Abstract
The blaNDM-1 gene encodes a carbapenemase that confers resistance to almost all β-lactams, including last-resort carbapenems. This is increasingly reported worldwide in nosocomial and community-acquired Gram-negative bacteria. Acinetobacter baumannii is an important opportunistic pathogen that is considered an intermediate reservoir for the blaNDM-1 gene. In this species, the blaNDM-1 gene is located within the Tn125 composite transposon. The mechanism driving the mobility of Tn125 has not yet been elucidated. Here we experimentally demonstrated the transposition of Tn125 in A. baumannii. Systematic 3-bp duplication of the target site, being the signature of transposition, was evidenced. The target site consensus sequence for Tn125 transposition was found to be GC enriched at the duplicated 3 bp and AT rich in the vicinity. Transposition frequency was not influenced by temperature changes or by exposure to subinhibitory concentrations of various antibiotics. This work is the first direct evidence of the functionality of a composite transposon in A. baumannii. It provides a mechanistic clue for the dissemination of the blaNDM-1 gene in Acinetobacter spp. and subsequently among Enterobacteriaceae.
INTRODUCTION
Carbapenemases are enzymes hydrolyzing most β-lactams, including penicillins, cephalosporins, and carbapenems. During the last decade, they have been increasingly reported worldwide in Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae. The spread of carbapenemases is of utmost importance for medicine, since carbapenems are last-resort antibiotics for treating the most severe and hospital-acquired infections. The class B New Delhi metallo-β-lactamase (NDM-1) is a broad-spectrum β-lactamase, hydrolyzing penicillins, cephalosporins, and carbapenems (1). NDM-1 was first identified in a Klebsiella pneumoniae isolate from a patient previously hospitalized in India in 2008 (2). Since then, it has been found mostly in Enterobacteriaceae and A. baumannii and to a lesser extend in P. aeruginosa (1, 3). The origin of the blaNDM-1 resistance gene and the mechanism(s) driving its mobility remains unknown. However, a current hypothesis suggests that the blaNDM-1 gene originates from an environmental bacterial progenitor species and that A. baumannii is an intermediate reservoir for the blaNDM-1 gene (4).
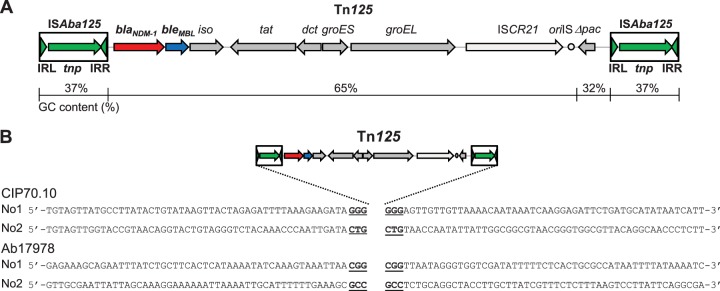
A. baumannii is an opportunistic human pathogen and a common cause of sepsis, pneumonia, urinary tract infection, and primary bacteremia. Multidrug-resistant A. baumannii isolates are of great concern (5, 6). In Acinetobacter spp., the blaNDM-1 gene is embedded in transposon Tn125 (4, 7, 8). Tn125 is a 10,099-bp composite transposon bracketed by two copies of the insertion sequence (IS) ISAba125 orientated in the same direction (Fig. 1A) (4, 7, 8). The 1,087-bp ISAba125 element belongs to the IS30 family and encodes a 322-amino-acid-long DDE-type transposase surrounded by imperfect terminal inverted repeat sequences (IRR [inverted repeat right] and IRL [inverted repeat left], sharing 20/26 nucleotide identity) (9, 10). The bleMBL gene, which encodes a 121-amino-acid-long protein conferring resistance to bleomycin, a glycopeptide antibiotic used as an antitumor agent, is located downstream of the blaNDM-1 gene (11). In addition, Tn125 comprises six genes encoding putative proteins (iso, tat, dct, groES, groEL, and Δpac [a truncated phospholipid acetyltransferase gene]), the ISCR21 element, and the putative oriIS sequence that defines the origin of replication of ISCR21 (7, 8). ISCR elements are peculiar ISs belonging to the IS91 family, likely mobilizing genes located at their left-hand extremity by a rolling-circle transposition process (12, 13).
FIG 1.
Transposition of Tn125. (A) Schematic representation of transposon Tn125. The total size is 10,099 bp. The ORFs are represented by arrows, and the lengths are to scale. The two ISAba125 elements bracketing Tn125 are indicated in green, each with the transposase (tnp) gene and the IRL and IRR inverted repeats. The blaNDM-1 gene is in red, the bleMBL gene is in blue, and the 7 other ORFs are in dark gray; the ISCR21 and the oriIS (circle) are in light gray. The GC content (%) for each of the following fragments is indicated: 37% for each ISAba125 (nucleotides 1 to 1087 and 9013 to 10099), 65% for the sequence encompassing blaNDM-1 to oriIS, and 32% for Δpac. (B) Characterization of four Tn125 transposition events in A. baumannii CIP70.10 and Ab17978 (recA mutant) strains. The duplicated 3-bp target sites are underlined. The surrounding 50 nucleotides, upstream and downstream of each transposon insertion, are shown. In A. baumannii CIP70.10, transposon Tn125 transposed between ORFs designated No1 (encoding proteins deposited under accession no. CRL92855.1 and CRL92856.1) or an ORF designated No2 (encoding an outer membrane protein A precursor [accession no. CRL95910.1); in A. baumannii Ab17978 (recA mutant), transposon Tn125 transposed in the ORF designated No1 (encoding a hypothetical protein [accession no. AKQ26110.1]) or in the ORF designated No2 (encoding a CinA-like protein [accession no. KNZ37258.1]).
Current observations suggest that the blaNDM-1 gene originates from an unknown environmental bacterial progenitor species and is integrated into the chromosome of Acinetobacter spp. The blaNDM-1-bearing Tn125 transposon was likely subsequently built from such Acinetobacter spp. and then transferred onto broad-host-range plasmids, followed by horizontal transfer to Enterobacteriaceae and P. aeruginosa. This hypothesis is supported by a series of genetic features (4, 5), as follows. (i) The blaNDM-1 gene displays a higher GC percentage (62%) than that of the genome of Acinetobacter spp. (38% to 42%), arguing in favor of a phylogenetic distance between the progenitor species and Acinetobacter spp. (ii) ISCR21 may have mobilized a fragment encompassing the blaNDM-1 gene that displays a similar GC percentage from an unknown bacterial progenitor. (iii) ISAba125 has also been identified in Acinetobacter species isolates without physical association with the blaNDM-1 gene and shows a low GC content of 37%, consistent with a possible Acinetobacter species origin (4, 5).
A critical step in the dissemination process of the blaNDM-1 gene is the mobility of the Tn125 transposon in Acinetobacter spp. According to its genetic structure, it was presumed that Tn125 can move through transposition. Transposition is a catalytic process, driven by an element-specific transposase. During this process, the transposase generates a short direct repeat flanking the transposon in the target DNA, corresponding to a signature of the transposition event (10). Alternative mechanisms for Tn125 transfer might involve nonhomologous recombination or cointegration. Here, we show that the Tn125 transposon can efficiently transpose in A. baumannii and that the transposition frequency is not influenced by temperature changes or antibiotic pressure.
MATERIALS AND METHODS
Strains.
Transposition experiments were performed in A. baumannii CIP70.10 and Ab17978 (recA::Km) (14) reference strains. Plasmids were constructed in Escherichia coli TOP10 (Invitrogen).
Plasmid construction.
For pTOPO-Tn125, the Tn125 transposon was amplified from A. baumannii strain JH (8) together with 98 bp and 99 bp of the flanking genomic sequences present in upstream and downstream Tn125, respectively, with primers JHorfTn125-HindIII-F (5′-gatgataagcttTCAGCAATAAATTTGTCACCAGC-3′) and JHorfTn125-XbaI-R (5′-gatgattctagaCAAGCTGCTCAAGTTAAAGATCG-3′) (the HindIII and XbaI restriction sites are underlined, and the uppercase letters correspond to the open reading frame [ORF] identified in strain JH). This amplicon was subcloned into the HindIII and XbaI restriction sites of the pCR-BluntII-TOPO plasmid (Invitrogen). The integrity of both ISAba125 elements and of blaNDM-1 was confirmed by sequencing. pTOPO-zeodel-Tn125 was derived from pTOPO-Tn125, in which a frameshift in the zeocin resistance gene was generated by digestion at the unique FseI site, blunting, and self-ligation. The resulting zeocin resistance protein lacks the 32 C-terminal amino acids. For the pTOPO-shuttle-Tn125 plasmid, the A. baumannii-specific origin of replication was amplified from pWH1266 (a kind gift from P. Higgins) and subcloned into pTOPO-Tn125 between the BsrGI and HindIII restriction sites. Plasmid pTOPO-shuttle-Tn125 replicates in E. coli, from which it can be selected with 50 μg/ml zeocin, 100 μg/ml ampicillin, 25 μg/ml kanamycin, or 0.5 μg/ml imipenem (IPM), in A. baumannii CIP70.10, from which it can be selected with 25 μg/ml kanamycin, 200 μg/ml zeocin, or 1 μg/ml IPM, and finally in A. baumannii Ab17978 recA::Km, from which it can be selected with 10 μg/ml zeocin or 1 μg/ml IPM. Full sequences of the Tn125 plasmids are available upon request.
Transposition assays.
One-hundred-nanogram amounts of pTOPO-Tn125 or pTOPO-zeodel-Tn125 suicide plasmids were electroporated into 25 μl of electrocompetent A. baumannii cells with a MicroPulser (Bio-Rad). The bacteria were resuspended in 2 ml LB and incubated for 1 h 30 min at 37°C with agitation. A total of 100 μl was plated onto LB agar plates supplemented with 1 μg/ml IPM to select the transposition events. The transformation efficiency was determined with the highly similar pTOPO-shuttle-Tn125 plasmid (replicating in A. baumannii), which was electroporated and processed in parallel to the suicide plasmids. The transposition frequencies were calculated by dividing the number of transposition events by the number of transformed cells. Experiments were done in triplicate. The same procedure was used to address the effect antibiotics has on transposition, but cells were incubated for 3 h at 37°C after electroporation with the following antibiotics: the glycopeptide bleomycin (1 μg/ml and 5 μg/ml) (Molekula) or zeocin (4 μg/ml and 8 μg/ml), the fluoroquinolone ciprofloxacin (0.05 μg/ml and 1 μg/ml) (Sigma-Aldrich), the aminoglycoside kanamycin (1.5 μg/ml and 3 μg/ml) (Roth; ThermoFisher Scientific), and the carbapenem imipenem (0.125 μg/ml and 0.25 μg/ml) (Mylan). The MIC of bleomycin for Tn125-containing CIP70.10 was 10 μg/ml, while it was <1 μg/ml for the parental CIP70.10 strain.
Molecular characterization of transposition events.
The presence of the full-length Tn125 was confirmed by the amplification in 5′ of a 925-bp fragment spanning from ISAba125 to blaNDM-1 with primers 125-F (5′-ACACCATTAGAGAAATTTGC-3′) and NDM-R (5′-CGGAATGGCTCATCACGATC-3′) and in 3′ of a 1,326-bp fragment spanning from Δpac to ISAba125 with primers dpac-F (5′-CAACTGTGAGTCCTTTACTGAC-3′) and 125-R (5′-GCAAATTTCTCTAATGGTGT-3′). The integration sites of the transposition events were characterized by shotgun cloning. Total genomic DNA was digested with EcoRV and ligated into the EcoRV-digested and dephosphorylated pBSKS-kanR vector. The libraries were electroporated into E. coli TOP10 and plated onto LB agar containing 1 mg/liter IPM to select for blaNDM-1-positive clones, which were sequenced with the primer IS125tpase_NORF-R (5′-CTCACGATAGATCGTACTAGG-3′) to identify the genomic sequence upstream of Tn125. For each transposition event, a primer was designed to amplify a fragment spanning from the 3′ end of Tn125 to the genomic sequence downstream of Tn125. These PCR amplicons were sequenced with primer IS125tpase-CORF-F (5′-CATGTCACTGAATACTCGTCC-3′).
Determination of the AT and GC contents and pictogram of the target site consensus.
For the 27 transposition events characterized, the relative frequencies of each A and T, and G and C, for the region extending from 50 nucleotides upstream to 50 nucleotides downstream from the duplicated 3-bp target site were calculated and plotted onto a graph. The pictures of the relative frequencies of the bases at each position were generated with the Pictogram program (http://genes.mit.edu/pictogram.html).
Detection of ISAba125 in Acinetobacter species strains.
Strains were screened by PCR with primers Tn125-F (5′-TGTATATTTCTGTGACCCAC-3′) and Tn125-R (5′-GAAGGCGAATTCAAACATGAGGTGC-3′). A 255-bp product was amplified in the presence of ISAba125.
RESULTS
Transposition of Tn125 in A. baumannii.
To address the transposition of Tn125 in A. baumannii, the suicide plasmid pTOPO-Tn125 was transformed as the donor for the transposon in two A. baumannii recipient strains, CIP70.10 (recA wild type) and Ab17978 (recA mutant) (14). The pTOPO-Tn125 plasmid contains the entire Tn125 transposon, together with the flanking chromosomal sequences present in the Tn125-positive A. baumannii isolate JH (98 and 99 bp upstream and downstream of the transposon, respectively) (8). Four imipenem-resistant clones, two in A. baumannii CIP70.10 and two in A. baumannii Ab17978, were stepwise characterized as follows. First, they were tested for the loss of kanamycin and zeocin resistance markers, confirming that the donor plasmid did not integrate into the genome. Second, the presence of the full-length transposon Tn125 and the absence of the flanking sequences present in the donor plasmid were confirmed by PCR. Third, the genomic sequences flanking Tn125 were characterized in order to map the transposition sites and analyze the target site duplication. As shown in Fig. 1B, for each of the four studied clones, the entire Tn125 transposed into distinct loci of the A. baumannii chromosome. A 3-bp target site duplication was present in each case. Transposition of Tn125 conferred resistance to cephalosporins and carbapenems, with MIC values of ceftazidime, imipenem, and meropenem being >256, >32, and >32 μg/ml, respectively.
Transposition frequencies.
The transposition frequencies measured with pTOPO-Tn125 and pTOPO-zeodel-Tn125 as donors were 4.5 × 10−4 (±0.5 × 10−4) and 5.7 × 10−4 (±0.9 × 10−4) per transformed cell, respectively (Table 1, experiment 1). The frequencies with both donor plasmids were in the same range, excluding the potential influence of the zeocin marker gene, which confers resistance to the same class of molecules (bleomycin) as the bleMBL gene. Since the source of dissemination of the blaNDM-1 gene is likely the environment, in particularly in Asia (15, 16), where a variety of antibiotics has been widely identified (17, 18) and where temperature changes might influence the transposition frequency (19, 20), corresponding experiments were conducted. Incubation for 3 h with 1 or 5 μg/ml bleomycin, a glycopeptide antibiotic used as an antitumor agent, did not influence the transposition rate of Tn125 (Table 1, experiment 2). Similarly, the transposition rate was not influenced by incubation for 3 h at different temperatures (25°C, 30°C, 37°C, or 44°C) or by the presence of subinhibitory concentrations of structurally nonrelated antibiotics, such as fluoroquinolone (ciprofloxacin), aminoglycoside (kanamycin), glycopeptide (zeocin), and carbapenem (imipenem) (data not shown).
TABLE 1.
Transposition frequencies of Tn125 in A. baumannii CIP70.10
| Expta | Incubation drug (concn) | Transposition frequency (10−4)b |
|
|---|---|---|---|
| pTOPO-Tn125 | pTOPO-zeodel-Tn125 | ||
| 1 | None | 4.5 (±0.5) | 5.7 (±0.9) |
| 2 | None | 11.8 (±3.0) | 9.6 (±5.1) |
| Bleomycin (1 mg/liter) | 11.4 (±2.2) | 9.0 (±3.4) | |
| Bleomycin (5 mg/liter) | 7.6 (±3.9) | 4.6 (±1.8) | |
In experiment 1, the bacteria were incubated for 1 h 30 min at 37°C after electroporation and then plated on 1-mg/liter imipenem-containing plates. In experiment 2, the bacteria were incubated for 3 h at 37°C with the indicated concentration of bleomycin.
pTOPO-Tn125 or pTOPO-zeodel-Tn125 was transformed as the donor of Tn125. The transposition frequencies are expressed as the number of transposition events relative to the number of cells transformed. Experiments were performed in triplicate. Values in parentheses are standard deviations.
Target site specificity.
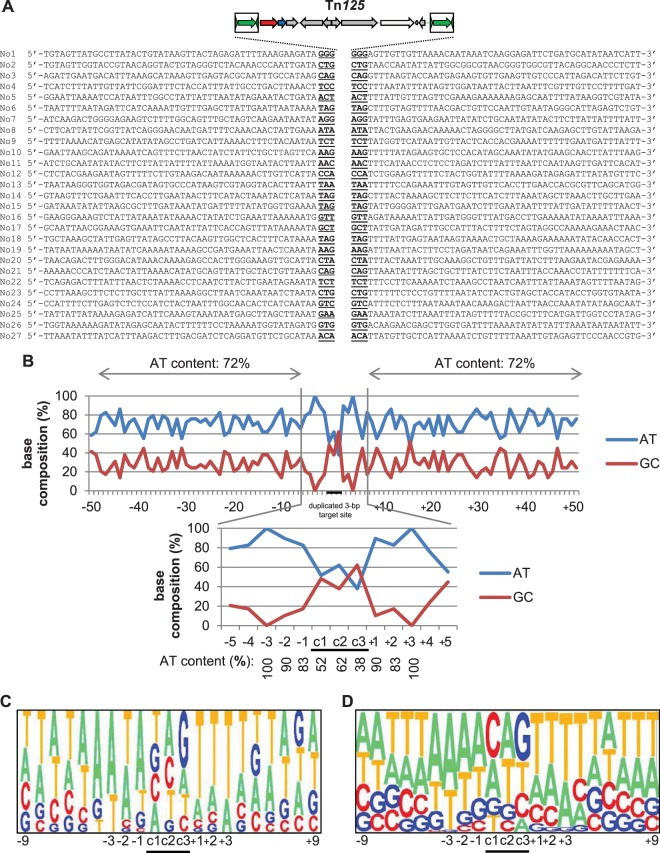
In order to determine a consensus target site for Tn125 transposition, 25 additional independent transposition events, in 25 independent isolates, were characterized in A. baumannii CIP70.10. Among the transposition events, 19 occurred within open reading frames (ORFs), 8 in direct orientation and 11 in reverse orientation compared to the disrupted ORF, and 8 outside of ORFs. For each transposition event, a systematic 3-bp duplication of the target site was evidenced (Fig. 2A). To further characterize the features of the target site, the surrounding genomic sequences of the 27 transposition events were aligned, from 50 bp upstream up to 50 bp downstream of Tn125. The mean AT content for the regions distal to the duplicated target site, from −50 to −4 bp and from +4 to +50 bp relative to it, was 72% on both sides (Fig. 2B, upper graph). Around the duplicated target site, the AT content increased, with 100% at positions −3 and +3 and 83% to 90% at positions −2, −1, +1, and +2 (Fig. 2B, lower graph). By analysis of the nucleotide composition, the −3 position was found to be predominantly an A (69%), and the +3 position was found to be predominantly a T (72%) (Fig. 2C). At the duplicated target site positions, named here c1, c2, and c3, the AT content was lower (38% to 62%) (Fig. 2B, lower graph). At c1, A, T, C, and G were equally represented (24% to 28%), while G was underrepresented at c2 (10% versus 38%, 24%, and 28% for A, T, and C, respectively) and predominant at c3 (48% versus 21%, 17%, and 14% for A, T, and C, respectively) (Fig. 2C). Further analysis of sequences flanking 74 ISAba125 elements retrieved from GenBank confirmed the consensus derived from the analysis of Tn125 transposition events (Fig. 2D). In conclusion, the target site specificity of Tn125 was driven mainly both by the 3 bp surrounding the duplicated target site, being AT rich and reaching 96% to 99% A or T at positions −3 and +3, and by the duplicated 3-bp target site itself, which was GC enriched, with a strong bias for nucleotide G at the position c3.
FIG 2.
Target site preferences of Tn125. (A) Molecular characterization of 27 transposition events of Tn125 in A. baumannii CIP70.10 (two of them are the same as in Fig. 1B). For each of them, the duplicated 3-bp target site is underlined. The surrounding 50 nucleotides upstream and downstream of the target sites are shown. (B) The 27 transposition sites of Fig. 2A were aligned, and the percentages of AT and GC at each position, from 50 nucleotides upstream to 50 nucleotides downstream of the target site, are shown on the graph. The 3 bp of the duplicated target site, named here c1, c2, and c3, are highlighted by a black bar. The AT percentages of regions spanning positions −50 to −4 and positions +4 to + 50 and those of the region spanning positions −3 to + 3 are indicated in the upper and lower graphs, respectively. (C) Pictogram showing the relative frequencies of each A, T, C, and G at the target site, deduced from the 27 experimental transposition events shown in panel A. (D) Pictogram showing the relative frequencies of each A, T, C, and G at the target site, deduced from 74 ISAba125 elements retrieved from GenBank (see Table 3 for references).
Distribution of ISAba125 in Acinetobacter spp.
In order to evaluate the distribution of ISAba125, its occurrence was evaluated among 17 A. baumannii and 16 Acinetobacter species clinical isolates. These strains were negative for the blaNDM-1 gene, and some of them carried previously characterized resistance genes to broad-spectrum β-lactams. ISAba125 was detected in 21 out of 33 strains (Table 2). In addition, the locations of 74 ISAba125 elements found in genomic sequences of 8 A. baumannii strains deposited in GenBank were mapped with respect to their target sites. Half of the ISAba125 elements were located within ORFs. For 6 of them, the IRR of ISAba125, which provides a −35 box promoter element orientated toward the flanking DNA, was found in proximity (less than 100 bp) of the ATG start codon of the adjacent ORFs (Table 3). In conclusion, ISAba125 was detected in more than half of the Acinetobacter species clinical isolates tested and potentially contributed to the genetic plasticity of those strains, either by disrupting ORFs or by potentially bringing promoter sequences to chromosomal genes.
TABLE 2.
Distribution of ISAba125 in Acinetobacter species isolates, detected by PCR
| Isolate | Speciesa | Resistance gene(s) characterizedb | ISAba125 | Reference or sourcec |
|---|---|---|---|---|
| Rem | A. baumannii | None | − | Collection |
| Son | A. baumannii | None | + | Collection |
| ROU | A. baumannii | None | − | Collection |
| 4547 | A. baumannii | None | + | Collection |
| MK8744 | A. baumannii | None | − | 33 |
| AS1 | A. baumannii | blaOXA-23 | + | 34 |
| 1279 Bahe | A. baumannii | blaOXA-51 oE | + | Collection |
| CLA-1 | A. baumannii | blaOXA-40 | + | Collection |
| 75510 | A. baumannii | blaIMP-4 | + | Collection |
| FER | A. baumannii | blaOXA-23 | − | Collection |
| PIN | A. baumannii | blaOXA-40 | + | Collection |
| 1637 | A. baumannii | armA | − | Collection |
| AP | A. baumannii | blaOXA-91, blaGES-14 | + | Collection |
| ELF | A. baumannii | blaOXA-40, blaTEM-1 | − | Collection |
| 614 | A. baumannii | blaOXA-23 | + | 34 |
| 133 | A. baumannii | blaOXA-58, blaOXA-64 | + | 35 |
| 5179 | A. baumannii | blaVEB-1a, blaSCO-1 | + | 36 |
| CGL-3 | A. haemolyticus | blaOXA-58 | + | Collection |
| 7446 | A. junii | blaOXA-58, blaPER-2, blaSCO-1 | + | 36 |
| 7368 | A. lwoffii | blaOXA-58, blaPER-2, blaSCO-1 | + | 36 |
| BER | A. radioresistens | blaOXA-105 | − | Collection |
| R864 | A. lwoffii | blaOXA-134 | + | Collection |
| 5400 | A. baylyi | blaPER-2, blaSCO-1 | + | 36 |
| 0551 | A. radioresistens | blaOXA-23 | + | Collection |
| CIP103788 | A. radioresistens | blaOXA-23, blaOXA-103 | + | Collection |
| CIP64.7 | A. genomospecies 6 | blaOXA-134 | + | Collection |
| 9905 | A. johnsonii | blaOXA-211 | + | 37 |
| CIP64.5 | A. junii | None | − | Collection |
| CIP107464 | A. gerneri | None | − | Collection |
| CIP107468 | A. bouvetii | None | − | Collection |
| CIP107469 | A. tandoii | None | − | Collection |
| CIP107470 | A. grimontii | None | + | Collection |
| CIP107472 | A. towneri | None | − | Collection |
A. baumannii is the clinically most important species.
blaOXA genes encode carbapenem-hydrolyzing class D oxacillinases, blaTEM-1 and blaSCO-1 encode penicillinases, blaVEB-1a and blaPER-2 encode extended-spectrum beta-lactamases (ESBLs), blaIMP-4 and blaGES-14 encode carbapenemases, and armA encodes a 16S rRNA methylase. oE, overexpression.
Collection, personal laboratory collection.
TABLE 3.
Position of 74 ISAba125 elements with respect to genomic ORFsa
| Strain | Accession no. | No. of ISAba125 copies |
Reference | |||
|---|---|---|---|---|---|---|
| Totalb | Within ORF | Between ORFs | Promoter providerc | |||
| NCGM 237 | AP013357 | 26 | 14 | 12 | 2 | 38 |
| LAC-4 | CP007712 | 14 | 9 | 5 | 1 | 39 |
| AbH12O-A2 | CP009534 | 3 | 2 | 1 | 0 | 40 |
| AB04-mff | CP012006 | 12d | 5 | 6 | 0 | 41 |
| ACICU | CP000863 | 7 | 2 | 5 | 2 | 29 |
| BJAB0715 | CP003847 | 8 | 3 | 5 | 0 | 30 |
| TTHO-4 | CP012608 | 4 | 2 | 2 | 0 | Unpublished |
| TCDC-AB0715 | CP002522 | 1 | 0 | 1 | 1 | 42 |
Sequences were retrieved from 8 A. baumannii complete genome sequences deposited in GenBank.
Number of ISAba125 copies found in each strain.
Among ISAba125 intergenic copies, those copies where the distance between the end of the IRR (which provides a −35 box promoter element) and the ATG of the following ORF was less than 100 bp.
Ten single ISAba125 elements and 1 composite transposon made of 2 ISAba125 copies.
DISCUSSION
This study actually corresponds to the first experimental demonstration of the functionality of the blaNDM-1-carrying Tn125 transposon. The ability of Tn125 to transpose in A. baumannii sustains the following model: A. baumannii is an intermediate reservoir for the blaNDM-1 gene and may contribute to further dissemination of the blaNDM-1 gene among species. Indeed, environmental A. baumannii is originally likely in close contact with a still unknown progenitor of the blaNDM-1 gene or with disseminated blaNDM-1-positive bacteria (15). It could then transfer resistance genes to Enterobacteriaceae in the environment or in humans.
The transposase directly influences the choice of the target site, with a preference for GC- or AT-rich DNA domains, local DNA structure, degree of supercoiling, replication or transcription level, and chromosome or plasmid location (10). The target site of Tn125 was AT rich when the 3 bp surrounding the duplicated target site was considered, reaching 99% A and 96% T at positions −3 and +3, respectively, and was enriched in GC within the 3-bp target site, which is duplicated upon transposition, with a strong preference for G at c3 (Fig. 2). In A. baumannii clinical isolates, the same features are present in the chromosomal regions flanking the Tn125 and TnaphA6 composite transposons, the most recently identified being composed of the aphA6 aminoglycoside resistance gene surrounded by two ISAba125 elements (8, 21). The preference for AT-rich regions, previously observed for ISAba1 (22), is consistent with the Acinetobacter species origin of ISAba125, whose genome is AT rich (58 to 62%). The frequencies of transposition of Tn125 observed here were several orders of magnitude higher than, for example, those of the Tn2006 composite transposon and the ISAba1 and IS1999 elements (22, 23); unfortunately, these values are difficult to compare due to different experimental setups.
Two ISs were previously shown to be functional for transposition in A. baumannii, namely, ISAba825 (IS982 family) (24) and ISAba1 (IS4 family) (22, 25). Here, we experimentally demonstrated the functionality of ISAba125 (IS30 family). Consistent with its ability to transpose, ISAba125 was described in association with increased resistance levels to β-lactams in Acinetobacter spp., for example, through duplication of the blaOXA-58 carbapenemase gene (26), disruption of the carO gene encoding an outer membrane protein acting in synergy with other resistance mechanisms (27), or insertion upstream of the blaADC cephalosporinase gene (28). In the sequenced genomes of strains listed in Table 3, ISAba125 was found upstream of the ampC gene in strain ACICU (29) or flanking the aphA6 gene conferring resistance to amikacin in strain BJAB0715 (30).
ISAba125 was present in more than half of the clinical isolates tested (Table 2). In a parallel study, analysis of 131 Acinetobacter spp. revealed that 5 strains had 1 copy, 34 strains had few (2 to 9) copies, 12 strains exhibited numerous (≥10) copies, and 80 strains contained no copy of ISAba125 (31). This high frequency and high copy number of ISAba125 in Acinetobacter spp. are indicative of frequent transposition events, lateral gene transfer within Acinetobacter spp., a replicative mechanism of transposition (as shown for the IS30 family), and the capacity of ISAba125 to expand within genomes, a general feature of ISs (9). Taken together, these characteristics are suggestive of an evolutionary role of ISAba125 customized for Acinetobacter spp., as proposed for ISAba1 in A. baumannii (32, 33).
This work demonstrates the ability of the blaNDM-1-carrying Tn125 transposon to transpose in A. baumannii. It underlines the importance of A. baumannii, which has been considered for years as playing a minor role in medical microbiology. A. baumannii is likely in close contact in the environment with the (still unknown) natural progenitor of the blaNDM-1 gene and may play a key role in the spread of the blaNDM-1 gene to clinically relevant bacterial species, in particular members of the Enterobacteriaceae.
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation (project FNS-31003A_163432) and by the University of Fribourg.
REFERENCES
- 1.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dortet L, Poirel L, Nordmann P. 2014. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int 2014:249856. doi: 10.1155/2014/249856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnin RA, Poirel L, Nordmann P. 2014. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol 9:33–41. doi: 10.2217/fmb.13.69. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Potron A, Poirel L, Nordmann P. 2015. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Pfeifer Y, Wilharm G, Zander E, Wichelhaus TA, Gottig S, Hunfeld KP, Seifert H, Witte W, Higgins PG. 2011. Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. J Antimicrob Chemother 66:1998–2001. doi: 10.1093/jac/dkr256. [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman's guide to bacterial insertion sequences. Microbiol Spectr 3:MDNA3-0030-2014. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 10.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dortet L, Nordmann P, Poirel L. 2012. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob Agents Chemother 56:1693–1697. doi: 10.1128/AAC.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett PM. 2008. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol 153(Suppl 1):S347–S357. doi: 10.1038/sj.bjp.0707607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aranda J, Bardina C, Beceiro A, Rumbo S, Cabral MP, Barbe J, Bou G. 2011. Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J Bacteriol 193:3740–3747. doi: 10.1128/JB.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Zhou Z, Jiang Y, Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother 66:1255–1259. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 17.Diwan V, Tamhankar AJ, Khandal RK, Sen S, Aggarwal M, Marothi Y, Iyer RV, Sundblad-Tonderski K, Stalsby-Lundborg C. 2010. Antibiotics and antibiotic-resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health 10:414. doi: 10.1186/1471-2458-10-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fick J, Soderstrom H, Lindberg RH, Phan C, Tysklind M, Larsson DG. 2009. Contamination of surface, ground, and drinking water from pharmaceutical production. Environ Toxicol Chem 28:2522–2527. doi: 10.1897/09-073.1. [DOI] [PubMed] [Google Scholar]
- 19.Nagel M, Reuter T, Jansen A, Szekat C, Bierbaum G. 2011. Influence of ciprofloxacin and vancomycin on mutation rate and transposition of IS256 in Staphylococcus aureus. Int J Med Microbiol 301:229–236. doi: 10.1016/j.ijmm.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsubo Y, Genka H, Komatsu H, Nagata Y, Tsuda M. 2005. High-temperature-induced transposition of insertion elements in Burkholderia multivorans ATCC 17616. Appl Environ Microbiol 71:1822–1828. doi: 10.1128/AEM.71.4.1822-1828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamidian M, Holt KE, Pickard D, Dougan G, Hall RM. 2014. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J Antimicrob Chemother 69:955–958. doi: 10.1093/jac/dkt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mugnier PD, Poirel L, Nordmann P. 2009. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J Bacteriol 191:2414–2418. doi: 10.1128/JB.01258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aubert D, Naas T, Heritier C, Poirel L, Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of β-lactam resistance genes. J Bacteriol 188:6506–6514. doi: 10.1128/JB.00375-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravasi P, Limansky AS, Rodriguez RE, Viale AM, Mussi MA. 2011. ISAba825, a functional insertion sequence modulating genomic plasticity and blaOXA-58 expression in Acinetobacter baumannii. Antimicrob Agents Chemother 55:917–920. doi: 10.1128/AAC.00491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo HY, Chang KC, Liu CC, Tang CY, Peng JH, Lu CW, Tu CC, Liou ML. 2014. Insertion sequence transposition determines imipenem resistance in Acinetobacter baumannii. Microb Drug Resist 20:410–415. doi: 10.1089/mdr.2014.0004. [DOI] [PubMed] [Google Scholar]
- 26.Evans BA, Hamouda A, Towner KJ, Amyes SG. 2010. Novel genetic context of multiple blaOXA-58 genes in Acinetobacter genospecies 3. J Antimicrob Chemother 65:1586–1588. doi: 10.1093/jac/dkq180. [DOI] [PubMed] [Google Scholar]
- 27.Mussi MA, Limansky AS, Viale AM. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob Agents Chemother 49:1432–1440. doi: 10.1128/AAC.49.4.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopes BS, Amyes SG. 2012. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J Med Microbiol 61:1103–1108. doi: 10.1099/jmm.0.044156-0. [DOI] [PubMed] [Google Scholar]
- 29.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJ, Sicheritz-Ponten T, De Bellis G, Visca P, Cassone A, Carattoli A. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother 52:2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu L, Yan Z, Zhang Z, Zhou Q, Zhou J, Wakeland EK, Fang X, Xuan Z, Shen D, Li QZ. 2013. Complete genome analysis of three Acinetobacter baumannii clinical isolates in China for insight into the diversification of drug resistance elements. PLoS One 8:e66584. doi: 10.1371/journal.pone.0066584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon EJ, Goussard S, Touchon M, Krizova L, Cerqueira G, Murphy C, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P. 2014. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3′)-VI. mBio 5:e01972-14. doi: 10.1128/mBio.01972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segal H, Garny S, Elisha BG. 2005. Is IS(ABA-1) customized for Acinetobacter? FEMS Microbiol Lett 243:425–429. doi: 10.1016/j.femsle.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Heritier C, Poirel L, Nordmann P. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect 12:123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 34.Mugnier PD, Poirel L, Naas T, Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis 16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figueiredo S, Poirel L, Papa A, Koulourida V, Nordmann P. 2009. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob Agents Chemother 53:4045–4047. doi: 10.1128/AAC.00292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirel L, Corvec S, Rapoport M, Mugnier P, Petroni A, Pasteran F, Faccone D, Galas M, Drugeon H, Cattoir V, Nordmann P. 2007. Identification of the novel narrow-spectrum β-lactamase SCO-1 in Acinetobacter spp. from Argentina. Antimicrob Agents Chemother 51:2179–2184. doi: 10.1128/AAC.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figueiredo S, Bonnin RA, Poirel L, Duranteau J, Nordmann P. 2012. Identification of the naturally occurring genes encoding carbapenem-hydrolysing oxacillinases from Acinetobacter haemolyticus, Acinetobacter johnsonii, and Acinetobacter calcoaceticus. Clin Microbiol Infect 18:907–913. doi: 10.1111/j.1469-0691.2011.03708.x. [DOI] [PubMed] [Google Scholar]
- 38.Tada T, Miyoshi-Akiyama T, Shimada K, Shimojima M, Kirikae T. 2014. Dissemination of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and emergence of OXA-72 carbapenemase coproducers in Japan. Antimicrob Agents Chemother 58:2916–2920. doi: 10.1128/AAC.01212-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ou HY, Kuang SN, He X, Molgora BM, Ewing PJ, Deng Z, Osby M, Chen W, Xu HH. 2015. Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci Rep 5:8643. doi: 10.1038/srep08643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merino M, Poza M, Roca I, Barba MJ, Sousa MD, Vila J, Bou G. 2014. Nosocomial outbreak of a multiresistant Acinetobacter baumannii expressing OXA-23 carbapenemase in Spain. Microb Drug Resist 20:259–263. doi: 10.1089/mdr.2013.0127. [DOI] [PubMed] [Google Scholar]
- 41.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CC, Lin YC, Sheng WH, Chen YC, Chang SC, Hsia KC, Liao MH, Li SY. 2011. Genome sequence of a dominant, multidrug-resistant Acinetobacter baumannii strain, TCDC-AB0715. J Bacteriol 193:2361–2362. doi: 10.1128/JB.00244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]