Abstract
A carbohydrate esterase called glucuronoyl esterase (GE) was discovered 10 years ago in a cellulolytic system of the wood-rotting fungus Schizophyllum commune. Genes coding for GEs were subsequently found in a number of microbial genomes, and a new family of carbohydrate esterases (CE15) has been established. The multidomain structures of GEs, together with their catalytic properties on artificial substrates and positive effect on enzymatic saccharification of plant biomass, led to the view that the esterases evolved for hydrolysis of the ester linkages between 4-O-methyl-d-glucuronic acid of plant glucuronoxylans and lignin alcohols, one of the crosslinks in the plant cell walls. This idea of the function of GEs is further supported by the effects of cloning of fungal GEs in plants and by very recently reported evidence for changes in the size of isolated lignin-carbohydrate complexes due to uronic acid de-esterification. These facts make GEs interesting candidates for biotechnological applications in plant biomass processing and genetic modification of plants. This article is a brief summary of current knowledge of these relatively recent and unexplored esterases.
INTRODUCTION
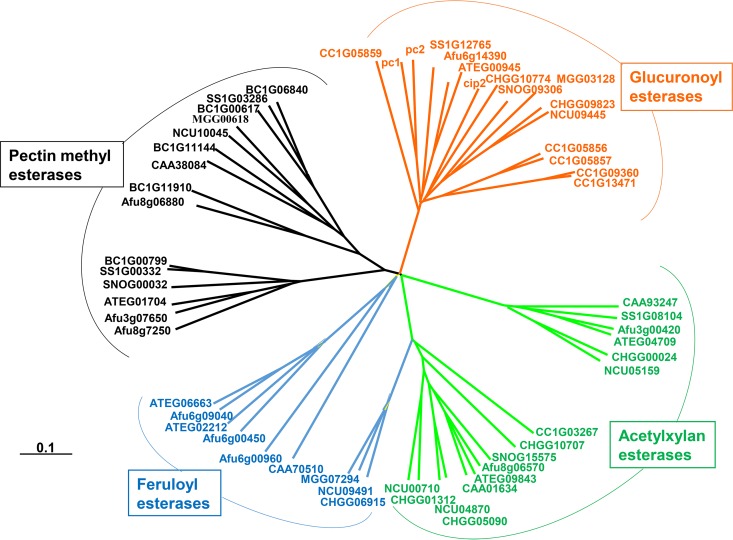
One of the crosslinks in plant cell walls contributing to their recalcitrance is the ester linkage between 4-O-methyl-d-glucuronic acid (MeGlcA) residues of xylans and hydroxyl groups of lignin alcohols (1–4). This linkage does not survive alkaline pretreatments of plant biomass; therefore, it contributes to enzymatic resistance of only plant material pretreated under nonalkaline conditions, such as steam explosion. Ten years ago, we published the first evidence for the existence of an esterase that could cleave such linkages. The enzyme, found in the cellulolytic system of the wood-rotting fungus Schizophyllum commune, was capable of hydrolyzing various synthetic unnatural esters of 4-O-methyl-d-glucuronic acid (MeGlcA) and glucuronic acid (GlcA) and was named glucuronoyl esterase (GE) (5). The sequence of the first gene coding for GE was identified in Trichoderma reesei (Hypocrea jecorina) (6). The T. reesei GE gene was found to be identical with gene cip2 of unknown function described earlier by Genencor (7) and was claimed by the authors to code for a protein stimulating enzymatic saccharification of plant cell walls (8). Cip2, containing a carbohydrate binding module (CBM), CBM1 (9, 10), is a protein inducible by cellulose and sophorose. Occurrence of similar genes in fungal and bacterial genomes resulted in the introduction of new carbohydrate esterase (CE) family CE15 (6) in the carbohydrate-active enzyme (CAZy) classification (11). The GE sequences were unique and phylogenetically distant from those of other carbohydrate esterases, acetylxylan esterases (CE1 and CE5), feruloyl esterases (CE1 and the putative CE3), and pectin methyl esterases (CE8) (Fig. 1) (12). GEs not only appear to be inducible constituents of plant cell wall-degrading enzyme systems but also are frequently constituents of bi- or multimodular enzymes. Some of them, such as GE from T. reesei and Podospora anserina, contain a family 1 carbohydrate binding module, CBM1 (6, 13). Phanerochaete chrysosporium produces two forms, one without and one with the CBM1 module (12). The catalytic domains of the Ph. chrysosporium GEs are almost identical; however, expression of their genes appears to be mediated by different forms of regulatory control (12). Later studies reported that the genome of this white-rot fungus and also the genome of its close relative Phanerochaete carnosa each contain three GE genes, two of which code for CBM-containing enzymes (14, 15). However, the majority of the genomes of white-rot fungi contain two GE genes whereas the genomes of brown-rot fungi contain usually only one CE15 gene (15, 16). It is also worth mention that not all fungal genomes contain GE genes (12) and that basidiomycetes have on average more genes in CE15 than do aspergilli (16). Further biochemical studies are needed to understand the significance of this uneven GE gene distribution in microbial wood decay. Such studies will probably show that, although they belong to the same CE family, the enzymes may differ in catalytic properties and physiological function.
FIG 1.
Phylogenetic tree of confirmed and putative GEs, acetylxylan esterases, feruloyl esterases, and pectin methyl esterases, constructed by rearrangement of the published data (9). The protein sequences are marked by accession numbers given in databases.
GEs are also present in cellulosomes, such as the enzyme from Ruminococcus flavefaciens. In this bacterium, GE occurs in a bifunctional enzyme in combination with a catalytic module of an acetylxylan esterase (17). In Teredinibacter turnerae, a shipworm gut bacterium, GE is connected with endo-β-1,4-xylanase of glycoside hydrolase (GH) family 11 (18). In both cases, the two enzymes are tightly functionally bound. These data support the view that GEs play an important role in microbial breakdown of plant cell walls.
STRUCTURE AND PROPOSED MODE OF ACTION
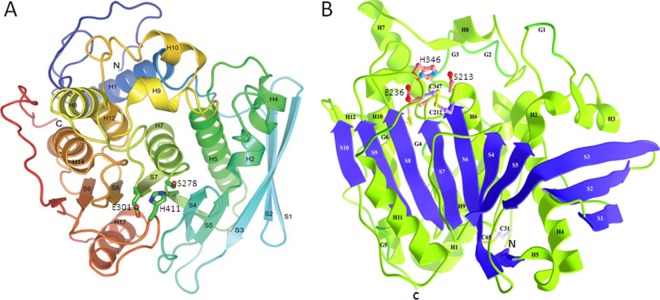
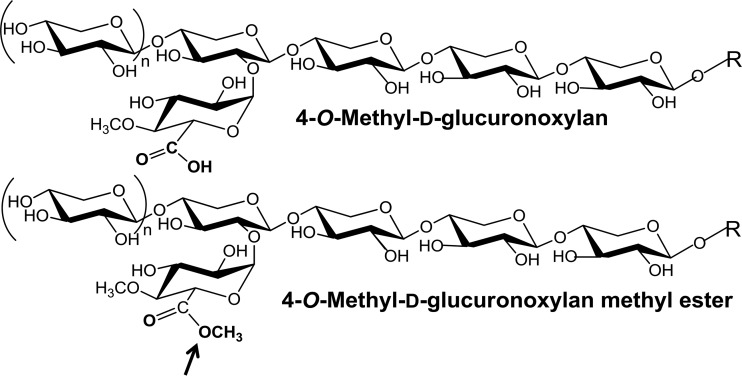
The first three-dimensional (3D) structure of GE was elucidated on the catalytic domain of the T. reesei GE (Cip2), which was homologously overexpressed using a cellobiohydrolase promoter and purified from the growth medium (6, 19). The structure has an α/β-hydrolase fold with an overall αβα-sandwich architecture, as shown in Fig. 1 (19). The twisted β-sheet is sandwiched between two layers of α-helices with the catalytic triad Ser-His-Glu exposed on the protein surface. A similar 3D structure was reported for GE from Myceliophthora thermophila (earlier assigned as Sporotrichum thermophile) (20) (Fig. 1). A mutation of catalytic serine to alanine in M. thermophila GE abolished the enzyme activity (21) but did not affect its active site architecture (20). The mutant was also successfully crystallized with methyl 4-O-methyl-d-glucopyranuronate, an artificial GE substrate (5), to reveal the topology of the active site (20). In contrast to the majority of serine type esterases, glutamic acid replaces aspartic acid in known GEs. The active site on the surface of the protein has implications for the ability of the enzyme to hydrolyze the ester bonds between large molecules in the plant cell walls. This is supported by experimental evidence indicating that GEs can de-esterify the methyl ester of beech wood glucuronoxylan, a synthetic substrate with methyl-esterified uronic acids linked to the polymeric carbohydrate chain (Fig. 2) (22). The de-esterification of this substrate by GEs can easily be followed by 1H-nuclear magnetic resonance (1H-NMR) spectroscopy (22). The GE-catalyzed hydrolysis of synthetic substrates shown in Fig. 3 demonstrates the tolerance of GEs for larger aryl propane structures on the side of lignin alcohols (Fig. 4) (23–25). It is worth noting that the esterification of glucuronoxylan with methanol reduces its solubility. This indicates that a similar esterification of the polysaccharide MeGlcA with aryl alkyl alcohols as “more natural” GE substrates would lead to water-insoluble products. These considerations suggest that, during microbial attack of plant cell walls, GEs most probably operate on a phase boundary between highly hydrated partially acetylated hemicellulose and hydrophobic lignin structures.
FIG 2.
3D structures of the catalytic domain of GE from T. reesei (19) (A) and GE from M. thermophila (20) (B). Both structures show the twisted β-sheet structure sandwiched between two layers of α-helixes. Strands are labeled with the letter “S” followed by a number, helices with the letter “H” followed by a number, C termini with “C,” and N termini with “N.” The catalytic triad Ser, His, and Glu, shown in balls and sticks (S278, H411, and E301 in T. reesei GE; S213, H346, and E236 in M. thermophila GE), is in both cases located on the surface of the enzyme.
FIG 3.
Fragment of alkali-extracted beech wood glucuronoxylan and its methyl ester serving as the substrate of GEs (22). The site of attack is marked by an arrow.
FIG 4.
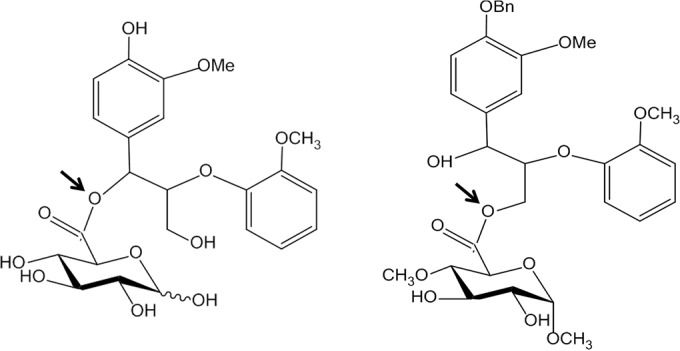
Esters of GlcA (left) and MeGlcA (right) methyl glycoside with the largest aryl propane structures found to be hydrolyzed by GEs so far (23–25). The sites of attack are marked by arrows.
GEs—ESTERASES WITH UNEXPLORED BIOTECHNOLOGICAL POTENTIAL
Surprisingly, a real physiological role of GEs in natural substrates was demonstrated for the first time just before submission of this article. A recombinant GE from Acremonium alcalophilum reduced the molecular mass of isolated lignin-carbohydrate complexes from spruce and birch, as shown by size exclusion chromatography (26). Simultaneous analyses of hydroxyl groups before and after GE treatment of subsequently phosphitylated derivatives of the complexes by 31P-NMR spectroscopy (27) showed an increase in the content of the carboxyl hydroxyl groups as a result of de-esterification of uronic acids (26). The enzymes certainly play an important role in plant cell wall degradation since their addition to a commercial cellulolytic system from Novozymes considerably enhanced the amount of liberated reducing sugars, including both pentoses and hexoses (25). With the Novozymes cellulase CellicCTec fortified with β-xylosidase, the presence of GE led to a 25% increase in the amount of pentoses released from a pretreated corn fiber (25). Additional indirect evidence for a possible role of GEs in plant cell wall degradation has so far been obtained by cloning fungal GE genes in plants (28, 29). Expression of a fungal GE in Arabidopsis or aspen (Populus tremula) alters plant cell wall composition and architecture, improves extractability of xylan from plant biomass, and enhances enzymatic cellulose digestibility (28, 29). These effects were ascribed to reduced cell wall ester crosslinks leading to increased lignin deposition. The greatest improvement in cell wall fractionation was achieved through expression of fungal GEs under the control of a developmentally regulated promoter, e.g., a promoter specifically active in the plant cell wall during secondary cell wall deposition (30). Such an approach applied to tobacco plant biomass resulted in a significant increase of the enzymatic saccharification yields and became a subject of a patent of FuturaGene Ltd. (Israel and Brazil), claiming again a decrease of lignin-hemicellulose ester crosslinks in the cell walls of plants expressing fungal GEs (30). One line of eucalyptus genetically modified by fungal GE was reported to produce much stronger wood (communicated by E. R. Gonzales, FuturaGene Brazil, at the 2nd Brazilian BioEnergy Science and Technology Conference, Campos de Jõrdao, Brazil, October 2014). To my knowledge, these are the first examples of biotechnological significance of these relatively very recent and not extensively studied microbial carbohydrate esterases. Many other possible applications, such as those involving their effects on delignification and bleaching of sulfite and hydrothermal pulps, have not been examined. Reverse and transesterification reactions, which could lead to high-value products derived from plant constituents, remain unexamined. Such catalytic properties should not be dismissed, because some types of deacetylases catalyze efficient transesterification reactions to carbohydrates in aqueous medium (31, 32).
SUBSTRATES TO PROMOTE RESEARCH OF GEs
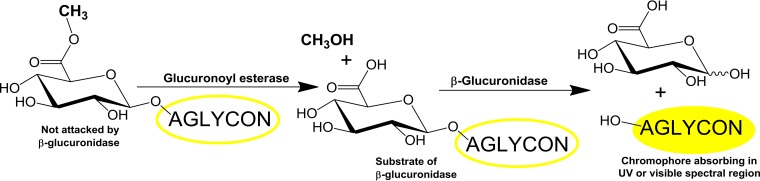
Studies of GEs are hampered by unavailability of proper substrates. Many synthetic substrates, such as alkyl and alkyl aryl alcohol esters of MeGlcA and GlcA or their glycosides used earlier (5, 12, 21, 33–35), are not commercially available. Natural substrates derived from plant cell wall material, such as suitable lignin-carbohydrate complexes, await their introduction. The isolation, purification, and characterization of such complexes, as well as the synthesis of their analogues, are not easy tasks (4, 26). Benzyl glucuronate, which is the only synthetic substrate commercially available, was recently suggested for both qualitative and quantitative GE assays, including a uronic acid dehydrogenase-coupled assay (36). The drawback of the coupled assay is the fact that uronic acid dehydrogenase accepts only the β-anomer of the de-esterified GlcA (37). Fortunately, there is continuous progress in methods to study GEs. We have introduced new β-glucuronidase-coupled assays using easily synthesized prochromogenic substrates (38). The assays are based on the facts that the GEs do not differentiate esters of α- or β-glucuronides (34) and that β-glucuronidases are available in the market. The substrates are methyl esters of commercially available 4-nitrophenyl and 5-bromo-4-chloro-3-indolyl β-glucuronides. They can be prepared simply by cation-exchanger-catalyzed esterification in dry methanol followed by evaporation of the solvent. β-Glucuronidases do not hydrolyze esterified glycosides. Thus, the coupling of the action of GE with β-glucuronidase leads to release of the chromophore aglycones (Fig. 5). The assays are suitable for microplate setup and also for high-throughput screenings of genomic libraries. Particularly convenient is the substrate with the indolyl aglycon, release of which leads to the blue indigo type product. The substrates enabled us to examine for the first time the presence of GE in commercial cellulolytic and hemicellulolytic preparations (38). However, as in the similar case of benzyl glucuronate, the lack of the 4-O-methyl group in GlcA decreases the affinity of the enzymes for the substrates (5, 12, 33–35). It is obvious that methyl esters of analogous glycosides of MeGlcA could serve as ideal substrates. However, their preparation requires a considerable amount of MeGlcA. Its chemical synthesis (39, 40) is certainly a better way to achieve larger quantities than enzymatic liberation from glucuronoxylan by α-glucuronidase.
FIG 5.
Scheme of the β-glucuronidase-coupled assay of glucuronoyl esterase on methyl esters of d-glucuronic acid β-glycosylated with chromophore aglycons. Thus far, the tested substrates have been found to contain 4-nitrophenol and 5-bromo-4-chloro-3-hydroxyindol as aglycons (38).
CONCLUSIONS
The number of papers dedicated to GEs since their discovery remains still very limited. However, the recent evidence that the addition of GEs to saccharification enzyme systems improves sugar yields from pretreated plant biomass (25, 30), as well as the observation that GEs are present in commercial enzyme preparations (38), underlines the need to elucidate further the role of GEs in microbial degradation of plant cell walls and other biotechnological potential, particularly in biorefinery and saccharification processes. This is stressed by the recent observation that lignin-hemicellulose esters are present in both hardwood and softwood (26). As indicated by patent literature on GEs (30), new opportunities can be foreseen in the area of genetic modification of plants by expression of microbial GE genes. However, further progress in this area is dependent on the availability of new simple and sensitive assays for screening of genomic libraries for GEs with specific properties. Such substrates are not commercially available, and those which have so far been suggested suffer from the lack of the 4-O-methyl group on the uronic acid moiety recognized by GEs. Therefore, the synthesis of esters of MeGlcA instead of GlcA, and their commercialization, is an interesting challenge for organic chemists.
ACKNOWLEDGMENTS
I am grateful to colleagues at home and abroad who contributed to current knowledge of glucuronoyl esterases.
Support by the grant from the Slovak Research and Development Agency APVV-0602-12 is greatly appreciated.
REFERENCES
- 1.Das NN, Das SC, Mukherjee AK. 1984. On the ester linkage between lignin and 4-O-methyl-d-glucurono-d-xylan in jute fiber (Corchorus capsularis). Carbohydr Res 127:345–348. doi: 10.1016/0008-6215(84)85369-0. [DOI] [Google Scholar]
- 2.Watanabe T, Koshijima T. 1988. Evidence for an ester linkage between lignin and glucuronic acid in lignin-carbohydrate complexes by DDQ-oxidation. Agric Biol Chem 52:2953–2955. doi: 10.1080/00021369.1988.10869116. [DOI] [Google Scholar]
- 3.Jeffries T. 1990. Biodegradation of lignin-carbohydrate complexes. Biodegradation 1:163–176. doi: 10.1007/BF00058834. [DOI] [Google Scholar]
- 4.Balashkin MY, Capanema EA, Chang H. 2007. MWL fraction with a high concentration of lignin-carbohydrate linkages: isolation and 2D NMR spectroscopic analysis. Holzforschung 61:1–7. doi: 10.1515/HF.2007.001. [DOI] [Google Scholar]
- 5.Spániková S, Biely P. 2006. Glucuronoyl esterase—novel carbohydrate esterase produced by Schizophyllum commune. FEBS Lett 580:4597–4601. doi: 10.1016/j.febslet.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 6.Li X-L, Spániková S, deVries RP, Biely P. 2007. Identification of genes encoding microbial glucuronoyl esterases. FEBS Lett 581:4029–4035. doi: 10.1016/j.febslet.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 7.Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, Meerman HJ, Mitchell T, Mitchinson C, Olivares HA, Teunissen PJM, Yao J, Ward M. 2003. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem 278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- 8.Foreman P, Goedegebuur F, Van Solingen P, Ward M. February 2010. Novel Trichoderma genes. European patent 1 627 049 B1.
- 9.Guillén D, Sánchez S, Rodríguez-Sanoja R. 2010. Carbohydrate-binding domains: multiplicity of biological roles. Appl Microbiol Biotechnol 85:1241–1249. doi: 10.1007/s00253-009-2331-y. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert HJ, Knox JP, Boraston AB. 2013. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol 23:669–677. doi: 10.1016/j.sbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duranová M, Spániková S, Wösten HOB, Biely P, de Vries RP. 2009. Two glucuronoyl esterases of Phanerochaete chrysosporium. Arch Microbiol 191:133–140. doi: 10.1007/s00203-008-0434-y. [DOI] [PubMed] [Google Scholar]
- 13.Katsimpouras C, Bénarouche A, Navarro D, Karpuss M, Dimarogona M, Berrin J-G, Christakopoulos P, Topakas E. 2014. Enzymatic synthesis of model substrates recognized by glucuronoyl esterases from Podospora anserina and Myceliophthora thermophila. Appl Microbiol Biotechnol 98:5507–5516. doi: 10.1007/s00253-014-5542-9. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H, MacDonald J, Syed K, Salamov A, Hori C, Aerts A, Henrissat B, Wiebenga A, VanKuyk PA, Barry K, Lindquist E, LaButti K, Lapidus A, Lucas S, Coutinho P, Gong Y, Samejima M, Mahadevan R, Abou-Zaid M, de Vries RP, Igarashi K, Yadav JS, Grigoriev IV, Master ER. 2012. Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P. chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC Genomics 13:444. doi: 10.1186/1471-2164-13-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hori C, Gaskell J, Igarashi K, Samejima M, Hibbett D, Henrissat B, Cullen D. 2013. Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia 105:1412–1427. doi: 10.3852/13-072. [DOI] [PubMed] [Google Scholar]
- 16.Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR. 2014. Plant-Polysaccharide-degrading enzymes from basidiomycetes. Microbiol Mol Biol Rev 78:614–649. doi: 10.1128/MMBR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aurilia V, Martin JC, McCrae SI, Scott KP, Rincon MT, Flint HJ. 2000. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology 146:1391–1397. doi: 10.1099/00221287-146-6-1391. [DOI] [PubMed] [Google Scholar]
- 18.Yang JC, Madupu R, Durkin AS, Ekborg NA, Pedamallu CS, Hostetler JB, Radune D, Toms BS, Henrissat B, Coutinho PM, Schwarz S, Field L, Trindade-Silva AE, Soares CA, Elshahawi S, Hanora A, Schmidt EW, Haygood MG, Posfai J, Benner J, Madinger C, Nove J, Anton B, Chaudhary K, Foster J, Holman A, Kumar S, Lessard PA, Luyten YA, Slatko B, Wood N, Wu B, Teplitski M, Mougous JD, Ward N, Eisen JA, Badger JH, Distel DL. 2009. The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (shipworms). PLoS One 4:e6085. doi: 10.1371/journal.pone.0006085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pokkuluri PR, Duke NEC, Wood SJ, Cotta MA, Li X-L, Biely P, Schiffer M. 2011. Structure of the catalytic domain of glucuronoyl esterase, Cip2 from Hypocrea jecorina. Proteins 79:2588–2592. doi: 10.1002/prot.23088. [DOI] [PubMed] [Google Scholar]
- 20.Charavgi M-D, Dimarogona M, Topakas E, Christakopoulos P, Chrysina ED. 2013. The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst. Acta Cryst D 69:63–73. doi: 10.1107/S0907444912042400. [DOI] [PubMed] [Google Scholar]
- 21.Topakas E, Moukouli M, Dimarogona M, Vafiadi C, Christakopoulos P. 2010. Functional expression of a thermophilic glucuronoyl esterase from Sporotrichum thermophile: identification of the nucleophilic serine. Appl Microbiol Biotechnol 87:1765–1772. doi: 10.1007/s00253-010-2655-7. [DOI] [PubMed] [Google Scholar]
- 22.Biely P, Malovíková A, Uhliariková I, Li X-L, Wong DWS. 2015. Glucuronoyl esterases are active on the polymeric substrate methyl esterified glucuronoxylan. FEBS Lett 589:2334–2339. doi: 10.1016/j.febslet.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 23.Nylander F, Sunner H, Olsson L, Christakopoulos P, Westman G. 2016. Synthesis and enzymatic hydrolysis of a diaryl benzyl ester model of a lignin-carbohydrate complex (LCC). Holzforschung 70:385–391. doi: 10.1515/hf-2014-0347. [DOI] [Google Scholar]
- 24.Li K, Helm RF. 1995. Synthesis and rearrangement reactions of ester-linked lignin-carbohydrate model compounds. J Agric Food Chem 43:2098–2103. doi: 10.1021/jf00056a026. [DOI] [Google Scholar]
- 25.d'Errico C, Börjesson J, Ding H, Krogh KBRM, Spodsberg Madsen N, Nygaard R, Monrad R. 2016. Improved biomass degradation using fungal glucuronoyl esterases – hydrolysis of natural corn fiber substrate. J Biotechnol 219:117–123. doi: 10.1016/j.jbiotec.2015.12.024. [DOI] [PubMed] [Google Scholar]
- 26.Bååth JA, Giummarella N, Klaubauf S, Lawoko M, Olsson L. 26 July 2016. A glucuronoyl esterase from Acremonium alcalophilum cleaves native lignin-carbohydrate esters bonds. FEBS Lett doi: 10.1002/1873-3468.12290. [DOI] [PubMed] [Google Scholar]
- 27.King AWT, Zoia L, Filpponen I, Olszewska A, Haibo XIE, Kilpeläinen I, Argyropoulos DS. 2009. In situ determination of lignin phenolics and wood solubility in imidazolium chlorides using 31P NMR. J Agric Food Chem 57:8236–8243. doi: 10.1021/jf901095w. [DOI] [PubMed] [Google Scholar]
- 28.Tsai AY-L, Canam T, Gorzsas A, Mellerowicz EJ, Campbell MM, Master ER. 2012. Constitutive expression of a fungal glucuronoyl esterase in Arabidopsis reveals altered cell wall composition and structure. Plant Biotechnol J 10:1077–1087. doi: 10.1111/j.1467-7652.2012.00735.x. [DOI] [PubMed] [Google Scholar]
- 29.Latha Gandla M, Derba-Maceluch M, Liu X, Gerber L, Master ER, Mellerowicz EJ, Jönsson LJ. 2015. Expression of a fungal glucuronoyl esterase in Populus: effect on wood properties and saccharification efficiency. Phytochemistry 112:210–220. doi: 10.1016/j.phytochem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Abramson M, Shani Z, Shoseyov O. August 2013. Transgenic plants with improved saccharification yields and methods of generating same. US patent application 2013/0227724 A1.
- 31.Kremnický L, Biely P. 2005. Unique mode of acetylation of oligosaccharides in aqueous two-phase system by Trichoderma reesei acetyl esterase. J Mol Catal B Enzym 37:72–78. doi: 10.1016/j.molcatb.2005.09.011. [DOI] [Google Scholar]
- 32.Topakas E, Kyriakopoulos S, Biely P, Hirsch J, Vafiadi C, Christakopoulos P. 2010. Carbohydrate esterases of family 2 are 6-O-deacetylases. FEBS Lett 584:543–548. doi: 10.1016/j.febslet.2009.11.095. [DOI] [PubMed] [Google Scholar]
- 33.Spániková S, Poláková M, Joniak D, Hirsch J, Biely P. 2007. Synthetic esters recognized by glucuronoyl esterase from Schizophyllum commune. Arch Microbiol 188:185–189. doi: 10.1007/s00203-007-0241-x. [DOI] [PubMed] [Google Scholar]
- 34.Duranová M, Kolenová K, Hirsch J, Biely P. 2009. Fungal glucuronoyl esterases and their substrate uronic acid recognition. Biosci Biotechnol Biochem 73:2483–2487. doi: 10.1271/bbb.90486. [DOI] [PubMed] [Google Scholar]
- 35.d'Errico C, Jorgensen JO, Krogh KBRM, Spodsberg N, Madsen R, Nygaard Monrad R. 2015. Enzymatic degradation of lignin-carbohydrate complexes (LCCs): model studies using a fungal glucuronoyl esterase from Cerrena unicolor. Biotechnol Bioeng 112:914–922. doi: 10.1002/bit.25508. [DOI] [PubMed] [Google Scholar]
- 36.Sunner H, Charavgi M-D, Olsson L, Topakas E, Christakopoulos P. 2015. Glucuronoyl esterase screening and characterization assays utilizing commercially available benzyl glucuronic acid ester. Molecules 20:17807–17817. doi: 10.3390/molecules201017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkkinen T, Boer H, Janis J, Andberg M, Penttila M, Koivula A, Rouvinen J. 2011. Crystal structure of uronate dehydrogenase from Agrobacterium tumefaciens. J Biol Chem 286:27294–27300. doi: 10.1074/jbc.M111.254854.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraňová L, Puchart V, Biely P. 2016. β-Glucuronidase-coupled assays of glucuronoyl esterases. Anal Biochem 510:114–119. doi: 10.1016/j.ab.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Li K, Helm RF. 1995. A practical synthesis of methyl 4-O-methyl-α-d-glucopyranosiduronic acid. Carbohydr Res 273:249–253. doi: 10.1016/0008-6215(95)00087-A. [DOI] [Google Scholar]
- 40.Hirsch J, Langer V, Koós M. 2005. Synthesis and molecular structure of methyl 4-O-methyl-d-glucopyranuronate. Molecules 10:251–258. doi: 10.3390/10010251. [DOI] [PMC free article] [PubMed] [Google Scholar]