ABSTRACT
Aerobic anoxygenic phototrophic bacteria (AAPB) are thought to be important players in oceanic carbon and energy cycling in the euphotic zone of the ocean. The genus Citromicrobium, widely found in oligotrophic oceans, is a member of marine alphaproteobacterial AAPB. Nine Citromicrobium strains isolated from the South China Sea, the Mediterranean Sea, or the tropical South Atlantic Ocean were found to harbor identical 16S rRNA sequences. The sequencing of their genomes revealed high synteny in major regions. Nine genetic islands (GIs) involved mainly in type IV secretion systems, flagellar biosynthesis, prophage, and integrative conjugative elements, were identified by a fine-scale comparative genomics analysis. These GIs played significant roles in genomic evolution and divergence. Interestingly, the coexistence of two different photosynthetic gene clusters (PGCs) was not only found in the analyzed genomes but also confirmed, for the first time, to our knowledge, in environmental samples. The prevalence of the coexistence of two different PGCs may suggest an adaptation mechanism for Citromicrobium members to survive in the oceans. Comparison of genomic characteristics (e.g., GIs, average nucleotide identity [ANI], single-nucleotide polymorphisms [SNPs], and phylogeny) revealed that strains within a marine region shared a similar evolutionary history that was distinct from that of strains isolated from other regions (South China Sea versus Mediterranean Sea). Geographic differences are partly responsible for driving the observed genomic divergences and allow microbes to evolve through local adaptation. Three Citromicrobium strains isolated from the Mediterranean Sea diverged millions of years ago from other strains and evolved into a novel group.
IMPORTANCE Aerobic anoxygenic phototrophic bacteria are a widespread functional group in the upper ocean, and their abundance could be up to 15% of the total heterotrophic bacteria. To date, a great number of studies display AAPB biogeographic distribution patterns in the ocean; however, little is understood about the geographic isolation impact on the genome divergence of marine AAPB. In this study, we compare nine Citromicrobium genomes of strains that have identical 16S rRNA sequences but different ocean origins. Our results reveal that strains isolated from the same marine region share a similar evolutionary history that is distinct from that of strains isolated from other regions. These Citromicrobium strains diverged millions of years ago. In addition, the coexistence of two different PGCs is prevalent in the analyzed genomes and in environmental samples.
INTRODUCTION
Aerobic anoxygenic phototrophic bacteria (AAPB) are a widespread functional microbial group in the euphotic zone of the ocean and are thought to play important roles in the cycling of marine carbon and energy (1–10). AAPB can harvest light using bacteriochlorophyll a (BChl a) and various carotenoids to form a transmembrane proton gradient for the generation of ATP (1, 11). The photosynthetic process is performed by a series of photosynthetic operons, including bch, crt, puf, puh, and some regulatory genes, forming a highly conserved 40- to 50-kb photosynthetic gene cluster (PGC) (12, 13). The heart of anoxygenic photosynthesis is the reaction center, encoded by the puf and puh operons (14).
The genus Citromicrobium, belonging to the order Sphingomonadales in the class Alphaproteobacteria, is a member of the marine AAPB (15–17). Since the initial isolation of the type species, Citromicrobium bathyomarinum strain JF-1, from deep-sea hydrothermal vent plume waters in the Juan de Fuca Ridge (Pacific Ocean), dozens of Citromicrobium strains were isolated from a depth of 500 to 2,379 m near this region (17, 18). Genomic analyses showed that members of the genera Citromicrobium and Erythrobacter generally contained the shortest and simplest PGC structures among all known AAPB (16). Analysis of the C. bathyomarinum JL354 genome led to the discovery that two different PGCs could coexist in one bacterium, with one complete cluster and the other cluster incomplete (15, 16). Horizontal gene transfer (HGT) was detected to mediate the incomplete PGC acquisition, and multiple mechanisms mediating HGT were also found through studying its genome, including a gene transfer agent (GTA), prophage, integrative conjugative element (ICE), and the type IV secretion system (T4SS). Citromicrobium species may benefit from obtaining genes by HGT to compete and survive in natural environments. Additionally, genes encoding xanthorhodopsin, which is a light-driven proton pump like bacteriorhodopsin (19–21), but more effective at collecting light, are also found in citromicrobial genomes (22, 23).
A recent pyrosequencing analysis of pufM genes showed that Citromicrobium-like AAPB were mainly distributed in the oligotrophic ocean and were relatively more abundant in the upper twilight zone (150 to 200 m depth) than in the subsurface waters (5 m and 25 m) of the western Pacific Ocean (24). However, in this study, no sequences belonging to the incomplete PGC were detected, although the Citromicrobium-like pufM gene belonging to the complete PGC showed a high relative abundance.
Fortunately, a great number of citromicrobial strains have been isolated from the Mediterranean Sea, the South China Sea, and the South Atlantic Ocean (16, 25). The Mediterranean Sea is an almost completely enclosed sea connected with the eastern North Atlantic Ocean by the narrow Strait of Gibraltar. The South China Sea is the largest marginal sea in the western Pacific Ocean, extending from subtropical to tropical zones. The sampled area in the South Atlantic Ocean (13.857°S, 26.018°W) is a region of tropical open oceans. Although a great number of studies showed that microbes displayed biogeographic patterns in the ocean (26, 27), little is understood about the geographic isolation impact on the genome divergence of marine microbes.
The aims of this study were to (i) illustrate the evolutionary divergence of Citromicrobium genomes with identical 16S rRNA sequences but different geographical origins and (ii) demonstrate the prevalence of the coexistence of two PGCs in these strains and within the marine environment.
MATERIALS AND METHODS
Isolation of Citromicrobium strains.
Citromicrobium strains JL31 (24.458°N, 118.247°E), JL354 (21.684°N, 112.918°E), JL477 (22.167°N, 115.153°E), JL1351 (17.994°N, 120.287°E), and WPS32 (17.000°N, 115.000°E) were isolated from euphotic waters from the South China Sea on plates containing rich organic (RO) medium (Table 1) (17). Strain JL2201 was isolated from South Atlantic surface water (13.857°S, 26.018°W) on an RO medium plate (Table 1). Strains RCC1878 and RCC1885 were isolated from the Ionian Sea (at the middle of the Mediterranean Sea) surface water (34.133°N, 18.450°E) on two low-strength agar medium plates, and strain RCC1897 was isolated from western Mediterranean Sea (38.633°N, 7.917°E) water (Table 1) (25).
TABLE 1.
Genome information for the nine strains
| Strain | Genome size (Mb) | No. of contigs | G+C content (%) | No. of genes | No. of CDSsa | No. of COGs | No. of tRNAs | Sequencing coverage (×) | Isolation source | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| JL31 | 3.16 | 22 | 65.1 | 3,092 | 2,960 | 1,969 | 45 | 180 | South China Sea | LAIH00000000 |
| JL1351 | 3.16 | 17 | 65.1 | 3,090 | 2,961 | 1,981 | 45 | 155 | South China Sea | LAPR00000000 |
| WPS32 | 3.16 | 16 | 64.9 | 3,056 | 2,925 | 1,934 | 44 | 250 | South China Sea | LAPS00000000 |
| JL477 | 3.26 | 1 | 65.0 | 3,168 | 3,027 | 2,004 | 45 | 220 | South China Sea | CP011344 |
| JL354 | 3.27 | 68 | 65.0 | 3,208 | 3,137 | 2,010 | 45 | 26 | South China Sea | ADAE00000000 |
| JL2201 | 3.27 | 22 | 65.1 | 3,250 | 3,105 | 1,975 | 45 | 245 | South Atlantic Ocean | LARQ00000000 |
| RCC1878 | 3.28 | 14 | 64.8 | 3,194 | 3,061 | 1,966 | 45 | 205 | Mediterranean Sea | LBLZ00000000 |
| RCC1885 | 3.28 | 14 | 64.8 | 3,197 | 3,063 | 1,975 | 45 | 190 | Mediterranean Sea | LBLY00000000 |
| RCC1897 | 3.28 | 17 | 64.8 | 3,192 | 3,113 | 1,978 | 45 | 440 | Mediterranean Sea | LUGI01000000 |
CDSs, coding sequences.
Genome sequencing and assembly.
Whole-genome sequencing of Citromicrobium sp. strain JL354 was performed by 454 pyrosequencing, as previously reported (15, 16). The genomes of JL31, JL477, JL1351, JL2201, WPS32, RCC1878, RCC1885, and RCC1897 were obtained by an Illumina MiSeq system. Paired-end reads of average 250-bp length were assembled using the Velvet software (version 2.8) (28). The sequencing coverages ranged from 155× (JL1351) to 440× (RCC1897). The genome of strain JL477 has been completed, and the other seven genomes each possessed 14 to 22 contigs (Table 1).
Gene prediction and annotation.
The open reading frames (ORFs) were analyzed using a combination of Glimmer 3.02 (29) and GeneMark (30, 31). All predicted ORFs were then annotated using the NCBI Prokaryotic Genome Annotation Pipeline (32) and Rapid Annotations using Subsystems Technology (RAST) (33). rRNA identification was performed with the RNAmmer 1.2 software (34), and tRNAscan-SE (version 1.21) was used to identify the tRNA genes (35).
The genomic average nucleotide identity (ANI) was calculated by the JSpecies Online Service (http://jspecies.ribohost.com/jspeciesws) (36).
Core genome and pangenome analyses.
Orthologous clusters (OCs) were assigned by grouping all protein sequences from the nine genomes using OrthoMCL based on their sequence similarity (E value <10−5, >50% coverage) (37). The core and pangenomes were analyzed according to the method described by Tettelin et al. (38). The functional proteins were classified by comparison with the COG (Clusters of Orthologous Groups) databases (39).
SNP discovery.
Single-nucleotide polymorphisms (SNPs) were detected by sequence comparisons of the 9 Citromicrobium genomes using MUMmer (40). Because 8 out of 9 genomes were draft genomes, the positions of SNPs from all genomes relative to the sequence of strain JL477 were recorded. Paralogous genes and repeated regions were removed from our analysis. The synonymous SNPs of coding regions were used to roughly estimate the pairwise strain divergence time (41).
Sequence comparison.
The genome of JL477 was compared to the other eight citromicrobial genomes in silico using the BLAST Ring Image Generator (BRIG) software (42). Regions with nucleotide sequence similarity above 70% are shown on the map. Nine genetic islands (GIs) were identified from the comparative map. The upstream and downstream regions of each GI were then retrieved and manually searched for the presence of conserved regions or signature genes (such as tRNA). Some GIs and their flanking genes from different genomes were chosen for pairwise comparison (see Table S1 in the supplemental material). Although there were eight draft genomes with a number of contigs involved in analyses, the completeness for each genome was more than 99% as a result of the high genome sequencing coverage. Gaps between contigs usually were intergenic regions or did not contain more than three genes (see Table S1). All gene losses (especially more than a 10-kb fragment) occurred inside contigs but not between contigs (see Table S1).
Phylogenetic analysis.
All pufM gene sequences collected from the NCBI database, Citromicrobium genomes, and environmental samples were aligned using Clustal X, and phylogenetic trees were constructed using the maximum likelihood and neighbor-joining algorithms of the MEGA 6 software (43). The phylogenetic trees were supported by bootstrap for resampling test, with 100 and 1,000 replicates for the maximum likelihood and neighbor-joining algorithms, respectively.
Environmental sample collection.
Seawater samples were collected on board during a western Pacific Ocean cruise in July 2011. Seawater was collected at two stations (P3 [129.00°E, 14.00°N] and P10 [130.00°E, 2.00°N]) and five depths (5 m, 25 m, 75 m, 150 m, and 200 m). For each sample, 2 to 3 liters of seawater was prefiltered through a 20-μm-pore filter, and the microorganisms were then collected onto 0.22-μm-pore polycarbonate filters (Millipore). Nucleic acids were extracted using hot sodium dodecyl sulfate, phenol, chloroform, and isoamyl alcohol (24, 44). The high-quality DNA was stored at −20°C for future use.
Sequence generation and processing.
Considering that previous primers (2, 45) had five mismatches with pufM sequences belonging to the incomplete PGC in Citromicrobium, the following primer set was used to amplify the environmental DNA: pufM_Citro forward (5′-TACGGSAAYTTSTWCTAC-3′) and pufM_Citro reverse (5′-GCRAACCAGYANGCCCA-3′). High-throughput sequencing of the pufM gene (∼240 bp) was performed using Illumina MiSeq technology. The generated high-throughput sequence data were processed as described in Zheng et al. (24). Briefly, after quality control, all sequences were grouped into operational taxonomic units (OTUs) using a 6% cutoff. One representative sequence for each OTU was chosen to perform local BLAST against our pufM sequence database (for details, see Zheng et al. [24]).
Accession number(s).
The complete JL477 genome sequence is available under GenBank accession no. CP011344. Whole-genome sequences of strains JL31, JL354, JL1351, WPS32, JL2201, RCC1878, RCC1885, and RCC1897 are available under GenBank accession numbers LAIH00000000, ADAE00000000, LAPR00000000, LAPS00000000, LARQ00000000, LBLZ00000000, LBLY00000000, and LUGI01000000, respectively. All pufM gene representative sequences of Citromicrobium-related OTUs and each OTU's abundance are shown in Table S2 in the supplemental material.
RESULTS AND DISCUSSION
Overview of nine Citromicrobium strains.
Nine Citromicrobium strains were used to perform comparative genome analyses, which were isolated from the South China Sea (strains JL31, JL354, JL477, JL1351, and WPS32), the Mediterranean Sea (strains RCC1878, RCC1885, and RCC1897), and the tropical South Atlantic Ocean (strain JL2201) (Table 1). Although 1 to 2 base mismatches were found in the 16S rRNA sequences collected from the GenBank database (strains JL354, RCC1878, RCC1885, and RCC1897) or after Sanger sequencing (the other five strains), the 16S rRNA sequences (1,442 bp, one copy per genome) extracted from the nine genomes with high sequencing coverage were identical. The Sanger sequencing might induce some biases or mismatches during the 16S rRNA amplification and sequencing PCR steps.
The nine genomes displayed highly similar genomic characteristics in terms of genome size (from 3.16 to 3.28 Mb), G+C content (from 64.8 to 65.1%), gene number (from 3,056 to 3,250), COGs (from 1,934 to 2,010), and tRNA number (44 or 45) (Table 1).
Pan- and core genomes of the Citromicrobium strains.
Based on the total set of genes from the 9 sequenced strains, the Citromicrobium pangenome consisted of 3,546 predicted orthologous clusters (OCs), with a conserved core genome of 2,691 OCs. The cumulative length of the core genome was approximately 2.50 Mbp, which covered >75% of each genome. The flexible genome comprises 853 OCs, including 362 unique OCs and 490 OCs shared by more than one strain but not all strains.
The core genome is mainly involved in housekeeping functions and central metabolism, from the Calvin cycle to the tricarboxylic acid (TCA) cycle. Approximately 80% of the predicted core genes are assigned to COG functional categories. The predicted core genes contain a relatively high percentage of genes assigned to the following COG categories: general function prediction only (R), amino acid transport and metabolism (E), unknown function (S), translation, ribosomal structure, and biogenesis (J), energy production and conversion (C), lipid transport and metabolism (I), cell wall/membrane/envelope biogenesis (M), and inorganic ion transport and metabolism (P). Due to a larger fraction of putative or hypothetical genes, only 36.8% of the flexible genes are assigned to COG functional categories. Compared to the core genes, they include an overrepresentation of genes assigned to the following COG categories: general function prediction only (R), lipid transport and metabolism (I), replication, recombination and repair (L), intracellular trafficking, secretion, and vesicular transport (U), secondary metabolite biosynthesis, transport, and catabolism (Q), cell motility (N), transcription (K), and defense mechanisms (V). Most of flexible genes were sourced from the genetic island regions.
Genomic ANI.
The average nucleotide identity (ANI) shared between genome pairs ranged from 95.96% to 100% (see Table S3 in the supplemental material). Five genomes, JL31, JL1351, JL354, JL477, and JL2201, share more than 99.5% ANI between them but share lower values with three RCC strains (from 95.96 to 96.47%) and WPS32 (from 96.39 to 96.44%). The three RCC strains had the lowest percentages of all the genomes involved in pairwise comparisons (see Table S3).
Genome pairs JL31 and JL1351, JL31 and JL2201, JL1351 and JL2201, JL477 and JL354, RCC1878 and RCC1885, RCC1878 and RCC1897, and RCC1885 and RCC1897 showed strikingly high ANI (almost 100%) (see Table S3 in the supplemental material). Among them, genome pairs JL31 and JL1351, JL477 and JL354, RCC1878 and RCC1885, RCC1878 and RCC1897, and RCC1885 and RCC1897 showed high genomic percentages (>98.0%) involved in pairwise comparisons (see Table S3), indicating closer evolutionary relationships with each other.
The proposed cutoff of the ANI between two genome sequences for a species boundary is 95 to 96% (36). Five JL strains share 95.96 to ∼96.47% ANI with three RCC strains; therefore, three RCC strains isolated from the almost-enclosed Mediterranean Sea have a long divergence history from other strains and tend to evolve into a novel group. However, all nine Citromicrobium strains have identical 16S rRNA sequences. This emphasizes that traditional diversity studies, which classify sequences into operational taxonomic units based on the nucleotide sequence similarity, underestimate real environmental microbial information. The classification and diversity results based on environmental 16S rRNA could not link to in situ microbial functions (46, 47).
Comparison of nine genomes.
A comparison of all nine genomes (JL477 versus the others) showed high synteny of major regions and a significantly high level of sequence conservation (Fig. 1; see Table S1 in the supplemental material). DNA fragment insertions and deletions were detected in a genome comparison (one versus the other eight) (see Table S1).
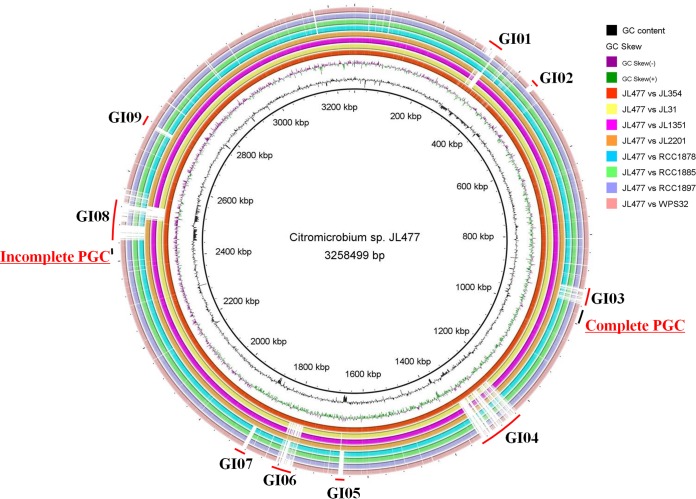
FIG 1.
Whole-genome map of Citromicrobium sp. JL477 compared with other eight Citromicrobium genomes. From the inner to outer circles: G+C content plot with a gray circle representing 50%, GC skew plot, Citromicrobium sp. JL354, Citromicrobium sp. JL31, Citromicrobium sp. JL1351, Citromicrobium sp. JL2201, Citromicrobium sp. RCC1878, Citromicrobium sp. RCC1885, RCC1897, and Citromicrobium sp. WPS32. Genomic island regions are indicated with red line on the outermost circle from GI01 to GI09. The complete and incomplete PGCs are labeled by the black line on the outermost circle.
Horizontal gene transfer (HGT) is common in bacteria, contributing to the genomic plasticity and possibly to environmental adaptation (48). To better understand genome plasticity and unique genome characteristics, nine specific genomic regions larger than 10 kb (except GI07, with 9.7 kb) in size were identified based on the comparative genome map (Fig. 1). They were absent or different in the corresponding regions of the eight other genomes (JL477 versus the others) and are here designated genomic islands (GIs) GI01 to GI09 (Fig. 1). These nine GIs each contribute approximately 12% of the genome size. Almost all the GIs were regarded as originating from HGT (gene gain and loss) mediated by the transposases, integrases, and conjugal transfer systems, and five of them (GI03, GI04, GI07, GI08, and GI09) were flanked by a tRNA gene. Previous studies showed that GIs frequently originate from integration events associated with tRNA-encoding genes (49–51). These nine GIs are scattered throughout the genomes, and their general features and sequence information are summarized in Table 2.
TABLE 2.
Detailed information for the nine GIs
| GI | Size (kb) | G+C content (%) | tRNA(s) | No. of genes | No. of transposases and integrases | No. of hypothetical proteins | Predicted function(s) |
|---|---|---|---|---|---|---|---|
| 01 | 36.7 | 60.66 | 34 | 0 | 10 | T4SS | |
| 02 | 11.4 | 66.80 | 16 | 1 | 4 | GTA | |
| 03 | 35.5 | 65.13 | tRNA-Ser-GGA | 36 | 1 | 10 | Flagellar biosynthesis |
| 04 | 101.1 | 62.14 | tRNA-Ser-GCT | 88 | 1 | 18 | Choline and betaine uptake, glycerolipid and glycerophospholipid metabolism, fatty acid metabolism, pyruvate metabolism |
| 05 | 11.3 | 52.08 | 0 | 0 | Unknown | ||
| 06 | 38.1 | 65.98 | 58 | 2 | 30 | Prophage | |
| 07 | 9.7 | 54.39 | tRNA-Pro-TGG, tRNA-Met-CAT | 4 | 1 | 2 | Unknown |
| 08 | 113.4 | 60.62 | tRNA-Met-CAT | 98 | 1 | 21 | ICE |
| 09 | 11.5 | 65.76 | tRNA-Ser-CGA | 3 | 0 | 0 | Flagellar hook-length control |
GI01 and T4SS.
GI01 mainly consists of a trb gene cluster, trbBCDEJLFGI, which is probably involved in the conjugal transfer of mobile genetic elements mediated by the type IV secretion system (T4SS) (51–53). In the genomes of strains JL354, RCC1878, RCC1885, and RCC1897, the highly homologous gene cluster (here denoted T4SS-I) is detected at the same chromosome position as in strain JL477 (Fig. 2A), which is flanked by putative genes for T4SS protease (traF), relaxase (virD2), and ATPase for transfer DNA (T-DNA) transfer (virD4) in the upstream regions and for genes associated with amino acids metabolism, transmembrane transport, and transcriptional regulation in the downstream region.
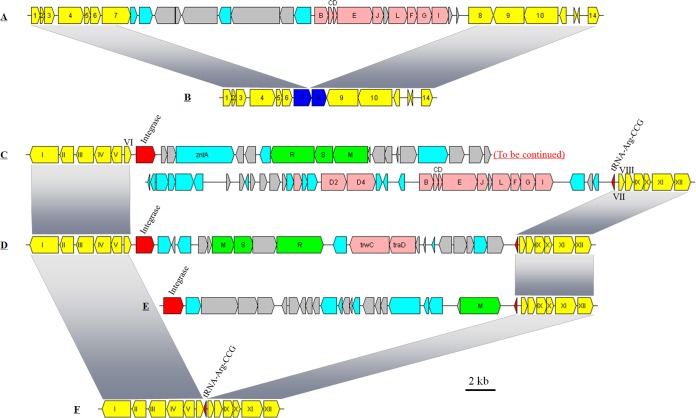
FIG 2.
Organization of GI01 structural genes in Citromicrobium genomes. (A) Structure found in strains JL477, JL354, RCC1878, RCC1885, and RCC1897. (B) Structure found in strains JL31, JL1351, and JL2201. (C) Structure found in strains JL31, JL1351, and JL2201. (D) Structure found in the three RCC strains. (E) Structure found in strain WPS32. (F) Structure found in strains JL477 and JL354. Yellow, conserved upstream and downstream genes (from 1 to 14, and from I to XII) of the GI01 gene cluster in Citromicrobium genomes; pink, trb gene cluster; red, tRNA or integrase; green, type I restriction-modification system; cyan, genes with other known functions; light gray, hypothetical genes. 1, type IV secretory pathway, protease TraF; 2, hypothetical protein; 3, membrane-bound lytic murein transglycosylase C precursor; 4, type IV secretory pathway, VirD2 components (relaxase); 5, hypothetical protein; 6, hypothetical protein; 7, coupling protein VirD4, ATPase required for T-DNA transfer; 8, asparagine synthetase (glutamine-hydrolyzing); 9, acylamino-acid-releasing enzyme; 10, TonB-dependent receptor; 11, RNA polymerase sigma-70 factor, extracytoplasmic factor (ECF) subfamily; 12, hypothetical protein; 13, hypothetical protein; 14, transcriptional regulator. I, cell division protein FtsH; II, ATPase, ParA family protein; III, butyryl-CoA dehydrogenase; IV, alpha-methylacyl-CoA racemase; V, enoyl-CoA hydratase; VI, ferrichrome-iron receptor; VII, hypothetical protein; VIII, hypothetical protein; IX, sterol desaturase family protein; X, hypothetical protein; XI, hypothetical protein; XII, hypothetical protein.
Interestingly, the same flanking gene organization was found in the genomes of strains JL31, JL1351, and JL2201 but with gene fragment loss in the middle of two genes (genes 7 and 8) (Fig. 2B). There is a 770-bp deletion in the latter part of gene 7 and a 670-bp deletion in the front part of gene 8, which indicates a large DNA fragment deletion in these three genomes.
In the genomes of strains JL31, JL1351, and JL2201, a trb gene cluster (here denoted T4SS-II) that is located in an integrase-mediated foreign DNA fragment was also found (Fig. 2C). The average nucleotide identity between T4SS-I and T4SS-II was low (<50%), indicating that the T4SS-II gene cluster was acquired via HGT mediated by the integrase. In addition, a three-gene cluster coding for the type I restriction-modification system (TIRS), which protects microbes from the foreign DNA (e.g., bacteriophage), was detected in the integrated DNA fragment. The inserted sequence is adjacent to the tRNA-CCG gene in these three genomes. In the genomes of strains RCC1878, RCC1885, RCC1897, and WPS32, HGT derived from integration events also occurred adjacent to the tRNA-CCG gene (Fig. 2D and E). However, different inserted gene clusters were found in these three genomes. A TIRS gene cluster was also observed in the genomes of strains RCC1878, RCC1885, and RCC1897, but TIRS gene clusters from three RCC strains share less than 50% nucleotide identity with strains JL31, JL1351, and JL2201 (Fig. 2D). Two genes homologous to trwC and traD, both involved in conjugative transfer, were found next to the TIRS in the inserted sequence of strains RCC1878, RCC1885, and RCC1897 (Fig. 2D). The inserted sequence flanking tRNA-CCG in the genome of strain WPS32 contains a few genes involved in restriction-modification and antirestriction, and several hypothetical genes (Fig. 2E). No inserted genes were found around tRNA-CCG in the genomes of JL477 and JL354 (Fig. 2F). This suggests that different foreign DNA fragments are independently integrated into the same tRNA gene, which contributes to bacterial genome evolution and species divergence (50). No trb gene cluster was found in the genome of strain WPS32.
GI02, containing a gene transfer agent.
A gene transfer agent (GTA) is an unusual bacteriophage-like element of genetic exchange that transfers a random host genomic DNA fragment (4 to 14 kb in size) between closely related bacteria (54, 55). Analysis of citromicrobial genomes found a GTA gene cluster present in all nine genomes at the same chromosome position in GI02 (see Fig. S1 in the supplemental material). The structure and composition of the GTA gene cluster and flanking genes are identical in all the genomes, except for that of strain WPS32 (see Fig. S1B). For example, an approximately 3-kb DNA fragment mediated by transposase is inserted in the front of GTA in all genomes but was absent in strain WPS32. We speculate that in this strain, the transposase, after acquisition, mediated the gene loss of the downstream region, including the ORFs of the GTA.
GI03, involved in flagella and motility.
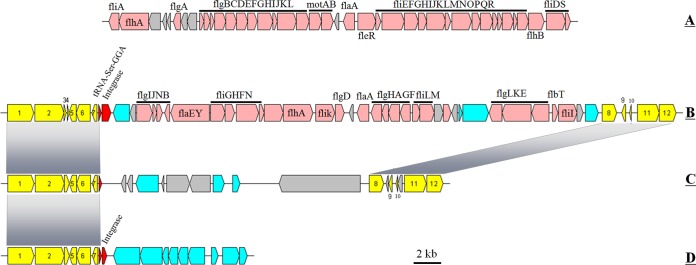
Flagella support marine bacterial motility and allow cells to move toward favorable living conditions in the environment, e.g., nutrient-rich and light (56–58). Sometimes, flagella also contribute to adhesion (56, 59). In some special cases, flagella might also provide an advantage for bacterial competition (56, 57). Two gene clusters for flagellar biosynthesis were found in the nine genomes (Fig. 3). The first cluster (flagella I) was common in all genomes (Fig. 3A), while the second (35.5 kb in size, flagella II) was detected in only five genomes, JL31, JL354, JL477, JL2201, and JL1351 (Fig. 3B). Flagella I mainly consists of two large gene clusters (flgBCDEFGHIJKL and fliEFGHIJKLMNOPQR), motAB, fliDS, and some regulatory genes (Fig. 3A). The organization of flagella II is irregular (Fig. 3B).
FIG 3.
Organization of flagellar and GI03 structural genes in Citromicrobium genomes. Yellow, conserved upstream and downstream genes (from 1 to 12) of the GI03 gene cluster in Citromicrobium genomes; pink, flagellar gene cluster; red, tRNA or integrase; cyan, genes with other known functions; light gray, hypothetical genes.
An integrase mediates the acquisition of the flagella II gene cluster, and the integration event occurs adjacent to the tRNA-GGA gene. In the other four genomes, the inserted DNA sequences were also found at the same position. In the three RCC genomes, a 17.1-kb inserted fragment was detected, and only a few genes could be annotated as encoding a known function (traG and DNA invertase) (Fig. 3C). In the WPS32 genome, genes involved in the serine-glyoxylate cycle and that were respiration related were found at the same position (Fig. 3D).
GI04.
GI04, the longest GI, at 101.1 kb in size, is mainly involved in choline and betaine uptake as well as the metabolism of glycerolipids, glycerophospholipids, fatty acids, and pyruvate. This large DNA fragment was integrated into JL354 and JL477 chromosomes via an integrase flanked by the tRNA-GCT gene. The other genomes, except for WPS32, have the same flanking genes with no GI04 sequences. GI04 contains 88 genes, 18 of which have unknown functions. The G+C content of GI04 (62.14%), is lower than the genomic G+C content.
In the WPS32 genome, an approximately 47.9-kb inserted DNA fragment was also found at the same chromosome position adjacent to the tRNA-GCT gene, and its acquisition was mediated by the integrase. It also contains several fatty acid metabolism-related genes but with significantly lower sequence identity (or different genes) than in strains JL477 and JL354.
GI05.
GI05 (approximately 11.3 kb) displayed the lowest G+C content (52.08%). Only found in the genomes of strains JL31, JL354, JL477, JL1351, and JL2201, it contains four genes, and its coding region represents less than 50% of its sequence. The only known function was a DNA polymerase of family B.
GI06, a Mu-like prophage.
In a previous study, we isolated one inducible bacteriophage from strain JL354, consisting of three parts: an early expression region and regions encoding heads and tails (60). Nearly identical prophage sequences were observed in the genomes of strains JL354, JL477, and JL2201. They share high levels of structural conservation and sequence identity and are defined here as prophage type I (see Fig. S2 in the supplemental material). In addition, another type of prophage (here defined as prophage type II) was found in strains JL2201, RCC1878, RCC1885, and RCC1897. The type II prophage has structure modules similar to those of type I (see Fig. S2), but they share significantly lower sequence identity.
All three type I prophages share the same upstream and downstream genes, and the first downstream gene is a transposase. Type II prophages integrated into the host chromosome in a different position with type I prophage. This indicates that these two types of prophages originate from different integration events.
Two types of prophages coexist in the genome of strain JL2201. The type I prophage in strain JL2201 is identical to the prophage found in strains JL354 and JL477. However, the type II prophage in strain JL2201 lost its early expression region but kept the structural genes encoding heads and tails (see Fig. S2 in the supplemental material). Interestingly, the structural genes form duplication (approximately 26 kb × 2) centers around the last gene with less than 93% nucleotide identity (see Fig. S2). The incomplete duplicated prophage sequence might contribute to an increase in viral particle production under the control of the early expression genes of prophage I in strain JL2201.
GI07.
GI07, located between the tRNA-TGG and tRNA-CAT genes, is the shortest (approximately 9.7 kb) among all GIs. Its G+C content (54.39%) is much lower than the genomic G+C content (64.8 to 65.1%). It contains three genes, an integrase, a hypothetical gene, and a reverse transcriptase, and the coding sequences represent approximately 50% of its length. The gene organization and composition of GI07 in the five JL genomes are identical.
In the three RCC strains, the inserted foreign DNA (approximately 11.3 kb with 60.67% G+C content) is after the tRNA-TGG gene. It consists of nine genes mainly involved in the type I restriction-modification system, flavodoxin reductase, and fatty acid metabolism. The corresponding region was not detected in the WPS32 genome.
GI08, involved in ICEs.
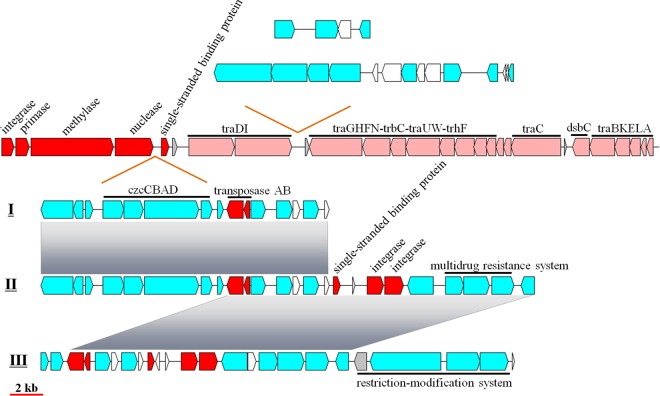
Integrative and conjugative elements (ICEs) are defined as self-transmissible mobile genetic elements with the capacity to integrate into and excise from a host chromosome (61, 62). The core ICEs are made up of three typical genetic modules: ICE integration and excision, ICE conjugation, and ICE regulation modules (62–64). ICEs integrate characteristics of both temperate bacteriophages (the front part) and conjugative plasmids (the latter part) (Fig. 4) (62, 65). ICEs have been reported to contain several intergenic hot spots where a diverse range of exogenous genes can be carried, including antibiotic or heavy-metal resistance genes (65). ICEs mediate HGT among prokaryotes and greatly facilitate microbial genome evolution and ecological fitness (61, 62).
FIG 4.
Structure and composition of ICEs. Two hot spots were detected in all ICEs. One contained three types of exogenous gene clusters (I, II, and III), and the other contained two types. Red, phage-related genes; pink, conjugation-related genes; cyan, genes with other known functions; white, hypothetical genes.
All analyzed genomes except that of strain WPS32 possess an ICE. Based on the structure and gene composition, the eight ICEs could be classified into three groups, namely, group 1 (JL477 and JL354), group 2 (JL1351, JL2201, and JL31), and group 3 (RCC1878, RCC1885, and RCC1897).
Two intergenic hot spots carrying exogenous genes were found in the eight genomes. The first one is located between genes encoding a nuclease and a single-stranded DNA binding protein. It also contains three different exogenous gene clusters (I, II, and III) corresponding to three types of ICEs (group 1, group 2, and group 3). Exogenous gene cluster I is present in the genomes of JL477 and JL354 (group 1) and mainly consists of heavy-metal resistance genes (czcCBAD) and their transcriptional regulator (merR), transposase AB, copper homeostasis, and antirestriction genes. Exogenous gene cluster II, found in group 2, contains all the genes of exogenous gene cluster I and also possesses several extra multidrug resistance genes, whose acquisition is mediated by two integrases. Interestingly, although no ICE was found in the genome of WPS32, this genome contains multidrug resistance genes and two integrases. Exogenous gene cluster III lost heavy-metal resistance genes but gained restriction-modification-related genes. This hot spot might carry a limited length of foreign genes, suggesting that these microbes might carefully select foreign genes that are optimally adapted to their environment.
The second hot spot, located between gene clusters traDI and traGHFN, mostly comprises peptidase and nuclease metabolism-related genes. In ICE group 1, it contains 14 genes with known functional genes for nucleases, helicases, ATPases, peptidases, and a transcriptional regulator. The group 2 ICE lost a large part of the conjugation module (traDI and traGHFN-trbC-traUW-trhF). The second hot spot in ICE group 3 is composed of only five genes related to peptidases, pyrophosphatases, and transcriptional regulators. Downstream of ICE group 3, an approximately 27-kb gene fragment mainly involved in fatty acid metabolism, butyrate metabolism, and branched-chain amino acid biosynthesis is absent in the genomes of three RCC strains and WPS32.
GI09.
GI09 is an approximately 11.5-kb fragment with a G+C content similar to that of the genome (65.76%). It contains three genes, a giant hypothetical gene (8.70 kb), a transcriptional regulator, and a histidine kinase, which together represent 97.14% of its sequence. GI09 is adjacent to a tRNA-CGA gene. The protein for the giant hypothetical gene product comprises three domains: an immunoglobulin beta-sandwich folding domain, a cadherin-like beta-sandwich domain, and an autotransporter beta-domain. Cadherins are suggestive of adhesion molecules that mediate Ca2+-dependent cell-cell junctions (66). Usually, bacteria or cells containing the same cadherins tend to preferentially aggregate.
GI09 was not detected at the same position in the genomes of strains RCC1878, RCC1885, and RCC1897. However, we found a remnant short sequence predicted to be a hypothetical gene (324 bp) that shares 91% (296/324) nucleotide identity with the giant hypothetical gene of strain JL477. This supports the hypothesis that three RCC strains lost the GI09 sequence.
Coexistence of two PGCs in genomes of Citromicrobium isolates.
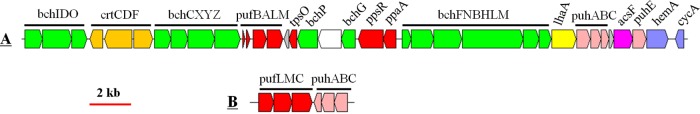
Interestingly, two different (one complete and one incomplete) PGCs were found in all nine genomes. The complete PGC consists of two conserved subclusters, crtCDF-bchCXYZ-pufBALM and bchFNBHLM-lhaA-puhABC (Fig. 5A). The complete PGC organization is identical in all nine genomes in terms of gene arrangement and composition. The incomplete PGC contains only the pufLMC and puhABC genes (Fig. 5B). The incomplete PGC, which was proved to be obtained by HGT (15, 16), is located at the same position in all the genomes and is flanked by respiratory complex I and coenzyme A (CoA) metabolism-related genes. This indicates that the ancestral Citromicrobium strains obtained the incomplete PGC before divergence. Both the complete and incomplete PGCs are close to the GI regions, creating conditions for gain and loss of phototrophic genes (Fig. 1).
FIG 5.
Structure and arrangement of two PGCs in Citromicrobium. (A) Complete PGC. (B) Incomplete PGC. Green, bch genes; red, puf and regulator genes; pink, puh genes; orange, crt genes; blue, hem and cyc genes; yellow, lhaA gene; blank, uncertain or unrelated genes; gray, hypothetical protein. The horizontal arrows represent putative transcripts.
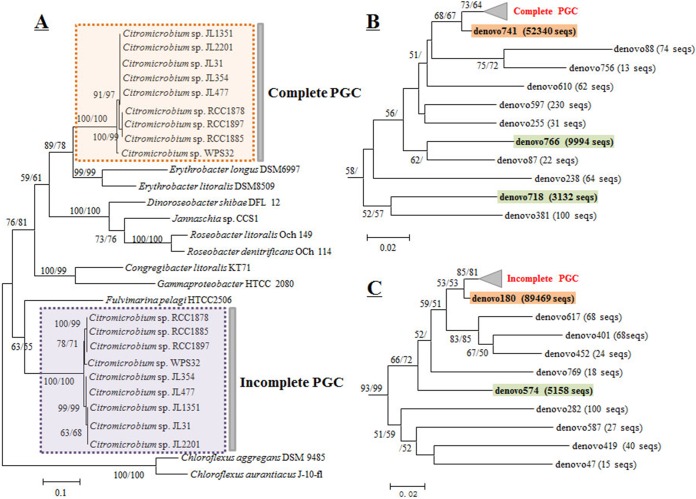
The pufM sequences from the complete PGC formed a clade close to that of Erythrobacter species also belonging to the order Sphingomonadales, alpha-IV subcluster (Fig. 6A). The pufM sequences from the incomplete PGC formed a distant clade branching with Fulvimarina pelagi HTCC 2506 (alpha-VI subcluster) (Fig. 6A). This phylogenetic placement is in agreement with our previous finding showing that the incomplete PGC genes might have been acquired from a Fulvimarina-related species (16).
FIG 6.
Neighbor-joining phylogenetic trees based on pufM gene sequences. (A) Phylogenetic tree containing pufM sequences of the nine isolates. (B) Partial tree containing environmental pufM sequences from the complete PGC. (C) Partial tree containing environmental pufM sequences from the incomplete PGC. Only bootstrap percentages (>50%) are shown (neighbor-joining/maximum likelihood).
In both pufM clades, the sequences could be grouped into three clusters: three RCC strains formed one cluster, WPS32 by itself was a second cluster, and the other five strains formed a third cluster (Fig. 6A).
Coexistence of two copies of pufM in Citromicrobium environmental sequences.
A total of 540,022 good-quality sequence reads were obtained from two stations at five depths (5, 25, 75, 150, and 200 m) using the revised primers (Table 3). A large proportion (29.8%) of pufM sequences having Citromicrobium as the closest relative were obtained. Among them, 66,182 and 95,052 sequences were classified into the complete and incomplete PGC clades, respectively.
TABLE 3.
Distribution and identity of environmental pufM sequences retrieved from sites P3 and P10 at different depths
| OTU ID | No. of environmental pufM sequences by site and depth (m) |
Identity (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P10 |
P3 |
Total | ||||||||||
| 5 | 25 | 75 | 150 | 200 | 5 | 25 | 75 | 150 | 200 | |||
| denovo180 | 820 | 1,517 | 7,885 | 1,830 | 15,898 | 5,382 | 14,689 | 5,287 | 10,405 | 25,756 | 89,469 | 99.6 |
| denovo574 | 17 | 301 | 1,956 | 15 | 81 | 2,255 | 126 | 47 | 308 | 52 | 5,158 | 94.3 |
| denovo741 | 721 | 460 | 3,645 | 982 | 10,915 | 783 | 9,503 | 3,423 | 7,093 | 14,815 | 52,340 | 99.1 |
| denovo766 | 76 | 586 | 2,950 | 36 | 147 | 5,586 | 73 | 23 | 419 | 98 | 9,994 | 91.4 |
| denovo718 | 11 | 109 | 662 | 10 | 9 | 645 | 44 | 1,629 | 3 | 10 | 3,132 | 88.4 |
| Total sequences | 62,205 | 61,988 | 42,912 | 72,509 | 78,655 | 35,183 | 73,232 | 37,557 | 31,088 | 44,693 | 540,022 | |
Eleven and 10 OTU (>10 sequences) were classified into the citromicrobial complete and incomplete PGC clades, respectively (Fig. 6B and C). All the environmental sequences differed from pufM sequences from the isolates. Five main OTUs (with more than 1,000 sequences) were retrieved, three (denovo741, denovo766, and denovo718) in the complete PGC clade and two (denovo180 and denovo574) in the incomplete PGC clade (Table 3). Interestingly, denovo741 and denovo180 showed similar positions in their phylogenetic trees (Fig. 6B and C). Their representative sequences shared 99.1% (230/232) and 99.6% (227/228) nucleotide identity with the pufM sequences belonging to the complete and incomplete PGCs of strain JL477, respectively. In addition, denovo741 and denovo180 demonstrated the same depth distribution pattern (Table 3). A similar situation was observed for denovo766 and denovo574, whose representative sequences shared 91.4% (212/232) and 94.3% (217/230) nucleotide identity, respectively (Table 3).
However, our analysis did not find an OTU corresponding to a copy of denovo718 in the incomplete PGC clade (Fig. 5B). This may suggest that some citromicrobial strains have lost the incomplete PGC or that denovo718 is a novel Citromicrobium relative.
SNPs.
The number of SNPs of the eight genomes relative to the complete genome of strain JL477 had a wide range. More than 84,000 SNPs were found in the genomes of strains RCC1878, RCC1885, RCC1897, and WPS32, while fewer than 200 SNPs were present in strains JL354, JL31, and JL1351. In the genome of JL2201, 1,603 SNPs were found, and most of them (1,379 SNPs) originated from the prophage I sequences, suggesting that viruses had much higher evolutionary rates. Approximately 90% of all SNPs are located in coding regions and are scattered throughout the genomes except in the genetic islands.
Based on the growth rate (0.72 to 2.13 day−1) of AAPB in the ocean (3), their generation time should be approximately 250 to 750 generations per year. The estimated divergence times based on the accumulation of synonymous mutations that excluded SNPs from GIs span a long history. The divergence times among JL477, JL31, JL1351, and JL354 are in century timescales, and these four strains diverge at a millennial timescale with JL2201. The three RCC strains and the WPS32 strain diverged from the five JL strains millions of years ago.
Geographic relationship.
The isolates used in the study originate from diverse geographic locations, including the Mediterranean Sea, the South China Sea, and the South Atlantic Ocean. Water from the Atlantic Ocean refilled the Mediterranean Sea through the Strait of Gibraltar 5.33 million years ago (67, 68). Before the water poured in, the Mediterranean almost entirely dried out as a result of the Messinian salinity crisis (67, 68). In other words, the modern Mediterranean Sea has an ∼5.33 million-year history. Microbes in the almost-enclosed Mediterranean Sea might have evolved their unique characteristics compared to those in the other open-ocean regions. That is consistent with the divergence time between three RCC strains and five JL strains.
Both phylogenies based on marker genes and comparisons of genome sequences revealed that strains from the same region (South China Sea or Mediterranean Sea) shared a similar evolutionary history and are distinct from those originating from other regions (South China Sea versus Mediterranean Sea). Geographic differences are partly responsible for driving the observed evolutionary divergences, and they allow microbes to diverge through local adaptation to specific environmental conditions (69–71). The divergence processes within species are traditionally considered microevolutionary. However, some specific events, such as viral infection, grazing, or extreme physical events, might contribute to unusual evolutionary diversification (e.g., strain WPS32).
HGT plays an important role in Citromicrobium genomic plasticity. Three integration events occurred, mediated by two types of prophages (JL477 and JL354; three RCC strains; JL2201), corresponding to the three marine regions from which the strains originated. Three of the nine strains were free of viral infection. Several genes preventing viral infection are detected in their GIs, suggesting that bacterium-phage interactions are actively ongoing in their environment.
A comparison of nine Citromicrobium genomes that share identical 16S rRNA sequences provides new insights into bacterial microevolution and divergence under different environments. The distribution of various genetic islands plays important roles in genomic plasticity and adaptability. The information gathered by comparing Citromicrobium genomes sheds new light on the evolution and environmental adaptations resulting from geographic isolation in Citromicrobium species.
Supplementary Material
ACKNOWLEDGMENTS
We thank the four anonymous reviewers for their useful comments and suggestions.
We declare no conflicts of interest.
Funding Statement
This work was supported by the NSFC project 41306126, the national key research programs (grants 2013CB955700 and 2016YFA0601400), the SOA project (grant GASI-03-01-02-05), CNOOC grants CNOOC-KJ125FZDXM00TJ001-2014 and CNOOC-KJ125FZDXM00ZJ001-2014, and the China National Marine Science Talent Training Base project (grant 2015Z01).
The work performed at the Station Biologique (Roscoff) is a contribution of the BOUM (Biogeochemistry from the Oligotrophic to the Ultraoligotrophic Mediterranean) experiment (http://www.com.univ-mrs.fr/BOUM/) of the French national LEFE-CYBER program, the European IP SESAME and the international IMBER project. The BOUM experiment was coordinated by the Institut National des Sciences de l'Univers (INSU) and managed by the Centre National de la Recherche Scientifique (CNRS).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02495-16.
REFERENCES
- 1.Kolber ZS, Gerald F, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblizek M, Rathgeber C, Falkowski PG. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 2.Béjà O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630–633. doi: 10.1038/415630a. [DOI] [PubMed] [Google Scholar]
- 3.Koblížek M, Mašín M, Ras J, Poulton AJ, Prášil O. 2007. Rapid growth rates of aerobic anoxygenic phototrophs in the ocean. Environ Microbiol 9:2401–2406. doi: 10.1111/j.1462-2920.2007.01354.x. [DOI] [PubMed] [Google Scholar]
- 4.Koblížek M. 2015. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev 39:854–870. doi: 10.1093/femsre/fuv032. [DOI] [PubMed] [Google Scholar]
- 5.Jiao N, Zhang Y, Zeng Y, Hong N, Liu R, Chen F, Wang P. 2007. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ Microbiol 9:3091–3099. doi: 10.1111/j.1462-2920.2007.01419.x. [DOI] [PubMed] [Google Scholar]
- 6.Yutin N, Suzuki MT, Teeling H, Weber M, Venter JC, Rusch DB, Béjà O. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ Microbiol 9:1464–1475. doi: 10.1111/j.1462-2920.2007.01265.x. [DOI] [PubMed] [Google Scholar]
- 7.Yurkov V, Csotonyi JT. 2009. New light on aerobic anoxygenic phototrophs, p 31–55. In Hunter N, Daldal F, Thurnauer MC, Beatty JT (ed), The purple phototrophic bacteria. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 8.Ritchie AE, Johnson ZI. 2012. Abundance and genetic diversity of aerobic anoxygenic phototrophic bacteria of coastal regions of the Pacific Ocean. Appl Environ Microbiol 78:2858–2866. doi: 10.1128/AEM.06268-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrera I, Borrego CM, Salazar G, Gasol JM. 2014. Marked seasonality of aerobic anoxygenic phototrophic bacteria in the coastal NW Mediterranean Sea as revealed by cell abundance, pigment concentration and pyrosequencing of pufM gene. Environ Microbiol 16:2953–2965. doi: 10.1111/1462-2920.12278. [DOI] [PubMed] [Google Scholar]
- 10.Stegman MR, Cottrell MT, Kirchman DL. 2014. Leucine incorporation by aerobic anoxygenic phototrophic bacteria in the Delaware estuary. ISME J 8:2339–2348. doi: 10.1038/ismej.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koblížek M, Béjà O, Bidigare RR, Christensen S, Benitez-Nelson B, Vetriani C, Kolber MK, Falkowski PG, Kolber ZS. 2003. Isolation and characterization of Erythrobacter sp. strains from the upper ocean. Arch Microbiol 180:327–338. doi: 10.1007/s00203-003-0596-6. [DOI] [PubMed] [Google Scholar]
- 12.Swingley WD, Sadekar S, Mastrian SD, Matthies HJ, Hao J, Ramos H, Acharya CR, Conrad AL, Taylor HL, Dejesa LC, Shah MK, O'huallachain ME, Lince MT, Blankenship RE, Beatty JT, Touchman JW. 2007. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J Bacteriol 189:683–690. doi: 10.1128/JB.01390-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Q, Zhang R, Koblížek M, Boldareva EN, Yurkov V, Yan S, Jiao N. 2011. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One 6:e25050. doi: 10.1371/journal.pone.0025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beatty JT. 1995. Organization of photosynthesis gene transcripts, p 1209–1219. In Blankenship RE, Madigan MT, Bauer CE (ed), Anoxygenic photosynthetic bacteria. Springer, Dordrecht, The Netherlands. [Google Scholar]
- 15.Jiao N, Zhang R, Zheng Q. 2010. Coexistence of two different photosynthetic operons in Citromicrobium bathyomarinum JL354 as revealed by whole-genome sequencing. J Bacteriol 192:1169–1170. doi: 10.1128/JB.01504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Q, Zhang R, Fogg PC, Beatty JT, Wang Y, Jiao N. 2012. Gain and loss of phototrophic genes revealed by comparison of two Citromicrobium bacterial genomes. PLoS One 7:e35790. doi: 10.1371/journal.pone.0035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yurkov VV, Krieger S, Stackebrandt E, Beatty JT. 1999. Citromicrobium bathyomarinum, a novel aerobic bacterium isolated from deep-sea hydrothermal vent plume waters that contains photosynthetic pigment-protein complexes. J Bacteriol 181:4517–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rathgeber C, Lince MT, Alric J, Lang AS, Humphrey E, Blankenship RE, Verméglio A, Plumley FG, Van Dover CL, Beatty JT, Yurkov V. 2008. Vertical distribution and characterization of aerobic phototrophic bacteria at the Juan de Fuca Ridge in the Pacific Ocean. Photosynth Res 97:235–244. doi: 10.1007/s11120-008-9332-z. [DOI] [PubMed] [Google Scholar]
- 19.Balashov SP, Imasheva ES, Boichenko VA, Antón J, Wang JM, Lanyi JK. 2005. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science 309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balashov S, Lanyi J. 2007. Xanthorhodopsin: proton pump with a carotenoid antenna. Cell Mol Life Sci 64:2323–2328. doi: 10.1007/s00018-007-7167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boeuf D, Audic S, Brillet-Guéguen L, Caron C, Jeanthon C. 2015. MicRhoDE: a curated database for the analysis of microbial rhodopsin diversity and evolution. Database (Oxford) 2015:bav080. doi: 10.1093/database/bav080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon S-K, Kim BK, Song JY, Kwak M-J, Lee CH, Yoon J-H, Oh TK, Kim JF. 2013. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol Evol 5:187–199. doi: 10.1093/gbe/evs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riedel T, Gómez-Consarnau L, Tomasch J, Martin M, Jarek M, González JM, Spring S, Rohlfs M, Brinkhoff T, Cypionka H, Goker M, Fiebig A, Klein J, Goesmann A, Fuhrman JA, Wagner-Dobler I. 2013. Genomics and physiology of a marine flavobacterium encoding a proteorhodopsin and a xanthorhodopsin-like protein. PLoS One 8:e57487. doi: 10.1371/journal.pone.0057487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Q, Liu Y, Steindler L, Jiao N. 2015. Pyrosequencing analysis of aerobic anoxygenic phototrophic bacterial community structure in the oligotrophic western Pacific Ocean. FEMS Microbiol Lett 362:fnv034. doi: 10.1093/femsle/fnv034. [DOI] [PubMed] [Google Scholar]
- 25.Jeanthon C, Boeuf D, Dahan O, Gall FL, Garczarek L, Bendif EM, Lehours A-C. 2011. Diversity of cultivated and metabolically active aerobic anoxygenic phototrophic bacteria along an oligotrophic gradient in the Mediterranean Sea. Biogeosciences 8:1955–1970. doi: 10.5194/bg-8-1955-2011. [DOI] [Google Scholar]
- 26.Agogué H, Lamy D, Neal PR, Sogin ML, Herndl GJ. 2011. Water mass-specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol 20:258–274. doi: 10.1111/j.1365-294X.2010.04932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso JG, Lauber CL, Walters WA, Berglyons D, Lozupone C, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salzberg SL, Delcher AL, Kasif S, White O. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res 26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borodovsky M, McIninch J. 1993. GENEMARK: parallel gene recognition for both DNA strands. Comput Chem 17:123–133. [Google Scholar]
- 31.Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angiuoli SV, Gussman A, Klimke W, Cochrane G, Field D, Garrity GM, Kodira CD, Kyrpides N, Madupu R, Markowitz V, Tatusova T, Thomson N, White O. 2008. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS 12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GS, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagesen K, Hallin P, Rødland E, Stærfeldt H, Rognes T, Ussery D. 2007. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:0955–0964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinklac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A 102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 40.Delcher AL, Salzberg SL, Phillippy AM. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics Chapter 10:Unit 10.13. [DOI] [PubMed] [Google Scholar]
- 41.Foster JT, Beckstrom-Sternberg SM, Pearson T, Beckstrom-Sternberg JS, Chain PS, Roberto FF, Hnath J, Brettin T, Keim P. 2009. Whole-genome-based phylogeny and divergence of the genus Brucella. J Bacteriol 191:2864–2870. doi: 10.1128/JB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuhrman JA, Comeau DE, Hagström Å, Chan AM. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol 54:1426–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yutin N, Suzuki MT, Béjà O. 2005. Novel primers reveal wider diversity among marine aerobic anoxygenic phototrophs. Appl Environ Microbiol 71:8958–8962. doi: 10.1128/AEM.71.12.8958-8962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajendhran J, Gunasekaran P. 2011. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res 166:99–110. doi: 10.1016/j.micres.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Delgado-Baquerizo M, Giaramida L, Reich PB, Khachane AN, Hamonts K, Edwards C, Lawton LA, Singh BK. 2016. Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J Ecol 104:936–946. doi: 10.1111/1365-2745.12585. [DOI] [Google Scholar]
- 48.Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 49.Hacker J, Hentschel U, Dobrindt U. 2003. Prokaryotic chromosomes and disease. Science 301:790–793. doi: 10.1126/science.1086802. [DOI] [PubMed] [Google Scholar]
- 50.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaneko T, Maita H, Hirakawa H, Uchiike N, Minamisawa K, Watanabe A, Sato S. 2011. Complete genome sequence of the soybean symbiont Bradyrhizobium japonicum strain USDA6T. Genes 2:763–787. doi: 10.3390/genes2040763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol 33:1210–1220. [DOI] [PubMed] [Google Scholar]
- 53.Lawley T, Klimke W, Gubbins M, Frost L. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett 224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 54.Marrs B. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc Natl Acad Sci U S A 71:971–973. doi: 10.1073/pnas.71.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang AS, Beatty JT. 2007. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol 15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Grossart H-P, Riemann L, Azam F. 2001. Bacterial motility in the sea and its ecological implications. Aquatic Microb Ecol 25:247–258. doi: 10.3354/ame025247. [DOI] [Google Scholar]
- 57.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 58.Stocker R. 2012. Marine microbes see a sea of gradients. Science 338:628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- 59.Svensson SL, Pryjma M, Gaynor EC. 2014. Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PLoS One 9:e106063. doi: 10.1371/journal.pone.0106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng Q, Zhang R, Xu Y, White RA III, Wang Y, Luo T, Jiao N. 2014. A marine inducible prophage vB_CibM-P1 isolated from the aerobic anoxygenic phototrophic bacterium Citromicrobium bathyomarinum JL354. Sci Rep 4:7118. doi: 10.1038/srep07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Böltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol 184:5158–5169. doi: 10.1128/JB.184.18.5158-5169.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burrus V, Marrero J, Waldor MK. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173–183. doi: 10.1016/j.plasmid.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 63.Ravatn R, Studer S, Springael D, Zehnder AJ, van der Meer JR. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J Bacteriol 180:4360–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hochhut B, Marrero J, Waldor MK. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol 182:2043–2047. doi: 10.1128/JB.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pokutta S, Weis WI. 2007. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol 23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 67.Krijgsman W, Langereis C, Zachariasse W, Boccaletti M, Moratti G, Gelati R, Iaccarino S, Papani G, Villa G. 1999. Late Neogene evolution of the Taza–Guercif Basin (Rifian Corridor, Morocco) and implications for the Messinian salinity crisis. Mar Geol 153:147–160. doi: 10.1016/S0025-3227(98)00084-X. [DOI] [Google Scholar]
- 68.Garcia-Castellanos D, Estrada F, Jiménez-Munt I, Gorini C, Fernández M, Vergés J, De Vicente R. 2009. Catastrophic flood of the Mediterranean after the Messinian salinity crisis. Nature 462:778–781. doi: 10.1038/nature08555. [DOI] [PubMed] [Google Scholar]
- 69.Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 70.Papke RT, Ramsing NB, Bateson MM, Ward DM. 2003. Geographical isolation in hot spring cyanobacteria. Environ Microbiol 5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- 71.Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JB. 2012. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10:497–506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.