Abstract
Members of the Erm methyltransferase family modify 23S rRNA of the bacterial ribosome and render cross-resistance to macrolides and multiple distantly related antibiotics. Previous studies have shown that the expression of erm is activated when a macrolide-bound ribosome stalls the translation of the leader peptide preceding the cotranscribed erm. Ribosome stalling is thought to destabilize the inhibitory stem-loop mRNA structure and exposes the erm Shine-Dalgarno (SD) sequence for translational initiation. Paradoxically, mutations that abolish ribosome stalling are routinely found in hyper-resistant clinical isolates; however, the significance of the stalling-dead leader sequence is largely unknown. Here, we show that nonsense mutations in the Staphylococcus aureus ErmB leader peptide (ErmBL) lead to high basal and induced expression of downstream ErmB in the absence or presence of macrolide concomitantly with elevated ribosome methylation and resistance. The overexpression of ErmB is associated with the reduced turnover of the ermBL-ermB transcript, and the macrolide appears to mitigate mRNA cleavage at a site immediately downstream of the ermBL SD sequence. The stabilizing effect of antibiotics on mRNA is not limited to ermBL-ermB; cationic antibiotics representing a ribosome-stalling inducer and a noninducer increase the half-life of specific transcripts. These data unveil a new layer of ermB regulation and imply that ErmBL translation or ribosome stalling serves as a “tuner” to suppress aberrant production of ErmB because methylated ribosome may impose a fitness cost on the bacterium as a result of misregulated translation.
INTRODUCTION
Macrolide antibiotics inhibit bacterial protein synthesis by binding to the ribosomal exit tunnel (1). The extensive use of macrolides in agribusiness and the medical community has accelerated the erosion of the efficacy of these drugs and the spread of transmissible resistant determinants (2–4). One of the major resistance mechanisms is caused by the members of the Erm methyltransferase family, which modify the single 23S rRNA nucleotide A2058 (Escherichia coli numbering) of the bacterial 50S ribosomal subunit and thereby reduce the drug-binding affinity. Dimethylation of A2058 (m62A2058) not only evokes resistance to the prototypic macrolide erythromycin (ERY) but also leads to cross-resistance to the structurally distinct lincosamides and streptogramins, which share the overlapping binding site (5). Erm enzymes are most prevalent in Gram-positive staphylococci, streptococci, enterococci, and clostridia but are increasingly found in Gram-negative bacteria of animal and human origins (4, 6–11). Previous studies on the ermCL-ermC operon have shown that ErmC methyltransferase translation is activated by macrolides when the antibiotic-bound ribosome stalls at a specific site in the ermCL leader peptide upstream of the cotranscribed ermC. The arrested ribosome is thought to induce a structural rearrangement of ermCL-ermC mRNA and allow the ribosome access to the Shine-Dalgarno (SD) sequence that would otherwise be occluded from translational initiation (12–14). Analogous leader peptide-dependent, ribosome-stalling-mediated translational regulation has also been proposed for other homologous erm systems (15–19) and in other ligand-dependent (20–23) and ligand-independent (24–29) bicistrons. In other cases, ribosome stalling in the leader sequence promotes downstream transcription by precluding termination factor binding or by melting of the termination mRNA structure (23, 30, 31).
Erm-directed resistance can either be constitutive or macrolide inducible, and the ribosome-stalling-based regulation belongs to the latter category (5, 32). The distributions of constitutive and inducible resistance in natural bacterial populations are not well documented. Nevertheless, in many clinical surveillance studies, the constitutive phenotype appears to be equally widespread, if not the most predominant type, in resistant isolates from patients (33–44). Insertions, duplications, deletions, and missense mutations within the leader regulatory region are commonly found in the constitutively expressed erm operons (33–43). The deletion of a substantial portion of the regulatory region could explain the overproduction of Erm enzyme because the inhibitory mRNA hairpin structure is removed. However, most naturally occurring mutations are more subtle and, in many cases, result in premature termination before translation reaches the ribosome-stalling site. These leader peptide mutants, which are presumably defective in ribosome stalling, remain responsive to macrolides that further upregulate the downstream erm expression. The mechanism underlying these apparently paradoxical phenomena has not been established.
We used the ermBL-ermB operon from methicillin-resistant Staphylococcus aureus CM05 as a model to investigate the inducible and constitutive resistance. The intergenic region of other homologous ermBL-ermB transcript is known to adopt secondary structure, which could mask the SD sequence of ermB (18) (Fig. 1A). Consistent with relevant clinical studies, we found that cells bearing truncated ErmBL (and consequently impaired in ribosome stalling) remain highly resistant via a previously underappreciated mechanism by which the antibiotic promotes the stability of ermB mRNA. We found that ermBL-ermB mRNA is more susceptible to RNase in the absence of ERY and that the nucleolytic site lies immediately downstream of the ermBL SD at a region coinciding with the reported consensus sequence of some RNase E substrates (45–47). Finally, we found that other cationic ribosome inhibitors, regardless of their ability to elicit ribosome stalling, can increase the mRNA abundance of a specific group of genes, most likely by enhancing mRNA stability. Our results reveal an alternative “off-target” resistance-inducing pathway and support the emerging idea that empirical antibiotic therapy can lead to unintended consequences by promoting mRNA stability, some of these mRNA coding products may be virulence factors.
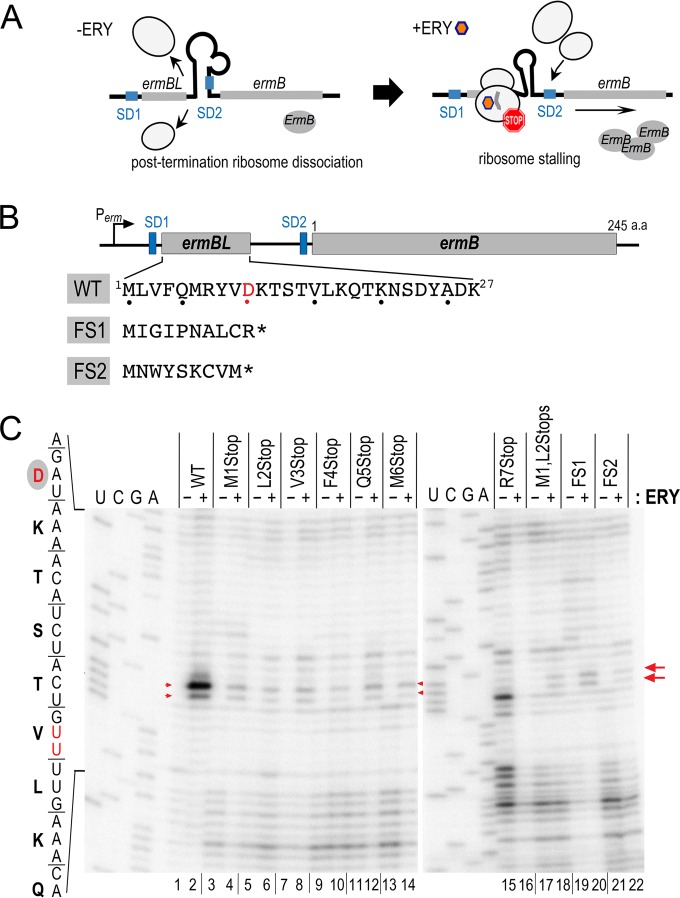
FIG 1.
Introducing premature ochre codons in ErmBLEF completely or severely impairs ribosome stalling in vitro. (A) A previously proposed model of ErmB upregulation by the erythromycin (ERY)-induced ErmBL ribosome stalling. ERY-bound ribosome stalls at the C terminus of ErmBL, causes structural rearrangement of the mRNA, and in turn exposes the Shine-Dalgarno (SD) sequence of downstream ermB for translational initiation. (B) Schematic diagram of the ermBLEF-ermB operon from S. aureus CM05. Protein sequences of wild-type (WT) ErmBLEF and the corresponding frameshift (FS) mutants are shown. The ribosome-stalling site D10 is highlighted in red. SD sequences are marked. An asterisk depicts a termination codon. (B) Mapping of the ERY-stalled ribosome on its mRNA template by toeprinting. Reverse transcriptase halts at a site 16 to 17 nt (red arrows) downstream of the P-site codon of a stalled ribosome, producing a truncated cDNA product that was analyzed using a denaturing sequencing gel. Weak background bands in the nonsense mutants are partially due to translational readthrough (see Fig. S3A in the supplemental material) and are partially due to reverse transcription pausing on the naked mRNA without added ribosome (see Fig. S3B in the supplemental material). The reason for the unusual primer extension signals in the R7Stop in the absence of ERY is unclear. ERY was used at 50 μM.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and reagents.
E. coli NM580 is a derivative of K-12 MG1655, which carries a mini-λRed recombinase and the counterselective markers comprising a kanamycin cassette and a ccdB toxin gene under the control of an arabinose-inducible promoter (a gift from N. Majdalani) (see Table S1 in the supplemental material) (48). Linear DNA templates bearing the Perm-ermBL-ermB′ or Perm-ermBL′ regions were PCR amplified using p494 and p495 as primers (see Table S2 in the supplemental material) and p381ermBL-B′ as the template. A 40-nucleotide (nt) homologous sequence flanking the kan-pBAD-ccdB cassette was incorporated into the 0.3- to 0.5-kb PCR products. After PCR purification, the DNA was recombined into the chromosomal lacZ locus of NM580 by inducing λRed recombinase using a temperature shift to 42°C. Successful recombinations were indicated by the loss of kanamycin resistance, and the identity of the recombinants was verified by DNA sequencing.
To construct p381ermBL-B′ and pTAermBL-ermB plasmids (see Table S1 in the supplemental material), the ermBL-ermB′ or ermBL-ermB region, including its native promoter, was amplified using p129+p151 or p805+p806 as a primer pair and the genomic DNA of S. aureus CM05 as the template. The ermBL-ermB′ fragment was cloned into the HindIII and BamHI sites of pRB381 (49) to create a lacZ translational fusion at codon 13 of ermB. The full-length ermB-ermBL fragment was cloned into pGEMT-Easy via TA cloning (Promega). Site-specific mutations in ermBL or ermB were introduced by QuikChange mutagenesis (Agilent). S. aureus cells were grown in tryptic soy broth (Difco), whereas E. coli strains were grown in Luria-Bertani (LB) broth (Fisher) at 37°C unless otherwise noted. DNA primers were obtained from IDT DNA. Antibiotics and chemicals were purchased from Sigma. Antibiotics were used at the following concentrations unless otherwise stated: ampicillin (100 μg/ml), kanamycin (50 μg/ml), erythromycin (10 to 200 μg/ml), clindamycin (100 to 1,500 μM), tylosin (0.25 to 3.5 mM), and rifampin (200 μg/ml). Serial titrations of antibiotics were performed to determine the optimal drug concentration for ermB-lacZ induction and toeprinting. The MIC of erythromycin was determined by Etest on the Mueller-Hinton agar (BD Difco) plates according to the manufacturer's instructions (bioMérieux).
In vitro translation and toeprinting analyses.
Linear DNA templates carrying the T7 promoter were programmed in 5 μl of cell-free PURExpress translation system (New England BioLabs) in the presence or absence of antibiotics. To detect translation products, the reactions were supplemented with 10 μCi of Tran35S-label (MP Biomedicals), incubated at 37°C for 1 h, precipitated with 4× volumes of acetone, resolved on 12% Bis-Tris SDS-PAGE gels, and autoradiographed (see Fig. S3A in the supplemental material). In the toeprinting experiments (50), the same translation reactions were assembled, except that the Tran35S-label was omitted. A PURExpress Δ ribosome translation mixture (NEB) without ribosome was programmed with appropriate DNA template and served as a toeprinting negative control. After incubation at 37°C for 15 min, primer extension was carried out at 37°C for 1 h using a 32P-labeled oligonucleotide that complemented a region 30 to 100 nt downstream of the ribosome-stalling site. The resulting cDNA was extracted once with phenol-chloroform (pH 6.8; Amresco) and was finally precipitated using 0.3 M sodium acetate (pH 5.2) and 3× volumes of isopropanol. The DNA pellet was washed with 70% ethanol, and the air-dried pellet was resuspended in 30 μl of formamide-containing loading buffer. DNA sequencing ladders were generated using a USB Thermo SEQ kit (Affymetrix). Five microliters of the toeprinting products and 1-μl portions of the ladders were heat denatured and resolved on 6% TBE (Tris-borate-EDTA)-urea polyacrylamide (19:1) sequencing gels and then scanned on a GE Typhoon phosphorimager. The intensity of m62A2058 signal was quantitated by ImageJ.
Northern blotting.
Oligonucleotides complementary to tRNAAsp, tRNAPro, and tRNALys (see Table S2 in the supplemental material) were biotinylated using a BrightStar psoralen-biotin kit (Ambion) under UV irradiation (365 nm) on ice for 45 min. The labeling efficiency was estimated by spotting serial diluted probes alongside the positive control provided in the kit, followed by signal detection with a BrightStar detection kit (Ambion). In vitro translation of the T7 -driven ermBL was carried out at 37°C for 1 h using a PURExpress kit (NEB). The translated products were resolved using neutral-pH 12% Bis-Tris SDS-PAGE (Invitrogen), and the tRNA-containing species were detected by chemiluminescence-based biotinylated oligonucleotide probe. Briefly, the translated products were transferred electrophoretically to a BrightStar-Plus nylon membrane (Ambion) using an Owl semidry HEP-1 transfer apparatus (Thermo Fisher) according to the manufacturer's protocol. Electrophoretic transfer was performed in 1× TBE buffer at 150 mA for 45 min, following UV cross-linking of the membrane inside a Stratalinker (Stratagene). Hybridization was performed at 45°C with biotinylated probes (1 pM final concentration), and washing steps were carried out using a Northern Max kit (Ambion). Finally, the hybridization signals were detected using a streptavidin-alkaline phosphatase-based BrightStar detection kit (Ambion) and by exposing the membrane to an autoradiography film (ISC BioExpress).
β-Galactosidase assay.
E. coli strains were grown in LB until reaching an optical density at 600 nm (OD600) of ≈0.3. Cultures were split into two portions; one portion was treated with antibiotics (erythromycin, 100 μg/ml; clindamycin, 230 μg/ml; tylosin, 800 μg/ml), and the second portion was supplemented with an equal volume of drug solvent. After treatment at 37°C for 30 min, 0.5-ml cell cultures were harvested in triplicate, and the β-galactosidase activity was measured in these samples according to standard protocols. Miller units were calculated by normalization to the cell density (OD600) (51). At least three independent biological replicates were performed.
Western blotting.
To determine the level of ErmB synthesis induction, E. coli XL1-Blue cells carrying the pTA derivatives (see Table S1 in the supplemental material) were grown and treated with antibiotics as described above. Portions (3 ml) of each culture were collected and resuspended in 1.5 ml of 20 mM Tris (pH 7.0). The suspensions were then divided in half. One portion was subjected to RNA isolation (see below), and the other portion was sonicated to obtain a crude cell lysate. Total soluble proteins (40 μg/lane) were resolved using 4 to 20% TGX SDS-PAGE (Bio-Rad), and the proteins were transferred to a nitrocellulose membrane using a Trans-Blot Turbo system (Bio-Rad). A 1/1,000 dilution of anti-ErmB (kindly provided by J. Rood) (52) and a 1/10,000 dilution of anti-RNAPα (NeoClone) were used for immunoblotting. The intensity of immunoblot bands was quantitated by using ImageJ.
RNA isolation and primer extension.
Total RNA was extracted using the hot phenol-SDS method (53) and an RNeasy kit (Qiagen). DNA contaminants were removed using two successive digestions with Turbo DNase I (Ambion), and RNA integrity was verified by nondenaturing agarose gel electrophoresis and ethidium bromide staining (54). Intact RNA was judged by determining the relative intensities of 23S and 16S rRNA bands, with a minimum accepted ratio of 1:1. A total of 250 ng of RNA input was used for primer extension (55); primer p1019 was used to detect m62A2058. To detect mRNA degradation intermediates, a final 6- to 8-ng/μl portion of total RNA and primers p700 or p299 was used for primer extension after normalization of the ermB mRNA between ERY-treated and untreated samples by quantitative reverse transcription-PCR (qRT-PCR; see Table S2 in the supplemental material) (55).
Rifampin chase and qRT-PCR.
E. coli NM580 derivatives were treated with antibiotics or mock solvent for 10 min. A final concentration of rifampin at 200 μg/ml or an equal volume of dimethyl sulfoxide was added to each culture. At time point zero (t0) and at subsequent time points (60, 90, 120, 150, 180, and 300 s), 1-ml samples of cells were subjected to hot-phenol-SDS RNA extraction (53) and RNeasy kit (Qiagen) purification. RT was performed using 5× iScript Supermix (Bio-Rad) and a 10-ng/μl concentration of DNase I-treated RNA. A “minus-RT” control was performed in parallel to ensure that the RNA was DNA-free. Quantitative PCR was performed in triplicate in 20-μl reaction mixtures containing 1× iTaq Universal SYBR green supermix (Bio-Rad), 0.4 μM concentrations of primers (see Table S2 in the supplemental material), and 1 μl of cDNA on a CFX96 real-time PCR instrument (Bio-Rad). Gene-specific primers were used (see Table S2 in the supplemental material), and 16S rRNA was used as an internal reference gene. Differences in mRNA levels were calculated using a published 2−ΔΔCT formula (56). mRNA half-lives were determined by fitting the data points to the equation y = a·e ^ (b·x), where y is the mRNA fraction, x is the time (in seconds), a is the initial number of mRNA, b is the decay constant, and e^ indicates exponential function; the fitted curve was used to calculate the half-life according to the equation t1/2 = x(1) − x(0.5).
RNA-Seq analysis.
Total RNA samples from three independent biological replicates were isolated as described above. RNA integrity after DNase I treatment was confirmed using a Bioanalyzer RNA 6000 Nano kit (Agilent). Four micrograms of RNA from each sample were subjected to rRNA depletion using a Ribo-Zero kit (Illumina) according to the manufacturer's protocol. RNA-Seq (transcriptome sequencing) was conducted at the Saint Louis University Biochemistry Genomics Core. Sequencing libraries were constructed using an Ion Total RNA-Seq kit (v2; Thermo Fisher) and were sequenced using an Ion Torrent Proton instrument (Life Technologies) with a mean read length of 101 bp and a minimum of 114× coverage. Alignment to the E. coli MG1655 reference genome (GenBank NC_000913.3) was performed using the TMAP aligner map4 algorithm (Life Technologies). Although Ion Torrent sequencing generates reads of different lengths, conventional RNA-Seq analyses typically use total reads (reads per kilobase per million/fragments per kilobase per million) to calculate expression levels under the assumption that all reads are the same length. To accurately measure expression levels, custom R scripts were used to calculate the total nucleotide coverage for each gene. The coverage values for all genes, expression ratios, standard errors, and P values are presented in Dataset S1 in the supplemental material. Data analyses are described in greater detail in reference 57. Functional groups of the differentially expressed genes with P ≤ 0.05 and 2-fold changes were classified by Panther GOC enrichment analysis.
Accession number(s).
Sequencing data were deposited in the NCBI database under accession number GSE80251.
RESULTS
Abrogating ErmBL-mediated ribosome stalling does not reduce ErmB expression.
The 27-amino-acid ErmBL leader peptide from S. aureus CM05 (GenBank accession number EF450709, referred to here as ErmBLEF) is highly conserved with Gram-positive bacterial homologs (Fig. 1B; see also Fig. S1 in the supplemental material) and only differs from the well-studied ErmBL at position 8 (Y8 instead of N8) (58, 59). An in vitro toeprinting assay was used to map the position of the stalled ribosome on ermBLEF mRNA in the presence of ERY. Consistent with previous in vitro results (58, 59), ERY arrested the ribosome with the D10 codon situated at the P-site (Fig. 1C; see also Fig. S2A in the supplemental material) and with a terminal aspartyl-tRNAAsp attached to the ErmBLEF nascent chain (see Fig. S2B in the supplemental material). Alanine mutagenesis showed that only residues R7, V9, D10, and K11 are critical for complete translation arrest (see Fig. S2A in the supplemental material). None of these alanine substitutions has been reported in the natural isolates; however, nonsense mutations, insertions, and deletions of ErmBL have been frequently found in clinical strains that exhibit high levels of multidrug resistance (34, 35, 37, 39, 40, 43, 60). Constitutive resistance derived from a complete loss of the ermBL regulatory region has been interpreted as a permanent disruption of the inhibitory mRNA structure that leads to downstream ErmB activation. The reasons underlying the hyper-resistance of less dramatic mutations, such as the introduction of a premature stop codon preceding the D10 ribosome-stalling sites, are poorly understood.
To investigate the consequences of introducing nonsense mutations and eliminating ErmBL translation, we replaced the first seven codons of ErmBLEF, one at a time, with an ochre codon that mimics the naturally occurring mutations (34, 37, 60). In principle, none of these mutants permits the ribosome to reach the ErmBLEF D10 stalling site. In the frameshift mutants FS1 and FS2, one or two adenine nucleotides were inserted immediately after the AUG initiation codon, which not only alters the sequence identity but also terminates the translation at positions 11 and 10, respectively (Fig. 1B). The M1L2Stop double mutant comprises an amber and an ochre codon, and V3Stop and Q5Stop are spontaneous ermBL mutations that are found in clinical isolates (34). In vitro toeprinting (Fig. 1C) demonstrated that the D10 toeprint signals were completely (R7Stop, lanes 15 and 16) or significantly diminished in all nonsense and frameshift mutants. The latter observation did not fully meet our initial expectation that the D10 toeprints would disappear completely in all early terminated ermBLEF mutants (Fig. 1C). Our subsequent investigations revealed that the residual background signals were derived in part from translational read-through past the stop codon (see Fig. S3A in the supplemental material) (61), and in some cases, resulted from the impeded reverse transcription on different mutated mRNA templates because toeprinting reactions that are programmed without a ribosome also produce weak signals (see Fig. S3B in the supplemental material).
To reassure ourselves that the mutations were defective in full-length translation of ErmBLEF, we examined the effects of ermBLEF nonsense mutations in vivo. Multidrug-resistant S. aureus CM05 carries Cfr RNA methyltransferase and the macrolide efflux pump proteins MefA and MsrA, which might mask the effect of ermBLEF-ermB. To avoid complications in interpreting the resistance phenotype, we either expressed the ermBLEF-ermB on a plasmid under the control of its native promoter or recombined the lacZ reporter alleles (ermBLEF-ermB′-lacZ or ermBLEF′-lacZ) to the native chromosomal lacZ locus of an E. coli surrogate host (Fig. 2). A similar approach is widely used to study antibiotic-induced resistance because E. coli cells and ribosomes are known to faithfully recapitulate peptide-dependent ribosome stalling in vivo and in vitro (14, 50, 58, 59, 62). Moreover, ermBL-ermB has been routinely found in many environmental and hospital E. coli strains (4, 6–10, 63, 64). By creating a translational fusion of the first nine codons of ermBLEF (excluding the D10 stalling site) directly to the lacZ, we confirmed that the nonsense mutations indeed abolish the translation of lacZ (as shown by a lack of β-galactosidase activity) with the exception of the M1Stop mutant. The mutant retains about 10% of the original β-galactosidase activity, presumably because L2 (UUG, 3% usage frequency in E. coli) acts as an alternate start codon (Fig. 2A). Furthermore, using the lacZ reporter fused to ermB, we found that the wild-type (WT) ermBLEF and M1Stop mutant moderately induced downstream ermB′-lacZ expression upon ERY exposure. In contrast, all other nonsense mutants demonstrated high basal expression of ermB′-lacZ without ERY treatment, and the levels were further elevated in the presence of ERY. Overall, ERY treatment increases ermB′-lacZ expression by ∼2-fold in the WT ermBLEF context and by ∼30% in the nonsense mutants (Fig. 2B), an induction level that is comparable to that of other erm systems (13, 14). These results indicate that the active translation of ermBLEF (WT and M1Stop) attenuates the capacity of ErmB synthesis, albeit ERY treatment can partially alleviate the repression.
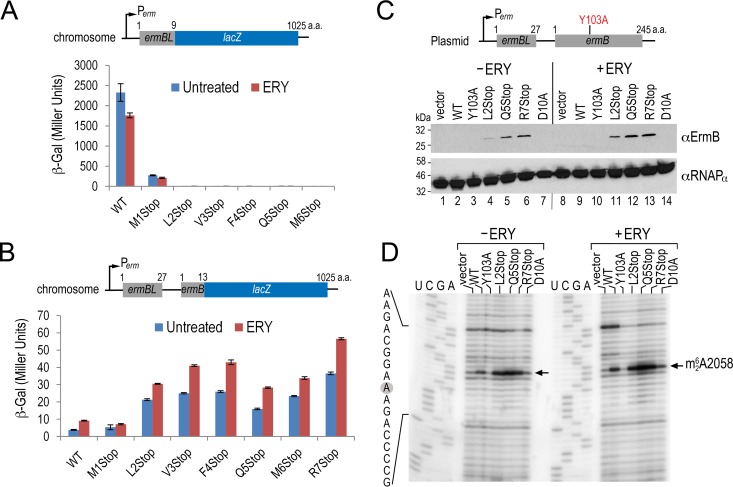
FIG 2.
ErmBLEF nonsense mutations result in a high basal and inducible expression of downstream ErmB that is consistent with the cellular concentrations of ErmB and the degree of ribosome methylation. (A) A β-galactosidase activity assay showing that nonsense mutations at residues L2 through R7 effectively shut down lacZ synthesis and revealing that L2 acts as an alternate start codon. β-Galactosidase activity (in Miller units) was conducted with E. coli bearing the chromosomal ermBL′-lacZ with or without 30 min of erythromycin (ERY) exposure. ERY was used at 100 μg/ml. (B) Results from a β-galactosidase assay showing that none of the premature nonsense mutations after codon M1 abolishes the downstream ermB′-lacZ expression. The β-galactosidase activity was determined as described in panel A except that chromosomal ermBL-ermB′-lacZ fusion was used. Error bars indicate standard deviations from three replicates. (C) Western blot analysis shows that ErmB overexpression remains inducible in response to ERY and that the degree of induction correlates with the lacZ reporter results. Log-phase cells with or without 30 min of ERY treatment were harvested and sonicated, and 40-μg portions of total soluble proteins were loaded per lane on the SDS-PAGE gel. Y103A is a catalytically inactive ErmB mutant. The alpha-subunit of RNA polymerase served as the loading control. A 1/1,000 dilution of anti-ErmB and a 1 1/10,000 dilution of anti-RNAPα were used for immunoblotting. (D) Results from a primer extension assay showing that the magnitude of A2058 methylation is consistent with the cellular levels of ErmB. Total RNAs were isolated from the same cells shown in panel C and used at 250 ng per lane. In primer extension, the reverse transcriptase halts at the methylated site and produces a truncated cDNA that is manifested by a strong signal at A2058.
To eliminate the possibility of artifacts associated with reporter mRNA and protein turnovers, we examined the expression of ErmB methyltransferase on a plasmid-borne ermBLEF-ermB in the E. coli background. An antibody against C. perfringens ErmB (52) was used to probe the expression level of ErmB. A catalytically inactive mutant of ErmB (Y103A, Y104 numbering in ErmC [65]) served as a control. Consistent with the β-galactosidase results, the basal levels of ErmB in the nonsense mutants were much greater than those in the ErmB (Y103A) mutant and WT in the absence of ERY (Fig. 2C, compare lanes 2 to 3 to lanes 4 to 6). The detection of ErmB in the WT and Y103A backgrounds was hampered by their low basal expression and in part by the low antibody titer. Nevertheless, we observed an ∼2-fold increase in ErmB induction in the nonsense mutants after 30 min of ERY exposure (compare lanes 4 to 6 to lanes 11 to 13). The expression level of ErmB was also consistent with the degree of in vivo ribosome methylation (Fig. 2D). Dimethylation of A2058 in 23S rRNA halts reverse transcription and produces a strong termination pause at this residue. We found that ERY treatment induces an ∼2-fold increase in methylation in the nonsense mutants and the WT, whereas the strains harboring the catalytically dead Y103A mutant and the empty vector did not undergo methylation.
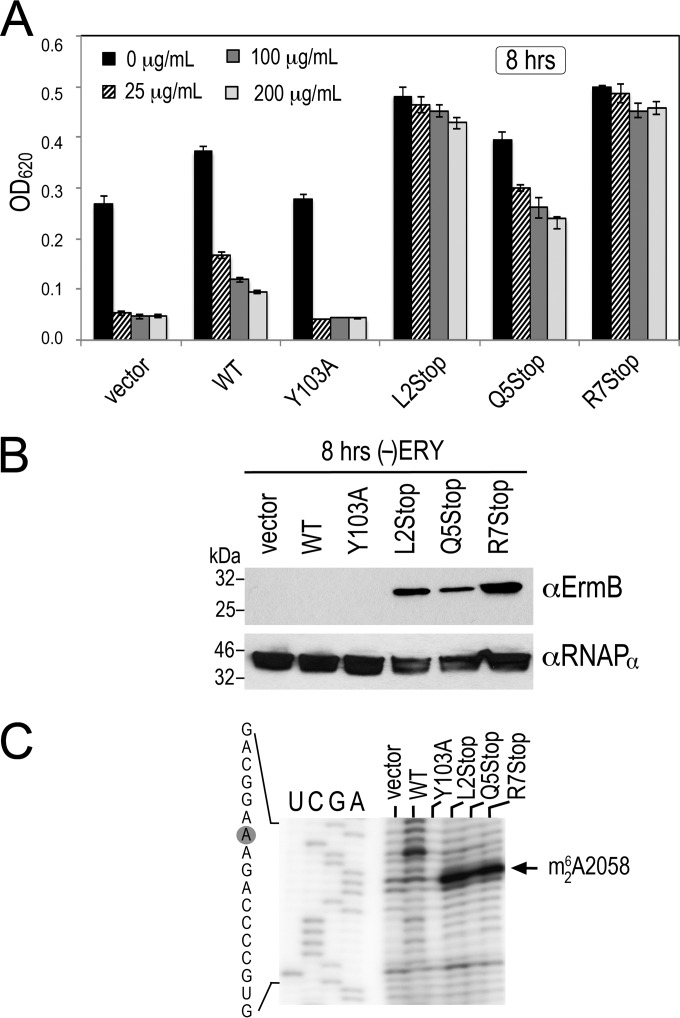
Ribosomal methylation appears to direct the hyper-resistance phenotypes of the ErmBLEF nonsense mutants. The same strains in Fig. 2C were treated with various concentrations of ERY. The MIC of ERY on the E. coli strain that we used was 46 to 64 μg/ml, as measured based on the Etest. The MIC of the ermBLEF-derived strains is >640 μg/ml. As expected, the vector and Y103A controls were extremely susceptible to subinhibitory doses of ERY, but the nonsense mutants were all resistant to high concentrations of ERY (Fig. 3A). The resistance phenotype was due to an increased level of ErmB (Fig. 3B) and an increase in ribosomal methylation (Fig. 3C). In these experiments, only untreated cells were analyzed because insufficient WT or control cells were recovered after 8 h of ERY inhibition. These data confirm that moderate 2- to 4-fold changes in ErmB expression can significantly affect bacterial resistance. Together, our results demonstrate that the ErmBLEF nonsense mutants are defective in ribosome stalling, but the mutations do not reduce ErmB expression. Rather, ErmB is constitutively expressed and is moderately inducible by ERY, thereby leading to higher resistance than that observed for WT ermBLEF-ermB.
FIG 3.
ErmBLEF nonsense mutations convert the susceptible WT into ERY-resistant cells. (A) Bacterial growth in the presence of various concentrations of erythromycin (ERY). Overnight E. coli LB cultures were diluted (1/100, normalized to an OD600 of 0.002) into fresh medium supplemented with ampicillin at 100 μg/ml to maintain the ermBL-ermB bearing plasmid. The cell density was recorded at 4, 8, and 24 h after inoculation. Only the 8-h dataset is shown. Error bars indicate standard deviations obtained from three independent experiments. (B) Detection of basal ErmB levels by immunoblotting after 8 h of growth in the absence of erythromycin (ERY). Total soluble proteins were extracted from the same plasmid-borne ermBL-ermB backgrounds shown in panel A. Each lane corresponds to 40 μg of total soluble proteins. Y103A is a catalytically inactive ErmB mutant. The alpha-subunit of RNA polymerase served as the loading control. A 1/1,000 dilution of anti-ErmB and a 1 1/10,000 dilution of anti-RNAPα were used for immunoblotting. (C) Results from a primer extension analysis showing the basal methylation of ribosomes (without ERY) after 8 h of growth. Total RNA was isolated from the same strain backgrounds shown in panels A and B. Each lane corresponds to 250 ng of RNA input.
Different ribosome-targeting antibiotics increase the steady-state level of ermB mRNA.
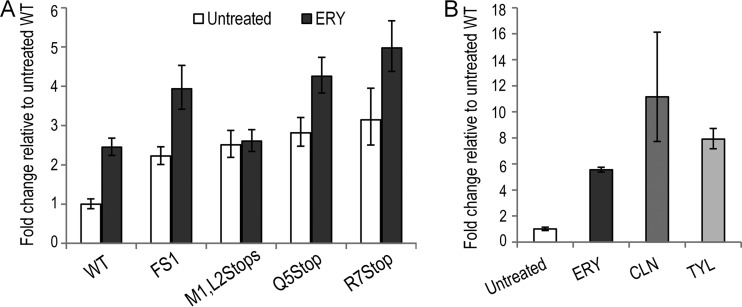
To understand how the ErmBLEF nonsense mutations stimulate ErmB overexpression, we performed qRT-PCR to determine whether the steady-state level of ermB mRNA was altered in different ermBLEF mutant E. coli backgrounds treated with or without ERY. Consistent with the results described earlier (Fig. 2), approximately 2- to 3-fold increases in the mRNA levels were observed in the mutants relative to the WT in the absence of ERY. In the presence of ERY, the mRNA levels of the mutants (except for M1L2Stop) were markedly elevated by an additional 1.5-fold (Fig. 4A). Interestingly, mRNA accumulation was also observed in cells that were exposed to other noninducers of the ErmBL-mediated ribosome stalling. We observed a similar trend of antibiotic-induced mRNA upregulation in response to clindamycin (CLN) and tylosin (TYL) treatments (Fig. 4B). CLN and TYL are ribosome inhibitors and, like ERY, are positively charged. However, CLN (a lincosamide) and TYL (a 16-membered macrolide) both fail to promote ErmBLEF-dependent ribosome stalling (see Fig. S4 in the supplemental material) (58). These results strongly suggest that ErmB upregulation can be separated from the ribosome-stalling pathway.
FIG 4.
Ribosome-targeting antibiotics increase steady-state ermB mRNA levels independently of ErmBLEF-mediated ribosome stalling. (A) Quantitative RT-PCR analysis demonstrated an increase in ermB mRNA abundance after ERY treatment in different ermBLEF mutant backgrounds. The fold change relative to the untreated WT is shown. (B) Quantitative RT-PCR analysis demonstrated that the noninducers of ErmBLEF-mediated ribosome stalling, clindamycin (CLN) and tylosin (TYL) (see Fig. S4 in the supplemental material) (58), upregulate ermB expression. Total RNAs were isolated from the WT ermBLEF-ermB strain treated with different antibiotics. The fold change relative to the untreated sample is shown. Error bars indicate the standard deviation of three replicates.
ERY promotes ermB mRNA stability.
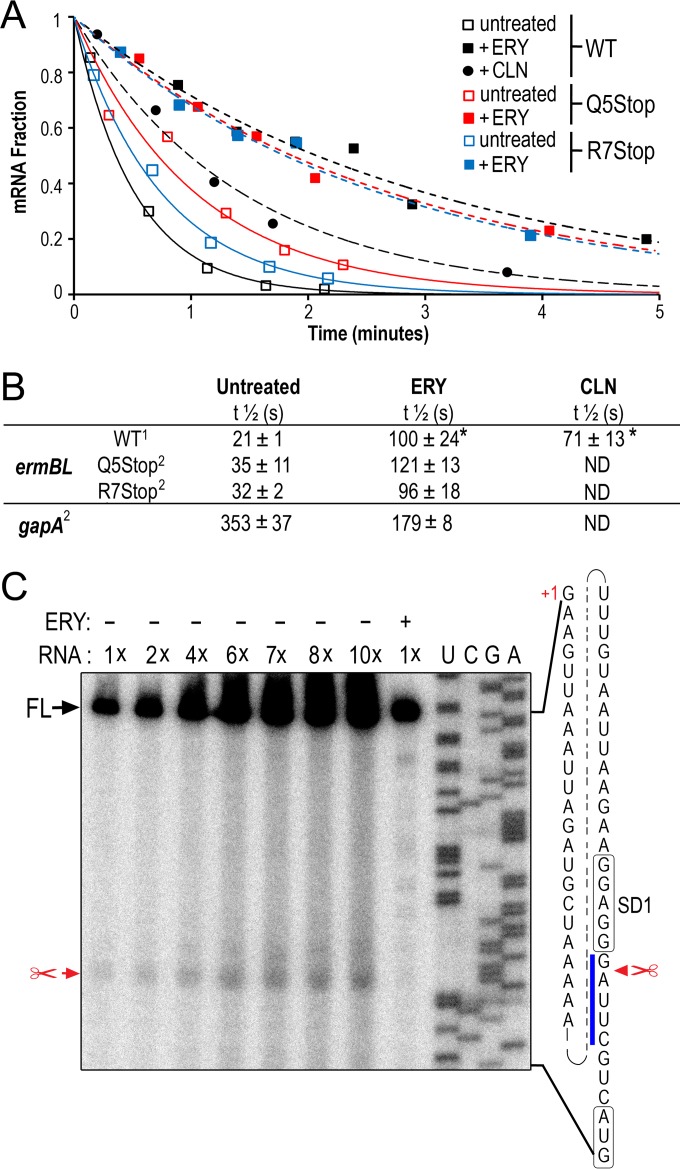
The observed increase in ermB mRNA abundance might be due to increased transcription initiation. Alternatively, ERY treatment and the ErmBLEF nonsense mutations might stabilize ermB mRNA. To distinguish between these alternatives, we performed a rifampin chase to measure the mRNA stability over time after the transcription inhibitor rifampin was added to the cell cultures. The vast majority of E. coli mRNAs exhibit half-lives of between 3 and 9 min (66); here, we found that the level of ermB mRNA decreased dramatically within the first minute after rifampin treatment (Fig. 5A). After fitting the data points, a half-life of 20 ± 1 s was observed for the untreated WT ermBLEF-ermB background from three independent experiments. The half-lives were extended by about 5- and 3.5-fold, respectively, after 10 min of exposure to ERY and CLN (Fig. 5B). The half-lives of ermB mRNA were slightly enhanced in the untreated Q5Stop and R7Stop mutant strains relative to the WT, and the mRNA was stabilized >3-fold after ERY treatment. In contrast, gapA, a housekeeping gene that has been frequently used as an internal reference for qRT-PCR (67, 68), showed an opposite effect; that is, the mRNA half-life was reduced 2-fold by ERY treatment (Fig. 5B). Because mRNA half-life is linked to degradation, we performed primer extension mapping with total cellular RNA isolated from WT ermBLEF-ermB cell cultures to identify degradation intermediates of the ermBLEF-ermB transcript. The steady-state level of ermB mRNA in untreated cells was lower than that in ERY-treated cells (Fig. 4A); therefore, we calibrated the signals by titrating the amount of RNA inputs from the untreated cells. Consistent with previous findings (19), the transcription start site of ermBLEF-ermB was mapped at a guanine located 50 nt upstream of the AUG start codon (Fig. 5C). A processed ermBLEF-ermB mRNA intermediate was detected at a site one nucleotide downstream of the predicted core SD sequence (69). Close inspection showed that the sequences flanking the cleavage site conspicuously resembled the previously reported RNase E target sequence ([A/G]AUU[A/U/C] (45–47). RNase E cleavage sites often occur in A/U-rich single-stranded regions; however, no definitive consensus has yet been identified (66, 70). Nevertheless, the results of the primer extension experiment implied that the 5′ end of the ermBLEF-ermB transcript is a substrate of RNase E when the ERY is omitted, which may account for the shorter mRNA half-life (Fig. 5B). The data also support the notion that ribosome-targeting antibiotics can enhance mRNA stability. However, it remains unclear whether the effect is direct or indirect and whether the phenomenon can be generalized to mRNAs other than the ermBLEF-ermB transcript.
FIG 5.
Antibiotics prolong ermB mRNA half-life. (A) The mRNA decay curves of ErmBLEF WT, Q5Stop, and R7Stop background strains after 10 min of exposure to erythromycin (ERY) and clindamycin (CLN). A rifampin chase experiment was performed in E. coli cells bearing the chromosomally located ermBL-ermB′. (B) Summary of the ermB mRNA half-lives determined from panel A. An asterisk (*) indicates statistical significance (t test P ≤ 0.05). Superscript numbers: 1, standard deviations obtained from three independent biological replicates; 2, means and standard deviations obtained from two replicates. ND, not determined. (C) Reverse transcription mapping shows that mRNA cleavage downstream of the ermBLEF SD is reduced in the presence of erythromycin (ERY). A 1× RNA input equals 4 μg of RNA template in primer extension. The start codon and SD of ermBLEF are boxed. The RNase E-targeting sequence is denoted by a blue line. FL, full-length cDNA. The “+1” indicates the transcription start site of ermBLEF-ermB.
ERY and CLN upregulate a specific subset of genes.
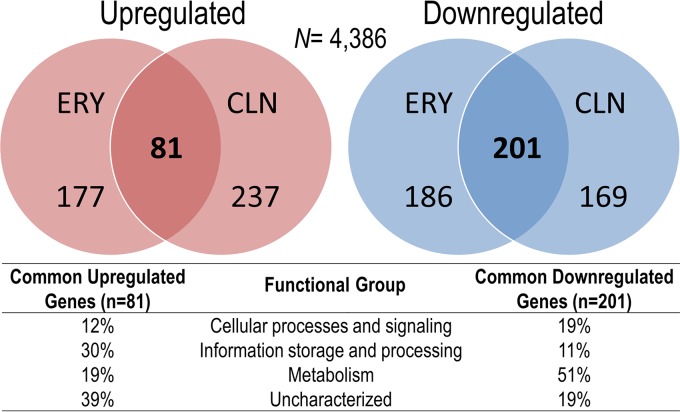
To determine the global effects of antibiotics, we performed an RNA-Seq analysis in E. coli (ermBL-ermB::lacZ) cells to measure changes in gene expression in response to ERY and CLN treatments. We found ca. 15% of the genes in each antibiotic treatment exhibited significant (P ≤ 0.05, 2-fold cutoff) increased or decreased mRNA levels (Fig. 6; see Dataset S1 in the supplemental material). Of note, a 2.3-fold increase (P = 0.007) in the ermBLEF-ermB′-lacZ transcript was observed in ERY-treated cells, validating our earlier results (Fig. 2 and 4A). Nearly half of the genes from each treatment were coregulated by both ERY and CLN. Of those, 30% of coupregulated genes are involved in information storage and processing, but many genes (30%) remain uncharacterized. In addition, half of the codownregulated genes are involved in cellular metabolism (Fig. 6; see Dataset S1 in the supplemental material). The expression patterns of 17 actively transcribed genes were further verified by qRT-PCR and were proven to follow the same profiles as the RNA-Seq data (see Fig. S5A in the supplemental material). Low concentrations of antibiotics can activate or repress the transcription of genes that are not their direct targets (71–76). We performed a rifampin chase to compare the mRNA decay rate in ERY-treated and untreated cells to examine whether the changes in mRNA abundance were caused by the altered stability. We found that 5 of 6 ERY-upregulated genes in ERY-treated cells exhibited significantly longer half-lives than those in untreated cells (see Fig. S5B in the supplemental material). The results suggest that the gene (deaD) whose mRNA half-life remained unaltered might be under transcriptional activation; however, most of the upregulated genes exhibited slowed mRNA decay when subjected to ERY treatment.
FIG 6.
Macrolide and lincosamide differentially regulate specific gene subsets. Genome-wide transcriptome analysis shows that ERY and CLN coregulate the expression of a subset of genes that is enriched in information storage and processing and metabolic pathways. Venn diagrams show genes that exhibited a 2-fold increase or decrease in nucleotide coverage density (P ≤ 0.05). Complete and sorted lists of the up- and downregulated genes are provided in Dataset S1 in the supplemental material.
DISCUSSION
The molecular- and atomic-level descriptions of macrolide-induced ribosome stalling have been elucidated in great detail (12–17, 19, 58, 59, 77–81); however, the relationship between ribosome stalling and the cellular levels of Erm methyltransferase (and thus bacterial resistance) has not been entirely consistent with clinical findings, wherein inducible constitutive resistance is commonly found in strains bearing ribosome-stalling-dead leader peptides. Our in vitro and in vivo analyses of the ErmBLEF nonsense mutants unequivocally demonstrate that increased mRNA stability could account for the observed ErmB overproduction, that distant macrolide relatives also promote the stabilization of the ermBLEF-ermB transcript, and that antibiotic exposure exerts a protective role on mRNA decay.
The translational attenuation model of erm regulation has been largely inferred from the ermCL-ermC system, for which the mRNA structural rearrangements in the presence or absence of ERY have been mapped in vivo and in vitro (13, 82). A similar conformational switch has been detected in ermBL-ermB mRNA (18). However, the two operons differ from each other in at least three aspects. First, the activation of ermC expression is induced by a narrow spectrum of macrolides via ribosome stalling and is induced by telithromycin via a frameshifting mechanism (77, 83). In contrast, ermB expression is induced by macrolides, lincosamides, and streptogramins (MLS) (18, 19, 84, 85) (Fig. 4B) and by the latest generation of macrolides, termed ketolides (58, 59). In this case, only macrolides and ketolides can act as an inducer of ErmBL-dependent ribosome stalling. It is unclear how lincosamides and streptogramins upregulate ermB expression because the two drugs do not appear to cause mRNA conformational changes (18) or induce ErmBL-dependent ribosome stalling (see Fig. S4 in the supplemental material).
Second, the synthesis of ErmC is strictly dependent on the translation of ermCL because an introduction of an ochre codon at the second position of ErmCL completely abolishes inducibility and ErmC production (12). ermBL-ermB nonsense mutations preceding the ErmBL D10 stalling sites are found in hyper-resistant clinical isolates (34, 37, 60), and ErmB overexpression has been observed when residues that are critical for ribosomes stalling (D10 and K11) are replaced with a termination codon (18). Intriguingly, the mRNAs from D10(UAA) and K11(UAA) mutants are processed in a distinctive manner in that the level of cleaved mRNA intermediates is increased after ERY treatment in the K11(UAA)-bearing cells but not in the D10(UAA) cells. The reason for this difference is unknown, but the high basal expression of ermB in these mutants has been interpreted as a structural disruption of the inhibitory stem-loop when the ribosome is paused at the tenth and eleventh termination codons, which are embedded inside the predicted hairpin structure (18). The ErmBL codons M1 through M6 are located outside the stem-loop structure (18); thus, the ochre codon we introduced (Fig. 2) and the previously reported spontaneous mutations V3(UAA) and Q5(UAA) (34) are unlikely to cause drastic changes within the presumptive mRNA hairpin, although some of these nonsense mutations may alter mRNA stability. For instance, we found that in ERY-free cells, the half-lives of Q5Stop and R7Stop are slightly longer than that of WT ermBLEF (Fig. 5B). Likewise, previous ermBL-ermB mRNA structural probing was conducted in the B. subtilis host and in the WT ermBL-ermB context (18); it is possible that ermB SD2 is unmasked in the nonsense mutants, which could explain the high basal expression of ErmB in the absence of ERY inducer. Nevertheless, we found that the early translational termination of ErmBLEF results in constitutive resistance and correlates with ErmB production and the extent of ribosome methylation. These data strongly suggest that ribosome stalling is not the sole determinant of the inducible resistance; the slowing down of mRNA turnover might represent an alternative pathway of conferring bacterial resistance.
Third, the improvement of mRNA stability upon ERY treatment has been previously reported for ermCL-ermC (86) and ermBL-ermB (18). Active ermCL translation is required for the observed stabilization, and stalling of the ribosome has been proposed to physically protect the mRNA from RNase action. The same interpretation has been posited for other ermBL-ermB system (18). In contrast, our data show that increased mRNA stability is not due to the translation of ermBLEF and ribosome stalling. Of note, the eighth position in the ErmBLEF that we used is a tyrosine rather than an asparagine (as used in reference 18; see Fig. S1 in the supplemental material). In addition, under our experimental settings, we did not detect the same processed intermediates possibly due to differences in the mRNA degradation enzymes between B. subtilis and E. coli (87). More strikingly, Min et al. (18) only observed mRNA intermediates in cells that had been treated with ERY; in that study, the processing sites were mapped at the edges of the ERY-stalled ribosome. In contrast, we detected an intermediate that might be a substrate of E. coli RNase E in cells without ERY treatment, and this intermediate is absent in ERY-treated cells (Fig. 5C). The lack of an mRNA decay product in the ERY-treated cells provides an explanation for the observed increase in the steady-state mRNA level. The mechanism by which ERY reduces mRNA degradation has yet to be determined. It is possible that ERY prevents mRNA decay by altering the mRNA conformation through direct RNA binding. Although macrolide binding to other structured RNAs has not been reported, the direct binding of aminoglycosides to other viral RNAs, ribozymes, and synthetic riboswitches has been described (88–92). Alternatively, ERY might indirectly stabilize mRNA by influencing the expression of protein factors and regulatory sRNAs that are responsible for the activity and expression of RNase. Finally, the possibility that ERY may directly inhibit nucleolytic activity or inhibit the binding of RNase to the mRNA cannot be ruled out.
The nonsense ErmBLEF mutations increase A2058 dimethylation, but the modification is relatively low in the WT ErmBLEF and in M1Stop, which remain active in terms of translation and ribosome stalling (Fig. 2A and B). Based on our results, translation of ermBLEF and ribosome stalling appear to suppress ErmB expression, whereas disrupting these functions results in “uncontrollable” ErmB expression. The ribosome-stalling mechanism thus may be a negative regulator to ensure that only an appropriate amount of ErmB is synthesized because methylated ribosome is known to compromise bacterial fitness by perturbing translational activity (93). Furthermore, our observation that ERY and CLN both have the ability to preferentially upregulate gene expression (Fig. 6; see Dataset S1 in the supplemental material) highlights an unexpected role of antibiotics in linking mRNA metabolism to resistance and underscores a need to examine the pleiotropic effects of antibiotic therapy.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Dale Dorsett for RNA-Seq data analyses, Nadim Majdalani for NM580, and Julian Rood for anti-ErmB. We thank Gaurab Kc, Suoami Abuirqeba, and Mariah Hassert for constructing and analyzing some of the ErmBL mutants.
This study was supported by the National Institutes of Health (grant R00GM094212 to M.N.Y.), the Edward Mallinckrodt Jr. Foundation, and the Saint Louis University faculty start-up and President's Research Fund. M.N.Y. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. The funders had no role in study design, data collection and interpretation, or the decision to submit this work for publication.
Funding Statement
This work, including the efforts of Mee-Ngan F. Yap, was funded by Edward Mallinckrodt Jr. Foundation, Saint Louis University President's Research Fund, HHS|NIH (R00GM094212), and Pew Charitable Trusts (0002920).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01806-16.
REFERENCES
- 1.Wilson DN. 2014. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 2.Thomas F, Depledge M. 2015. Medicine ‘misuse’: implications for health and environmental sustainability. Soc Sci Med 143:81–87. doi: 10.1016/j.socscimed.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 3.Yap MN. 2013. The double life of antibiotics. Mo Med 110:320–324. [PMC free article] [PubMed] [Google Scholar]
- 4.Wendlandt S, Shen J, Kadlec K, Wang Y, Li B, Zhang WJ, Fessler AT, Wu C, Schwarz S. 2015. Multidrug resistance genes in staphylococci from animals that confer resistance to critically and highly important antimicrobial agents in human medicine. Trends Microbiol 23:44–54. doi: 10.1016/j.tim.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Weisblum B. 1995. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother 39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts MC. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett 282:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 7.Kappell AD, DeNies MS, Ahuja NH, Ledeboer NA, Newton RJ, Hristova KR. 2015. Detection of multi-drug resistant Escherichia coli in the urban waterways of Milwaukee, WI. Front Microbiol 6:336. doi: 10.3389/fmicb.2015.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Bonnin RA, Nordmann P. 2011. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother 55:4224–4229. doi: 10.1128/AAC.00165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta A, Vlamakis H, Shoemaker N, Salyers AA. 2003. A new Bacteroides conjugative transposon that carries an ermB gene. Appl Environ Microbiol 69:6455–6463. doi: 10.1128/AEM.69.11.6455-6463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother 43:2823–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park AK, Kim H, Jin HJ. 2010. Phylogenetic analysis of rRNA methyltransferases, Erm and KsgA, as related to antibiotic resistance. FEMS Microbiol Lett 309:151–162. doi: 10.1111/j.1574-6968.2010.02031.x. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau D. 1985. Induction of ermC requires translation of the leader peptide. EMBO J 4:533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayford M, Weisblum B. 1989. Conformational alterations in the ermC transcript in vivo during induction. EMBO J 8:4307–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vazquez-Laslop N, Thum C, Mankin AS. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Ramu H, Vazquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, Micura R, Mankin AS. 2011. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell 41:321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 16.Sothiselvam S, Liu B, Han W, Ramu H, Klepacki D, Atkinson GC, Brauer A, Remm M, Tenson T, Schulten K, Vazquez-Laslop N, Mankin AS. 2014. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci U S A 111:9804–9809. doi: 10.1073/pnas.1403586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramu H, Mankin A, Vazquez-Laslop N. 2009. Programmed drug-dependent ribosome stalling. Mol Microbiol 71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 18.Min YH, Kwon AR, Yoon EJ, Shim MJ, Choi EC. 2008. Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob Agents Chemother 52:1782–1789. doi: 10.1128/AAC.01376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horinouchi S, Byeon WH, Weisblum B. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol 154:1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrod R, Lovett PS. 1997. Leader peptides of inducible chloramphenicol resistance genes from gram-positive and gram-negative bacteria bind to yeast and Archaea large subunit rRNA. Nucleic Acids Res 25:1720–1726. doi: 10.1093/nar/25.9.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Z, Rogers EJ, Lovett PS. 1993. Peptidyl transferase inhibition by the nascent leader peptide of an inducible cat gene. J Bacteriol 175:5309–5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovett PS, Rogers EJ. 1996. Ribosome regulation by the nascent peptide. Microbiol Rev 60:366–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R. 2016. Term-Seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 352:aad9822. doi: 10.1126/science.aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba S, Ito K. 2012. Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol Cell 47:863–872. doi: 10.1016/j.molcel.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 25.Butkus ME, Prundeanu LB, Oliver DB. 2003. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J Bacteriol 185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakatogawa H, Ito K. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell 7:185–192. doi: 10.1016/S1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 27.Nakatogawa H, Ito K. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108:629–636. doi: 10.1016/S0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 28.Yap MN, Bernstein HD. 2009. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell 34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii E, Chiba S, Hashimoto N, Kojima S, Homma M, Ito K, Akiyama Y, Mori H. 2015. Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc Natl Acad Sci U S A 112:E5513–E5522. doi: 10.1073/pnas.1513001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. 2005. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell 19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Park SY, Cromie MJ, Lee EJ, Groisman EA. 2010. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell 142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chancey ST, Zahner D, Stephens DS. 2012. Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol 7:959–978. doi: 10.2217/fmb.12.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon AR, Min YH, Yoon EJ, Kim JA, Shim MJ, Choi EC. 2006. ErmK leader peptide: amino acid sequence critical for induction by erythromycin. Arch Pharm Res 29:1154–1157. doi: 10.1007/BF02969307. [DOI] [PubMed] [Google Scholar]
- 34.Min YH, Kwon AR, Yoon JM, Yoon EJ, Shim MJ, Choi EC. 2008. Molecular analysis of constitutive mutations in ermB and ermA selected in vitro from inducibly MLSB-resistant enterococci. Arch Pharm Res 31:377–380. doi: 10.1007/s12272-001-1167-8. [DOI] [PubMed] [Google Scholar]
- 35.Min YH, Jeong JH, Choi YJ, Yun HJ, Lee K, Shim MJ, Kwak JH, Choi EC. 2003. Heterogeneity of macrolide-lincosamide-streptogramin B resistance phenotypes in enterococci. Antimicrob Agents Chemother 47:3415–3420. doi: 10.1128/AAC.47.11.3415-3420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malhotra-Kumar S, Mazzariol A, Van Heirstraeten L, Lammens C, de Rijk P, Cornaglia G, Goossens H. 2009. Unusual resistance patterns in macrolide-resistant Streptococcus pyogenes harbouring erm(A). J Antimicrob Chemother 63:42–46. doi: 10.1093/jac/dkn432. [DOI] [PubMed] [Google Scholar]
- 37.Rosato A, Vicarini H, Leclercq R. 1999. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J Antimicrob Chemother 43:559–562. doi: 10.1093/jac/43.4.559. [DOI] [PubMed] [Google Scholar]
- 38.Domelier AS, van der Mee-Marquet N, Arnault L, Mereghetti L, Lanotte P, Rosenau A, Lartigue MF, Quentin R. 2008. Molecular characterization of erythromycin-resistant Streptococcus agalactiae strains. J Antimicrob Chemother 62:1227–1233. doi: 10.1093/jac/dkn388. [DOI] [PubMed] [Google Scholar]
- 39.Min YH, Yoon EJ, Kwon AR, Shim MJ, Choi EC. 2011. Alterations in regulatory regions of erm(B) genes from clinical isolates of enterococci resistant to telithromycin. Arch Pharm Res 34:2149–2154. doi: 10.1007/s12272-011-1219-4. [DOI] [PubMed] [Google Scholar]
- 40.Vicarini H, Rosato A, Leclercq R. 1997. Analysis of regulatory region of ermAM genes in streptococci and enterococci highly resistant to macrolides and lincosamides. Adv Exp Med Biol 418:495–498. doi: 10.1007/978-1-4899-1825-3_118. [DOI] [PubMed] [Google Scholar]
- 41.de Vries LE, Christensen H, Agerso Y. 2012. The diversity of inducible and constitutively expressed erm(C) genes and association to different replicon types in staphylococci plasmids. Mob Genet Elements 2:72–80. doi: 10.4161/mge.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Millan L, Goni P, Cerda P, Rubio MC, Gomez-Lus R. 2007. Novel 10-bp deletion in the translational attenuator of a constitutively expressed erm(A) gene from Staphylococcus epidermidis. Int Microbiol 10:147–150. [PubMed] [Google Scholar]
- 43.Deng F, Shen J, Zhang M, Wu C, Zhang Q, Wang Y. 2015. Constitutive and inducible expression of the rRNA methylase gene erm(B) in Campylobacter. Antimicrob Agents Chemother 59:6661–6664. doi: 10.1128/AAC.01103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doktor SZ, Shortridge V. 2005. Differences in the DNA sequences in the upstream attenuator region of erm(A) in clinical isolates of Streptococcus pyogenes and their correlation with macrolide/lincosamide resistance. Antimicrob Agents Chemother 49:3070–3072. doi: 10.1128/AAC.49.7.3070-3072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehretsmann CP, Carpousis AJ, Krisch HM. 1992. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev 6:149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- 46.Kaberdin VR. 2003. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res 31:4710–4716. doi: 10.1093/nar/gkg690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richards J, Belasco JG. 2016. Distinct requirements for 5′-monophosphate-assisted RNA cleavage by Escherichia coli RNase E and RNase G. J Biol Chem 291:5038–5048. doi: 10.1074/jbc.M115.702555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Battesti A, Majdalani N, Gottesman S. 2015. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci U S A 112:5159–5164. doi: 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruckner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187–192. doi: 10.1016/0378-1119(92)90048-T. [DOI] [PubMed] [Google Scholar]
- 50.Davis AR, Gohara DW, Yap MNF. 2014. Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci U S A 111:15379–15384. doi: 10.1073/pnas.1410356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 52.Farrow KA, Lyras D, Polekhina G, Koutsis K, Parker MW, Rood JI. 2002. Identification of essential residues in the Erm(B) rRNA methyltransferase of Clostridium perfringens. Antimicrob Agents Chemother 46:1253–1261. doi: 10.1128/AAC.46.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuang SE, Daniels DL, Blattner FR. 1993. Global regulation of gene expression in Escherichia coli. J Bacteriol 175:2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. 2010. Nondenaturing agarose gel electrophoresis of RNA. Cold Spring Harb Protoc 2010:pdb.prot5445. doi: 10.1101/pdb.prot5445. [DOI] [PubMed] [Google Scholar]
- 55.Carey MF, Peterson CL, Smale ST. 2013. The primer extension assay. Cold Spring Harb Protoc 2013:164–173. doi: 10.1101/pdb.prot071902. [DOI] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Wu Y, Gause M, Xu D, Misulovin Z, Schaaf CA, Mosarla RC, Mannino E, Shannon M, Jones E, Shi M, Chen WF, Katz OL, Sehgal A, Jongens TA, Krantz ID, Dorsett D. 2015. Drosophila Nipped-B mutants model Cornelia de Lange syndrome in growth and behavior. PLoS Genet 11:e1005655. doi: 10.1371/journal.pgen.1005655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arenz S, Ramu H, Gupta P, Berninghausen O, Beckmann R, Vazquez-Laslop N, Mankin AS, Wilson DN. 2014. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun 5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gupta P, Liu B, Klepacki D, Gupta V, Schulten K, Mankin AS, Vazquez-Laslop N. 2016. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat Chem Biol 12:153–158. doi: 10.1038/nchembio.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Isogai N, Urushibara N, Kawaguchiya M, Ghosh S, Suzaki K, Watanabe N, Quinones D, Kobayashi N. 2013. Characterization of Enterococcus faecium with macrolide resistance and reduced susceptibility to quinupristin/dalfopristin in a Japanese hospital: detection of extensive diversity in erm(B)-regulator regions. Microb Drug Resist 19:298–307. doi: 10.1089/mdr.2012.0176. [DOI] [PubMed] [Google Scholar]
- 61.Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. 2000. A posttranslational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J 19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A 107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brisson-Noel A, Arthur M, Courvalin P. 1988. Evidence for natural gene transfer from gram-positive cocci to Escherichia coli. J Bacteriol 170:1739–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts MC, Soge OO, No DB. 2011. Characterization of macrolide resistance genes in Haemophilus influenzae isolated from children with cystic fibrosis. J Antimicrob Chemother 66:100–104. doi: 10.1093/jac/dkq425. [DOI] [PubMed] [Google Scholar]
- 65.Schluckebier G, Zhong P, Stewart KD, Kavanaugh TJ, Abad-Zapatero C. 1999. The 2.2-Å structure of the rRNA methyltransferase ErmC′ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J Mol Biol 289:277–291. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- 66.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci U S A 99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yap MN, Bernstein HD. 2013. Mutations in the Escherichia coli ribosomal protein L22 selectively suppress the expression of a secreted bacterial virulence factor. J Bacteriol 195:2991–2999. doi: 10.1128/JB.00211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McLaughlin JR, Murray CL, Rabinowitz JC. 1981. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem 256:11283–11291. [PubMed] [Google Scholar]
- 70.Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Masse E, Lafontaine DA. 2012. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci U S A 109:E3444–E3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davies J, Spiegelman GB, Yim G. 2006. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 72.Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. 2002. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A 99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin JT, Connelly MB, Amolo C, Otani S, Yaver DS. 2005. Global transcriptional response of Bacillus subtilis to treatment with subinhibitory concentrations of antibiotics that inhibit protein synthesis. Antimicrob Agents Chemother 49:1915–1926. doi: 10.1128/AAC.49.5.1915-1926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ng WL, Kazmierczak KM, Robertson GT, Gilmour R, Winkler ME. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J Bacteriol 185:359–370. doi: 10.1128/JB.185.1.359-370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsui WH, Yim G, Wang HH, McClure JE, Surette MG, Davies J. 2004. Dual effects of MLS antibiotics: transcriptional modulation and interactions on the ribosome. Chem Biol 11:1307–1316. doi: 10.1016/j.chembiol.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 76.Seyedsayamdost MR. 2014. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A 111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gupta P, Kannan K, Mankin AS, Vazquez-Laslop N. 2013. Regulation of gene expression by macrolide-induced ribosomal frameshifting. Mol Cell 52:629–642. doi: 10.1016/j.molcel.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vazquez-Laslop N, Klepacki D, Mulhearn DC, Ramu H, Krasnykh O, Franzblau S, Mankin AS. 2011. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci U S A 108:10496–10501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. 2010. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J 29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arenz S, Meydan S, Starosta AL, Berninghausen O, Beckmann R, Vazquez-Laslop N, Wilson DN. 2014. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol Cell 53:446–452. doi: 10.1016/j.molcel.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arenz S, Bock LV, Graf M, Innis CA, Beckmann R, Grubmuller H, Vaiana AC, Wilson DN. 2016. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat Commun 7:12026. doi: 10.1038/ncomms12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Narayanan CS, Dubnau D. 1985. Evidence for the translational attenuation model: ribosome-binding studies and structural analysis with an in vitro runoff transcript of ermC. Nucleic Acids Res 13:7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bailey M, Chettiath T, Mankin AS. 2008. Induction of erm(C) expression by noninducing antibiotics. Antimicrob Agents Chemother 52:866–874. doi: 10.1128/AAC.01266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leclercq R, Courvalin P. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob Agents Chemother 46:2727–2734. doi: 10.1128/AAC.46.9.2727-2734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shaw JH, Clewell DB. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol 164:782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bechhofer DH, Dubnau D. 1987. Induced mRNA stability in Bacillus subtilis. Proc Natl Acad Sci U S A 84:498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohanty BK, Kushner SR. 2016. Regulation of mRNA decay in bacteria. Annu Rev Microbiol 70:25–44. doi: 10.1146/annurev-micro-091014-104515. [DOI] [PubMed] [Google Scholar]
- 88.Beilstein K, Wittmann A, Grez M, Suess B. 2015. Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth Biol 4:526–354. doi: 10.1021/sb500270h. [DOI] [PubMed] [Google Scholar]
- 89.Chen D, Murchie AI. 2014. An aminoglycoside sensing riboswitch controls the expression of aminoglycoside resistance acetyltransferase and adenyltransferases. Biochim Biophys Acta 1839:951–958. doi: 10.1016/j.bbagrm.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 90.Jia X, Zhang J, Sun W, He W, Jiang H, Chen D, Murchie AI. 2013. Riboswitch control of aminoglycoside antibiotic resistance. Cell 152:68–81. doi: 10.1016/j.cell.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 91.Pitt SW, Zhang Q, Patel DJ, Al-Hashimi HM. 2005. Evidence that electrostatic interactions dictate the ligand-induced arrest of RNA global flexibility. Angew Chem Int Ed Engl 44:3412–3415. doi: 10.1002/anie.200500075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roth A, Breaker RR. 2013. Integron attI1 sites, not riboswitches, associate with antibiotic resistance genes. Cell 153:1417–1418. doi: 10.1016/j.cell.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta P, Sothiselvam S, Vazquez-Laslop N, Mankin AS. 2013. Deregulation of translation due to posttranscriptional modification of rRNA explains why erm genes are inducible. Nat Commun 4:1984. doi: 10.1038/ncomms2984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.