ABSTRACT
Chromosomal integration of heterologous metabolic pathways is optimal for industrially relevant fermentation, as plasmid-based fermentation causes extra metabolic burden and genetic instabilities. In this work, chromosomal integration was adapted for the production of mevalonate, which can be readily converted into β-methyl-δ-valerolactone, a monomer for the production of mechanically tunable polyesters. The mevalonate pathway, driven by a constitutive promoter, was integrated into the chromosome of Escherichia coli to replace the native fermentation gene adhE or ldhA. The engineered strains (CMEV-1 and CMEV-2) did not require inducer or antibiotic and showed slightly higher maximal productivities (0.38 to ∼0.43 g/liter/h) and yields (67.8 to ∼71.4% of the maximum theoretical yield) than those of the plasmid-based fermentation. Since the glycolysis pathway is the first module for mevalonate synthesis, atpFH deletion was employed to improve the glycolytic rate and the production rate of mevalonate. Shake flask fermentation results showed that the deletion of atpFH in CMEV-1 resulted in a 2.1-fold increase in the maximum productivity. Furthermore, enhancement of the downstream pathway by integrating two copies of the mevalonate pathway genes into the chromosome further improved the mevalonate yield. Finally, our fed-batch fermentation showed that, with deletion of the atpFH and sucA genes and integration of two copies of the mevalonate pathway genes into the chromosome, the engineered strain CMEV-7 exhibited both high maximal productivity (∼1.01 g/liter/h) and high yield (86.1% of the maximum theoretical yield, 30 g/liter mevalonate from 61 g/liter glucose after 48 h in a shake flask).
IMPORTANCE Metabolic engineering has succeeded in producing various chemicals. However, few of these chemicals are commercially competitive with the conventional petroleum-derived materials. In this work, chromosomal integration of the heterologous pathway and subsequent optimization strategies ensure stable and efficient (i.e., high-titer, high-yield, and high-productivity) production of mevalonate, which demonstrates the potential for scale-up fermentation. Among the optimization strategies, we demonstrated that enhancement of the glycolytic flux significantly improved the productivity. This result provides an example of how to tune the carbon flux for the optimal production of exogenous chemicals.
INTRODUCTION
Mevalonate pathways, important in the production of isoprenoids, are broadly present in eukaryotes, archaea, and some prokaryotes (1–3). Besides a precursor for the production of isoprenoids (2, 3), mevalonate has been used in cosmetics and as a building block for the production of sustainable polymers (4). Recently, it has been demonstrated that mevalonate could be transformed, by simple chemical steps, into a branched lactone β-methyl-δ-valerolactone (βMδVL), and its copolymerization with lactides results in a new class of polyesters with tunable mechanical properties (5). These potential applications motivate the development of efficient and scalable fermentation processes for mevalonate production (6, 7).
Since the discovery of mevalonate in 1956 (8, 9), its production by fermentation has been studied. In 1986, Tamura et al. utilized Lactobacillus heterohiochii growth on mevalonate as a quantitative determination of mevalonate production for screening mevalonate-producing organisms (10). However, the best organism identified, Saccharomycopsis fibuligera NRRL Y-7069, still showed a low mevalonate titer (0.9 g/liter). To improve mevalonate production, the 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) synthase (mvaS) and the HMG-CoA reductase (mvaE) genes from several different organisms were cloned into plasmids and transcriptionally controlled by inducible promoters (6). Heterologous expression from these plasmids in Escherichia coli resulted in high mevalonate titers (3.9 to ∼14.6 g/liter) and yields (0.195 to ∼0.411 g/g of glucose) (5–7).
Although plasmid-based production of mevalonate achieved high yields, the disadvantages of plasmid-based fermentation pose significant challenges for large-scale fermentation (11–15). First, the replication of plasmid and expression of the genes residing on the plasmid result in extra metabolic burden, which reduces the allocation of nutrient and energy resources for the desired product (12). Second, no mechanism ensures the distribution of plasmids equally into daughter cells, potentially causing segregational instability. This might lead to plasmid-free cells after several generations of cell division (14), which diminishes both productivity and yield.
With the advancement of technologies for chromosome manipulation (16–19), chromosomal integration of desired genes is considered the preferable alternative to plasmid-based fermentation to ensure stability. However, compared to plasmid-based overexpression, tuning the expression level of the heterologous genes remains a key challenge for chromosomal integration. To resolve this problem, different strategies have been reported to enhance the expression of desired genes by modifying the promoter (20) or increasing the gene copy number on the chromosome (11, 18). These successful strategies have broadened the application of chromosomal integration in the biotechnology industry.
Another disadvantage is that the current engineered mevalonate pathways were mainly modulated by inducible promoters that require the expensive inducer isopropyl-β-d-thio-galactoside (IPTG). Scaled-up application of these inducible promoters is prevented not only by inducer cost but also by transcriptional heterogeneity (21, 22). Alternative promoters, such as temperature-sensitive, anaerobic-inducible, or constitutive promoters, are also applied in metabolic engineering (23–26). Although timing of the expression of desired pathway genes can be achieved by the temperature-sensitive and anaerobic-inducible promoters, constitutive promoters could reduce the operational difficulty.
Glucose is widely used as the primary feedstock in the fermentation industry. Since the glycolysis pathway converts glucose to the precursors of many desired chemicals, enhancement of the glycolytic flux, in principle, has the potential to improve productivity for many fermentation products. Although the regulation of glycolysis remains unclear, a previous study has demonstrated that enhancing the consumption of ATP by expression of the F1 subunits of the H+-ATP synthase increases the glycolytic flux by 1.7 times (27). Similar strategies, for example, deletion of the atpF and atpH genes, encoding ATP synthase B and delta chains, respectively, have been applied to improve the production of acetate and pyruvate (28–30).
In this study, the gene cluster for mevalonate production was integrated into the chromosome of E. coli. To enhance the expression of this gene cluster, a high-strength and constitutive expression promoter, M1-93, was employed (20). The engineered strains required neither antibiotic nor inducer. Further improvement of the productivity was achieved by deletion of the atpFH genes to enhance the glycolytic flux. Finally, fed-batch fermentation results showed that after integrating two copies of the mevalonate pathway genes (atoB-mvaS-mvaE) at different loci of the E. coli chromosome and deleting the atpFH and sucA genes, the engineered strain CMEV-7 exhibited a high titer (30 g/liter), a high productivity (∼1.01 g/liter/h), and a high production yield (86.1% of the maximum theoretical yield), which demonstrates great potential for industrially relevant production.
MATERIALS AND METHODS
Strains.
E. coli strain BW25113 was used for the chromosomal integration and plasmid comparison experiments (31). E. coli DH10B was used for the plasmid construction (32). Based on the improved mevalonate-producing plasmid pMEV-7 (5), the inducible promoter PLlacO1 was replaced by the constitutive promoter M1-93 (20), and this new mevalonate biosynthetic operon together with an antibiotic resistance gene (upstream of the mevalonate operon) was integrated into the chromosome of BW25113 at different loci by lambda Red-mediated homologous recombination (16). To integrate the mevalonate biosynthetic pathway into the chromosome and replace the adhE gene, the mvaE gene from pMEV-7 was PCR amplified, first using the mvaEF and mvaER1 primers (Table 1) and then using the mvaEF and mvaER2 primers to introduce the 50 nucleotides of DNA homologous to the downstream of the adhE gene. The new mvaE DNA fragment plus 50 nucleotides of homologous region were cloned into pMEV-7 digested by BamHI/XbaI to replace the original mvaE gene, generating plasmid pMEV-10. The chloramphenicol resistance gene (cm) from pLysS, followed by the M1-93 promoter sequence, were constructed first using the CmF and CmR primers to amplify the cm gene, and then using the CmF, Pm1, Pm2, and Pm3 primers to add the M1-93 promoter sequence. Part of the atoB open reading frame (ORF) region from pMEV-7 was amplified using the atoF and atoR primers. Then, the DNA fragments containing the cm gene, followed by the M1-93 promoter and part of the atoB gene, were connected by overlap PCR (33), first using the CmLig1 and atoR primers and then using the CmLig2 and atoR primers to introduce 60 nucleotides of DNA homologous to the upstream region of the adhE gene together with the XhoI site. This new DNA fragment was digested by XhoI/NdeI and introduced into pMEV-10 to replace the original PLlacO1 promoter and part of the atoB ORF region, generating plasmid pMEV-11. Finally, the cm-M1-93-atoB-mvaS-mvaE fragment from plasmid pMEV-11 was electroporated into competent BW25113 with pKD46 for homologous recombination to generate strain CMEV-1 (Table 1). A similar method was used to integrate the mevalonate biosynthetic pathway with the kanamycin resistance gene (kan) into the chromosome and replace the lactate dehydrogenase gene (ldhA), generating strain CMEV-2.
TABLE 1.
Primers used in this study
| Primer name | Primer sequence (5′ to 3′) | Location |
|---|---|---|
| mvaEF | CGGTAAGGATCCAGGAGAAATTAACTATGAAATTTTACGAG | Forward primer to amplify the mvaE gene plus BamHI site |
| mvaERadhE1 | CGTTTATGTTGCCAGACAGCGCTACTGATTAATCCCGATTTTCATCTTTTGATTG | Reverse primer to amplify the mvaE gene plus 28 nt of DNA homologous to the downstream of the adhE gene |
| mvaERadhE2 | CTTTTCTAGAATCGGCATTGCCCAGAAGGGGCCGTTTATGTTGCCAGACAGCGCTACTGA | 50 nt of DNA homologous to the downstream of the adhE gene plus XbaI site |
| CmF | TCGTCTTCACCTCGAGGTAGTCAATAAACCGGTAAACCAGC | Forward primer to amplify the cm gene |
| CmR | ACGTTGTTATCTCTTGTCAACACCGCCAGAGATAACCTGTGACGGAAGATCACTTCGCAG | Reverse primer to amplify the cm gene plus 35 nt of the M1-93 promoter sequence |
| Pm1 | TTATCTCTGGCGGTGTTGACAAGAGATAACAACGTTGATATAATTGAGCC | Primer to amplify the M1-93 promoter |
| Pm2 | GACAAGAGATAACAACGTTGATATAATTGAGCCCGTATTGTTAGCATGTACGTTTAAAC | Primer to amplify the M1-93 promoter |
| Pm3 | AGTTAATTTCTCCTGTTTAAACGTACATGCTAACAATACGGGCTCAATTATATCAACGTT | Reverse primer to amplify the M1-93 promoter |
| atoF | GTTTAAACAGGAGAAATTAACTATGAAAAATTGTGTCATCGTCAGTGC | 22 nt of the M1-93 sequence plus the forward sequence to amplify the atoB gene |
| atoR | AATCCCCATATGATAACCATGGGTGGCGCACATC | Reverse primer to amplify the first 480 bp of the atoB gene, including NdeI site |
| CmLig1 | TCTAGTTGTGCAAAACATGCTAATGTAGCCACCAAGTAGTCAATAAACCGGTAAACCAGC | 35 nt of DNA homologous to the upstream of the adhE gene plus forward sequence to amplify the cm gene |
| CmLig2 | TCTTCACCTCGAGAAATTGCTATCATTCGTTATTGTTATCTAGTTGTGCAAAACATGC | 45 nt of DNA homologous to the upstream of the adhE gene plus XhoI site |
| atpF1 | CAATCCTCGGCCAGGCCATCGCGTTTGTCCTGTTTGTAGGCTGGAGCTGCTTCG | 34 nt of DNA homologous to the upstream of the atpF gene plus forward sequence to amplify the kan cassette |
| atpR1 | GTCTGCAAGGCGCTCAAGACGACCGCGTACGCTGCATTCCGGGGATCCGTCGACC | Reverse sequence to amplify the kan cassette plus 35 nt of DNA homologous to the downstream of the atpH gene |
| atpF2 | GTGAATCTTAACGCAACAATCCTCGGCCAGGCCATCGCGTTTGTCCTGTTTGTAG | Forward primer to add 50 nt of DNA homologous to the upstream of the atpF gene |
| atpR2 | TTAAGACTGCAAGACGTCTGCAAGGCGCTCAAGACGACCGCGTACGCTGCATTCC | Reverse primer to add 50 nt of DNA homologous to the downstream of the atpH gene |
To construct the atpFH deletion strain, the same homologous regions were chosen as described previously (30). The kan gene from pKD13, flanked by the homologous regions, was amplified, first using the atpF1 and atpR1 primers and then using the atpF2 and atpR2 primers. Then, the DNA fragment was electroporated into competent BW25113 with pKD46 to generate the ΔatpFH strain. Then, the ΔatpFH::kan fragment from the ΔatpFH mutant strain was transferred into CMEV-1 by P1 transduction to create strain CMEV-3 (34).
To delete the acetate fermentation pathways, the kan gene in CMEV-3 strain was eliminated by using pCP20, and then the pyruvate oxidase (poxB) and phosphate acetyltransferase (pta) genes in this strain were deleted by P1 transduction to generate CMEV-4 and CMEV-5, respectively. P1 phage of the ΔpoxB::kan (JW0855-1) or Δpta::kan (JW2294-1) mutant strains from CGSC were used for P1 transduction.
To generate CMEV-6, first a strain with two copies of mevalonate pathway was constructed as described below: strain CMEV-2 was used for P1 transduction of the mevalonate pathway at the ldhA locus into strain CMEV-1, and the kan gene was eliminated by using pCP20. Then, the atpFH genes in this new strain were deleted by P1 transduction.
To generate CMEV-7, after obtaining the strain with two copies of the mevalonate pathway in the construction of CMEV-6, the sucA gene was deleted by P1 transduction, and the kan gene was eliminated. Then, the atpFH genes were deleted. For the sucA deletion strains, all the media were supplemented with 5 mM succinate to prevent the spontaneous Succ+ mutation (28).
Growth condition and analysis.
The growth conditions were the same as the shake flask batch fermentation described previously (5), except that for chromosome-based mevalonate fermentation, neither antibiotic nor inducer was supplemented into the fermentation medium. Both shake flask and fed-batch fermentations were performed for ∼48 h at 30°C. For fed-batch fermentation, there was 4% initial glucose followed by the addition of 2% or 4% glucose at the indicated time. For fed-batch fermentation with different concentrations of yeast extract, the basic medium had the following composition: glucose, 40 g/liter; K2HPO4, 7.5 g/liter; citric acid monohydrate, 0.2 g/liter; MgSO4·7H2O, 2.0 g/liter; ferric ammonium citrate, 0.3 g/liter; thiamine hydrochloride, 0.008 g/liter; d+-biotin, 0.008 g/liter; nicotinic acid, 0.008 g/liter; pyridoxine, 0.032 g/liter; ampicillin, 0.1 g/liter; concentrated H2SO4, 0.8 ml/liter; disodium succinate, 0.8 g/liter; NH4Cl, 1 g/liter; and trace metal solution, 1 ml/liter (5). The pH was adjusted to 7.0 with NH4OH, and different concentrations (0, 0.5, 1, and 5 g/liter) of yeast extract were supplemented.
Fermentation samples were collected at ∼6- or ∼12-h intervals. For measurement of the optical density at 600 nm (OD600), 100 μl of the sample was mixed with 300 μl of 1 M HCl to eliminate the CaCO3 in the fermentation medium, and then an appropriate dilution with ultrapure water was made. For mevalonate concentration measurements, the fermentation samples were centrifuged at maximal speed for 5 min, and the supernatants were diluted 5- to 10-fold before measurement by an Agilent 1260 Infinity high-performance liquid chromatography (HPLC) equipped with an Aminex HPX-87H column (Bio-Rad, USA) and a refractive-index detector (5). The first derivative (mevalonate versus time) of each curve between two adjacent time points served as an estimate of local volumetric rate, from which the maximal value was taken as the maximal volumetric rate.
RESULTS
Chromosomal integration of the mevalonate pathway.
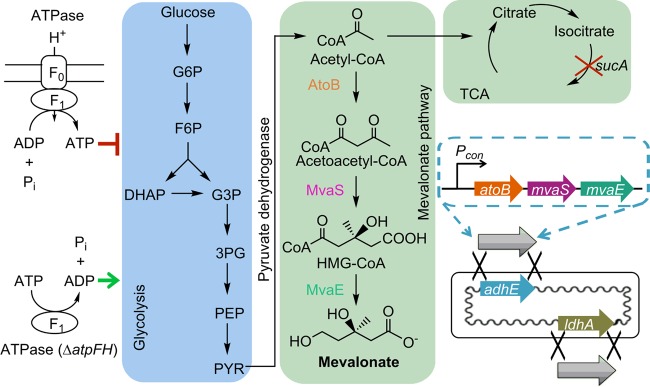
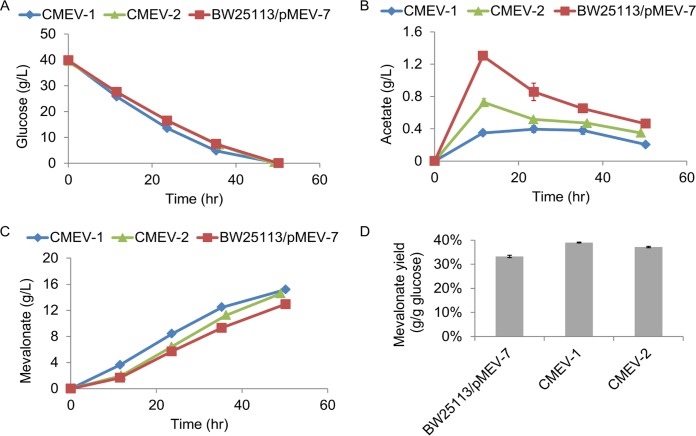
Previous work demonstrated that employing HMG-CoA synthase (MvaS) and HMG-CoA reductase (MvaE) from Lactobacillus casei could improve the production of mevalonate (5). Although this mevalonate production strategy was promising, the metabolic burden and genetic instability of plasmid prevented the industrial application (14). To circumvent such problems, these foreign genes were integrated into the chromosome of E. coli and controlled by the constitutive promoter M1-93, which has high promoter strength (Fig. 1) (20). Previously, the mevalonate pathway was assembled into a high-copy-number plasmid, pMEV-7, and controlled by the strong synthesized promoter PLlacO1 (5). To achieve comparable production of mevalonate after integrating the mevalonate pathway into the chromosome, we chose a constitutive promoter (M1-93) which is 5-fold stronger than the native Plac promoter. With these chromosomal constructs, neither antibiotic nor inducer was required during the fermentation. To prevent generation of the native fermentation products, chromosomal integration was performed to replace the adhE and ldhA genes, generating CMEV-1 and CMEV-2 strains, respectively (Table 2). Based on the constructed mevalonate pathway, the maximum theoretical yield is 0.548 g of mevalonate/g of glucose. Shake flask fermentation results showed that although the chromosome-based fermentation (CMEV-1 and CMEV-2) and the plasmid-based fermentation (BW25113/pMEV-7) exhibited similar consumption rates for glucose (Fig. 2A), CMEV-1 and CMEV-2 generated less acetate than BW25113/pMEV-7 (Fig. 2B), which likely accounts for the slightly higher mevalonate titers in strains CMEV-1 and CMEV-2 (Fig. 2C), since other by-products could barely be observed using HPLC. Consequently, the mevalonate yields (0.391 and 0.372 g/g of glucose) of the chromosome-based fermentation were slightly higher than that of the plasmid-based fermentation (0.332 g/g of glucose) (Fig. 2D).
FIG 1.
Diagram on chromosomal engineering for mevalonate production. Relevant metabolic reactions include glycolysis pathway, mevalonate pathway, and TCA cycle. Red line indicates that ATP inhibits glycolysis, while the green arrow represents that the ATP-limiting condition stimulates glycolysis. The red X represents deletion of the sucA gene, and the black Xs represent homologous recombination of pathway genes. Pi, inorganic phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde 3-phosphate; 3PG, 3-phosphoglyceric acid; PEP, phosphoenolpyruvic acid; PYR, pyruvate.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| BW25113 | Δ(araD-araB)567ΔlacZ4787(::rrnB-3) ΔlacZ4787(::rrnB-3) Δ(rhaD-rhaB)568 hsdR514 | CGSC |
| JW0855-1 | ΔpoxB::kan | CGSC |
| JW2294-1 | Δpta::kan | CGSC |
| CMEV-1 | BW25113 ΔadhE::cm-M1-93-atoB-mvaS-mvaE | This study |
| CMEV-2 | BW25113 ΔldhA::kan-M1-93-atoB-mvaS-mvaE | This study |
| CMEV-3 | BW25113 ΔadhE::cm-M1-93-atoB-mvaS-mvaE ΔatpFH::kan | This study |
| CMEV-4 | BW25113 ΔadhE::cm-M1-93-atoB-mvaS-mvaE ΔatpFH::FRT ΔpoxB::kan | This study |
| CMEV-5 | BW25113 ΔadhE::cm-M1-93-atoB-mvaS-mvaE ΔatpFH::FRT Δpta::kan | This study |
| CMEV-6 | BW25113 ΔadhE::cm-M1-93-atoB-mvaS-mvaE ΔldhA::FRT-M1-93-atoB-mvaS-mvaE ΔatpFH::kan | This study |
| CMEV-7 | BW25113 ΔadhE::cm-M1-93-atoB-mvaS-mvaE ΔldhA::FRT-M1-93-atoB-mvaS-mvaE ΔsucA ΔatpFH::kan | This study |
| Plasmids | ||
| pMEV-7 | pZE-PLlacO1-atoB-mvaS-mvaE | 5 |
| pKD13 | kan cassette | CGSC |
| pKD46 | λ Red recombinase expression | CGSC |
| pCP20 | FLP recombinase expression | CGSC |
| pMEV-10 | pZE-PLlacO1-atoB-mvaS-mvaE-50-nt homolog | This study |
| pMEV-11 | pZE-60 nt-homolog-cm-M1-93-atoB-mvaS-mvaE-50-nt homolog | This study |
FRT, Flp recombinase target; nt, nucleotide.
FIG 2.
Production of mevalonate after chromosomal integration of the mevalonate pathway. (A) Glucose concentration in the fermentation medium at indicated time. (B) Accumulation of acetate by-product. Mevalonate titers (C) and yields (D) achieved from plasmid-based fermentation (BW25113/pMEV-7), or after chromosomal integration of the mevalonate pathway (CMEV-1 and CMEV-2). Error bars represent the standard deviation (from three different cultures). The standard deviations are often small and masked by the data symbols.
Improvement of the production rate by atpFH deletion.
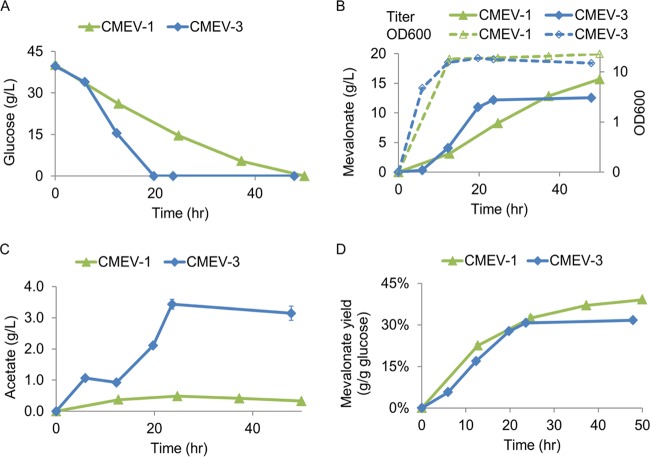
Previous work demonstrated that deleting the atpFH genes improved the production of acetate or pyruvate (28–30), and further deletion of acetate kinase (ackA) converted an acetate-producing strain (T36 in reference 28) to a pyruvate-producing strain (T38 in reference 29), which suggested that the accumulation of acetate in T36 was mainly through the following pathway: pyruvate → acetyl-CoA → acetyl-P → acetate (28, 29). These results indicate that an increase in the glycolytic flux by an atpFH deletion could improve the production of acetyl-CoA, the precursor of mevalonate synthesis. To confirm this and increase the productivity of mevalonate, the atpFH genes in CMEV-1 strain were deleted to generate CMEV-3. Shake flask fermentation showed that deletion of the atpFH genes resulted in an at least 2-fold increase in the glycolytic flux, as indicated by the much higher glucose consumption rate (Fig. 3A). Consequently, the increase in glycolytic flux accelerated the production of mevalonate (Fig. 3B). For the CMEV-1 strain, the mevalonate titer increased slowly during the entire fermentation process (Fig. 3B), and the maximal volumetric rate reached ∼0.43 g/liter/h. For the atpFH deletion strain CMEV-3, mevalonate started to accumulate during late-exponential phase and increased sharply until the carbon source has been completely depleted (Fig. 3A and B), and the maximal volumetric rate reached ∼0.92 g/liter/h. Compared to strain CMEV-1, the deletion of atpFH (CMEV-3) resulted in a 2.1-fold increase in the maximal productivity due to enhancement of the glycolytic flux. This result confirmed that improved glycolytic flux by deleting atpFH stimulates the production of acetyl-CoA and subsequently improves the productivity of mevalonate. This result also suggested that in CMEV-1, the carbon flux for mevalonate production is primarily restricted by the glycolysis pathway.
FIG 3.
Production of mevalonate after enhancing the glycolytic flux by deletion of the atpFH genes. (A) Glucose concentration in the fermentation medium. (B) Mevalonate concentration in the fermentation medium and the growth curve during the fermentation process. (C) Acetate concentration in the fermentation medium. (D) Mevalonate yield. Error bars are the standard deviations (SD) (from three different cultures). The standard deviations are often small and masked by the data symbols.
The acetate level increased from ∼0.33 to ∼3.14 g/liter in the atpFH deletion strain CMEV-3 (Fig. 3C). This most likely caused the relative lower mevalonate yield (Fig. 3D). To further improve the mevalonate yield in the atpFH deletion strain CMEV-3, bioengineering should be used to eliminate the generation of acetate. For this purpose, the poxB or pta gene for acetate synthesis in strain CMEV-3 was deleted to generate CMEV-4 or CMEV-5. Shake flask fermentation showed that deletion of the acetate synthetic pathways could neither decrease the acetate level nor improve the mevalonate yield (Table 3), which indicates that the acetate produced in CMEV-3 results from some other pathways.
TABLE 3.
Shake flask fermentationa
| Strain | Acetate (g/liter) | Mevalonate yield |
|
|---|---|---|---|
| g/g of glucose | % of theoreticalb | ||
| CMEV-3 | 3.14 ± 0.23 | 0.331 ± 0.006 | 60.4 ± 1.1 |
| CMEV-4 | 2.83 ± 0.15 | 0.332 ± 0.007 | 60.6 ± 1.3 |
| CMEV-5 | 6.35 ± 0.25 | 0.273 ± 0.005 | 49.8 ± 0.8 |
Data are indicated as means ± SD.
Maximum theoretical yield is 0.548 g of mevalonate per g of glucose.
Improvement of the production yield by enhancing the downstream pathway from acetyl-CoA to mevalonate.
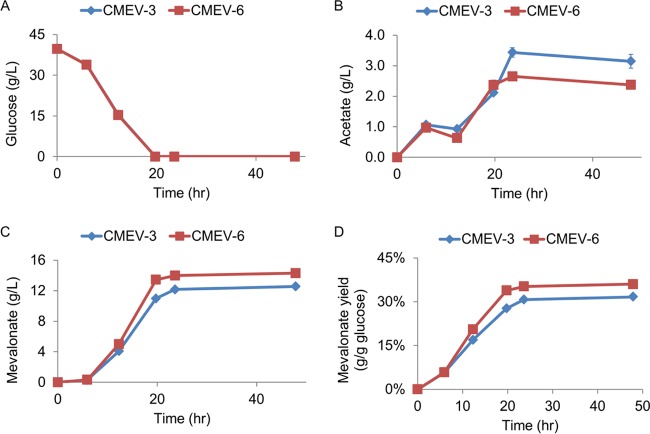
As deletion of the acetate fermentation pathway did not improve the mevalonate yield, an alternative strategy was to enhance the downstream enzyme(s) catalyzing the conversion of acetyl-CoA to mevalonate, which could competitively recruit more acetyl-CoA for mevalonate production (Fig. 1). To test this strategy, two copies of the atoB-mvaS-mvsE operon were integrated into the chromosome of E. coli, and the atpFH genes were deleted to generate CMEV-6 (Table 2). Shake flask fermentation results showed that CMEV-6 and CMEV-3 strains exhibited similar consumption rates for glucose (Fig. 4A), while strain CMEV-6 accumulated less acetate (Fig. 4B). Consequently, strain CMEV-6 produced more mevalonate than strain CMEV-3 (Fig. 4C). This result indicates that although two copies of the atoB-mvaS-mvsE operon did not further improve the glycolytic flux, it directed the redistribution of acetyl-CoA flux to synthesize more mevalonate, which decreased the accumulation of acetate.
FIG 4.
Enhancement of the downstream pathway from acetyl-CoA to mevalonate by integration of an extra copy of the atoB-mvaS-mvsE operon into the chromosome. (A to C) Glucose (A), acetate (B), and mevalonate (C) concentrations in the fermentation medium. (D) Mevalonate yield. Error bars are the SD (from three different cultures). The standard deviations are often small and masked by the data symbols.
Fed-batch fermentation study.
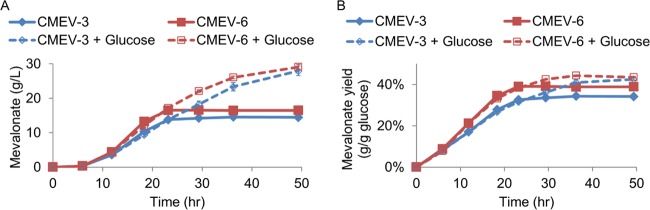
The production yield (Fig. 4D) of CMEV-3 and CMEV-6 stopped increasing after glucose was depleted within 24 h (Fig. 4A). To investigate whether the yield could further increase in the presence of glucose, fed-batch fermentations were performed by the addition of 2% glucose at 12-h intervals (Fig. 5). Under this condition, although the maximal volumetric rates did not increase, both CMEV-3 and CMEV-6 produced more mevalonate (27.9 g/liter and 29.1 g/liter, respectively; Fig. 5A), and the yield also increased to 0.425 and 0.435 g/g of glucose, respectively, after ∼49 h of fermentation (Fig. 5B). Compared to the fermentation results of CMEV-1 (Fig. 3), the deletion of atpFH (CMEV-3) improved both the productivity (from 0.43 to 0.92 g/liter/h) and the yield (from 0.391 to 0.425 g/g of glucose). Integrating another copy of the mevalonate pathway into the chromosome further improved the production yield in CMEV-6.
FIG 5.
Fed-batch fermentation by adding 2% glucose at 12-h intervals. (A and B) The mevalonate titer (A) and yield (B) comparison between CMEV-3 (CMEV-6) and CMEV-3 plus glucose (CMEV-6 + Glucose). CMEV-3 (CMEV-6) represents shake flask fermentation without adding glucose, while CMEV-3 plus glucose (CMEV-6 + Glucose) represents fed-batch fermentation with adding 2% glucose at 12-h intervals. Error bars are the SD. The standard deviations are often small and masked by the data symbols.
Further improvement of mevalonate yield by deletion of sucA.
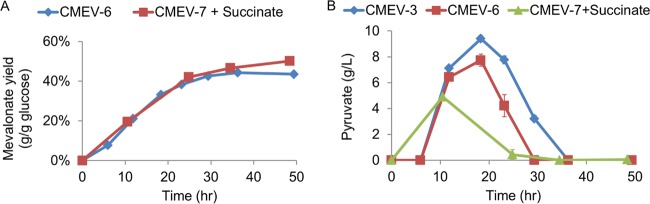
Since the entire atp operon deletion results in more carbon flux through the tricarboxylic acid (TCA) cycle (35), deletion of the sucA gene (encoding the E1 subunit of the 2-oxoglutarate decarboxylase) would disrupt the TCA cycle and redirect acetyl-CoA for mevalonate production, which is likely to improve the mevalonate yield. To confirm this, the sucA gene in CMEV-6 was deleted to generate CMEV-7 (see Materials and Methods). Fed-batch fermentations were performed by adding 4% glucose after 24 h and supplementing with 5 mM succinate. The fermentation results showed that by deleting the sucA gene, CMEV-7 exhibited higher yield than that of CMEV-6 (0.502 versus 0.435 g/g of glucose, respectively; P < 0.0001) (Fig. 6A), confirming that TCA cycle competes with the mevalonate pathway for acetyl-CoA.
FIG 6.
Fed-batch fermentation result of the sucA deletion strain. (A) The mevalonate yield comparison. (B) Pyruvate production. Error bars are the SD. The standard deviations are often small and masked by the data symbols.
In the atpFH deletion background, integrating another copy of the mevalonate pathway into the chromosome and deleting the sucA gene increased the production yield. However, the productivities of CMEV-3, CMEV-6, and CMEV-7 remained similar. Enhancement of the glycolytic flux resulted in accumulation of a metabolic intermediate pyruvate in strain CMEV-3, CMEV-6, and CMEV-7 during the fermentation processes (Fig. 6B), which is consistent with previous results of the acetate-producing strain T36 (29). This result indicates that in strains CMEV-3, CMEV-6, and CMEV-7, the limitation of productivity is probably located at the conversion of pyruvate into acetyl-CoA catalyzed by pyruvate dehydrogenase complex.
All the fermentation experiments described above were supplemented with 5 g/liter yeast extract, which may contribute to mevalonate production titer and yield. To investigate the effect of yeast extract on mevalonate production, an alternative fermentation medium was applied (see Materials and Methods and Table 4). Fed-batch fermentation results showed that without yeast extract, the mevalonate yield was still very high (86.1% of the maximum theoretical yield), but the mevalonate titer was lower (5.3 g/liter) probably due to the low biomass (OD600, 2.93). With 1 g/liter yeast extract, CMEV-7 exhibited a mevalonate titer similar to the plasmid-based fermentation result (Fig. 2C), although the OD600 only reached 6.37 (Table 4). The accumulation of mevalonate is not toxic to the E. coli, thus further increasing the concentration of yeast extract to 5 g/liter; this allowed for an increase in both the titer and yield, while reducing the acetate level. These results indicate that optimal concentrations of yeast extract could improve both titer and yield of mevalonate production. For the cost-effective production of mevalonate, adaptive evolution may be required to select a mevalonate-producing strain that can grow well without yeast extract and/or succinate (28).
TABLE 4.
Fed-batch fermentation with different concentrations of yeast extracta
| Yeast extract concn (g/liter) | OD600 | Acetate concn (g/liter) | Mevalonate concn (g/liter) | Yield (g/g of glucose) | % theoretical yield |
|---|---|---|---|---|---|
| 0 | 2.93 ± 0.04 | 1.05 ± 0.08 | 5.3 ± 0.3 | 0.472 ± 0.009 | 86.1 ± 1.7 |
| 0.5 | 4.61 ± 0.17 | 1.25 ± 0.28 | 8.8 ± 0.2 | 0.462 ± 0.016 | 84.3 ± 2.9 |
| 1 | 6.37 ± 0.59 | 1.08 ± 0.30 | 13.2 ± 1.1 | 0.453 ± 0.017 | 82.6 ± 3.2 |
| 5 | 11.61 ± 0.96 | 0.21 ± 0.10 | 23.3 ± 1.2 | 0.520 ± 0.026 | 94.9 ± 4.7 |
Data are indicated as means ± SD.
DISCUSSION
By chromosomal integration and modification of the endogenous/heterologous metabolic pathways, we engineered an E. coli strain for efficient mevalonate production in terms of titer, yield, and productivity. Since this engineered strain is plasmid free, and the heterologous pathway is controlled by a constitutive promoter, the production of mevalonate is genetically stable and inducer free, which has potential for scale-up fermentation (15, 36). We enhanced the glycolytic flux by deleting the atpFH genes to improve productivity. Previously, deletion of the atpFH genes has been applied to improve the production of acetate or pyruvate from glucose (28–30). However, to the best of our knowledge, this is the first report to improve the production of bioproducts beyond pyruvate and acetate using this strategy. Our result indicates that deletion of the atpFH genes could significantly increase the acetyl-CoA flux for biosynthesis of downstream products.
Compared to plasmid-based overexpression, chromosomal integration of the metabolic pathways for the desired chemicals has great advantages for industrial applications. Chromosomal integration of the mevalonate pathway requires no antibiotic to maintain the plasmid in the cell. Furthermore, a constitutive promoter was employed to control the expression of the mevalonate pathway (Fig. 1) without the use of any inducer. Although a high-copy-number plasmid (pMEV-7 in our case) may provide a high copy number of the atoB-mvaS-mvaE operon, replication of the plasmid and expression of the genes residing on the plasmid waste carbon and energy resources that can be used for the synthesis of mevalonate. To show this burden, we transformed pMEV-7 into strain CMEV-3, and shake flask fermentation showed that the atoB-mvaS-mvaE operon on pMEV-7 could not improve the mevalonate titer. In addition, we showed that enhancement of the downstream pathway by integration of another copy of the atoB-mvaS-mvsE operon into the chromosome of the CMEV-3 strain increased the mevalonate titer from 12.6 to 14.3 g/liter in the new strain, CMEV-6. These results indicate that enhancement of the downstream pathway for mevalonate production could increase the mevalonate titer if the atoB-mvaS-mvaE operon was integrated into the chromosome but could not increase the titer if the operon was cloned into a plasmid. This is probably because the metabolic burden of the high-copy-number plasmid pMEV-7 took carbon and energy resources away from mevalonate production.
Previous work demonstrated that the production of pyruvate or acetate could be improved by enhancement of the glycolytic flux (28–30). Since acetyl-CoA, the precursor of the mevalonate pathway (Fig. 1), resides in the main pathway for acetate fermentation, enhancement of the glycolytic flux is likely to enhance the production of acetyl-CoA and subsequently facilitate the downstream flux to any desired products derived from acetyl-CoA. To confirm this hypothesis and improve the production of mevalonate, the atpFH genes, encoding the b and c subunits of the Fo in membrane-bound (FoF1) H+-ATP synthase, were deleted. With deletion of the atpFH genes, both the glucose consumption and the productivity of mevalonate increased by at least 2-fold. However, the deletion of atpFH resulted in the accumulation of pyruvate at the early stages of fermentation. This indicates in the atpFH deletion background, the conversion of pyruvate to acetyl-CoA by the pyruvate dehydrogenase complex (PDC) is likely the rate-limiting step. Further improvement in the production of mevalonate would focus on increasing the flux through PDC. A previous study has shown that the protein level of PDC is transcriptionally regulated by Crp and Fur (37), so modification of these regulators may improve the acetyl-CoA flux. Since PDC catalyzes a redox reaction, another possibility is that imbalanced reducing equivalents also inhibit the reaction.
Compared to CMEV-1, deletion of the atpFH genes enhanced the productivity of mevalonate by ∼2.1-fold in CMEV-3. This indicates that in CMEV-1, the glycolysis pathway primary determines the productivity of mevalonate, while the downstream pathway (encoded by the atoB-mvaS-mvsE operon) has little effect on the synthetic rate. Additionally, for other acetyl-CoA-derived chemicals, if the engineered downstream metabolic pathway does not limit the productivity, enhancement of the glycolytic flux by deleting the atpFH genes has the potential to increase the productivity significantly. The disadvantage of this strategy is that the presence of native pathways in the host will compete with the desired pathway for acetyl-CoA or other intermediates along the glycolysis pathway, which might reduce the titer/yield. One possible solution is the deletion or downregulation of the genes in competitive pathways. Previous work has indicated that the TCA cycle is likely to be a competitive pathway (35, 38, 39), and this work showed that deletion of the sucA gene in the TCA cycle does improve mevalonate yield. An alternative solution applied here is the enhancement of the downstream pathway for the desired product, and although it could not eliminate the production of by-products, it did increase the carbon flux to the desired product.
ACKNOWLEDGMENTS
We thank Maria McClintock, Pooja Jambunathan, Qiuge Zhang, and Zhengyang Liu for discussions.
We declare no conflicts of interest.
This research was supported by a grant from the National Science Foundation through the Center for Sustainable Polymers (grant CHE-1413862).
REFERENCES
- 1.Kuzuyama T, Seto H. 2012. Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc Jpn Acad Ser B Phys Biol Sci 88:41–52. doi: 10.2183/pjab.88.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pitera DJ, Paddon CJ, Newman JD, Keasling JD. 2007. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng 9:193–207. doi: 10.1016/j.ymben.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 4.Beck ZQ, Eliot AC, Peres CM, Vaviline DV. April 2013. Utilization of phosphoketolase in the production of mevalonate, isoprenoid precursors, and isoprene. U.S. patent 20130089906 A1.
- 5.Xiong M, Schneiderman DK, Bates FS, Hillmyer MA, Zhang K. 2014. Scalable production of mechanically tunable block polymers from sugar. Proc Natl Acad Sci U S A 111:8357–8362. doi: 10.1073/pnas.1404596111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabata K, Hashimoto S. December 2005. Process for producing mevalonic acid. U.S. patent 20050287655 A1.
- 7.Beck ZQ, Miller MC, Peres CM, Primak YA, Pucci JP, Wells DH. November 2012. Production of mevalonate, isoprene, and isoprenoids using genes encoding polypeptides having thiolase, hmg-coa synthase and hmg-coa reductase enzymatic activities. WO patent 2012149469 A1.
- 8.Wright LD, Cresson EL, Skeggs HR, MacRae GDE, Hoffman CH, Wolf DE, Folkers K. 1956. Isolation of a new acetate-replacing factor. J Am Chem Soc 78:5273–5275. doi: 10.1021/ja01601a033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura G. 2004. Hiochic acid, a new growth factor for Lactobacillus homohiochi and Lactobacillus heterohiochi. J Gen Appl Microbiol 50:327–330. [PubMed] [Google Scholar]
- 10.Tamura G, Ando K, Kodama K, Arima K. 1968. Production of mevalonic acid by fermentation. Appl Microbiol 16:965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyo KE, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 12.Keasling JD. 2008. Synthetic biology for synthetic chemistry. ACS Chem Biol 3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 13.de la Cueva-Méndez G, Pimentel B. 2007. Gene and cell survival: lessons from prokaryotic plasmid R1. EMBO Rep 8:458–464. doi: 10.1038/sj.embor.7400957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friehs K. 2004. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol 86:47–82. [DOI] [PubMed] [Google Scholar]
- 15.Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas MA. 2016. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 34:652–664. doi: 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd D, Weiss DS, Chen JC, Beckwith J. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol 182:842–847. doi: 10.1128/JB.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu P, Yang F, Su T, Wang Q, Liang Q, Qi Q. 2015. A rapid and reliable strategy for chromosomal integration of gene(s) with multiple copies. Sci Rep 5:9684. doi: 10.1038/srep09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senecoff JF, Bruckner RC, Cox MM. 1985. The FLP recombinase of the yeast 2-micron plasmid: characterization of its recombination site. Proc Natl Acad Sci U S A 82:7270–7274. doi: 10.1073/pnas.82.21.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Tang J, Liu Y, Zhu X, Zhang T, Zhang X. 2012. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl Microbiol Biotechnol 93:2455–2462. doi: 10.1007/s00253-011-3752-y. [DOI] [PubMed] [Google Scholar]
- 21.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. 2005. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A 102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegele DA, Hu JC. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A 94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdez-Cruz NA, Caspeta L, Perez NO, Ramirez OT, Trujillo-Roldan MA. 2010. Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters. Microb Cell Fact 9:18. doi: 10.1186/1475-2859-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasr R, Akbari Eidgahi MR. 2014. Construction of a synthetically engineered nirB promoter for expression of recombinant protein in Escherichia coli. Jundishapur J Microbiol 7:e15942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nevoigt E, Kohnke J, Fischer CR, Alper H, Stahl U, Stephanopoulos G. 2006. Engineering of promoter replacement cassettes for fine-tuning of gene expression in Saccharomyces cerevisiae. Appl Environ Microbiol 72:5266–5273. doi: 10.1128/AEM.00530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim CG, Fowler ZL, Hueller T, Schaffer S, Koffas MA. 2011. High-yield resveratrol production in engineered Escherichia coli. Appl Environ Microbiol 77:3451–3460. doi: 10.1128/AEM.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J Bacteriol 184:3909–3916. doi: 10.1128/JB.184.14.3909-3916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Causey TB, Zhou S, Shanmugam KT, Ingram LO. 2003. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: homoacetate production. Proc Natl Acad Sci U S A 100:825–832. doi: 10.1073/pnas.0337684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Causey TB, Shanmugam KT, Yomano LP, Ingram LO. 2004. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc Natl Acad Sci U S A 101:2235–2240. doi: 10.1073/pnas.0308171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Eiteman MA, Altman R, Altman E. 2008. High glycolytic flux improves pyruvate production by a metabolically engineered Escherichia coli strain. Appl Environ Microbiol 74:6649–6655. doi: 10.1128/AEM.01610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grenier F, Matteau D, Baby V, Rodrigue S. 2014. Complete genome sequence of Escherichia coli BW25113. Genome Announc 2(5):e01038-14. doi: 10.1128/genomeA.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durfee T, Nelson R, Baldwin S, Plunkett G III, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csörgő B, Pósfai G, Weinstock GM, Blattner FR. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol 190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pont-Kingdon G. 2003. Creation of chimeric junctions, deletions, and insertions by PCR. Methods Mol Biol 226:511–516. [DOI] [PubMed] [Google Scholar]
- 34.Thomason LC, Costantino N, Court DL. 2001. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1.17. doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 35.Jensen PR, Michelsen O. 1992. Carbon and energy metabolism of atp mutants of Escherichia coli. J Bacteriol 174:7635–7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woolston BM, Edgar S, Stephanopoulos G. 2013. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng 4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Gosset G, Barabote R, Gonzalez CS, Cuevas WA, Saier MH Jr. 2005. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J Bacteriol 187:980–990. doi: 10.1128/JB.187.3.980-990.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cress BF, Toparlak OD, Guleria S, Lebovich M, Stieglitz JT, Englaender JA, Jones JA, Linhardt RJ, Koffas MA. 2015. CRISPathBrick: modular combinatorial assembly of type II-A CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Synth Biol 4:987–1000. doi: 10.1021/acssynbio.5b00012. [DOI] [PubMed] [Google Scholar]
- 39.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA. 2011. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng 13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]