Editor: In a recent AIDS Research and Human Retroviruses publication by C.N. Roy et al. [Intersubtype Genetic Variation of HIV-1 Tat Exon 1, 2015;33(6):1215–1225], the authors report the identification of 30 codon sites under positive selection in the first exon of the HIV-1 tat gene. Unfortunately, they have not considered the presence of overlapping coding sequences in HIV-1, invalidating most of their positive selection analysis.
Conventional phylogenetic selection analyses compare the rate of synonymous codon substitution (dS) with the rate of nonsynonymous codon substitution (dN). A ratio of dN/dS exceeding 1 indicates positive selection, as the rate of amino acid change is greater than the rate of (ostensibly) neutral evolution. Selection acting on overlapping coding sequences can invalidate such analyses, as nonsynonymous sites in one overlapping reading frame will correspond to synonymous sites in another.1
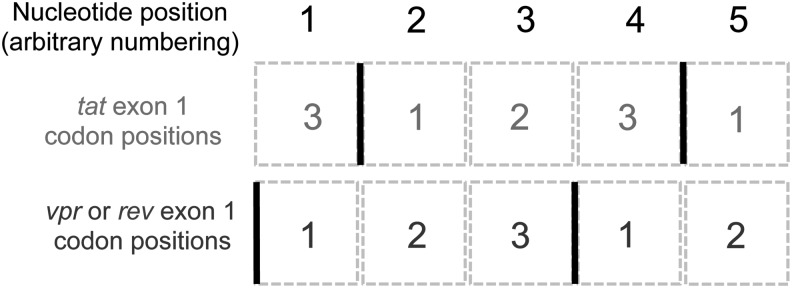
In the HIV-1 genome, the first seven codons at the 5′ end of tat exon 1 overlap with the 3′ end of the vpr gene, while the last 26 codons overlap with rev exon 1 (HXB2 reference sequence; www.hiv.lanl.gov/content/sequence/HIV/MAP/landmark.html). Thus, the first nucleotide position in vpr/rev codons corresponds to the third nucleotide position in tat codons, as shown in Fig. 1. Substitutions in the first nucleotide positions in the vpr/rev codons are likely to be nonsynonymous, meaning purifying selection acting on vpr and rev would reduce the substitution rate at these positions. This corresponds to a decreased substitution rate at the third nucleotide position in the tat codons, which are likely to be synonymous, therefore inflating the dN/dS ratio for tat and resulting in the mistaken impression of positive selection. For this reason, the analysis described by Roy et al. is suitable only for the region of tat between codons 8 and 46 that are not overlapping with other genes. Restricting the analysis to these residues eliminates the majority (19 of 30) of the sites identified by Roy et al. as under positive selection.
FIG. 1.
Overlapping codons in the HIV-1 tat and vpr/rev genes. Boxes represent nucleotide positions and solid black lines show the boundaries between codons. Numbers within boxes represent nucleotide positions within each codon triplet. It can be seen that the nucleotide occupying the first position in vpr/rev codons is the same as the nucleotide at the third codon position in tat.
A number of approaches to studying selection specifically in overlapping coding sequences have been developed,1–3 though none are practical for routine analyses. Investigators undertaking phylogenetic selection analyses with conventional codon models must be aware of the bias introduced by ignoring overlapping coding sequences in virus or bacterial genomes. The scope of such work should be limited to regions of genes in which there is no overlap.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Sabath N, Landan G, and Graur D: A method for the simultaneous estimation of selection intensities in overlapping genes. PLoS One 2008;3:e3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hein J. and Stovlbaek J: A maximum-likelihood approach to analyzing nonoverlapping and overlapping reading frames. J Mol Evol 1995;40:181–189 [DOI] [PubMed] [Google Scholar]
- 3.Pedersen AM. and Jensen JL: A dependent-rates model and an MCMC-based methodology for the maximum-likelihood analysis of sequences with overlapping reading frames. Mol Biol Evol 2001;18:763–76 [DOI] [PubMed] [Google Scholar]