Abstract
A total of 431 Pseudomonas aeruginosa clinical isolates were collected from 29 general hospitals in South Korea in 2015. Antimicrobial susceptibility was tested by the disk diffusion method, and MICs of carbapenems were determined by the agar dilution method. Carbapenemase genes were amplified by PCR and sequenced, and the structures of class 1 integrons surrounding the carbapenemase gene cassettes were analyzed by PCR mapping. Multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) were performed for strain typing. Whole-genome sequencing was carried out to analyze P. aeruginosa genomic islands (PAGIs) carrying the blaIMP-6, blaIMP-10, and blaGES-24 genes. The rates of carbapenem-nonsusceptible and carbapenemase-producing P. aeruginosa isolates were 34.3% (148/431) and 9.5% (41/431), respectively. IMP-6 was the most prevalent carbapenemase type, followed by VIM-2, IMP-10, and GES-24. All carbapenemase genes were located on class 1 integrons of 6 different types on the chromosome. All isolates harboring carbapenemase genes exhibited genetic relatedness by PFGE (similarity > 80%); moreover, all isolates were identified as sequence type 235 (ST235), with the exception of two ST244 isolates by MLST. The blaIMP-6, blaIMP-10, and blaGES-24 genes were found to be located on two novel PAGIs, designated PAGI-15 and PAGI-16. Our data support the clonal spread of an IMP-6-producing P. aeruginosa ST235 strain, and the emergence of IMP-10 and GES-24 demonstrates the diversification of carbapenemases in P. aeruginosa in Korea.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic pathogen that causes various nosocomial infections, including sepsis, pneumonia, and urinary tract infections (1). Treatment of P. aeruginosa infections is often difficult because of the intrinsic drug resistance of P. aeruginosa and the ability of this pathogen to acquire genes for antimicrobial resistance determinants (2). Carbapenems are used as last-resort drugs for the treatment of infections caused by multidrug-resistant P. aeruginosa, due to their high affinity for penicillin-binding proteins, stability to various β-lactamases, and ability to easily pass through the bacterial outer membrane. However, the increasing use of carbapenems has resulted in the emerging phenomenon of carbapenem resistance (3).
P. aeruginosa can become resistant to carbapenems by reduced permeability of the outer membrane due to loss of substrate-specific outer membrane porin OprD, frequently accompanied by AmpC hyperproduction and overexpression of efflux systems (4), along with acquisition of genes encoding carbapenemases (5). While the molecular mechanism of carbapenem resistance is geographically variable, diverse classes of carbapenemase have been increasingly identified in P. aeruginosa, including class A (KPC and GES variants), class B (IMP, VIM, and NDM metallo-β-lactamases [MBLs]), and class D (OXA variants) (6). The MBLs, in particular IMP and VIM enzymes, are the most widespread carbapenemases in P. aeruginosa (7), with IMP-6 exclusively present in South Korea (8).
Carbapenemase genes are found often on integrons in P. aeruginosa clinical isolates. Integrons are prevalent in Gram-negative clinical isolates and offer the ability to capture and excise gene cassettes, frequently including antibiotic resistance determinants, to the bacterial host (9). However, integrons accomplish mobility only when they are associated with a specific mobile genetic element, such as a transposon, conjugative plasmid, or genomic island (10). The genomic islands in P. aeruginosa are mostly integrative and conjugative elements (ICEs) and are named according to their characteristics, i.e., P. aeruginosa pathogenicity island or the name of their host, i.e., Liverpool epidemic strain genomic islands or, more broadly, i.e., P. aeruginosa genomic island (PAGI) (11). Thus far, 14 PAGIs have been identified (12). These genomic islands typically carry genes for integration, transfer, and maintenance, which confer self-transferability to the composite ICE, along with genes conferring metabolic, virulence, and heavy metal resistance capability, which offer extra benefit to the bacterial host. Although PAGIs associated with antimicrobial resistance are rare, some examples have been identified, e.g., derivatives of PAGI-1 and PAGI-2 (13).
Two nucleotide sequence-based bacterial typing techniques, pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST), are considered to be the gold standards for use in epidemiological studies. The PFGE provides fingerprints of entire bacterial genomes by restriction enzyme mapping to determine epidemiological relationships of an outbreak in detail, while the MLST method gives strain lineages based on DNA fragments of seven housekeeping genes which allow fewer and larger groups feasible for global comparison (14). Two international high-risk P. aeruginosa clones, sequence type 111 (ST111) and ST235, are responsible for the dissemination of carbapenemase genes worldwide (15), with GES-type carbapenemase-producing P. aeruginosa ST235 in Spain (16) and IMP-6-producing P. aeruginosa ST235 in South Korea (8). Meanwhile, ST175 and ST244 carrying genes that confer resistance to carbapenems have also been reported to be responsible for regional dissemination; as a result, it could be considered that they were associated with high-risk clones (17). Together with PFGE and MLST, relatively high-cost whole-genome sequencing (WGS) is now commonly used for epidemiological studies expecting an incomparable power of discrimination (14).
The aims of this study were to investigate the prevalence of carbapenemase genes in P. aeruginosa clinical isolates recovered from general hospitals in South Korea in 2015 and to determine the genetic environments surrounding the carbapenemase genes and epidemiological characteristics of carbapenemase-producing P. aeruginosa (CP-PA) clinical isolates.
MATERIALS AND METHODS
Bacterial strains.
A total of 431 nonduplicated P. aeruginosa clinical isolates were collected from 29 general hospitals located in 11 cities and provinces in South Korea between June and August 2015. The isolates were recovered from blood (n = 33), respiratory specimens (n = 180), urine (n = 93), pus (n = 97), and other specimens (n = 28). Bacterial species were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI MS) using a Bruker MALDI MS instrument (Bruker, Billerica, MA). Species identification was made only when the log score of the species was above 2.0; otherwise, partial sequences of the 16S rRNA gene were analyzed.
Antimicrobial susceptibility testing.
Antimicrobial susceptibilities were tested by the disk diffusion method by following the Clinical and Laboratory Standards Institute guidelines (18). Briefly, disks (Oxoid Ltd., Basingstoke, UK) containing the following antimicrobial agents were used: piperacillin, ceftazidime, cefepime, aztreonam, gentamicin, tobramycin, amikacin, ciprofloxacin, meropenem, imipenem, and colistin. The MICs of meropenem and imipenem were determined by the agar dilution method on Mueller-Hinton agar (Becton, Dickinson and Company, Sparks, MD). P. aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as control strains.
Identification of carbapenem resistance determinants.
The genes encoding IMP-, VIM-, and NDM-type MBLs, KPC- and GES-like serine β-lactamases (19), and OprD outer membrane porin were amplified by PCR in carbapenem-nonsusceptible P. aeruginosa clinical isolates. PCR amplicons were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced using an Applied Biosystems 3730xl DNA analyzer (Applied Biosystems, Foster City, CA) to identify the variant types of each carbapenemase family. Sequences were compared with those in GenBank (http://www.ncbi.nlm.nih.gov/GenBank) using the BLAST network service.
Integron mapping and sequencing.
The structures of the class 1 integrons carrying the carbapenemase gene cassettes were determined by PCR mapping and sequencing using the primers listed in Table 1.
TABLE 1.
Primers used in this study for integron characterization
| Target | Primer | Nucleotide sequence (5′–3′) | Positiona | Reference |
|---|---|---|---|---|
| intI | INT1-mF | GCCTGTTCGGTTCGTAAGCT | 908/927 | This study |
| INT1-F | CCAAGCTCTCGGGTAACATC | 462/481 | ||
| INT1-R | CATGAAAACCGCCACTGC | 997/1014 | ||
| qacEΔ1 | qacEΔ1-F | GAAAGGCTGGCTTTTTCTTG | 3/22 | This study |
| qacEΔ1-R | GCAGCGACTTCCACGATG | 311/328 | ||
| ereA | ereA2-F | TTGAGCGATTTTCGGATACC | 269/288 | This study |
| ereA2-R | GGCATGAATCCTCCTTACCA | 1071/1090 | ||
| qac | qac-F | CAATCTTTGGCGAGGTCATC | 29/48 | 8 |
| qac-R | CGCTGACCTTGGATAGCAG | 307/325 | ||
| catB3 | catB3-F | AAGGCAAGCTGCTTTCTGAG | 29/48 | 21 |
| catB3-R | AACGATAGCGTAAGGCTCCA | 440/459 | ||
| aadA1 | aadA1-F | ACATCATTCCGTGGCGTTAT | 284/303 | This study |
| aadA1-R | AGGTTTCATTTAGCGCCTCA | 489/508 | ||
| aacA4 | aacA4-F | TGACCTTGCGATGCTCTATG | 12/31 | This study |
| aacA4-R | CTGGCGTGTTTGAACCATGT | 470/489 | ||
| aacA7 | aacA7-F | CAGGCCTGTTGAAACTACCG | 21/40 | 22 |
| aacA7-R | CTTGAGCAACCTCCGTGAAT | 414/433 | ||
| blaOXA | OXA-1F | TATCTACAGCAGCGCCAGTG | 54/73 | 21 |
| OXA-1R | TGCACCAGTTTTCCCATACA | 635/654 | ||
| OXA-2F | CGATAGTTGTGGCAGACGAA | 128/147 | ||
| OXA-2R | CTTGACCAAGCGCTGATGT | 564/582 | ||
| tniC | TniC-R | TTTCCGAGCGAACAGTCGCT | 229/248 | 22 |
| blaIMP | IMP-1F | AAGGCGTTTATGTTCATACTTCG | 95/117 | 19 |
| IMP-1R | TTTAACCGCCTGCTCTAATGTAA | 677/699 | ||
| blaVIM | VIM-2F | ATCATGGCTATTGCGAGTCC | 46/65 | 19 |
| VIM-2R | ACGACTGAGCGATTTGTGTG | 775/794 | ||
| blaKPC | KPC-F | GTCACTGTATCGCCGTCTAGTTC | 3/25 | 19 |
| KPC-R | TGGTGGGCCAATAGATGATT | 919/938 | ||
| blaGES | GES-F | ATGCGCTTCATTCACGCAC | 1/19 | This study |
| GES-R | CTATTTGTCCGTGCTCAGGA | 864/845 | ||
| blaNDM | NDM-F | GCCCAATATTATGCACCCGG | 9/28 | 19 |
| NDM-R | CTCATCACGATCATGCTGGC | 649/668 | ||
| oprD | oprD-F | GGAACCTCAACTATCGCCAAG | −120/−99 | This study |
| oprD-R | GTTGCCTGTCGGTCGATTAC | 17/1328 |
Coordinates refer to the first base of each gene.
MLST.
PCR amplification and sequencing of partial fragments of seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE) were performed, and the experimentally determined nucleotide sequences of both strands were compared to preexisting sequences in the MLST database to assign allele numbers and STs (http://pubmlst.org/paeruginosa).
PFGE.
Agarose plugs containing XbaI-digested genomic DNA from P. aeruginosa clinical isolates were prepared and DNA fragments were separated for 20 h at 6 V/cm at 11°C using a CHEF-DRII system (Bio-Rad, Hercules, CA). Pulsing was carried out with initial and final pulse times of 0.5 s and 30 s, respectively. A lambda ladder (Bio-Rad) was used as a DNA size marker. Band patterns were analyzed with UVIband/Map software (UVItech Ltd., Cambridge, UK) and used to generate dendrograms based on the unweighted pair group method using arithmetic averages from the Dice coefficient.
Southern blot and hybridization.
Southern blotting was performed to determine the locations of the carbapenemase genes. Briefly, I-CeuI or S1 nuclease-digested DNA was blotted onto nylon membranes (Zeta-Probe blotting membranes; Bio-Rad) and hybridized with probes specific for the carbapenemase genes or 16S rRNA. The probes were obtained via PCR amplification as described above. Probe labeling, hybridization, and detection were performed with a digoxigenin DNA labeling and detection kit (Roche Diagnostics, Indianapolis, IN).
WGS.
For bacterial whole-genome sequencing (WGS), single-molecule real-time (SMRT) sequencing was carried out on a PacBio RSII instrument (Pacific Biosciences, Menlo Park, CA). Genomic DNA was extracted from the P. aeruginosa strains using a Wizard genomic DNA purification kit (Promega, Madison, WI). After extraction, DNA shearing was performed using a g-TUBE apparatus (Covaris, Inc., Woburn, MA), and the fragments were purified by using 0.45× the final volume of washed Agencourt AMPure XP magnetic beads (Beckman Coulter Inc., Brea, CA). SMRTbell template libraries were subsequently prepared, and adapter ligation was performed, followed by exonuclease digestion of incompletely ligated products. Reads with lengths that were less than 50 bp were filtered out after acquisition of the sequencing data; the minimum polymerase read quality was set to 0.75. De novo genome assembly of PacBio SMRT reads was performed with the PacBio SMRT analysis software suite (version 2.3.0) (20). Briefly, the hierarchical genome assembly process was used with default parameters and a seed read length cutoff of 6 kb. Following assembly, individual contigs with duplicate sequences on their 5′ and 3′ ends were manually trimmed. Similarly, overlapping sequences were manually trimmed and joined based on sequence similarity to form circular fragments. Following chromosome and plasmid circularization, the sequences were polished using Quiver. In this program, the raw reads are mapped back to the chromosome and plasmid sequences to validate the assembly and resolve any remaining sequence errors. The annotations of coding sequences, tRNA sequences, and rRNA sequences were performed using the NCBI Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/books/NBK174280).
Accession number(s).
The nucleotide sequence data reported in this paper are available in the GenBank nucleotide database under accession numbers KX196167 (PAGI-16 carrying the blaIMP-6 gene), KX196168 (PAGI-15 carrying the blaGES-24 gene), and KX196169 (PAGI-16 carrying the blaIMP-10 gene).
RESULTS
Antimicrobial resistance profiles of the P. aeruginosa isolates.
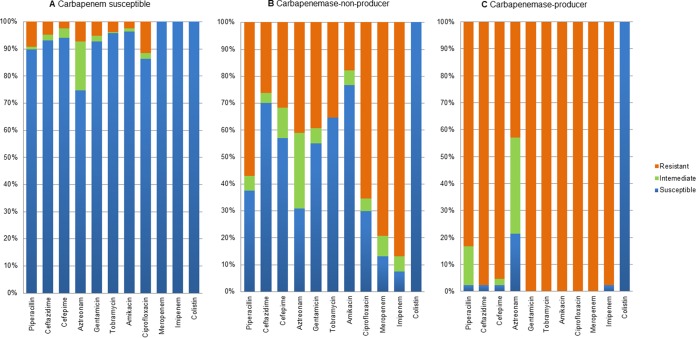
Of the 431 P. aeruginosa clinical isolates examined, 283 (65.7%) were found to be susceptible to carbapenems. The remaining 148 isolates (34.3%) that were nonsusceptible to imipenem and/or meropenem were classified by PCR into two groups: carbapenemase-producing P. aeruginosa (CP-PA) (n = 41), whose members carried the blaIMP, blaVIM, or blaGES gene, and non-carbapenemase-producing P. aeruginosa (NCP-PA; n = 107), whose members did not. Twenty isolates (18.7%) in the NCP-PA group had an OprD porin loss. The vast majority (40/41) of the clinical isolates in the CP-PA group exhibited an extensively drug-resistant phenotype and were resistant to all drugs tested except colistin (Fig. 1). Moreover, the CP-PA group showed extremely high rates of nonsusceptibility to piperacillin (97.6%, compared to 62.6% in the NCP-PA group and 10.2% in the carbapenem-susceptible group), ceftazidime (97.6%, compared to 29.9% and 7.1%, respectively), ciprofloxacin (100%, compared to 70.1% and 13.8%, respectively), and amikacin (100%, compared to 23.4% and 3.5%, respectively).
FIG 1.
Antimicrobial susceptibilities of P. aeruginosa clinical isolates. (A) Carbapenem-susceptible isolates (n = 283); (B) carbapenem-nonsusceptible non-carbapenemase producers (n = 107); (C) carbapenem-nonsusceptible carbapenemase producers (n = 41).
Presence of the carbapenemase gene and its correlation with carbapenem susceptibility.
The most prevalent gene, blaIMP-6, was identified in 36 isolates; moreover, another subtype, blaIMP-10, was identified in one isolate. P. aeruginosa clinical isolates harboring blaIMP-6 exhibited higher resistance to meropenem (MICs, 64 to >256 μg/ml; MIC50, >256 μg/ml; and MIC90, >256 μg/ml) than to imipenem (MICs, 16 to 256 μg/ml; MIC50, 16 μg/ml; and MIC90, 32 μg/ml). Isolate BP14, carrying blaIMP-10, had similar carbapenem MICs: for meropenem, >256 μg/ml, and for imipenem, 16 μg/ml. The products encoded by blaIMP-6 and blaIMP-1 differ by one amino acid, with the former having a Ser216-Gly substitution compared with the latter. Similarly, the product encoded by blaIMP-10 exhibits a single Val49-Phe substitution compared with the product encoded by blaIMP-1.
The blaVIM-2 gene was identified in three isolates. These three isolates had meropenem MICs ranging from 16 to 128 μg/ml and imipenem MICs ranging from 16 to 256 μg/ml. The blaGES-24 gene was identified in one isolate, and the isolate showed resistance to meropenem (MIC, 128 μg/ml) and imipenem (MIC, 64 μg/ml). Of note, this strain had a truncated OprD by a novel insertion sequence, ISPA67. Considering the relatively low carbapenem MICs for 20 P. aeruginosa strains having only the porin loss, 1 to 32 μg/ml (MIC50, 8 μg/ml, and MIC90, 16 μg/ml) for meropenem and 4 to 32 μg/ml (MIC50, 16 μg/ml, and MIC90, 32 μg/ml) for imipenem, the elevated carbapenem MICs were mostly by GES-24. GES-24 differs from GES-1 by two substitutions, Met62-Thr and Gly170-Ser, and the latter change is the same as in the carbapenem-hydrolyzing GES-5.
Strain typing for the CP-PA isolates.
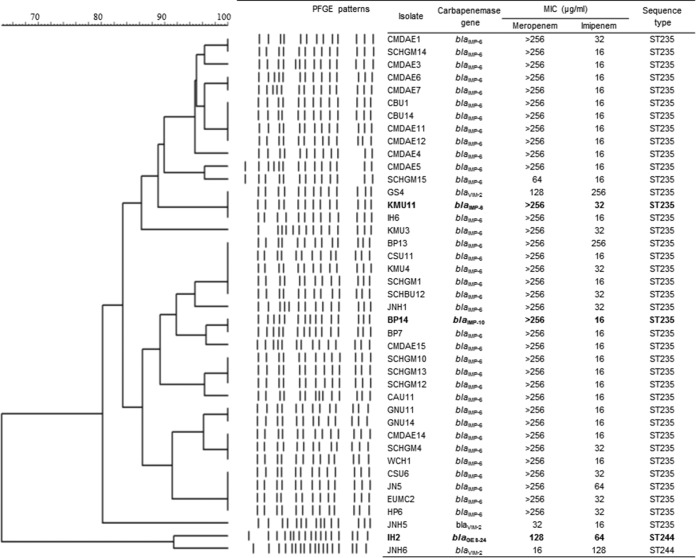
All CP-PA isolates were identified as ST235 (allele profile, 38-11-3-13-1-2-4), with the exception of two isolates: isolate JNH6, which carried blaVIM-2, and isolate IH2, which carried blaGES-24. These two isolates were identified as ST244 (17-5-12-3-14-4-7), which is very different from ST235 (see Table S1 in the supplemental material). The ST235 CP-PA isolates shared more than 80% similarity as determined by PFGE analysis, and the remaining two ST244 isolates also exhibited close relatedness (similarity, >90%). The ST235 and ST244 isolates did not show significant relatedness (similarity, <70%) to each other (Fig. 2).
FIG 2.
Characteristics of chromosome-encoded carbapenemase-producing P. aeruginosa clinical isolates. Strains which were subjected to whole-genome sequencing are shown in bold.
Genetic contexts of the class 1 integrons.
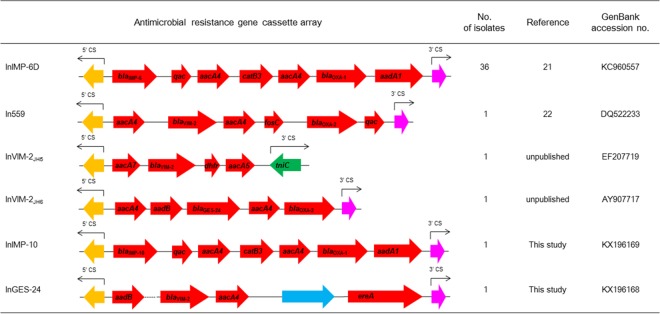
The blaIMP-6, blaIMP-10, blaVIM-2, and blaGES-24 genes were located on class 1 integrons as gene cassettes, and the structures determined by PCR mapping are indicated in Fig. 3. The blaIMP-6 gene in all 36 isolates was carried by InIMP-6D (intI1-blaIMP-6-qac-aacA4-catB3-aacA4-blaOXA-1-aadA1). This class 1 integron was identified in 2008 in one P. aeruginosa ST235 clinical isolate from South Korea (21). The blaIMP-10 gene in isolate BP14 composed a new class 1 integron, InIMP-10 (intI1-blaIMP-10-qac-aacA4-catB3-aacA4-blaOXA-1-aadA1). Between the integron integrase coding gene and the gene cassettes, an integron-associated 136-bp attI recombination site was identified.
FIG 3.
Schematic representation of class 1 integrons carrying the carbapenemase gene cassettes. Yellow arrows, integrase; red arrows, genes for resistance; magenta arrows, qacEΔ1; green arrow, tniC gene; blue arrow, a gene for tyrosine recombinase XerD; dotted line, unknown nucleotide sequences.
The blaVIM-2 gene cassette was found in three different integrons: In559 (intI1-aacA7-blaVIM-2-dhfa-aadA5), InVIM-2JH5 (intI1-aadB-blaVIM-2-aacA4-orf-ereA), and InVIM-2JH6 (intI1-aacA4-blaVIM-2-aacA4-fosC-blaOXA-2-qac). In559 was first identified in P. aeruginosa ST235 clinical isolates from Russia in 2002 (22), and InVIM-2JH5 and InVIM-2JH6 class 1 integrons were first found in P. aeruginosa and Pseudomonas putida isolates, respectively, from South Korea (unpublished data; GenBank accession numbers EF207719 for InVIM-2JH5 and AY907717 for InVIM-2JH6). Notably, In559 was distinct from the other class 1 integrons that culminate with qacΕΔ1 in that the Tn5090 tniC gene (also called tniR of Tn402) for the 3′ conserved sequence (CS) indicates a distinct evolutionary path driving the excision and addition of gene cassettes (23). Isolate IH2 was found to harbor a novel integron, InGES-24 (intI1-aacA4-aadB-blaGES-24-aacA4-blaOXA-2).
Two novel genomic islands, PAGI-15 and PAGI-16.
By Southern blotting using I-CeuI macrorestriction-digested fragments, the blaIMP-6, blaIMP-10, blaVIM-2, and blaGES-24 genes were verified to be chromosome carried in all CP-PA isolates (data not shown). Probes specific for the carbapenemase genes did not bind to any S1 nuclease-treated plasmids. To further characterize the genomic environments of the class 1 integrons, WGS was conducted using the PacBio RSII platform for three P. aeruginosa isolates: KMU11 of blaIMP-6, BP14 of blaIMP-10, and IH2 of blaGES-24.
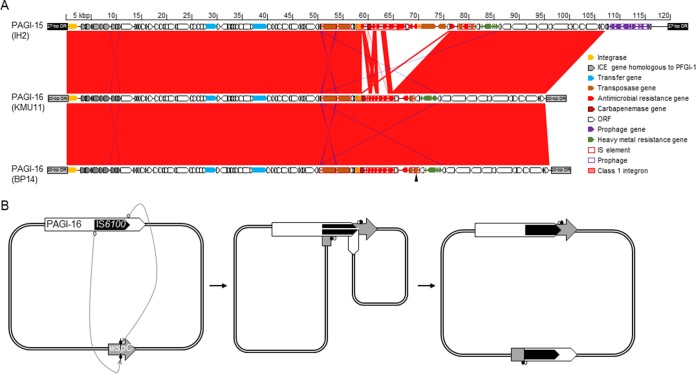
The draft genome sequences of KMU11 were composed of nine contigs, and three (a total of 6.9 Mb in size) of those carried the genes for ribosomal proteins indicating that they were carried on a chromosome. The class 1 integron InIMP-6D was found on a chromosome in a 1.6-Mb contig possessing together the genes encoding ribosomal proteins S6, S9, S15, S18, S20, L9, L13, L21, L25, L27, and L31. Moreover, the integron was in a novel genomic island, PAGI-16, which was 95,029 bp long. PAGI-16 was integrated at the end of the tRNAGly gene, like the other genomic islands found in P. aeruginosa (11), and flanked at its end by 20-bp direct repeats (DR), GATTCCCTTCACCCGCTCCA (Fig. 4A). PAGI-16 in isolate KMU11 had a G+C content of 61.4% and contained 99 open reading frames (ORFs), including a cluster of nine genes originating from the clc element, which is also found in PFGI-1 of Pseudomonas fluorescens (24). The clc element integrates through the attB site using a bacteriophage P4-like integrase; this site is situated in between two tandem tRNAGly genes in the 3′ end (11). The cluster also contains 10 genes associated with genome instability, such as genes for integrases and transposases and insertion sequences; eight genes for antimicrobial resistance, i.e., two copies of aacA4 plus aadA1, catB3, cmx, sul1, and blaOXA-1, together with blaIMP-6, which are mostly found in the class 1 integron between bp 57464 and 77106; and genes encoding hypothetical proteins, which comprise the rest of the sequences.
FIG 4.
Schematic representation of genomic islands in strains IH2, KMU11, and BP14. (A) The sequences of genomic islands of P. aeruginosa isolates were aligned using BLASTN and compared using Artemis Comparison Tool (32). The conserved regions (>95% identity) are indicated in red. Arrowhead, location of chromosomal inversion due to duplication-insertion of IS6100. (B) Schematic mechanism for the large chromosomal inversion mediated by duplication of IS6100. In a chromosome carrying PAGI-16, catalyzed 3′-OH groups of IS6100 (white circle) targeted the 5′ phosphate in the wspC gene (black circle) located elsewhere. The replication broke the cointegration, leading to the duplication in the opposite direction of IS6100 and a chromosomal inversion. The final chromosomal structure became 5′PAGI-16-IS6100-5′ΔwspC and 3′PAGI-16-IS6100 (opposite direction)-3′ΔwspC. White arrow, PAGI-16; black arrow, IS6100; gray arrow, wspC.
Isolate BP14 had a 7.1-Mb circularized chromosome without any plasmid, and the class 1 integron InIMP-10 was found within a 69,208-bp 5′ portion of a genomic island sharing 99.99% nucleic acid identity with PAGI-16 in isolate KMU11. This portion was integrated at the end of the tRNAGly gene, and the 20-bp DR GATTCCCTTCACCCGCTCCA was present in the 5′ end as in KMU11. PAGI-16 was ended by IS6100, and a truncated wspC, encoding a biofilm formation methyltransferase WspC, was found downstream (Fig. 4A). The 1,269-bp wspC gene encodes the biofilm formation methyltransferase WspC, and the left 619 bp was found 3.3 Mb away from the portion together with the rest of PAGI-16 in the opposite direction. The 26,701-bp 3′ portion of PAGI-16 in isolate BP16 was identical to that in KMU11, and the 20-bp DR sequence was found, indicating chromosomal inversion by a duplication-insertion of IS6100 (25). All of PAGI-16 in isolate BP14 had the same G+C content, 61.4%.
IH2 had a 7.3-Mb circularized chromosome devoid of any plasmid. In the chromosome, the class 1 integron InGES-24 was located within a 118,715-bp genomic island designated PAGI-15. The location of PAGI-15 was the same as that of PAGI-16, next to the tRNAGly gene, but the DR was 27 bp, AGGGTTCGATTCCCTTCGCCCGCTCCA (Fig. 4A). The G+C content of the PAGI-16 was 61.3%, slightly lower than that of the PAGI-15. This genomic island shared the backbone with PAGI-16, presenting 99.99% nucleic acid identity excluding the class 1 integron and a short putative prophage possessing nine ORFs located in the 3′ end of the genomic island (Fig. 4A). PAGI-16 contained 116 putative ORFs, mostly shared with PAGI-15 except 11 antimicrobial resistance genes, two copies of aacA4, aadB, strA, strB, tet(G), cmx, two copies of sul1, blaOXA-2, and blaGES-24.
DISCUSSION
A nationwide survey in 2009 reported that the carbapenem resistance rate in P. aeruginosa was 35.8% (138/386) and 8.0% (31/386) of the strains produced carbapenemases IMP-6 (n = 30) and VIM-2 (n = 1) (8). In this surveillance, a similar rate of CP-PA was found, 9.5% (41/431). Among CP-PA strains, prevalence of blaIMP-6-producing P. aeruginosa has been detected exclusively in South Korea. IMP-6 production by the bacterial host results in a higher MIC for meropenem than for imipenem. This is due to the Ser216-Gly substitution, which results in increased hydrolysis of meropenem compared with that of imipenem (26). Interestingly, meropenem is highly recommended for treating Gram-negative bacterial infections, rather than imipenem (27), and meropenem usage is increasing faster than that of imipenem (28). It is likely that antibiotic selective pressure leads to the predominance of IMP-6 in P. aeruginosa.
Clonal diversity of the epidemic isolates, which consisted mostly of ST235 isolates presenting indistinguishable PFGE patterns, allowed us to understand that the dissemination of IMP-6 was mostly by clonal dissemination of the P. aeruginosa ST235 isolates possessing the gene. The gene cassette array added more evidence for that, showing all IMP-6-positive strains possessed an identical class 1 integron, InIMP-6D (intI1-blaIMP-6-qac-aacA4-catB3-aacA4-blaOXA-1-aadA1). Interestingly, the gene cassette array of this class 1 integron resembled that of the integron intI1-blaIMP-6-qac-aacA4-blaOXA-1-aadA1 in epidemic strains of P. aeruginosa isolated in 2009 (8) (Fig. 3).
The second most prevalent carbapenemase was VIM-2, although it was identified in only three isolates. The blaVIM-2 gene was sporadically identified to be associated with unrelated integrons in P. aeruginosa of two different STs, ST235 and ST244, each with different PFGE patterns (Fig. 2). These results suggest that blaVIM-2 dissemination was due not to clonal spread but rather to an occasional occurrence. Interestingly, In559, one of the three class 1 integrons carrying the blaVIM-2 gene cassette, had a distinct 3′ end, matured by tniC of Tn5090 instead of the usual 3′ CS with qacEΔ1. This finding likely indicates that In559 has been developed through a different evolutionary scheme used by Tn5090. A similar blaVIM-2 gene cassette-carrying integron, which was identified as an ancestral class 1 integron 3′ ended by the tniC gene, was identified in a P. aeruginosa strain isolated in India in 2003 (23). The precise epidemiological relationship of the unusual integrons in our strain and in the Indian strain is unclear, since no ST information could be obtained for the case.
This surveillance study enabled the first observation of IMP-10 and GES-24, indicating diversification of carbapenemases in P. aeruginosa in South Korea. The blaIMP-10-associated integron has previously been identified in P. aeruginosa in Japan (29). We initially suspected that the first identification of blaIMP-10 reflected a traveler carrier; however, after inspecting the genomic environment of this gene, we concluded that this gene appearance was a result of spontaneous nucleotide substitutions independently from blaIMP-1, a G640A substitution resulting in Gly216-Ser mutation for IMP-6 in PAGI-16 of KMU11 and G145T leading to Val49-Phe mutation in IMP-10 in PAGI-16 of BP14. Indeed, this conclusion is supported by two additional pieces of evidence. First, InIMP-6D and InIMP-10 have the same genetic context, with the exception of the MBL gene cassettes (blaIMP-6 and blaIMP-10) between the 5′ CS and the 3′ CS (Fig. 3). Second, PAGI-16, which was found to carry the blaIMP-6 gene (GenBank accession number KX196167), and PAGI-16, which was found to carry the blaIMP-10 gene (GenBank accession number KX196169), exhibited a high degree of homology based on WGS results and shared most of the conserved regions (95% identity) (Fig. 4A).
So far, six GES β-lactamases, namely, GES-2, -4, -5, -6, -14, and -18, have been shown to present detectable carbapenemase activity (6). This activity is predominantly a factor of substitutions of amino acid residues 104 and 170. The GES-24 enzyme has Gly170, like GES-5, which results in high affinity for carbapenems and high turnover rates due to the low rate constants for acylation and deacylation (30). The GES-5 producers Klebsiella pneumoniae and E. coli have been found in South Korea (31), but the genes were on very different integrons. The genes for GES-24 in Enterobacter cloacae and Acinetobacter baumannii from Japan are available in GenBank (last updated on 13 May 2016), but to the best of our knowledge, this is the first description of GES-24 in P. aeruginosa.
Interestingly, PAGI-15 and PAGI-16 were found to differ only by their integrons. It is likely that the two genomic islands derived from a common origin. They once possessed a backbone possessing a clc-like element, into which two unrelated integrons were later integrated. Moreover, PAGI-16 in one isolate was found to have split into two parts by large chromosomal inversion that resulted in duplication-insertion of the IS6100 gene in the opposite direction (Fig. 4B). Similar IS6100-mediated large chromosomal inversions have been found in P. aeruginosa clinical isolates and are likely to be involved in phenotypic adaptations of the different strains to the environment (25).
PAGI-15 and -16 in this study were found in P. aeruginosa belonging to ST244 and ST235. These genomic islands have shared origin evidenced by the same backbone in spite of the distinct STs of the host P. aeruginosa. Interestingly, a genomic island having a same backbone is found also in the chromosome of P. aeruginosa ST395 isolated in France in 1997 (GenBank accession number CP013993). The genomic island has a class 1 integron possessing one gene cassette, aadB. This highly mobile genomic island is likely to have a key role for capturing and disseminating the multiple antibiotic resistance genes in P. aeruginosa belonging to various STs, similar to the case with PAGI-1 and -2 (13).
In conclusion, our data indicate that IMP-6 is highly prevalent in CP-PA ST235 isolates. Moreover, our results clearly demonstrate that WGS methodology can be used to identify the genomic environments associated with resistance determinants, which promises to shed light on many epidemiological questions regarding the mechanism of dissemination of resistance determinants.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Research Program funded by the Korea Centers for Disease Control and Prevention (2015E4400200).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01601-16.
REFERENCES
- 1.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morita Y, Kimura N, Mima T, Mizushima T, Tsuchiya T. 2001. Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO1. J Gen Appl Microbiol 47:27–32. doi: 10.2323/jgam.47.27. [DOI] [PubMed] [Google Scholar]
- 3.Zavascki AP, Carvalhaes CG, Picão RC, Gales AC. 2010. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther 8:71–93. doi: 10.1586/eri.09.108. [DOI] [PubMed] [Google Scholar]
- 4.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crespo MP, Woodford N, Sinclair A, Kaufmann ME, Turton J, Glover J, Velez JD, Castañeda CR, Recalde M, Livermore DM. 2004. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-beta-lactamase, in a tertiary care center in Cali, Colombia. J Clin Microbiol 42:5094–5101. doi: 10.1128/JCM.42.11.5094-5101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diene SM, Rolain JM. 2014. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect 20:831–838. doi: 10.1111/1469-0691.12655. [DOI] [PubMed] [Google Scholar]
- 7.Pasteran F, Faccone D, Gomez S, De Bunder S, Spinelli F, Rapoport M, Petroni A, Galas M, Corso A, Pseudomonas aeruginosa KPC Group 2012. Detection of an international multiresistant clone belonging to sequence type 654 involved in the dissemination of KPC-producing Pseudomonas aeruginosa in Argentina. J Antimicrob Chemother 67:1291–1293. doi: 10.1093/jac/dks032. [DOI] [PubMed] [Google Scholar]
- 8.Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. 2011. Dissemination of IMP-6 metallo-β-lactamses-producing Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother 66:2791–2796. doi: 10.1093/jac/dkr381. [DOI] [PubMed] [Google Scholar]
- 9.Boucher Y, Labbate M, Koenig JE, Stokes HW. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol 15:301–309. doi: 10.1016/j.tim.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Hall RM. 2012. Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann N Y Acad Sci 1267:71–78. doi: 10.1111/j.1749-6632.2012.06588.x. [DOI] [PubMed] [Google Scholar]
- 11.Kung VL, Ozer EA, Hauser AR. 2010. The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev 74:621–641. doi: 10.1128/MMBR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silveira MC, Albano RM, Asensi MD, Carvalho-Assef AP. 2016. Description of genomic islands associated to the multidrug-resistant Pseudomonas aeruginosa clone ST277. Infect Genet Evol 42:60–65. doi: 10.1016/j.meegid.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 13.Roy Chowdhury P, Scott M, Worden P, Huntington P, Hudson B, Karagiannis T, Charles IG, Djordjevic SP. 2016. Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol 6:pii=150175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foxman B, Zhang L, Koopman JS, Manning SD, Marrs CF. 2005. Choosing an appropriate bacterial typing technique for epidemiologic studies. Epidemiol Perspect Innov 2:10. doi: 10.1186/1742-5573-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. 2014. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseuomonas aeruginosa collected during 2009-11 in 14 European and Mediterranean countries. J Antimicrob Chemother 69:1804–1814. doi: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 16.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. 2009. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 53:4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 21-22:41–59. [DOI] [PubMed] [Google Scholar]
- 18.CLSI. 2016. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Document M100-S25. CLSI, Wayne, PA. [Google Scholar]
- 19.Hong JS, Kim JO, Lee H, Bae IK, Jeong SH, Lee K. 2015. Characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa in Korea. Infect Chemother 47:33–40. doi: 10.3947/ic.2015.47.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner CP, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 21.Bae IK, Suh B, Jeong SH, Wang KK, Kim YR, Yong D, Lee K. 2014. Molecular epidemiology of Pseudomonas aeruginosa clinical isolates from Korea producing β-lactamases with extended-spectrum activity. Diagn Microbiol Infect Dis 79:373–377. doi: 10.1016/j.diagmicrobio.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Edelstein MV, Skleenova EN, Shevchenko OV, D'souza JW, Tapalski DV, Azizov IS, Sukhorukova MV, Pavlukov RA, Kozlov RS, Toleman MA, Walsh TR. 2013. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: a longitudinal epidemiological and clinical study. Lancet Infect Dis 13:867–876. doi: 10.1016/S1473-3099(13)70168-3. [DOI] [PubMed] [Google Scholar]
- 23.Toleman MA, Vinodh H, Sekar U, Kamat V, Walsh TR. 2007. blaVIM-2-harboring integrons isolated in India, Russia, and the United States arise from an ancestral class 1 integron predating the formation of the 3′ conserved sequence. Antimicrob Agents Chemother 51:2636–2638. doi: 10.1128/AAC.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavrodi DV, Loper JE, Paulsen IT, Thomashow LS. 2009. Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol 9:8. doi: 10.1186/1471-2180-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coyne S, Courvalin P, Galimand M. 2010. Acquisition of multidrug resistance transposon Tn6061 and IS6100-mediated large chromosomal inversions in Pseudomonas aeruginosa clinical isolates. Microbiology 156:1448–1458. doi: 10.1099/mic.0.033639-0. [DOI] [PubMed] [Google Scholar]
- 26.Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. 2001. Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother 45:1343–1348. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joly-Guillou ML, Kempf M, Cavallo JD, Chomarat M, Dubreuil L, Maugein J, Muller-Serieys C, Roussel-Delvallez M. 2010. Comparative in vitro activity of meropenem, imipenem and piperacillin/tazobactam against 1071 clinical isolates using 2 different methods: a French multicentre study. BMC Infect Dis 10:72. doi: 10.1186/1471-2334-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo RN, Dong YH, Liu JP, Chang CH, Shau WY, Lai MS. 2011. Predicting healthcare utilization using a pharmacy-based metric with the WHO's Anatomic Therapeutic Chemical algorithm. Med Care 49:1031–1039. doi: 10.1097/MLR.0b013e31822ebe11. [DOI] [PubMed] [Google Scholar]
- 29.Iyobe S, Kusadokoro H, Takahashi A, Yomoda S, Okubo T, Nakamura A, O'Hara K. 2002. Detection of a variant metallo-beta-lactamase, IMP-10, from two unrelated strains of Pseudomonas aeruginosa and an Alcaligenes xylosoxidans strain. Antimicrob Agents Chemother 46:2014–2016. doi: 10.1128/AAC.46.6.2014-2016.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frase H, Shi Q, Testero SA, Mobashery S, Vakulenko SB. 2009. Mechanistic basis for the emergence of catalytic competence against carbapenem antibiotics by the GES family of β-lactamases. J Biol Chem 284:29509–29513. doi: 10.1074/jbc.M109.011262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Hong SG, Bae IK, Kang JR, Jeong SH, Lee W, Lee K. 2011. Emergence of Escherichia coli sequence type ST131 carrying both the blaGES-5 and blaCTX-M-15 genes. Antimicrob Agents Chemother 55:2974–2975. doi: 10.1128/AAC.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.