Abstract
BEL-1 is an acquired class A extended-spectrum β-lactamase (ESBL) found in Pseudomonas aeruginosa clinical isolates from Belgium which is divergent from other ESBLs (maximum identity of 54% with GES-type enzymes). This enzyme is efficiently inhibited by clavulanate, imipenem, and moxalactam. Crystals of BEL-1 were obtained at pH 5.6, and the structure of native BEL-1 was determined from orthorhombic and monoclinic crystal forms at 1.60-Å and 1.48-Å resolution, respectively. By soaking native BEL-1 crystals, complexes with imipenem (monoclinic form, 1.79-Å resolution) and moxalactam (orthorhombic form, 1.85-Å resolution) were also obtained. In the acyl-enzyme complexes, imipenem and moxalactam differ by the position of the α-substituent and of the carbonyl oxygen (in or out of the oxyanion hole). More surprisingly, the Ω-loop, which includes the catalytically relevant residue Glu166, was found in different conformations in the various subunits, resulting in the Glu166 side chain being rotated out of the active site or even in displacement of its Cα atom up to approximately 10 Å. A BEL-1 variant showing the single Leu162Phe substitution (BEL-2) confers a higher level of resistance to CAZ, CTX, and FEP and shows significantly lower Km values than BEL-1, especially with oxyiminocephalosporins. BEL-1 Leu162 is located at the beginning of the Ω-loop and is surrounded by Phe72, Leu139, and Leu148 (contact distances, 3.5 to 3.9 Å). This small hydrophobic cavity could not reasonably accommodate the bulkier Phe162 found in BEL-2 without altering neighboring residues or the Ω-loop itself, thus likely causing an important alteration of the enzyme kinetic properties.
INTRODUCTION
Among Gram-negative bacteria, the production of one or several β-lactamase(s) represents the major mechanism of resistance to β-lactam antibiotics (1). Extended-spectrum β-lactamases (ESBLs) are enzymes which confer resistance not only to penicillins and narrow-spectrum cephalosporins but also to expanded-spectrum cephalosporins, such as cefotaxime, ceftriaxone, ceftazidime, and cefepime. ESBL production in clinically relevant opportunistic pathogens, such as Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter baumannii (2), is now relatively common, and such isolates are globally spread, representing an important issue for successful antimicrobial chemotherapy (1, 2).
Besides the TEM- and SHV-derived variants, which acquired ESBL properties by means of specific mutations, an increasing number of β-lactamases have been reported which showed native ESBL properties, such as the widespread CTX-M-type enzymes, but also the less frequently observed PER-, VEB-, and GES-type enzymes. More recently, a novel and divergent ESBL was identified in a Pseudomonas aeruginosa clinical isolate from Belgium that shared at most 50% identity with GES-type enzymes and showed interesting biochemical properties, such as inhibition by moxalactam and low affinity for ceftazidime, as well as relatively low turnover rates for most substrates (3). Even more recently, a variant exhibiting a unique substitution (Leu162Phe in BEL-2) was identified and showed significantly different activity on ceftazidime, resulting from much lower Km values for expanded-spectrum cephalosporins (4).
In this work, we describe the crystal structure of the native BEL-1 ESBL and of its acyl-enzyme complexes with imipenem and moxalactam, the latter representing the first observation of this substrate in a class A β-lactamase, providing insight into the specific structural features that could account for the enzyme properties and the importance of position 162 (where the single substitution occurring in BEL-2 is observed) in the binding and recognition of β-lactam substrates.
MATERIALS AND METHODS
We use the term “native” BEL-1, although most of the subunits contain a citrate molecule in the active site, to make a distinction from the imipenem- and moxalactam-bound forms of the enzyme. The nomenclature of BEL-1 used here refers to the residue numbering in the PDB coordinates and follows the BEL-1 sequence [UniProt Q3SAW3_PSEAI]. When the consensus numbering of class A β-lactamases [ABL numbering] is used, the position of corresponding BEL-1 residues is provided.
Purification of BEL-1.
A near-homogeneity BEL-1 enzyme preparation was purified from a 2-liter culture of Escherichia coli BL21(DE3)/pET-BEL-1 grown for 24 h in autoinducing medium ZYP-5052. The expression plasmid pET-BEL-1 was obtained by cloning the blaBEL-1 open reading frame (ORF), amplified by PCR, into the NdeI and BamHI cloning sites of pET-9a plasmid, as previously described (4). The culture was centrifuged (10,000 × g, 10°C, 30 min), and the clarified supernatant was concentrated using an Amicon YM30 membrane (Amicon model 2000 concentrator; Millipore Corp., Carlsbad, CA) and desalted using a HiPrep desalting column (GE Healthcare, Uppsala, Sweden) against 30 mM morpholineethanesulfonic acid (MES) buffer, pH 6.2 (buffer M). The resulting sample was loaded on an XK 16/20 column packed with 25 ml of CM Sepharose FF (GE Healthcare) equilibrated with buffer M and the proteins eluted using a linear NaCl gradient (0 to 0.3 M in 150 ml). The β-lactamase-containing fractions were pooled, desalted as before in buffer M, and loaded on a Resource S column (column volume, 1 ml; GE Healthcare). Elution was achieved with a linear NaCl gradient in buffer M (0 to 1 M in 20 ml). Resulting fractions were then concentrated to 10 mg/ml in buffer M containing 0.16 M NaCl using Amicon Ultra ultrafiltration devices (10-kDa-molecular-mass cutoff; Millipore, Bedford, MA). This protocol yielded ∼10 mg of >98% pure BEL-1 enzyme per liter of culture. Electrospray ionization mass spectrometry (ESI-MS) analysis, performed as previously described (41), confirmed the authenticity of the BEL-1 preparation, with an excellent agreement between the observed mass (28,691.0) and the theoretical value (28,688.5; after the removal of a 20-amino-acid signal peptide). A higher-mass species was also observed (28,873.0) which likely corresponds to an alternate cleavage of the N-terminal signal sequence occurring between residues 18 and 19 (theoretical mass, 28,887.7).
Enzyme kinetics and thermal unfolding experiments.
The kinetic parameters for the inactivation of BEL-1 by moxalactam were determined using the reporter substrate method as previously described (42). The rate of inactivation (k) of BEL-1 (final concentration, 1.2 nM) was determined in the presence of various concentrations of moxalactam (final concentration, 20 to 80 μM) and 220 μM cephalothin (reporter substrate) in 50 mM HEPES (pH 7.5) buffer. Nonlinear regression analysis of the dependence of k over [moxalactam] was performed using Sigma Plot and the following equation: , where [I] is the moxalactam concentration and K* is equal to ; is the Km value of the reporter substrate; K is equal to k−1/k+1. The conformational stabilities of BEL-1 and BEL-2 (purified as previously described [4]) were compared using thermal unfolding experiments by monitoring the far-UV circular dichroism (CD) signal of a 20 μM enzyme solution in 50 mM HEPES (pH 7.5) buffer using a Jasco J-815 spectropolarimeter. The protein samples were heated from 20 to 90°C at a rate of 0.55°C/min, and data were interpreted as previously described (43). Under these conditions, unfolding of the β-lactamases was irreversible.
Crystallization of BEL-1.
Crystallization trials were performed using the sitting-drop method (96-well CrystalEX plates; Corning) (5) and the commercially available Crystal Screen I and II (Hampton Research, Aliso Viejo, CA). Drops consisting of 2 μl protein solution (10 mg/ml) and 2 μl reservoir solution were equilibrated at room temperature (20°C) against a 100-μl reservoir volume. Small, needle-shaped crystals appeared in 3 weeks with Crystal Screen I condition 40 (0.1 M sodium citrate tribasic dihydrate, pH 5.6, 20% [wt/vol] polyethylene glycol [PEG] 4000, 20% [vol/vol] 2-propanol), which provided a basis for further optimization of the crystallization conditions. Using 24-well sitting-drop plates (Cryschem plate; Hampton Research), protein drops with variable protein/precipitant volume ratios (1:1 and 1:2) were equilibrated over a 700- or 800-μl reservoir volume. Protein concentration ranged from 5.2 to 10 mg/ml, PEG 4000 concentration from 5 to 20%, wt/vol, and 1-propanol, 2-propanol, 2-butanol, or methyl-pentanediol (MPD) concentrations from 12 to 25%, vol/vol. Crystals suitable for diffraction were obtained in drops prepared by mixing 4 μl protein solution (5.2 mg/ml) with 2 μl two different precipitant solutions. The one composed of 0.1 M sodium citrate, pH 5.6, 15% (wt/vol) PEG 4000, and 15% (vol/vol) 2-propanol, equilibrated over an 800-μl reservoir volume (solution A), provided orthorhombic crystals in space group P21212. Monoclinic P21 crystals were obtained from a precipitant solution containing 0.1 M sodium citrate, pH 5.6, 20% (wt/vol) PEG 4600, and 20% (vol/vol) MPD, equilibrated over an 800-μl reservoir volume (solution B).
Crystals of the adducts with imipenem and moxalactam were obtained by soaking experiments (30- to 60-min soaking times, respectively) using 1 to 2 mM final substrate concentrations (resuspended in precipitant solution) in the crystal drop in both cases. The BEL-1–imipenem complex was obtained in the monoclinic form, while the BEL-1–moxalactam complex was obtained in the orthorhombic form (Table 1).
TABLE 1.
Data collection, refinement and ligand validation statistics
| Parameter | Value(s) for BEL-1 type: |
|||
|---|---|---|---|---|
| Native (orthorhombic form) | Native (monoclinic form) | Imipenem complex | Moxalactam complex | |
| PDB code | 5EOE | 5EOO | 5EPH | 5EUA |
| X-ray source | ESRF ID14-1 | ESRF ID29 | ESRF ID29 | DLS I04-1 |
| Wavelength (Å) | 0.934 | 0.976 | 0.976 | 0.920 |
| Data collection temp (K) | 100 | 100 | 100 | 100 |
| Space group (no.) | P21212 (18) | P21 (4) | P21 (4) | P212121 (19) |
| Cell dimensions (Å) | a = 100.27, b = 120.92, c = 53.41 | a = 54.84, b = 94.69, c = 103.68, β = 92.64 | a = 53.98, b = 95.86, c = 103.42, β = 91.97 | a = 70.53, b = 80.91, c = 86.64 |
| Subunit/asymmetric unit (no.) | 2 | 4 | 4 | 2 |
| Matthews coefficient (Å3 Da−1) | 2.66 | 2.11 | 2.10 | 2.20 |
| Solvent content (%) | 53.87 | 47.79 | 41.46 | 44.21 |
| Resolution limita (Å) | 29.25–1.60 (1.69–1.60) | 51.78–1.48 (1.56–1.48) | 51.68–1.79 (1.89–1.79) | 33.58–1.85 (1.95–1.85) |
| No. of reflectionsa | ||||
| Measured | 1,177,930 (169,097) | 389,425 (57,732) | 223,222 (33,531) | 264,536 (40,492) |
| Unique | 86,406 (12,467) | 154,141 (22,851) | 91,836 (13,854) | 42,901 (6,182) |
| Completenessa (%) | 99.9 (99.9) | 88.0 (89.7) | 93.2 (96.4) | 99.7 (100.0) |
| Rmergea,b (%) | 7.6 (39.7) | 14.9 (38.0) | 8.4 (38.8) | 8.0 (42.7) |
| Multiplicitya | 13.6 (13.6) | 2.5 (2.5) | 2.4 (2.4) | 6.2 (6.5) |
| I/σ(I)a | 25.0 (6.9) | 4.3 (2.2) | 7.5 (2.7) | 12.6 (3.8) |
| Wilson B factor (Å2) | 14.11 | 12.68 | 16.44 | 19.68 |
| Rcrysta,b (%) | 14.2 (18.4) | 16.3 (23.1) | 16.4 (23.1) | 16.6 (22.9) |
| Rfreea,b (%) | 16.7 (22.1) | 18.7 (24.5) | 20.2 (29.5) | 21.5 (26.7) |
| No. of: | ||||
| Protein atoms | 4009 | 7951 | 7862 | 3936 |
| Ligand atoms | 86 | 75 | 93 | 104 |
| Water molecules | 800 | 944 | 685 | 305 |
| Avg B factor (Å2) | 14.59 | 15.98 | 20.74 | 27.59 |
| RMSD | ||||
| Bond length (Å) | 0.013 | 0.013 | 0.017 | 0.015 |
| Bond angle (°) | 1.677 | 1.646 | 1.767 | 1.697 |
| Ramachandran plot residues (%) | ||||
| Most favored regions | 98.7 | 98.9 | 98.7 | 98.8 |
| Additionally allowed regions | 1.3 | 1.1 | 1.3 | 1.2 |
| Disallowed regions | 0.0 | 0.0 | 0.0 | 0.0 |
| Ligand | ||||
| LLDFc | 1.02, 4.06, 0.10, 4.88f | 0.04, 2.26g | ||
| RSRd | 0.13, 0.11, 0.10, 0.12f | 0.12, 0.21g | ||
| RSCCe | 0.95, 0.96, 0.96, 0.94f | 0.91, 0.79g | ||
Data in parentheses refer to results for the highest-resolution shell.
Rmerge = ΣhΣi|Ii,h − Īh|/ΣhΣiIi,h × 100. Rcryst(Rfree) = Σh||Fh,obs| − |Fh,calc|/Σh||Fh,obs| × 100.
LLDF, local ligand density fit as reported by PDB validation.
RSR, real space R value as reported by PDB validation.
RSCC, real space density correlation coefficient as reported by PDB validation.
Values for ligand in chains A, B, C, and D.
Values for ligand in chains A and B.
Data collection and processing.
Before data collection, all crystals were transferred to a cryoprotectant solution (30%, wt/vol, PEG-4000/4600, 15%, vol/vol, 2-propanol, 20%, vol/vol, MPD, and 0.1 M sodium citrate, pH 5.6) and flash frozen in liquid nitrogen. Diffraction data on the orthorhombic crystal of the native enzyme were collected (100 K; wavelength, 0.983 Å; Δφ, 0.5°) at the European Synchrotron Radiation Facility (ESRF) on beamline ID14-1, equipped with an ADSC Quantum 210R detector. Data collection on the monoclinic crystal of the native enzyme and the complex with imipenem were carried out at the ESRF beamline ID29 (100 K; wavelength, 0.976 Å; Δφ [rotation angle for data collection], 0.5°) equipped with a Pilatus 6M detector. Diffraction data on BEL-1 in complex with moxalactam were collected (100 K; wavelength, 0.920 Å; Δφ, 0.2°) at the Diamond Light Source (DLS) beamline I04-1 equipped with a Pilatus 2M detector. Data were processed using the program MOSFLM (6) and scaled with SCALA (7) from the CCP4 suite (8). Data collection parameters and data reduction statistics are reported in Table 1.
Structure determination and refinement.
The BEL-1 structure was obtained by molecular replacement using the structure of the class A ESBL Toho-1 (44% identity with BEL-1) as a search model (PDB code 1IYS; water molecules and ions were omitted) (9). The program MOLREP (10) provided an evident solution for the positioning of the two and four BEL-1 subunits found in the asymmetric unit of the orthorhombic and monoclinic crystal forms, respectively. Isotropic refinement of the four structures was carried out with the program REFMAC5 (11) from the CCP4 suite. Between refinement cycles, the model was subjected to manual rebuilding using Coot (12). Upon completion of the protein models of the native enzyme, inspection of the Fourier difference map clearly demonstrated the presence of citrate anions in both the enzyme subunits in the orthorhombic form and in three out of the four subunits in the monoclinic form. The imipenem and moxalactam inhibitors are bound in all BEL-1 subunits.
Water molecules were added using the standard procedure within the ARP/wARP suite (13). In the last cycle of refinement, hydrogen atoms have been added in calculated positions, contributing to the Fc calculation. The stereochemical quality of the refined model was assessed using the program PROCHECK (14) and Coot. Refinement statistics are reported in Table 1. The ligand placement was checked in the final models through the calculation of the omit maps and validated during the deposition in the Protein Data Bank. Figures were prepared using the molecular graphics software CCP4mg (15).
Accession number(s).
Coordinates and structure factors of native BEL-1 and its acyl-enzyme complexes with imipenem and moxalactam have been deposited into the Protein Data Bank under codes 5EOE (BEL-1 orthorhombic form), 5EOO (BEL-1 monoclinic form), 5EPH (BEL-1–imipenem complex), and 5EUA (BEL-1–moxalactam complex).
RESULTS AND DISCUSSION
BEL-1 overall fold.
BEL-1 crystals display a high propensity to polymorphism. The native enzyme was indeed obtained in both orthorhombic (P21212 space group) and monoclinic (P21 space group) forms. Similarly, the imipenem and moxalactam acyl-enzyme complexes crystallized in the monoclinic P21 and orthorhombic P212121 space groups, respectively (Table 1). In the orthorhombic form P21212, the final model consisted of two complete, independent BEL-1 molecules related by almost exact noncrystallographic 2-fold symmetry. The asymmetric unit of the monoclinic crystals consists of two of such dimers. However, analysis of the protein interface by the PISA software (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) indicates that the dimeric arrangement observed in the crystals does not represent a stable quaternary structure and is not maintained in solution. Similar unstable dimeric arrangements also have been observed in the crystal structures of other class A enzymes, like PER-2 (PDB code 4D2O) (16) and SED-1 (PDB code 3BFE) (L. Pernot et al., unpublished data).
The four BEL-1 crystal structures presented here are determined at high resolution, and the electron density is of very good quality, allowing the tracing of the chains from Asp21 (two N-terminal residues, Gln19-Ala20, of the mature protein are missing) to His283, except in some portions of the Ω loop (residues 150 to 172) and the C termini of a few subunits (see “Acyl-enzyme complexes of BEL-1,” below).
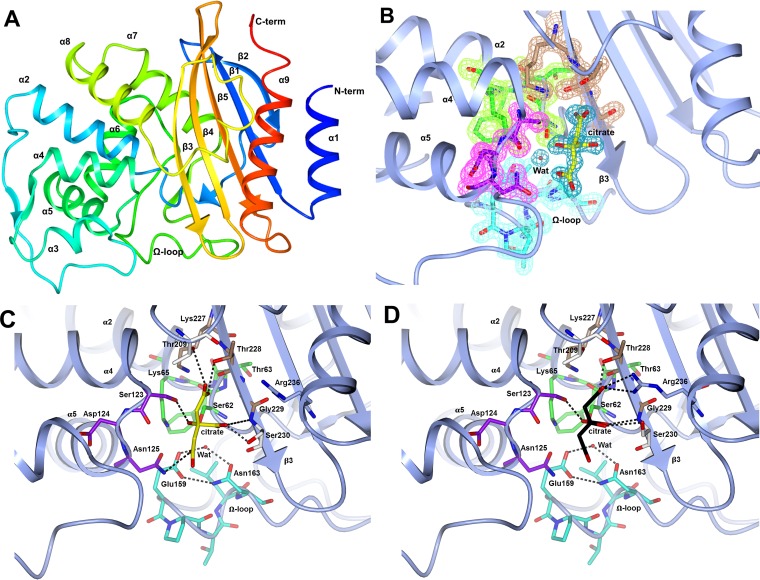
The tertiary structure of BEL-1 displays the characteristic fold of class A β-lactamases (17) (Fig. 1A), consisting of a five-stranded antiparallel β-sheet (β2, β1, β5, β4, and β3) sandwiched on one side by the N- and C-terminal helices α1 and α9 and on the other by helices α2 to α8, as shown in Fig. 1A. The active site is defined by the interface between the β-sheet and the large α2-α8 domain.
FIG 1.
(A) Overall fold of BEL-1. The polypeptide chain is rainbow colored, starting from the N terminus (blue) to the C terminus (red). The secondary-structure elements are labeled. (B) The four conserved motifs in BEL-1 are shown as sticks. Motif 1 (green) is located at the beginning of helix-2, motif 2 (purple) is located in the loop between α4 and α5, motif 3 (brown) is on β3, and motif 4 (cyan) is on the Ω-loop. The citrate molecule is shown as yellow sticks. The Fo-Fc Fourier difference map, corresponding to the four motifs and citrate, is shown as a wire contoured at 1.2 σ in colors related to motifs 1 to 4 (pale-green, pink, pale brown, and cyan, respectively), while that of citrate is colored blue. (C) The H-bonding scheme involving the citrate atoms is shown together with the water molecule involved in the deacylation of the reacted substrate (Wat). This binding of the citrate anion was observed in both subunits of the orthorhombic BEL-1 crystals. (D) Alternate binding of the citrate anion and relative interaction network, as observed in subunits A and C of the monoclinic BEL-1 crystals (in subunit B, citrate was modeled in two alternative conformations, omitted for clarity, whereas in subunit D it was not detected). Arg236 was observed in two alternative orientations, but only the one involved in citrate binding is shown here for clarity.
Helix α6 is linked to helix α7 by the large Ω-loop that covers the active site. Another large loop (BEL-1 residues 79 to 98) linking α2 to α3 constitutes part of the active-site channel. The initial turn of helix α2 contains the active-site motif 1 (S70xxK), bearing the catalytically active serine residue (BEL-1 residues 62 to 65).
All of the subunits in all crystal structures display the same conformation except, as already mentioned, for residues in some Ω-loops that are not visible due to the high flexibility of this region. No significant structural differences could be observed between independent enzyme molecules found in the different crystal asymmetric units. Their root mean square deviations (RMSDs) calculated on Cα atoms range between 0.26 Å and 0.66 Å.
BEL-1 active site and interaction with citrate.
Figure 1B shows the electron density 2Fo-Fc map in the BEL-1 (orthorhombic form) active site where a citrate molecule from the crystallization buffer is bound. Besides motif 1 (S70xxK; BEL-1 residues Ser62-Thr63-Phe64-Lys65), the electron density of the conserved motifs 2 (S130xN; BEL-1 residues Ser123-Asp124-Asn125) and 3 (K234xG; BEL-1 residues Lys227-Thr228-Gly229) is also shown. Other residues, including the catalytically important Glu166, are located in the omega loop and constitute the floor of the active site (BEL-1 residues Glu159, Pro160, Thr161, Leu162, and Asn163) (Fig. 1B). These residues have been shown to be involved in substrate recognition in class A β-lactamases (18). Figure 1C shows the H-bonding network involving the citrate anion and the residues in the BEL-1 active site. The citrate oxygen atoms (in the orthorhombic form) interact with the side chains of catalytically relevant residues of the three conserved motifs and with the side chains of BEL-1 residues Thr209, Ser230, and Arg236 (in the monoclinic form only). This positively charged area in the active-site cavity is important to accommodate the carboxylated group of β-lactamase substrates, as it has been shown in the structures of various class A β-lactamase acyl-enzyme complexes (e.g., TEM-1 and imipenem (19).
The binding of polycarboxylates is not uncommon for β-lactamases, as citrate has already been observed bound to TEM-72 (20) and to KPC-2 (21), to the class C CMY-2 (PDB entry 1ZC2) (C. Bauvois et al., unpublished data), to a β-lactamase/d-Ala-d-Ala-carboxypeptidase from Yersinia pestis (PDB: 3RJU) (Y. Kim et al., unpublished data), and even to the metallo-β-lactamase BcII (PDB code 1MQO) (I. Garcia-Saez et al., unpublished data). Tartrate has been found bound to OXA-46 (22). Interestingly, in the last example, the tartrate binding can be observed only when the conserved Lys75 (motif 1) is not carbamylated. This finding supports the hypothesized preferential inhibition of class A β-lactamases, rather than other serine-β-lactamases, by polycarboxylate molecules. The possibility of inhibiting β-lactamases by using polycarboxylates has been explored recently (23).
Citrate anions are found bound in both independent subunits of the orthorhombic form of BEL-1, while citrate is present only in three out of four BEL-1 subunits of the monoclinic form, where it adopts a different orientation that displaces the Wat molecule H bonded to Glu166 and Asn170 (BEL-1 residues 159 and 163) in the orthorhombic form.
The Nδ atoms of Lys73 (BEL-1 residue 65), in motif 1, establish a hydrogen bond with the catalytic Ser70 (BEL-1 residue 62) on the same motif, with Ser130 and Asn132 of motif 2 (BEL-1 residues 123 and 125) and Glu166 on the Ω-loop (not shown in Fig. 1C).
The two residues Glu166 and Asn170 establish a hydrogen bonding network with Wat (Fig. 1C), the nucleophile in the second step of the catalyzed reaction (24). The other highly conserved water molecule, which usually occupies the oxyanion hole between helix α2 and strand β3, bridging Ser70 and Thr235, is not present in this BEL-1 structure, being displaced by the carboxylate group of citrate. Upon binding, the citrate anions displace at least four water molecules seen in the active site of the unbound subunit.
Acyl-enzyme complexes of BEL-1.
The structure of the acyl-enzyme form of BEL-1 resulting from binding of imipenem and moxalactam, two β-lactam compounds which efficiently inactivated BEL-1, have been obtained at a resolution of 1.79 and 1.85 Å, respectively (Table 1). In both adducts, the hydrolyzed form of the inhibitor could be modeled in all subunits.
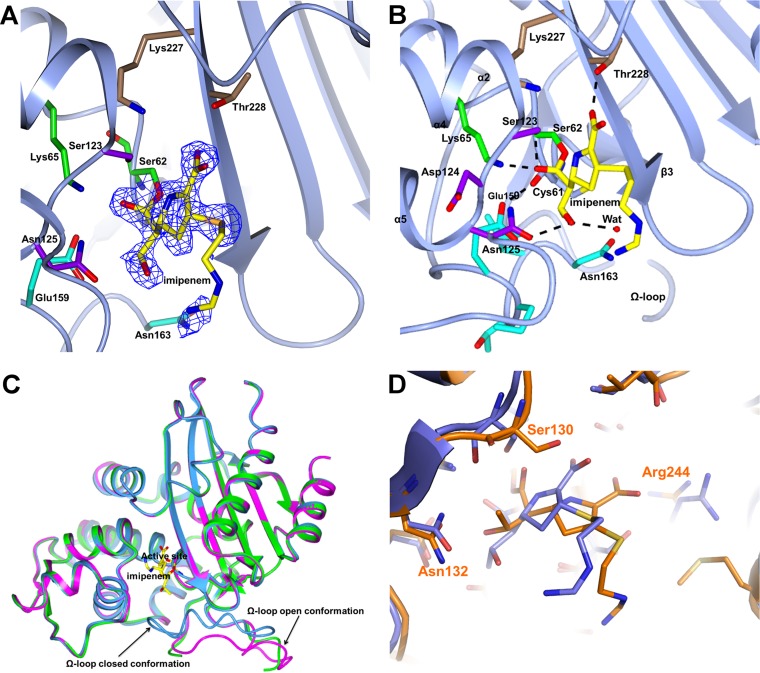
In the BEL-1–imipenem complex, obtained in the monoclinic crystal form (Fig. 2), the inhibitor adopts similar conformations in all subunits except for the imino-methylamino-ethyl chain, which extends out from the cavity and is disordered. Beyond the covalent bond to Ser70 (BEL-1 Ser62), the imipenem carboxylate group is H bonded to Thr235 (BEL-1 Thr228), although this interaction involves a single oxygen atom (O31) of the substrate carboxylate group, and the 6-α-hydroxyethyl substituent is H bonded to Asn132 (BEL-1 Asn125). A comparison with the TEM-1–imipenem acyl-enzyme complex highlights some differences (19) (Fig. 2D). In TEM-1, the imipenem carboxylate is located close to and interacts with Ser235 and Arg244 residues, but sits approximately 2.8 Å away from that position in the BEL-1 complex and only interacts with the Thr235 (228) side chain hydroxyl group and backbone carbonyl. Interestingly, the carbonyl oxygen of the TEM-1-bound imipenem is located in the oxyanion hole of the enzyme active site, while it is located outside the BEL-1 complex. Indeed, the O6 carbonyl oxygen does not occupy the oxyanion hole but is H bonded to Lys73 (BEL-1 Lys65) in a conformation that would not allow inhibitor hydrolysis. In addition, the C6-α-hydroxyethyl group also shows a different orientation in both enzymes, resulting from the rotation of C5-C6 of the molecule. In the TEM-1 complex, the deacylation water molecule, although displaced by the hydroxyl group of the C6 imipenem substituent, is visible and interacts with Glu166, Asn170, and the hydroxyl oxygen of the substrate C6 substituent. In the BEL-1 complex, this water molecule could not be observed, and the hydroxyl group of this C6 substituent points towards the active site. The binding of imipenem in BEL-1 seems to modify the H-bonding pattern linking the active site with residues in the Ω-loop, causing disorder in the loop that is fully visible only in subunit D, while 2 to 10 residues are missing from the other subunits (missing residues are Thr164 and Asn165 in subunit A, Glu159 to Asn165 in subunit B, and Thr164 to Pro167 in subunit C). Figure 2B shows that Glu166, located at the Ω-loop, is H bonded to carbonyl O of Cys69 (2.9 Å), suggesting its protonated state. Asn170 has moved away from the cavity, and the water molecule bridging the two residues in the native form of the enzyme (Fig. 1D) disappeared. The Ω-loop in the D subunit has an open conformation, unobserved so far in class A enzymes, pointing away from the active-site crevice as shown in Fig. 2C. The open conformation of the Ω-loop, although not fully traceable, is also seen in the unbound subunit D of the BEL-1 native monoclinic form. The Glu166-Wat-Asn170-H-bonded triad shown in Fig. 1C is disrupted in all subunits of imipenem-bound BEL-1.
FIG 2.
(A) Electron density from the final omit map corresponding to imipenem covalently bound to Ser62 is shown as a blue wire contoured at 2.7 σ. The BEL-1 residues corresponding to the four motifs described in the legend to Fig. 1 are shown as sticks following the same color code. The traced portion of the open, disordered Ω-loop is also shown. (B) H bonds involving imipenem within the BEL-1 active site. The residue colors follow the above-described scheme. The Glu159 (ABL Glu166) side chain is now H bonded to the carbonyl oxygen of Cys61 (ABL Cys68), and Asn163 (ABL Asn170) has moved away from the active site. (C) The two observed conformations of the Ω-loop shown by superimposing the citrate-bound orthorhombic form of BEL-1 (closed conformation; light blue ribbon) with subunit D of the unbound monoclinic BEL-1 form (green ribbon; Ω-loop partially traced) and with the imipenem-bound BEL-1 (magenta ribbon; complete open conformation). The imipenem molecule is shown as yellow sticks to locate the BEL-1 active site. (D) Comparison of the active site of the imipenem-bound structures of BEL-1 (blue) and TEM-1 (orange), showing the different position and orientation of the imipenem molecule in both enzymes.
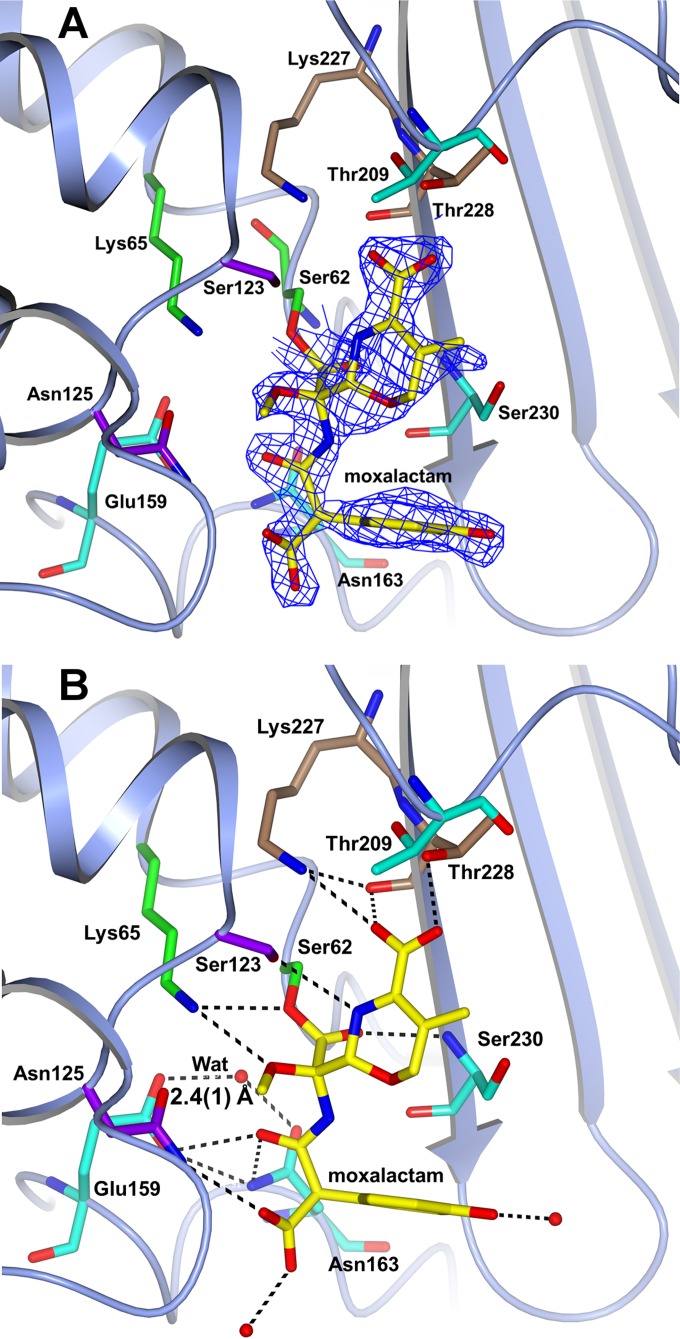
The two independent subunits of the moxalactam-bound complex of BEL-1 (orthorhombic form P212121) are identical except in the Ω-loop region, which is observed in subunit A but highly disordered in subunit B (missing residues are Glu159 to Asp169). Moxalactam, bearing a C7-α-methoxy substituent granting the compound some level of stability to β-lactamase-mediated hydrolysis, is covalently bound to the Ser62 side chain in both subunits, but the 1-methyl-5-thiotetrazole group of moxalactam is not visible and appears to have been cleaved upon rearrangement of moxalactam electronic structure due to the acylation reaction leading to a 3′-methylene species (25), as already observed in reported AmpC-moxalactam complex structures (26, 27) (Fig. 3). Although moxalactam is surely bound to subunit B (the open β-lactam ring and the oxa-azaciclo-carboxyl moiety are clearly visible in the electron density map), less defined electron density was observed for the carboxy-hydroxyphenyl portion of the molecule, preventing the moxalactam molecule from being fully modeled in this subunit. For this reason, the following description refers to moxalactam bound in BEL-1 subunit A.
FIG 3.
(A) Electron density from the final omit map corresponding to moxalactam covalently bound to Ser62 is shown as a blue wire contoured at 3 σ. The BEL-1 residues corresponding to the four motifs described in the legend to Fig. 1 are shown as sticks by following the same color code. (B) H bonds involving moxalactam within the BEL-1 active site. The residue colors follow the above-described scheme. The short H-bond distance between the carboxylate of Glu159 and the deacylating water (Wat) is indicated.
The hydrolyzed moxalactam molecule interacts with many conserved residues of the site, as reported in Fig. 3B. The C8 carbonyl oxygen is found in the oxyanion hole formed by the backbone N atoms of Ser70 (BEL-1 Ser62) and Ser237 (BEL-1 Ser230). The C7 α-methoxy oxygen makes an H bond to Lys73, while the C4 carboxylate group is H bonded (with both oxygen atoms) to the side chains of residues Lys234 and Thr235 (Fig. 3B).
In contrast to the BEL-1–imipenem complex, in which the Glu166 residue was observed in a significantly different position compared to that in the structure of the native protein, residues Asn159 and Glu166 were observed in a native-like conformation, and the deacylated water was located at 3.2 (± 0.1) Å from the moxalactam C28 atom of the scissile Ser70-moxalactam bond.
The inactivation of BEL-1 was investigated by kinetic methods, which confirmed that BEL-1 acylation by moxalactam is relatively fast (k+2/K, 1.9 × 103 M−1 · s−1) but that deacylation takes place slowly (k+3, 3.2 × 10−3 s−1). The slow deacylation of a β-lactamase inhibitor has been observed for avibactam bound to the class A CTX-M-15 enzyme. In this case, it has been demonstrated that deacylation occurs by a recyclization mechanism and not by the intervention of the deacylating water (28–30). The atomic-resolution crystal structure of the CTX-M-15–avibactam complex (29) shows a protonated Glu166 carboxylate oxygen forming a short low-barrier H bond to the deacylating water that acts as an H bond acceptor. In such an arrangement, the lone pair of that water is not productively oriented toward the scissile acyl carbon. At the resolution of the present BEL-1–moxalactam structure, hydrogen atoms cannot be observed. However, Glu166 establishes a very short H bond to the deacylating water (2.4 Å) (Fig. 3B), suggesting the presence of a low-barrier H bond, similar to that observed in the CTX-M-15–avibactam complex and the same nonproductive orientation of the water lone pairs, possibly explaining the kinetic data on moxalactam.
Structural comparison of native BEL-1 and its complexes with other class A β-lactamases.
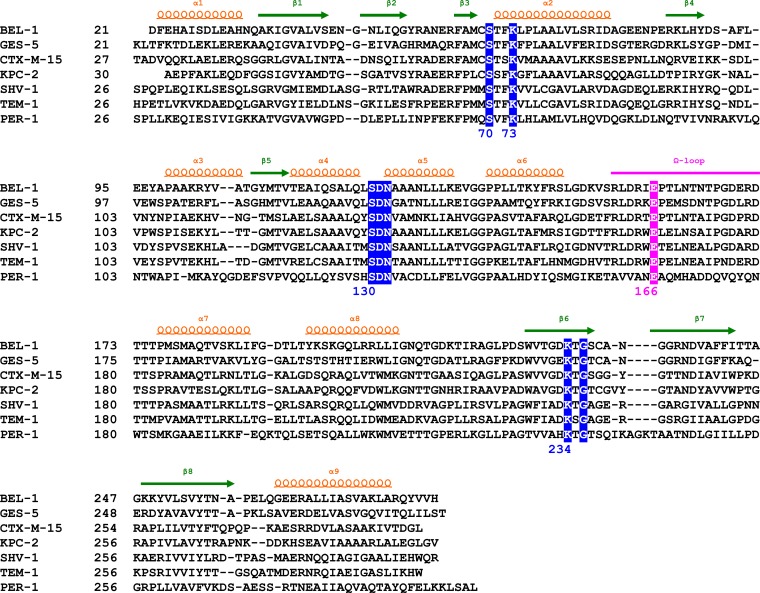
The comparison of the BEL-1 crystal structure with other class A β-lactamases clearly demonstrates the high structural conservation that characterizes the enzymes belonging to this class despite low sequence identity (ranging from 26% to 54% for PER-1 and GES-1; the sequence alignments are reported in Fig. 4).
FIG 4.
Structure-based sequence alignment of representative class A β-lactamases of known three-dimensional structure (PDB codes used to generate the structural alignment are the following: GES-5, 4GNU; CTX-M-15, 4HBT; KPC-2, 2OV5; SHV-1, 1SHV; TEM-1, 1BTL; PER-1, 1E25). The secondary-structure elements of BEL-1 are shown above the sequences, and residues of the conserved motifs of class A β-lactamases are shaded in blue (consensus numbering is reported under the sequences). The omega-loop (magenta), bearing the catalytically relevant Glu166 residue, is also indicated.
Least-squares superimposition of the BEL-1 structure with representatives of TEM, SHV, CTX-M, PER, SED, PenP, KPC, and GES subfamilies yields RMSDs on Cα ranging between 0.96 Å (GES) and 1.75Å (PER-1). The largest structural deviations occur in the two terminal helices (α1 and α9), in the short loop between strands β1 and β2, in helix α8 and the loop connecting this helix with strand β3, and in the short loop between β4 and β5. The three conserved motifs overlap almost perfectly in all of the crystal structures of the above-described class A enzymes.
Surprisingly, a disulfide bond between Cys69 and Cys238 (BEL-1 residues 61 and 231) is clearly visible in the BEL-1 crystal structure, as indicated by the continuous electron density between the sulfur atoms of the two cysteine residues. This disulfide bond was previously observed in GES-type β-lactamases and in carbapenemases, such as SME-1, NMC-A, and KPC-type enzymes (21, 31–33). The superimposition of disulfide-containing carbapenemases (including GES-2, SME-1, NMC-A, and KPC-2) and a number of penicillinases and cephalosporinases which do not have a similar disulfide bridge (such as TEM-1, in which a disulfide bridge is found between residues 77 and 123 instead) shows that in the disulfide-containing enzymes, helix α2 is positioned slightly closer to the β-domain than in the other enzymes. This highly conserved bond in the active site is considered a defining difference between carbapenemases and penicillinases in class A enzymes (34), and until now, its presence has been observed in all subtypes of class A β-lactamases which successfully evolved carbapenemase activity (such as in NMC-A and some GES-type enzymes). It is therefore surprising that this disulfide bond is present in BEL-1, which is unable to hydrolyze carbapenem substrates and is instead readily inactivated by imipenem (3) and moxalactam.
A structural feature conserved in class A β-lactamases is the cis conformation of the Glu166-Pro167 peptide bond. This configuration is stabilized by two hydrogen bonds between Glu166 and Asn136 favoring the configuration of the Ω-loop and the correct placement of Glu166 and Asn170. These two residues establish an H-bonding network involving their side chains and the catalytic water molecule for the deacylation step and contribute to its activation (Fig. 1D). The relevance of the H-bonding network in the catalysis and inhibition mechanism of class A enzymes has been demonstrated (29, 35). The arrangement of the Ω-loop is also stabilized by a salt bridge between Arg164 and Asp179 (BEL-1 residues 157 and 172). These four residues (Arg164, Glu166, Pro167, and Asp179) are conserved in all sequences of class A β-lactamases considered in the present alignment (Fig. 4) except for the PER-1 enzyme, in which residues Arg164, Pro167, and Asp179 are replaced by Ala164, Ala167, and Asn179, respectively. The crystal structure of PER-1 clearly shows that the Glu166-Ala167 peptide bond adopts a trans-conformation (36). Moreover, the replacement of the conserved Arg164 (BEL-1 residue 157) by an alanine in PER-1 results in the loss of the salt bridge interaction that contributes to stabilize this loop in other class A enzymes. Despite this, the present structures unequivocally show that the Ω-loop exists in different conformations in the unbound form (subunit D of the BEL-1 monoclinic form, in which the omega loop is disordered), in the citrate-bound form, or in subunit D of the imipenem-bound complex (Fig. 3C). In the latter, a Glu166-Pro167 trans-peptide bond is clearly observed.
Comparison with the class A CTX-M-15, TEM-1 GES-2, KPC-2, SHV-1, and Toho-1 native enzymes shows that they all have the same Ω-loop conformation as that seen in the citrate-bound subunits of BEL-1. Thus, it is possible that the shift between the cis- and trans-Glu159-Pro160 peptide bond in BEL-1 can be due either to spontaneous variability of the loop or to crystal-packing effects and that the native conformation of the Ω-loop is needed to bind the noncovalent weak inhibitor citrate, whereas the covalent inhibitors imipenem and moxalactam bind the enzyme independently of the conformation. The superimposition of BEL-1 with different class A enzymes also shows that the conformation of the peripheral region of the active site is affected by the variable conformation of the loop between strand β5 and helix α9 and the first two turns of the same helix. The initial part of helix α9 (residues 262 to 268 of BEL-1) forms a wall of the active-site cavity. The arrangement of this loop in BEL-1 shows a remarkable matching with the corresponding loop in GES-2, and it is also similar to PER-1, TEM-1, and SHV-1. The corresponding residues in Toho-1 and KPC-2 instead are shifted toward the β3-β4 loop (maximal displacement of about 6 Å), generating a greater steric hindrance in the peripheral region of the active site, especially in Toho-1. The major hindrance in Toho-1 and KPC-2 with respect to BEL-1 and GES-2 is due not only to the different arrangement of this loop but also to its amino acid composition. The presence in this site of an arginine (Arg274) and of a histidine (His274) in Toho-1 and KPC-2, respectively, narrows the active-site crevice compared to that of BEL-1 and GES-2, where a glycine (Gly263) and an alanine (Ala264) are found in the corresponding positions.
Functional implications and relevance of position 162.
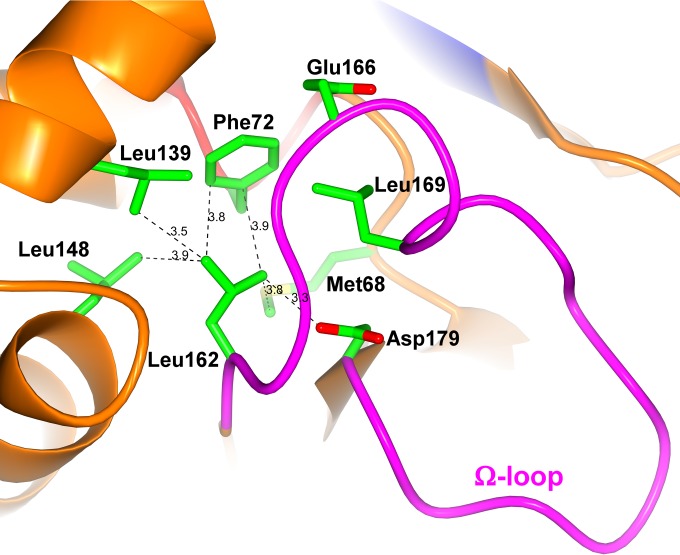
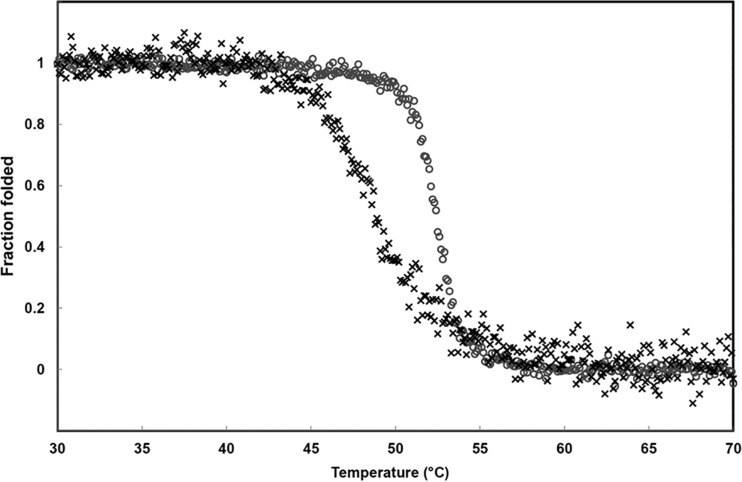
Residue 162 (BEL-1 position 155), located at the beginning of the Ω-loop and relatively close to catalytically relevant Glu166, seems to determine important functional differences between the two BEL-type variants detected so far, BEL-1 and BEL-2, which differ by a single-residue substitution at this position (Leu162 to Phe in BEL-2). Indeed, BEL-1 and BEL-2 show significantly different kinetic parameters for the hydrolysis of β-lactam substrates, with an overall decrease of both turnover rates (kcat) and Michaelis constants (Km) for BEL-2 (4). As a consequence, the production of BEL-2 in both E. coli laboratory strains and P. aeruginosa clinical isolates translated as a higher level of resistance to oxyimino-cephalosporins, in particular ceftazidime (4). The analysis of the BEL-1 structure provides a rationale for these differences. Leu162 (in BEL-1) is found in a hydrophobic pocket of limited dimensions, and its side chain interacts via van der Waals contacts with Met68, Phe72, Leu139, Leu148, and Asp179 (BEL-1 residues 60, 64, 132, 141, and 172) (Fig. 5). Its replacement with a larger Phe residue, whose side chain is approximately 2 Å longer than that of a Leu residue, is expected to strongly affect the position of the side chains of neighboring residues, as well as on the orientation and flexibility of the whole Ω-loop itself. The increased flexibility of regions defining the active site could determine both an increased enzyme activity and overall loss of stability. This is illustrated by the fact that many mutations conferring ESBL properties in class A enzymes are associated with decreased protein stability, which could be restored by remote substitutions, such as the so-called suppressor Met182Thr substitution occurring in TEM-type ESBLs (37). The study of both BEL-1 and BEL-2 conformational stability, determined following the circular dichroism signal in thermal denaturation experiments (Fig. 6), confirmed the loss of stability associated with the substitution (apparent melting point [Tm] values were the following: BEL-1, 52.4 ± 0.2°C; BEL-2, 48.8 ± 0.2°C). The observed Tm values were similar to that previously measured for another class A β-lactamase, TEM-1 (38). Interestingly, a similar decrease (3 to 4°C) of the Tm value was also observed, with some TEM-1 variants showing ESBL activity (e.g., with TEM-19, carrying the Gly238Ser substitution). Although BEL-1 is already an ESBL, the Leu162Phe substitution confers a higher level of activity on some oxyimino cephalosporins, particularly ceftazidime. Thus, a common strategy of class A β-lactamases to achieve this is to accumulate substitutions that enlarge the active site but often decrease the enzyme stability, unless a compensatory suppressor mutation (e.g., Met182Thr) can be introduced (39). Overall, these data indicate that the accommodation of the larger Phe162 residue in BEL-2 destabilizes the whole enzyme structure. This in turn is associated with greater flexibility, allowing a better accommodation of β-lactam substrates (lower Km values), but it could negatively affect turnover rates, considering that the position of Glu166 might be significantly altered, being unable to carry out efficient deacylation of the acyl-enzyme complex.
FIG 5.
Hydrophobic pocket hosting the side chain of the Ω-loop residue Leu155 (green sticks, Leu162) is shown.
FIG 6.
Thermal unfolding transition curves of purified BEL-1 (○) and BEL-2 (×) β-lactamases followed by circular dichroism.
Concluding remarks.
The crystal structure of BEL-1 shows a conformational variability of the functionally relevant Ω-loop, which is uncommon in class A β-lactamases. Surprisingly, this conformational variability is observed in the crystal structures of the imipenem- and moxalactam-bound BEL-1, while in the native citrate-bound structure, this loop is observed in a conformation similar to that of other class A β-lactamase structures. The conformational variability also appears to be linked to the slow deacylation rate observed for covalently inhibited BEL-1 (both imipenem and moxalactam behave as inhibitors of this enzyme), as the open conformation of the Ω-loop destroys the Glu166-Wat-Asn170 H-bonded triad that is needed to properly orient the water molecule prior to the deacylation reaction.
In addition, the presence of a disulfide bond between Cys69 and Cys238, as commonly found in class A carbapenemases, indicates that this structural element is not responsible per se for carbapenemase activity in the latter, as recently discussed (40).
Finally, the structures of BEL-1 also provide a rationale to explain the drastic alteration of the activity profile of BEL-2, a Leu162Phe variant. In this variant, an alteration of the Ω-loop structure is very likely, allowing both better recognition of bulkier substrates (BEL-2 has much lower Km values overall for many β-lactams) and, although turnover rates overall are lower with BEL-2, a better catalytic efficiency, especially for ceftazidime.
REFERENCES
- 1.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livermore DM. 2008. Defining an extended-spectrum β-lactamase. Clin Microbiol Infect 14(Suppl 1):S3–S10. [DOI] [PubMed] [Google Scholar]
- 3.Poirel L, Brinas L, Verlinde A, Ide L, Nordmann P. 2005. BEL-1, a novel clavulanic acid-inhibited extended-spectrum β-lactamase, and the class 1 integron In120 in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3743–3748. doi: 10.1128/AAC.49.9.3743-3748.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Docquier JD, De Luca F, Verlinde A, Ide L, Rossolini GM, Nordmann P. 2010. BEL-2, an extended-spectrum β-lactamase with increased activity toward expanded-spectrum cephalosporins in Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:533–535. doi: 10.1128/AAC.00859-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benvenuti M, Mangani S. 2007. Crystallization of soluble proteins in vapor diffusion for X-ray crystallography. Nat Protoc 2:1633–1651. doi: 10.1038/nprot.2007.198. [DOI] [PubMed] [Google Scholar]
- 6.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 8.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibuka AS, Ishii Y, Galleni M, Ishiguro M, Yamaguchi K, Frere JM, Matsuzawa H, Sakai H. 2003. Crystal structure of extended-spectrum β-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry 42:10634–10643. doi: 10.1021/bi0342822. [DOI] [PubMed] [Google Scholar]
- 10.Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 11.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langer G, Cohen SX, Lamzin VS, Perrakis A. 2008. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 15.McNicholas S, Potterton E, Wilson KS, Noble MEM. 2011. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr Sect D 67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruggiero M, Kerff F, Herman R, Sapunaric F, Galleni M, Gutkind G, Charlier P, Sauvage E, Power P. 2014. Crystal structure of the extended-spectrum β-lactamase PER-2 and insights into the role of specific residues in the interaction with β-lactams and β-lactamase inhibitors. Antimicrob Agents Chemother 58:5994–6002. doi: 10.1128/AAC.00089-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matagne A, Lamotte-Brasseur J, Frere JM. 1998. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J 330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee S, Pieper U, Kapadia G, Pannell LK, Herzberg O. 1998. Role of the ω-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry 37:3286–3296. doi: 10.1021/bi972127f. [DOI] [PubMed] [Google Scholar]
- 19.Maveyraud L, Mourey L, Kotra LP, Pedelacq JD, Guillet V, Mobashery S, Samama JP. 1998. Structural basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A β-lactamases from the antibiotic-resistant bacteria. J Am Chem Soc 120:9748–9752. doi: 10.1021/ja9818001. [DOI] [Google Scholar]
- 20.Docquier JD, Benvenuti M, Calderone V, Rossolini GM, Mangani S. 2011. Structure of the extended-spectrum β-lactamase TEM-72 inhibited by citrate. Acta Crystallogr F Struct Biol Cryst Commun 67:303–306. doi: 10.1107/S1744309110054680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrella S, Ziental-Gelus N, Mayer C, Renard M, Jarlier V, Sougakoff W. 2008. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A β-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob Agents Chemother 52:3725–3736. doi: 10.1128/AAC.00163-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Docquier JD, Benvenuti M, Calderone V, Giuliani F, Kapetis D, De Luca F, Rossolini GM, Mangani S. 2010. Crystal structure of the narrow-spectrum OXA-46 class D β-lactamase: relationship between active-site lysine carbamylation and inhibition by polycarboxylates. Antimicrob Agents Chemother 54:2167–2174. doi: 10.1128/AAC.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck J, Vercheval L, Bebrone C, Herteg-Fernea A, Lassaux P, Marchand-Brynaert J. 2009. Discovery of novel lipophilic inhibitors of OXA-10 enzyme (class D β-lactamase) by screening amino analogs and homologs of citrate and isocitrate. Bioorg Med Chem Lett 19:3593–3597. doi: 10.1016/j.bmcl.2009.04.149. [DOI] [PubMed] [Google Scholar]
- 24.Matagne A, Lamotte-Brasseur J, Dive G, Knox JR, Frere JM. 1993. Interactions between active-site-serine β-lactamases and compounds bearing a methoxy side chain on the alpha-face of the β-lactam ring: kinetic and molecular modelling studies. Biochem J 293:607–611. doi: 10.1042/bj2930607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishikawa J, Watanabe F, Shudou M, Terui Y, Narisada M. 1987. Proton NMR study of degradation mechanisms of oxacephem derivatives with various 3′-substituents in alkaline solution. J Med Chem 30:523–527. doi: 10.1021/jm00386a014. [DOI] [PubMed] [Google Scholar]
- 26.Patera A, Blaszczak LC, Shoichet BK. 2000. Crystal structures of substrate and inhibitor complexes with AmpC β-lactamase: possible implications for substrate-assisted catalysis. J Am Chem Soc 122:10504–10512. doi: 10.1021/ja001676x. [DOI] [Google Scholar]
- 27.Trehan I, Beadle BM, Shoichet BK. 2001. Inhibition of AmpC β-lactamase through a destabilizing interaction in the active site. Biochemistry 40:7992–7999. doi: 10.1021/bi010641m. [DOI] [PubMed] [Google Scholar]
- 28.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahiri SD, Mangani S, Durand-Reville T, Benvenuti M, De Luca F, Sanyal G, Docquier JD. 2013. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob Agents Chemother 57:2496–2505. doi: 10.1128/AAC.02247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sougakoff W, L'Hermite G, Pernot L, Naas T, Guillet V, Nordmann P, Jarlier V, Delettre J. 2002. Structure of the imipenem-hydrolyzing class A β-lactamase SME-1 from Serratia marcescens. Acta Crystallogr D Biol Crystallogr 58:267–274. doi: 10.1107/S0907444901019606. [DOI] [PubMed] [Google Scholar]
- 32.Frase H, Smith CA, Toth M, Champion MM, Mobashery S, Vakulenko SB. 2011. Identification of products of inhibition of GES-2 β-lactamase by tazobactam by x-ray crystallography and spectrometry. J Biol Chem 286:14396–14409. doi: 10.1074/jbc.M110.208744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. 2007. Crystal structure of KPC-2: insights into carbapenemase activity in class A β-lactamases. Biochemistry 46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaren P, Maveyraud L, Raquet X, Cabantous S, Duez C, Pedelacq JD, Mariotte-Boyer S, Mourey L, Labia R, Nicolas-Chanoine MH, Nordmann P, Frere JM, Samama JP. 1998. X-ray analysis of the NMC-A β-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity. J Biol Chem 273:26714–26721. doi: 10.1074/jbc.273.41.26714. [DOI] [PubMed] [Google Scholar]
- 35.Lamotte-Brasseur J, Dive G, Dideberg O, Charlier P, Frere JM, Ghuysen JM. 1991. Mechanism of acyl transfer by the class A serine β-lactamase of Streptomyces albus G. Biochem J 279:213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tranier S, Bouthors AT, Maveyraud L, Guillet V, Sougakoff W, Samama JP. 2000. The high resolution crystal structure for class A β-lactamase PER-1 reveals the bases for its increase in breadth of activity. J Biol Chem 275:28075–28082. doi: 10.1074/jbc.M003802200. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Minasov G, Shoichet BK. 2002. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol 320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 38.Raquet X, Vanhove M, Lamotte-Brasseur J, Goussard S, Courvalin P, Frère JM. 1995. Stability of TEM β-lactamase mutants hydrolyzing third generation cephalosporins. Proteins Gen 23:63–72. doi: 10.1002/prot.340230108. [DOI] [PubMed] [Google Scholar]
- 39.Huang W, Palzkill T. 1997. A natural polymorphism in β-lactamase is a global suppressor. Proc Natl Acad Sci U S A 94:8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frère JM, Sauvage E, Kerff F. 2016. From “an enzyme able to destroy penicillin” to carbapenemases: 70 years of β-lactamase misbehaviour. Curr Drug Targets 17:974–982. doi: 10.2174/1389450116666151001112859. [DOI] [PubMed] [Google Scholar]
- 41.Docquier JD, Lamottte-Brasseur J, Galleni M, Amicosante G, Frère JM, Rossolini GM. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J Antimicrob Chemother 51:257–266. [DOI] [PubMed] [Google Scholar]
- 42.De Meester F, Joris B, Reckinger G, Bellefroid-Bourguignon C, Frère JM, Waley SG. 1987. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem Pharmacol 36:2393–2403. [DOI] [PubMed] [Google Scholar]
- 43.Borgianni L, Vandenameele J, Matagne A, Bini L, Bonomo RA, Frère JM, Rossolini GM, Docquier JD. 2010. Mutational analysis of VIM-2 reveals an essential determinant for metallo-β-lactamase stability and folding. Antimicrob Agents Chemother 54:3197–3204. doi: 10.1128/AAC.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]