in mice. Alcoholism, clinical and experimental research 30, 407–413, doi: 10.1111/j.1530-0277.2006.00063.x (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazak R., Walterova D. & Kren V. Silybin and silymarin–new and emerging applications in medicine. Current medicinal chemistry 14, 315–338 (2007). [DOI] [PubMed] [Google Scholar]
- Li D. et al. Silymarin attenuates airway inflammation induced by cigarette smoke in mice. Inflammation 38, 871–878, doi: 10.1007/s10753-014-9996-9 (2015). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. Silibinin protects murine fibroblast L929 cells from UVB-induced apoptosis through the simultaneous inhibition of ATM-p53 pathway and autophagy. The FEBS journal 280, 4572–4584, doi: 10.1111/febs.12426 (2013). [DOI] [PubMed] [Google Scholar]
- Dai J. P. et al. Identification of 23-(s)-2-amino-3-phenylpropanoyl-silybin as an antiviral agent for influenza A virus infection in vitro and in vivo. Antimicrobial agents and chemotherapy 57, 4433–4443, doi: 10.1128/AAC.00759-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L. et al. WNT/beta-catenin signaling regulates cigarette smoke-induced airway inflammation via the PPARdelta/p38 pathway. Laboratory investigation; a journal of technical methods and pathology 96, 218–229, doi: 10.1038/labinvest.2015.101 (2016). [DOI] [PubMed] [Google Scholar]
- Kabeya Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal 19, 5720–5728, doi: 10.1093/emboj/19.21.5720 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. H. et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336, 225–228, doi: 10.1126/science.1218395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. et al. Autophagy inhibits reactive oxygen species-mediated apoptosis via activating p38-nuclear factor-kappa B survival pathways in oridonin-treated murine fibrosarcoma L929 cells. The FEBS journal 276, 1291–1306, doi: 10.1111/j.1742-4658.2008.06864.x (2009). [DOI] [PubMed] [Google Scholar]
- Kroemer G., Marino G. & Levine B. Autophagy and the integrated stress response. Molecular cell 40, 280–293, doi: 10.1016/j.molcel.2010.09.023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. Palmitate promotes autophagy and apoptosis through ROS-dependent JNK and p38 MAPK. Biochemical and biophysical research communications 463, 262–267, doi: 10.1016/j.bbrc.2015.05.042 (2015). [DOI] [PubMed] [Google Scholar]
- Pattingre S., Bauvy C. & Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. The Journal of biological chemistry 278, 16667–16674, doi: 10.1074/jbc.M210998200 (2003). [DOI] [PubMed] [Google Scholar]
- Tang D. et al. Endogenous HMGB1 regulates autophagy. The Journal of cell biology 190, 881–892, doi: 10.1083/jcb.200911078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toklu H. Z. et al. Silymarin, the antioxidant component of Silybum marianum, prevents sepsis-induced acute lung and brain injury. The Journal of surgical research 145, 214–222, doi: 10.1016/j.jss.2007.03.072 (2008). [DOI] [PubMed] [Google Scholar]
- Choi Y. H. et al. Silibinin attenuates allergic airway inflammation in mice. Biochemical and biophysical research communications 427, 450–455, doi: 10.1016/j.bbrc.2012.07.112 (2012). [DOI] [PubMed] [Google Scholar]
- Boeckenholt C., Begrow F. & Verspohl E. J. Effect of silymarin and harpagoside on inflammation reaction of BEAS-2B cells, on ciliary beat frequency (CBF) of trachea explants and on mucociliary clearance (MCC). Planta medica 78, 761–766, doi: 10.1055/s-0031-1298374 (2012). [DOI] [PubMed] [Google Scholar]
- Green D. R., Galluzzi L. & Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333, 1109–1112, doi: 10.1126/science.1201940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N. & Virgin H. W. Autophagy in immunity and inflammation. Nature 469, 323–335, doi: 10.1038/nature09782 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Kang R., Zeh H. J. 3rd, Lotze M. T. & Tang D. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy 9, 451–458, doi: 10.4161/auto.23691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. P. et al. Autophagic proteins regulate cigarette smoke-induced apoptosis: protective role of heme oxygenase-1. Autophagy 4, 887–895 (2008). [DOI] [PubMed] [Google Scholar]
- An C. H. et al. TLR4 deficiency promotes autophagy during cigarette smoke-induced pulmonary emphysema. American journal of physiology. Lung cellular and molecular physiology 303, L748–757, doi: 10.1152/ajplung.00102.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M. L. et al. Rescue of dysfunctional autophagy attenuates hyperinflammatory responses from cystic fibrosis cells. Journal of immunology 190, 1227–1238, doi: 10.4049/jimmunol.1201404 (2013). [DOI] [PubMed] [Google Scholar]
- Jessop F., Hamilton R. F., Rhoderick J. F., Shaw P. K. & Holian A. Autophagy deficiency in macrophages enhances NLRP3 inflammasome activity and chronic lung disease following silica exposure. Toxicology and applied pharmacology, doi: 10.1016/j.taap.2016.08.029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu G., Dull R. O., Schwartz D. E. & Hu G. Autophagy in pulmonary macrophages mediates lung inflammatory injury via NLRP3 inflammasome activation during mechanical ventilation. American journal of physiology. Lung cellular and molecular physiology 307, L173–185, doi: 10.1152/ajplung.00083.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Sun Q., Mi R. & Zhang H. Avian influenza A virus H5N1 causes autophagy-mediated cell death through suppression of mTOR signaling. Journal of genetics and genomics = Yi chuan xue bao 38, 533–537, doi: 10.1016/j.jgg.2011.10.002 (2011). [DOI] [PubMed] [Google Scholar]
- Sun Y. et al. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Science signaling 5, ra16, doi: 10.1126/scisignal.2001931 (2012). [DOI] [PubMed] [Google Scholar]
- Chen Z. H. et al. Autophagy Is Essential for Ultrafine Particle-Induced Inflammation and Mucus Hyperproduction in Airway Epithelium. Autophagy 0, doi: 10.1080/15548627.2015.1124224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. 14, 15-epoxyeicosatrienoic acid suppresses cigarette smoke condensate-induced inflammation in lung epithelial cells by inhibiting autophagy. American journal of physiology. Lung cellular and molecular physiology, ajplung 00161 02016, doi: 10.1152/ajplung.00161.2016 (2016). [DOI] [PubMed] [Google Scholar]
- Fujii S. et al. Insufficient autophagy promotes bronchial epithelial cell senescence in chronic obstructive pulmonary disease. Oncoimmunology 1, 630–641, doi: 10.4161/onci.20297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Molecular and cellular biology 30, 3275–3285, doi: 10.1128/MCB.00248-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Harper R., Barchowsky A. & Di Y. P. Identification of multiple MAPK-mediated transcription factors regulated by tobacco smoke in airway epithelial cells. American journal of physiology. Lung cellular and molecular physiology 293, L480–490, doi: 10.1152/ajplung.00345.2006 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G. L. & Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298, 1911–1912, doi: 10.1126/science.1072682 (2002). [DOI] [PubMed] [Google Scholar]
- Huang C., Jacobson K. & Schaller M. D. MAP kinases and cell migration. Journal of cell science 117, 4619–4628, doi: 10.1242/jcs.01481 (2004). [DOI] [PubMed] [Google Scholar]
- Renda T. et al. Increased activation of p38 MAPK in COPD. The European respiratory journal 31, 62–69, doi: 10.1183/09031936.00036707 (2008). [DOI] [PubMed] [Google Scholar]
- Mehra D., Geraghty P. M., Hardigan A. A. & Foronjy R. A comparison of the inflammatory and proteolytic effects of dung biomass and cigarette smoke exposure in the lung. PloS one 7, e52889, doi: 10.1371/journal.pone.0052889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffey K., Reynolds S., Plumb J., Kaur M. & Singh D. Increased phosphorylated p38 mitogen-activated protein kinase in COPD lungs. The European respiratory journal 42, 28–41, doi: 10.1183/09031936.00170711 (2013). [DOI] [PubMed] [Google Scholar]
- Kim Y. C. & Guan K. L. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 125, 25–32, doi: 10.1172/JCI73939 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. R. et al. A Non-canonical MEK/ERK Signaling Pathway Regulates Autophagy via Regulating Beclin 1. Journal of Biological Chemistry 284, 21412–21424, doi: 10.1074/jbc.M109.026013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N., Athonvarangkul D., Mishall P., Sahu S. & Singh R. Autophagy proteins regulate ERK phosphorylation. Nat Commun 4, doi: Artn 279910.1038/Ncomms3799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.