Abstract
Despite remarkable advances in our understanding of the drivers of human malignancies, new targeted therapies often fail to show sufficient efficacy in clinical trials. Indeed, the cost of bringing a new agent to market has risen substantially in the last several decades, in part fuelled by extensive reliance on preclinical models that fail to accurately reflect tumour heterogeneity. To halt unsustainable rates of attrition in the drug discovery process, we must develop a new generation of preclinical models capable of reflecting the heterogeneity of varying degrees of complexity found in human cancers. Patient-derived tumour xenograft (PDTX) models prevail as arguably the most powerful in this regard because they capture cancer’s heterogeneous nature. Herein, we review current breast cancer models and their use in the drug discovery process, before discussing best practices for developing a highly annotated cohort of PDTX models. We describe the importance of extensive multidimensional molecular and functional characterisation of models and combination drug–drug screens to identify complex biomarkers of drug resistance and response. We reflect on our own experiences and propose the use of a cost-effective intermediate pharmacogenomic platform (the PDTX-PDTC platform) for breast cancer drug and biomarker discovery. We discuss the limitations and unanswered questions of PDTX models; yet, still strongly envision that their use in basic and translational research will dramatically change our understanding of breast cancer biology and how to more effectively treat it.
Keywords: breast cancer, drug discovery, biomarker discovery, high-throughput screening, patient-derived tumour xenografts, targeted therapies, pharmacogenomics
Introduction
Breast cancer (herein BC) is not a single disease, but is instead a collection of diseases that have distinct histopathological features and genetic and genomic variability linked to diverse prognostic outcomes. Recent research has highlighted this heterogeneity and defined ten molecular subtypes based on copy number and gene expression data from over 2000 patient tumours (Curtis et al. 2012). Coupled with advances in our understanding of intertumour heterogeneity, large scale genomics projects such as The Cancer Genome Atlas (Chang et al. 2013) and METABRIC (Pereira et al. 2016) have led to unprecedented annotation of the drivers of BC. It is hoped that these advances will help improve patient stratification for targeted therapy based on the molecular underpinnings of individual cancer samples, paving the way towards personalised cancer treatment. However, despite the remarkable success of many such targeted agents, most investigational agents fail to show significant efficacy in clinical trials. Consequently, the oncological drug space suffers from 88% attrition between Phase I agents and market approval (Hutchinson & Kirk 2011). In cases where agents are initially efficacious, responses can be fleeting and the development of drug resistance is often seen as an inevitable consequence of cancer’s heterogeneity (Aparicio & Caldas 2013). Our reliance on preclinical models, unable to reflect this heterogeneity is therefore likely to underpin failures of the drug development framework (Cassidy et al. 2015).
Realising these limitations, the scientific community has been driven to create novel preclinical models that are able to recapitulate the complexity of human cancers. Many have turned to patient-derived tumour xenografts (PDTXs) (Whittle et al. 2015), which retain the complex heterogeneity of their originating tumour samples (DeRose et al. 2011, Cassidy et al. 2015, Eirew et al. 2015). PDTX models of BC resemble primary tumours across the genomic, epigenomic and transcriptomic landscape and are stable across multiple passages (Marangoni et al. 2007, Kabos et al. 2012, Eirew et al. 2015, Bruna et al. 2016). As preclinical models, PDTXs can be used to predict clinical trial responses (Gao et al. 2015); however, there use in the discovery phase itself has thus far been limited.
The drug discovery process is almost exclusively split between rational design, based on structural biology of the target protein and high-throughput screening (HTS). HTS strategies typically rely on increasingly simplified biological models, such as cancer cell lines containing reporter constructs for the pathway of interest and highly complex compound libraries (Kenny et al. 2015). Although PDTX models are undoubtedly more biologically relevant than cancer cell lines, they are limited by low throughput and high establishment costs (Siolas & Hannon 2013, Whittle et al. 2015).
Herein, we discuss the role of PDTX models in the BC drug discovery process. We begin by considering currently available models of BC and their uses in the drug discovery process before presenting the argument for increased use of models accurately reflecting the complexity of human malignancies. This complexity brings specific considerations, particularly in the need for high-throughput drug combination screens and deep genomic characterisation of models to enable biomarker discovery. We conclude by reflecting on our own experiences in developing an integrated pharmacogenomic pipeline for breast cancer drug discovery using PDTX cells (or PDTCs).
Preclinical models of breast cancer
BC is a collection of diseases with distinct biological traits and clinical outcomes. Thus, no individual model would be expected to completely recapitulate human BC in its entirety. Nevertheless, multiple models of BC have been established over the years, both patient derived and artificially engineered. For a full overview of these models, including their respective limitations, the reader is directed to an excellent review by Vargo-Gogola and Rosen (Vargo-gogola & Rosen 2007). In this section, we consider the models most often used in basic research and how these have fared in the drug discovery process.
Breast cancer cell lines
BC cell lines have found extensive use in the investigation of proliferation, apoptosis, migration and the tumour-initiating cell (TIC) phenomenon. The first BC cell line capable of surviving in culture for longer than 2 months was isolated in Detroit in 1970 and named MCF-7 (Soule et al. 1973). This oestrogen receptor alpha (ER)-positive luminal cell line has been heavily relied on in the study of tamoxifen resistance, leading to predictive biomarkers of resistance in patients (Ross-innes et al. 2012). Together, MDA-MB-231 (a triple-negative cell line), T-47D (a luminal cell line) and MCF-7 account for more than two-thirds of all abstracts mentioning BC cell lines (Lacroix & Leclercq 2004). Experiments in cell lines were crucial in the development of one of the first targeted therapeutic agents launched in 1998 – the anti-HER2 Herceptin, a humanised antibody that binds to the ectodomain of HER2 (Carter et al. 1992), has demonstrated a remarkable clinical impact on HER2-positive BC. Cell lines have also helped to elucidate the mechanisms of primary and acquired resistance to Herceptin and are still being used for a significant proportion of BC research today. These early successes supported the use of cancer cell lines for both drug development and biomarker discovery (Heiser et al. 2012).
The artificial 2D system of in vitro culture has many drawbacks, and several attempts have been made to increase the relevance of these incredibly tractable models. A seminal paper published in 2003 by Al-Hajj and coworkers demonstrated the presence of TICs in pleural effusions from BC patients, which later were shown to be maintained in suspension as 3D spheroids called mammospheres (Al-Hajj et al. 2003, Dontu et al. 2003). Accumulating evidence has supported the use of this system to better understand the biology of specific facets of BC, drug resistance and metastasis (Reya et al. 2001, Polyak 2002, Wicha et al. 2006). Mammosphere cultures have also been used to unravel molecular mechanisms of signalling networks, for example, those underlying the apparently paradoxical role of transforming growth factor beta (TGFβ) in BC (Bruna et al. 2012). The authors further identified, using these 3D mammosphere cultures, that TGFβ BC subtype’s specific regulatory networks are dictated by epigenomic landscapes (Tufegdzic-Vidakovic et al. 2015). However, as yet, no significant progress has been made in drug discovery using these, or other, complex 3D culture systems.
The tumour microenvironment (TME), comprising the extracellular matrix (ECM) and stromal and immune infiltrates, has significant bearing on the course of tumour development (Straussman et al. 2012, Junttila & de Sauvage 2013). Bissell and coworkers pioneered methods to model the microenvironment in 3D-cultured BC cell lines (Lee et al. 2007). By profiling gene expression patterns, Kenny and coworkers were able to show that a panel of 27 BC cell lines more accurately reflected human tumours when cultured on recombinant basement membrane (rBM) (Kenny et al. 2007), though cells in vivo are subject to a plethora of ECM-derived signalling gradients not easily recapitulated in vitro (Cassidy 2014). The growth of BC cell lines as xenografts allows investigation of the tumour–stromal interactions seen in vivo. For example, Kitamura and coworkers have recently reported a CCL2-induced chemokine cascade that promotes metastasis to the lung in tail-vein-injected murine cell line E0771-LG (derived from a spontaneous medullary breast adenocarcinoma of C57BL/6 background) through the recruitment of metastasis-associated macrophages (Kitamura et al. 2015). By including stromal and immune components, such syngeneic cell-line xenografts can have substantial utility as preclinical models and in the drug development process.
Breast cancer mouse models
Despite not always exhibiting typical histopathological phenotypes seen in human BC, genetically engineered mouse (GEM) models have been used extensively to investigate tumour initiation and progression. GEM models generally fall into three distinct histopathological categories: those closely resembling non-GEM tumours, those with unique transgene-specific phenotypes and those that resemble human malignancies (Cardiff et al. 2000). The choice of gene promoter and the mechanism of induction greatly influence the histological phenotype of the resulting tumour, and this needs to be taken into consideration for all GEM studies (Cardiff et al. 2000).
GEM models driven by the mouse mammary tumour virus (MMTV) promoter were used to characterise the effects of several now widely accepted oncogenes and tumour suppressors in BC (including tumour suppressors Pten, Brca1 and Trp53 and oncogenes Erbb2, Myc and Ccnd1) (Vargo-gogola & Rosen 2007). When combined with advanced intravital imaging, GEM models have also been used to elucidate the precise role of macrophages in BC metastasis. Jeffrey Pollard’s Lab has relied heavily on these models to show that the purported metastasis-associated macrophages are active promoters of the metastatic cascade rather than bystanders (Wyckoff et al. 2007). Like syngeneic models of BC, GEM models have the advantage of including native stromal compartments of a malignancy. In this regard, they may outperform many patient-derived models (see the ‘Strategies for breast cancer drug discovery’ section below) in the drug discovery process. However, a major limitation of GEM models is their tendency to form ER tumours suggesting that specific drug discovery processes are more suitable for other platforms (Medina & Thompson 2000).
Heterogeneity within a clonally expanding tumour is a consequence of intertumour heterogeneity (intrinsic molecular and cellular load) and clonal evolution upon selective pressures, which may also occur in spatially distinct tumour compartments. A recent study has uncovered a network of inter-clonal cooperation maintaining intratumour heterogeneity in a Wnt-driven MMTV GEM BC model (Cleary et al. 2014). The authors simulated targeted therapy by removing Wnt1 and found that relapsing basal populations recruited heterologous Wnt-producing luminal cells to restore cooperation. Alternatively, tumours evolved to rescue Wnt pathway activation through some other mutational event. In each case, drug resistance occurred from the cancer cell autonomous compartment in a fashion that could not be predicted easily. If such inter-clonal cooperation exists in human BC, this would underline the need for polyclonal preclinical models. However, it is worth noting that Wnt is thought to play relatively minor role in human BC (Curtis et al. 2012).
In vitro patient-derived models of breast cancer
Realising the importance of the cancer cell autonomous compartment in driving therapeutic responses, many researchers have turned to organoid cultures to study a wide variety of processes involved in the development and disease. Beginning in 2009, the Clevers lab showed that single Lgr5+ intestinal stem cells (ISCs) could build crypt-villus structures in vitro without a supporting mesenchymal niche (Sato et al. 2009). Subsequent research by this lab has identified culture conditions for normal and malignant pancreatic (Huch et al. 2013a) and liver (Huch et al. 2013b) organoids, amongst other tissue types. Organoids are generally genomically stable over long-term passage (Huch et al. 2015), though it is unclear whether mixed organoid cultures of primary tumours can truly recapitulate the complex clonal heterogeneity seen in vivo.
Recently, a biobank of 20 human colorectal carcinoma (CRC) organoids was established and characterised by exome sequencing, RNA expression analysis and high-throughput drug screening (van de Wetering et al. 2015). The authors show that CRC organoids largely recapitulate most features of the originating tumour sample, and the biobank captures most of the mutational and expression landscapes observed in large CRC studies. The authors screened these cultures using an 83 compound library to identify the molecular signatures associated with drug responses (van de Wetering et al. 2015). Although this study represented an important step forward for the field, cancers exist as communities of competing and cooperating clones surrounded by infiltrating stromal and soluble growth factors (Tabassum & Polyak 2015), each of which contributes to intratumour heterogeneity and therefore treatment response. It is therefore likely that in vitro co-culture techniques would have to be of exquisite complexity to maintain cancer cells in a niche as suitable as that found in a murine host.
Patient-derived tumour xenografts
Arguably the model best reflecting the complexity of human malignancies is the patient-derived tumour xenograft (PDTX). In this model, BC clinical samples are implanted and propagated in highly immunodeficient mice, typically NSG (NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ) or NRG (NOD.Cg-Rag1tm1Mom IL2rgtm1Wjl/SzJ) strains. Early BC PDTX studies suffered from low transplantation efficiencies and consequently a limited diversity of models (Vargo-gogola & Rosen 2007, Siolas & Hannon 2013); for example, one study reported only three ER+ models in a cohort of 32 stably transplantable PDTXs (Zhang et al. 2013). Clearly, to be useful as preclinical models, the early bias towards aggressive triple-negative BCs (TNBCs) has to be overcome. In this context, a new protocol involving intraductal injection of cells in female mice has been developed with the hope to dramatically increase engraftment rates especially in less-aggressive tumour samples (Sflomos et al. 2016).
PDTXs reflect the originating sample’s morphological and molecular features, and these remained stable throughout serial passaging (Bergamaschi et al. 2009, DeRose et al. 2011, Bruna et al. 2016). Remarkably, PDTXs are also a community of clones of varying degrees of complexity to that found in the clinical population (Eirew et al. 2015, Bruna et al. 2016). Moreover, our recent observations also showed that most of the clonal composition of a given BC-originating sample prevailed in the matched PDTX.
The most obvious limitation of the PDTX model is the lack of a functional immune system. Recent studies have highlighted both the essential role of the immune system in tumour progression (Gajewski et al. 2013) and the utility of targeted therapies designed to activate immune components (Gubin et al. 2014). The necessity of using severely immunocompromised mice as host animals severely limits the investigation of such therapies in PDTX models. Likewise, the tumour microenvironment in the PDTX model typically lacks patient-matched stromal cells, which can confer resistance to cytotoxic and targeted therapies (Straussman et al. 2012). We, and others, have found that human stromal cells are replaced by murine equivalents upon engraftment in the mouse, suggesting that implanted human cells retain the ability to recruit murine accessory cells to their niche (Bruna et al. 2016). However, it should be noted that differences exist between ligand repertoires of human and murine fibroblasts (Mestas & Hughes 2004). Clearly, stromal architecture and activity are mimicked in the murine host; however, it is currently unclear how this reflects the human TME with regards to supporting tumour growth and development.
The advantages of the PDTX over traditional BC models have been discussed extensively both here and elsewhere (Siolas & Hannon 2013, Whittle et al. 2015). However, as with any model system, we must be clear of their limitations and learn to interpret preclinical findings within this context. For a detailed discussion of the limitation of the PDTX model, we direct the reader to a prior review by the authors Cassidy et al. (2015). Although the tumour’s stromal compartments are undoubtedly important in drug response, the most important factor is undoubtedly the composition of the cancer cell autonomous compartment. Thus, PDTX models are likely the most powerful models currently available in the BC drug discovery process, and every effort should be made to adapt their use to high-throughput screens.
Recently, a PDTX-based drug screening program of unprecedented scale was reported (Gao et al. 2015). A large collection (n = 1075) of molecularly annotated PDTXs derived from the most common adult cancer types was shown to capture the genomic and transcriptomic features of tumours seen in the clinical population as a whole. The majority of PDTXs in this collection were treated with a variety of targeted compounds in a strategy dubbed ‘1 × 1 × 1’ for ‘one animal per model per treatment’. This approach mimics the reality of human clinical trials, which do not allow for technical or biological replicates. One of the key findings of this study was that a population of PDTXs mimicked the spectrum of human clinical responses, reinforcing the translatability of these models to predict population-based drug responses. Moreover, known mechanisms of resistance were identified by this strategy; for example, three PDTXs treated with encorafenib developed resistance through BRAF amplification – a clinically relevant resistance mechanism (Shi et al. 2014). It follows that there is a strong rationale for performing drug screens in PDTX models to investigate population-based treatment responses.
Strategies for breast cancer drug discovery
Despite increasing knowledge of the molecular structure of target proteins and a corresponding increase in the ability to design candidate therapeutics in silico, HTS are still essential in the drug discovery process. In order to reduce variability in HTS results, isogenic preclinical models have long been preferred in this regard. However, we have seen in the previous section how such in vitro models are not the most faithful reflections of the biological reality, and several steps can be taken to incorporate patient-derived material into this process (Fig. 1). In this section, we consider the impact of tumour heterogeneity on treatment response and explore at which points PDTX models may be introduced into the drug discovery process.
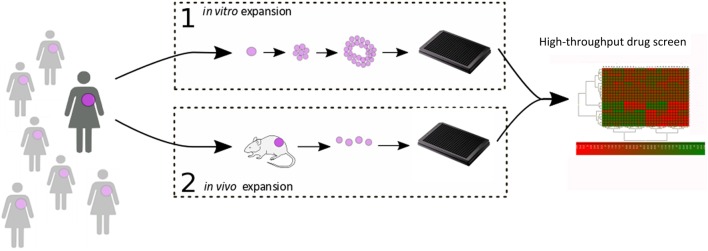
Figure 1.
High-throughput drug screen using patient-derived material. Figure 1 highlights high-throughput screening approaches using patient tumour material. (1) represents in vitro culture of tumour explants (for example as organoids/tumoroids). (2) represents the integrated PDTX:PDTC platform developed by our lab. In this strategy, patient tumour material is passaged and maintained in the murine host, and patient-derived tumour cells (PDTCs) are periodically dissociated for short-term ex vivo culture and high-throughput drug screens.
Tumour heterogeneity
Complex patterns of inter- and intra-tumour heterogeneity are a defining feature of human malignancy. Although the past few years have seen an unprecedented increase in research into tumour heterogeneity, we are far from a complete understanding. Indeed, current opinions suggest that cancer is better seen as community of co-existing and co-operating cells than a disease of a specific cell type (Tabassum & Polyak 2015). The origins of this heterogeneity are diverse, but Darwinian evolution of clonal populations has seen the most research in the context of treatment response. Moreover, a varying degree of intratumour heterogeneity exists in triple-negative BCs at diagnosis (Shah et al. 2012). Aside from the prognostic features of specific rare subclones (Diaz et al. 2012), there is an association between clonal diversity and treatment resistance for at least some tumour types – notably ovarian (Bashashati et al. 2013) and oesophageal (Maley et al. 2006). Basal-like TNBCs have previously been linked with shorter disease-free survival compared with non-basal-like TNBCs and tend to be associated with a higher clonal diversity (Shah et al. 2012, Pereira et al. 2016). Together with the clinical observation that targeted therapies often fail to show a sustained and complete response (Aparicio & Caldas 2013), it is apparent that intratumour heterogeneity plays a crucial role in treatment responses. Recently, spatial and temporal heterogeneity, with a mark remodelling of tumour clonal architecture, has been observed in response to aromatase inhibition in BC tumours (Miller et al. 2016). Thus, for a more effective oncological drug development process, we should ensure that preclinical models preserve the clonal architecture of the originating tumour sample, and second, that combinatorial drug treatment regimens are investigated to simultaneously target multiple clonal populations and extend time to recurrence.
In the context of targeting multiple specific clonal populations within a tumour, the use of patient-derived preclinical models retaining cancer’s polyclonal architecture is essential. Several groups have attempted to define the clonal structure within PDTX models through mutational clustering by population- and single cell-based computational approaches (Fischer et al. 2014, Eirew et al. 2015). We have recently quantified heterogeneity in a large biobank of BC PDTXs using statistical tools such as the MATHs score (Mroz & Rocco 2013) and PyClone (Roth et al. 2014). PDTXs display a range of heterogeneity similar to that found in the clinical population. PDTXs also preserve most of the originating sample’s intratumour clonal architecture (Eirew et al. 2015, Bruna et al. 2016). Clonal changes occur to some extent, which were more prominent upon initial engraftment into the mouse than serial passaging, yet these rarely contained breast cancer driver genes (Bruna et al. 2016, Pereira et al. 2016). In line with previous hypothesis that specific genetic alterations act as markers of fitness and drive evolutionary trajectories, we observed that clonal dynamics are replicated in different mice concurrently engrafted with spatially separated biopsies of the same sample (Eirew et al. 2015, Bruna et al. 2016). These features position PDTXs as the only preclinical models, currently available, which are able to mimic the intratumour heterogeneity found in the cancer of origin (Cassidy et al. 2015). However, further studies are needed to unravel whether the patterns of clonal trajectories seen in PDTX models resemble those seen in the patient.
Combination drug screens
Resistance to therapy can occur because of differing sensitivity to targeted agents or differing reliance on oncogenic signalling pathways between clonal populations of the same tumour. Although it is often the case that resistant populations exist in a tumour before treatment, acquisition of new mechanisms of resistance can occur as a consequence of tumour evolution. For example, Hata and coworkers have recently shown that acquired resistance of epidermal growth factor receptor (EGFR)-mutant non-small-cell lung cancers to anti-EGFR therapy can occur through the genetic evolution of initially EGFRT790M-negative drug-tolerant cells (Hata et al. 2016). These de novo mutant cells had a distinct phenotype from pre-existing resistant EGFRT790M populations, and drug sensitivity could be restored by co-treatment with Navitoclax, a BCL-2 inhibitor. Thus, tumours are dynamic, constantly evolving populations, and successful combination therapies may be hard to predict from single genomic characterisation.
In a large PDTX study referenced earlier (see the ‘Patient-derived tumour xenografts’ section above), combination therapy resulted in longer disease-free survival compared with single agent therapy (Gao et al. 2015). A good example of this is the combination of CDK4/6 inhibitor (LEE011) and PI3K inhibitor (BYL179) in BC models, which showed increased efficacy relative to single-agent treatment. Similar results were seen in melanoma PDTXs with LEE011 and Encorafenib. Although the benefits of using combination therapy to reduce proliferation and apoptosis are well explored, another advantage is delaying the onset of drug resistance. Significantly, some PDTXs treated with LEE011 and Encorafenib combination failed to develop resistance under continuous treatment for up to 200 days. In another combination study conducted by Xu and coworkers, a synergistic effect was observed with MK-8869 and MK-2206 (mTOR and AKT inhibitor, respectively) in two BC PDTXs with high levels of AKT phosphorylation and loss of PTEN expression. MK-2206 was shown to inhibit AKT activation induced by MK-8869, suggesting that combination therapy is essential in tumours with cross-talking signalling pathways (Xu et al. 2013). These and other studies strengthen the opinion that drug combinations are essential for more efficacious treatments.
Because PDTX models of BC preserve the clonal architecture of the originating tumour sample, they are greatly suited to study drug–drug combination screens. Although known co-operating signalling pathways (such as MET amplification and EGFR mutation in non-small-cell lung cancer) (Bhang et al. 2015) may be simulated in cell line models, the use of PDTX models that mimic human cancer’s complexity and dynamics ensures that our ability to identify novel, clinically relevant, targeted strategies is maximised.
Predictive biomarker identification
Recent work has highlighted the differential prognostic outcome of mutations in the same gene between patients belonging to different integrative subtypes (Pereira et al. 2016). The complex intertumour heterogeneity seen in BC means that drug screens are unlikely to identify novel therapies effective across all patients. Thus, it is essential that we develop large biobanks of PDTX models and deeply characterise them across genomic, transcriptomic and epigenomic spaces. It has been suggested that PDTX models are derived for each patient seen in the clinic and that these personalised PDTX models could be used to inform treatment decisions on a case-by-case basis (Malaney et al. 2014). However, this view needs to be taken with caution as there are still many unanswered questions that require further investigation. In the interim, efforts should be focussed on developing robust predictive biomarkers of both drug sensitivity and resistance to extrapolate results from a limited number of models to the BC population as a whole. It is our view that this stratified approach to treatment will be more effective in the near term.
Single-gene mutations are perhaps most widely adopted as clinical biomarkers of response (Garnett et al. 2012). As an example, mutations in EGFR, FLT3 and PIK3CA predict to some extent the efficacy of targeted therapies directed against the respective mutant proteins (O’Farrell et al. 2003, Lynch et al. 2004). In other instances, single-gene biomarkers can give insight into the interplay between the drug’s mechanism of action and the tumour’s genetic makeup. For example, TP53 is an important mediator of apoptosis and cell cycle arrest through its protein product p53. Inactivation of TP53 through mutation confers resistance to Nutlin-3a, an inhibitor of the MDM2 E3-ligase, which negatively regulates p53 protein levels (Vassilev et al. 2004, Garnett et al. 2012). Similarly, RB1 loss and high p16Ink4a levels confer resistance to Palbociclib, a selective CDK4/6 inhibitor showing promising clinical results in ER + BC (O’Leary et al. 2016). Mutations in the ligand-binding domain of ESR1 itself have been found in several cases of metastatic ER + BC after treatment with antiestrogen therapy (Robinson et al. 2013, Toy et al. 2013), but Fulvestrant is potentially effective in such ESR1-mutant cells (Li et al. 2013, Merenbakh-Lamin et al. 2013, Toy et al. 2013). Clearly in cases such as these, biomarkers of drug response consisting of single mutational events are relatively easily understood and implication in the clinic can be swift. Unfortunately, single-gene drug response associations are uncommon, suggesting complex molecular circuits underlie sensitivity to therapy.
Poly(adenosine diphosphate-ribose) polymerase (PARP) inhibitors in ovarian cancer patients have provided one of the best examples to date of stratification based on biomarkers of drug response. PARP inhibitors exert their cytotoxic effect by modulating the repair of DNA damage (Lord & Ashworth 2012). Although originally developed as chemosensitisers, these compounds induce synthetic lethality in tumour cells from patients carrying germline loss-of-function mutations in DNA damage pathway tumour suppressor genes BRCA1 and BRCA2 (Lord & Ashworth 2012). Several evidence suggests that the benefit of PARP inhibitors is not restricted to germline BRCA1/2 mutants and could be associated with a wider range of disrupted mechanisms. Hence, efforts have focussed on establishing a refined BRCAness signature (molecular signatures that mimic the phenotype seen in a BRCA1/2 germline loss-of-function mutation context) that can better identify patients who would benefit from PARP inhibition treatments (Turner et al. 2004, Konstantinopoulos et al. 2010).
Identification of complex genomic/epigenomic correlates of drug sensitivity requires unbiased and extensive genomic profiling of resistant and sensitive models (Fig. 2). For example, by applying elastic net regression (Zou & Hastie 2005) to genomewide expression data, Garnett and coworkers were able to identify cooperative interactions associated with drug resistance in the NCI60 cell line panel (Garnett et al. 2012). Interestingly, the authors observed several instances where transcriptional features correlated with drug sensitivity more than mutational events. For example, Lapatinib (an EGFR/ERBB2 inhibitor for HER2-positive BC) sensitivity unsurprisingly correlated with ERBB2 expression and mutation status, but the strongest correlate was expression of the matrix metalloproteinase MMP28. Likewise, together with BRAF mutation, sensitivity to RAF or MEK1/2 inhibitors was recurrently associated with 67 features, including expression of the MAP kinase signalling regulators SPRY2 and DUSP4/6 (Hanafusa et al. 2002, Garnett et al. 2012).
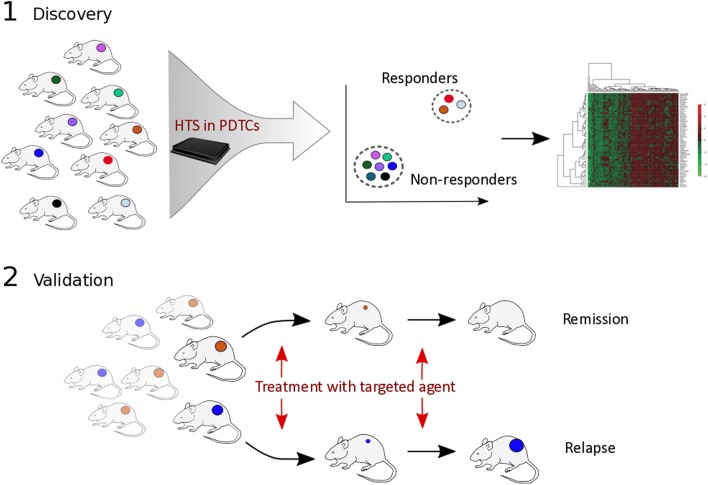
Figure 2.
Biomarker discovery using PDTX models. Figure 2 highlights an unbiased approach for biomarker discovery. (1) a mixed cohort of PDTX models is screened with multiple compounds affecting different members of the same signalling pathway and are subsequently clustered based on responders and non-responders. Genomic correlates of drug response are computed before validation in vivo (2).
Clearly, biomarkers of treatment response and the emergence of resistance are essential to relate the findings from a limited biobank of PDTX models to the BC population as a whole. This becomes vastly more difficult when attempting to systematically identify correlates of sensitivity from combination drug screens across models that are themselves dynamic, heterogeneous and constantly evolving. Thus, it is essential that PDTX biobanks are characterised and annotated across genomic, epigenomic, transcriptomic and proteomic spaces and that bioinformatics pipelines are incredibly robust and reproducible across labs and through time.
An integrated pharmacogenomic platform for drug discovery
The previous two sections have described the key features of an ideal BC drug screening program. Over the last 5 years, our lab has developed a successful, robust and reliable framework for the generation of breast cancer explants annotated with extensive molecular data (Bruna et al. 2016). Each model has been extensively characterised across genomic, transcriptomic and epigenomic spaces showing that these resemble the originating cancer. The extensive data generated represents a valuable resource to the scientific community, which can be easily browsed using a purpose-built web portal (http://caldaslab.cruk.cam.ac.uk/bcape).
During this time, we have refined our tumour engraftment protocol and now have a large biobank of BC PDTX samples that comprises almost equal percentages of originating primary and metastatic tumour samples. Modifications in the implantation protocol have included, but are not limited to dissociation of tumour samples to single cells with or without in vitro culture before implantation and alteration in implantation substrate. Interestingly, speed of implantation after collection in theatre (30–180 min) has been perhaps our most important protocol refinement to date, translating into significantly increased engraftment efficiencies and consequently a more thorough representation of all BC subtypes across the BC PDTX biobank, although with a bias towards ER-negative and poorer prognosis ER-positive tumours (Bruna et al. 2016).
A significant limitation of PDTXs as a preclinical platform is the fact that in vivo studies are not well suited for high-throughput drug screening due to financial and animal welfare reasons. We have recently reported a method for isolating single-cell suspensions from PDTX tumours for short-term cultured assays (Bruna et al. 2016). These PDTX cells (or PDTCs) resemble the originating sample across genomic, epigenomic and transcriptomic landscapes and show similar intratumour clonal architectures. This PDTX/PDTC platform ensures the maintenance of the human BC in vivo in the mouse, while allowing for ex vivo short-term cultures for single and combination HTS. As a proof of principle, a selection of 22 PDTXs were screened for over 100 different compounds relevant to cancer treatment and the observed drug responses highly correlated across technical and biological replicates. Similarly, compounds with similar target specificities or mechanism of action also had similar responses across all models tested. For example, similar responses were observed in 14 out of the 19 models tested with PARP inhibitor BMN-673 and Cisplatin (a DNA cross-linking agent), both of which exert their effects by increasing the frequency of mis-repaired double-strand breaks in the absence of effective homologous recombination. Also, inhibitors of the PI3K-AKT-mTOR pathway shared a similar pattern of response across all samples tested. To extend these observations, we computed the correlation scores of drug responses for all pair of compounds affecting the same pathway and observed that most compounds with overlapping specificity had similar responses, supporting the biological robustness of our data (Bruna et al. 2016). Recognising the importance of combination therapies in achieving long-lasting responses (Friedman et al. 2015), we also tested and validated the PDTX/PDTC platform in a HT combinatorial drug screen with standard-of-care chemotherapeutic agents (Cisplatin and Paclitaxel) and six clinically relevant BC-targeted compounds. Remarkably, 33 of 40 (82.5%) drug responses tested ex vivo in PDTCs were recapitulated in vivo using PDTXs. We propose the use of the PDTX/PDTC platform as a resourceful intermediate in the drug discovery process before in vivo testing using PDTXs (Bruna et al. 2016).
Finally, to extrapolate our data to the BC population as a whole and to aid in patient stratification during therapy, we sought to identify biomarkers of resistance and response in our models. Analysis of the multidimensional data generated for each model was performed to identify known and novel biomarkers of drug response. Significantly, the PDTX/PDTC platform could identify known mechanisms of drug response and resistance. However, the heterogenous nature of plausible biomarkers of drug response spread across the models tested suggests that integrated genomic data will be a stronger predictor of drug response than single-gene biomarkers.
It is our belief that the integrated pharmacogenomics PDTX/PDTC platform will show the greatest utility in preclinical studies and will considerably shorten the time of testing new drugs in patients (Bruna et al. 2016). This approach extends the predictive value of PDTX models in oncological drug development by allowing HT screening of hundreds of compounds, as well as reducing considerably the financial costs and number of animals used in such studies.
Conclusions and future directions
The oncological drug space suffers from an unsustainable rate of attrition, in part attributed to preclinical models that fail to accurately represent the complexity of BC. BC PDTX models broadly maintain the heterogeneity of their originating patient tumour; however, historically they have suffered from considerable engraftment bias towards TNBCs. Although the lower-fidelity BC models (cell lines etc.) have long been used in the drug discovery process, the inherent low throughput of PDTX models has also limited their adoption. In the context of clonal diversity and intertumour heterogeneity, it is essential that we adapt the models best reflecting their originating sample to the drug discovery process. Here, we have considered BC models currently used in the drug discovery process and reflected on our own experiences in adapting BC PDTX models to HTS in our integrated PDTX/PDTC platform (Bruna et al. 2016). Efforts such as these can only be achieved if the establishment of PDTX models is shared across large collaborative networks such as the EuroPDX consortium (Hidalgo et al. 2014) or in the context of large pharmaceutical companies. Alternatively, the process of establishing and maintaining large cohorts of PDTX models might be best suited to pharmaceutical companies (Gao et al. 2015).
The current generation of PDTX models is unlikely to be their final iteration. It is important to stress areas where improvements can be made and emphasise that no single model system can represent fully the complexity of a human malignancy. Ideally, the next generation of PDTX models would incorporate tumour extrinsic compartments of the microenvironment specific to the patient, either in the form of matched patient peripheral blood leukocytes or matched patient fibroblasts. These humanised PDTX models (huPDTX) would significantly increase the utility of the model and perhaps decrease the selective pressures seen on engraftment. However, the generation of huPDTX models is fraught with difficulties, and technical challenges are yet to be overcome (Cassidy et al. 2015). An intermediate solution to screen targeted therapies with an essential immune component could be to isolate and expand patient-derived peripheral blood leukocytes and incorporate them into complex short-term co-cultures with PDTCs. The use of checkpoint inhibitors in BC is of paramount interest (Ali et al. 2015) and as such this strategy could be of substantial utility in future drug screens.
Footnote
This paper is part of a thematic review section on hormone-dependent cancers. The Guest Editor for this section was Wayne Tilley.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
Funding
This work was supported by Cancer Research UK (grant number RG84936).
Author contribution statement
J W C and A B wrote the manuscript. A S B and W G contributed to structure, content and editing of the manuscript. All authors approved the final version of the manuscript.
Acknowledgements
The authors are particularly grateful to Prof C Caldas and Dr O M Rueda for valuable discussion and expertise. Establishment of their Patient-Derived Tumour Xenograft models would not have been possible without the dedicated work of the Cambridge Breast Unit, Cambridge Institute Biological Resources Unit and their collaborators in EuroPDX consortium (http://www.EuroPDX.eu). Additionally, they are particularly grateful to the patients who donated tissues to further our research and to Cancer Research UK for support.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. 2003. Prospective identification of tumorigenic breast cancer cells. PNAS 100 3983–3988. ( 10.1073/pnas.0530291100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali HR, Glont SS-E, Blows FM, Provenzano E, Dawson S-JS, Liu B, Hiller L, Dunn J, Poole CJ, Bowden S, et al. 2015. PD-L1 protein expression in breast cancer is rare, enriched in basal-like tumours and associated with infiltrating lymphocytes. Annals of Oncology 26 1488–1493. ( 10.1093/annonc/mdv192) [DOI] [PubMed] [Google Scholar]
- Aparicio S, Caldas C. 2013. The implications of clonal genome evolution for cancer medicine. New England Journal of Medicine 368 842–851. ( 10.1056/NEJMra1204892) [DOI] [PubMed] [Google Scholar]
- Bashashati A, Ha G, Tone A, Ding J, Prentice LM, Roth A, Rosner J, Shumansky K, Kalloger S, Senz J, et al. 2013. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. Journal of Pathology 231 21–34. ( 10.1002/path.4230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi A, Hjortland GO, Triulzi T, Sørlie T, Johnsen H, Ree AH, Russnes HG, Tronnes S, Mælandsmo GM, Fodstad O, et al. 2009. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Molecular Oncology 3 469–482. ( 10.1016/j.molonc.2009.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhang HC, Ruddy DA, Krishnamurthy Radhakrishna V, Caushi JX, Zhao R, Hims MM, Singh AP, Kao I, Rakiec D, Shaw P, et al. 2015. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nature Medicine 21 440–448. ( 10.1038/nm.3841) [DOI] [PubMed] [Google Scholar]
- Bruna A, Greenwood W, Le Quesne J, Teschendorff A, Miranda-Saavedra D, Rueda OM, Sandoval JL, Vidakovic AT, Saadi A, Pharoah P, et al. 2012. TGFβ induces the formation of tumour-initiating cells in claudinlow breast cancer. Nature Communications 3 1055 ( 10.1038/ncomms2039) [DOI] [PubMed] [Google Scholar]
- Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, Pogrebniak K, Sandoval JL, Cassidy JW, Tufegdzic-Vidakovic A, et al. 2016. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 167 260–274.e22. ( 10.1016/j.cell.2016.08.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, Rehm S, Russo J, Tavassoli FA, Wake LM, et al. 2000. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene 19 968–988. ( 10.1038/sj.onc.1203277) [DOI] [PubMed] [Google Scholar]
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. 1992. Humanization of an anti-p185HER2 antibody for human cancer therapy. PNAS 89 4285–4289. ( 10.1073/pnas.89.10.4285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy JW. 2014. Nanotechnology in the regeneration of complex tissues. Bone and Tissue Regeneration Insights 5 25–35. ( 10.4137/BTRI.S12331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy JW, Caldas C, Bruna A. 2015. Maintaining heterogeneity in patient derived tumour xenografts. Cancer Research 75 1–6. ( 10.1158/0008-5472.CAN-15-0727) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Creighton CJ, Davis C, Donehower L, Drummond J, Wheeler D, Ally A, Balasundaram M, Birol I, Butterfield YSN, et al. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genetics 45 1113–1120. ( 10.1038/ng.2764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary AS, Leonard TL, Gestl SA, Gunther EJ. 2014. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 508 113–117. ( 10.1038/nature13187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486 346–352. ( 10.1038/nature10983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose YS, Wang G, Lin Y-C, Bernard PS, Buys SS, Ebbert MTW, Factor R, Matsen C, Milash BA, Nelson E, et al. 2011. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nature Medicine 17 1514–1520. ( 10.1038/nm.2454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. 2012. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486 537–540. ( 10.1038/nature11219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. 2003. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes & Development 17 1253–1270. ( 10.1101/gad.1061803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. 2015. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518 422–426. ( 10.1038/nature13952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Vázquez-García I, Illingworth CJR, Mustonen V. 2014. High-definition reconstruction of clonal composition in cancer. Cell Reports 7 1740–1752. ( 10.1016/j.celrep.2014.04.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AA, Amzallag A, Pruteanu-malinici I, Baniya S, Frederick T, Lawrence DP, Haber DA, Flaherty KT. 2015. Landscape of targeted anti-cancer drug synergies in melanoma identifies a novel BRAF-VEGFR/PDGFR combination treatment. PLoS ONE 10 e0140310 ( 10.1371/journal.pone.0140310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF, Schreiber H, Fu Y-X. 2013. Innate and adaptive immune cells in the tumor microenvironment. Nature Immunology 14 1014–1022. ( 10.1038/ni.2703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, et al. 2015. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nature Medicine 21 1318–1325. ( 10.1038/nm.3954) [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Liu Q, et al. 2012. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483 570–575. ( 10.1038/nature11005.Systematic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber W-J, et al. 2014. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515 577–581. ( 10.1038/nature13988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Nishida E. 2002. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nature Cell Biology 4 850–858. ( 10.1038/ncb867) [DOI] [PubMed] [Google Scholar]
- Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HC, Krishnamurthy Radhakrishna V, et al. 2016. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nature Medicine 22 262–269. ( 10.1038/nm.4040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser LM, Sadanandam A, Kuo W-L, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, et al. 2012. Subtype and pathway specific responses to anticancer compounds in breast cancer. PNAS 109 2724–2729. ( 10.1073/pnas.1018854108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo M, Amant F, Biankin AV, Budinska E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Maelandsmo GM, et al. 2014. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discovery 4 998–1013. ( 10.1158/2159-8290.CD-14-0001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJM, Van De Wetering M, Sojoodi M, Li VSW, Schuijers J, Gracanin A, et al. 2013a. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO Journal 32 2708–2721. ( 10.1038/emboj.2013.204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, Van Es JH, Van De Wetering M, Li VSW, Hamer K, Sasaki N, Finegold MJ, Haft A, et al. 2013b. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494 247–250. ( 10.1038/nature11826.In) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, et al. 2015. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160 299–312. ( 10.1016/j.cell.2014.11.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Kirk R. 2011. High drug attrition rates--where are we going wrong? Nature Reviews. Clinical Oncology 8 189–190. ( 10.1038/nrclinonc.2011.34) [DOI] [PubMed] [Google Scholar]
- Junttila MR, de Sauvage FJ. 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501 346–354. ( 10.1038/nature12626) [DOI] [PubMed] [Google Scholar]
- Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, Manuel CA, Edgerton SM, Harrell JC, Elias A, et al. 2012. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Research and Treatment 135 415–432. ( 10.1007/s10549-012-2164-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-hoff MH, Petersen OW, et al. 2007. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Molecular Oncology 1 84–96. ( 10.1016/j.molonc.2007.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny HA, Lal-nag M, White EA, Shen M, Chiang C, Mitra AK, Zhang Y, Curtis M, Schryver EM, Bettis S, et al. 2015. Impact of high-throughput screening in biomedical research. Nature Communications 6 1–10. ( 10.1038/ncomms7220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Qian B, Soong D, Cassetta L, Noy R, Sugano G, Kato Y, Li J, Pollard JW. 2015. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. Journal of Experimental Medicine 212 1043–1059. ( 10.1084/jem.20141836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N, Levine DA, Cannistra SA. 2010. Gene expression profile of BRCA ness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. Journal of Clinical Oncology 28 3555–3561. ( 10.1200/JCO.2009.27.5719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G. 2004. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Research and Treatment 83 249–289. ( 10.1023/B:BREA.0000014042.54925.cc) [DOI] [PubMed] [Google Scholar]
- Lee GY, Kenny PA, Lee EH, Bissell MJ. 2007. Three-dimensional culture models of normal and malignant breast epithelial cells. Nature Methods 4 359–365. ( 10.1038/nmeth1015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, et al. 2013. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Reports 4 1116–1130. ( 10.1016/j.celrep.2013.08.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. 2012. The DNA damage response and cancer therapy. Nature 481 287–294. ( 10.1038/nature10760) [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. 2004. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. New England Journal of Medicine 350 2129–2139. ( 10.1056/NEJMoa040938) [DOI] [PubMed] [Google Scholar]
- Malaney P, Nicosia SV, Davé V. 2014. One mouse, one patient paradigm: new avatars of personalized cancer therapy. Cancer Letters 344 1–12. ( 10.1016/j.canlet.2013.10.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, Paulson TG, Blount PL, Risques R-A, Rabinovitch PS, et al. 2006. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nature Genetics 38 468–473. ( 10.1038/ng1768) [DOI] [PubMed] [Google Scholar]
- Marangoni E, Vincent-Salomon A, Auger N, Degeorges A, Assayag F, de Cremoux P, de Plater L, Guyader C, De Pinieux G, Judde J-G, et al. 2007. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clinical Cancer Research 13 3989–3998. ( 10.1158/1078-0432.CCR-07-0078) [DOI] [PubMed] [Google Scholar]
- Medina D, Thompson HJ. 2000. A comparison of the salient features of mouse, rat, and human mammary tumorigenesis. In Methods in Mammary Gland Biology and Breast Cancer Research, pp 31–36. Eds Ip MM, Asch BB. Boston, MA, USA: Springer; ( 10.1007/978-1-4615-4295-7_3) [DOI] [Google Scholar]
- Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, Yelensky R, Brown M, Miller VA, Sarid D, et al. 2013. D538G mutation in estrogen receptor-α: a novel mechanism for acquired endocrine resistance in breast cancer. Cancer Research 73 6856–6864. ( 10.1158/0008-5472) [DOI] [PubMed] [Google Scholar]
- Mestas J & Hughes CCW 2004. Of mice and not men: differences between mouse and human immunology. Journal of Immunology 172 2731–2738. ( 10.4049/jimmunol.172.5.2731) [DOI] [PubMed] [Google Scholar]
- Miller CA, Gindin Y, Lu C, Griffith OL, Griffith M, Shen D, Hoog J, Li T, Larson DE, Watson M, et al. 2016. Aromatase inhibition remodels the clonal architecture of estrogen-receptor-positive breast cancers. Nature Communications 7 12498 ( 10.1038/ncomms12498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz EA, Rocco JW. 2013. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncology 49 211–215. ( 10.1016/j.oraloncology.2012.09.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell A-M, Abrams TJ, Yuen HA, Ngai TJ, Louie SG, Yee KWH, Wong LM, Hong W, Lee LB, Town A, et al. 2003. SU11248 is a novel FLT3 tyrosine kinase inhibitor with potent activity in vitro and in vivo. Blood 101 3597–3605. ( 10.1182/blood-2002-07-2307) [DOI] [PubMed] [Google Scholar]
- O’Leary B, Finn RS, Turner NC. 2016. Treating cancer with selective CDK4/6 inhibitors. Nature Reviews Clinical Oncology 13 417–430. ( 10.1038/nrclinonc.2016.26) [DOI] [PubMed] [Google Scholar]
- Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut S-J, et al. 2016. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nature Communications 7 11908 ( 10.1038/ncomms11908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K. 2002. Molecular alterations in ductal carcinoma in situ of the breast. Current Opinion in Oncology 14 92–96. ( 10.1097/00001622-200201000-00016) [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. 2001. Stem cells, cancer, and cancer stem cells. Nature 414 105–111. ( 10.1038/35102167) [DOI] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, et al. 2013. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nature Genetics 45 1446–1451. ( 10.1038/ng.2823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-innes CS, Stark R, Teschendorff AE, Holmes KA, Raza H, Dunning MJ, Brown GD, Gojis O, Ellis IO, Andrew R, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481 389–393. ( 10.1038/nature10730.Differential) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, Ha G, Aparicio S, Bouchard-Cote A, Shah SP. 2014. PyClone: statistical inference of clonal population structure in cancer. Nature Methods 11 396–398. ( 10.1038/nmeth.2883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. 2009. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459 262–265. ( 10.1038/nature07935) [DOI] [PubMed] [Google Scholar]
- Sflomos G, Dormoy V, Metsalu T, Jeitziner R, Battista L, Scabia V, Raffoul W, Delaloye J-F, Treboux A, Fiche M, et al. 2016. A preclinical model for ERα-positive breast cancer points to the epithelial microenvironment as determinant of luminal phenotype and hormone response. Cancer Cell 29 407–422. ( 10.1016/j.ccell.2016.02.002) [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. 2012. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486 395–399. ( 10.1038/nature10933) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. 2014. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discovery 4 1–15. ( 10.1158/2159-8290.CD-13-0642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Hannon GJ. 2013. Patient derived tumor xenografts: transforming clinical samples into mouse models. Cancer Research 73 5315–5319. ( 10.1158/0008-5472.CAN-13-1069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule HD, Vazquez J, Long A, Albert S, Brennan M. 1973. A human cell line from a pleural effusion derived from a breast carcinoma 1,2. Journal of the National Cancer Institute 51 1409–1416. [DOI] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. 2012. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487 500–504. ( 10.1038/nature11183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabassum DP, Polyak K. 2015. Tumorigenesis: it takes a village. Nature Reviews Cancer 15 473–483. ( 10.1038/nrc3971) [DOI] [PubMed] [Google Scholar]
- Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, et al. 2013. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nature Genetics 45 1439-1445. ( 10.1038/ng.2822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufegdzic-Vidakovic A, Rueda OM, Vervoort SJ, Batra AS, Goldgraben MA, Uribe-Lewis S, Greenwood W, Coffer PJ, Bruna A. 2015. Context-specific effects of TGF- b/SMAD3 in cancer are modulated by the epigenome article context-specific effects of TGF- b/SMAD3 in cancer are modulated by the epigenome. CellReports 13 2480–2490. ( 10.1016/j.celrep.2015.11.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. 2004. Hallmarks of ‘BRCAness’ in sporadic cancers. Nature Reviews Cancer 4 814–819. ( 10.1038/nrc1457) [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161 933–945. ( 10.1016/j.cell.2015.03.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargo-gogola T, Rosen JM. 2007. Modelling breast cancer: one size does not fit all. Nature Reviews Cancer 7 659–672. ( 10.1038/nrc2193) [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Klein C, Fotouhi N, Liu EA. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303 844–848. ( 10.1126/science.1092472) [DOI] [PubMed] [Google Scholar]
- Whittle JR, Lewis MT, Lindeman GJ, Visvader JE. 2015. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Research 17 17 ( 10.1186/s13058-015-0523-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicha MS, Liu S, Dontu G. 2006. Cancer stem cells: an old idea – a paradigm shift. Cancer Research 66 1883–1890. ( 10.1158/0008-5472.CAN-05-3153) [DOI] [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li J, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. 2007. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Research 67 2649–2657. ( 10.1158/0008-5472.CAN-06-1823) [DOI] [PubMed] [Google Scholar]
- Xu S, Li S, Guo Z, Luo J, Ellis MJ, Ma CX. 2013. Combined targeting of mTOR and AKT is an effective strategy for basal-like breast cancer in patient-derived xenograft models. Molecular Cancer Therapeutics 12 1665–1675. ( 10.1158/1535-7163.MCT-13-0159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano M, et al. 2013. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Research 73 4885–4897. ( 10.1158/0008-5472.CAN-12-4081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. 2005. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society 67 301–320. ( 10.1111/j.1467-9868.2005.00503.x) [DOI] [Google Scholar]



 This work is licensed under a
This work is licensed under a