Abstract
Topical antimicrobials are often employed for decolonization and infection prevention and may alter the endogenous microbiota of the skin. The objective of this study was to compare the microbial communities and levels of richness and diversity in community-dwelling subjects and intensive care unit (ICU) patients before and after the use of topical decolonization protocols. We enrolled 15 adults at risk for Staphylococcus aureus infection. Community subjects (n = 8) underwent a 5-day decolonization protocol (twice daily intranasal mupirocin and daily dilute bleach-water baths), and ICU patients (n = 7) received daily chlorhexidine baths. Swab samples were collected from 5 anatomic sites immediately before and again after decolonization. A variety of culture media and incubation environments were used to recover bacteria and fungi; isolates were identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry. Overall, 174 unique organisms were recovered. Unique communities of organisms were recovered from the community-dwelling and hospitalized cohorts. In the community-dwelling cohort, microbial richness and diversity did not differ significantly between collections across time points, although the number of body sites colonized with S. aureus decreased significantly over time (P = 0.004). Within the hospitalized cohort, richness and diversity decreased over time compared to those for the enrollment sampling (from enrollment to final sampling, P = 0.01 for both richness and diversity). Topical antimicrobials reduced the burden of S. aureus while preserving other components of the skin and nasal microbiota.
INTRODUCTION
Nosocomial infections pose significant clinical and financial burdens to patients and health care systems (1, 2). Colonization with potential pathogens serves as an endogenous source of infection (e.g., central line-associated bloodstream infection with Staphylococcus aureus) (3). For hospitalized patients, particularly those in intensive care units (ICUs), decolonization with topical antimicrobials, including chlorhexidine and/or mupirocin, has been demonstrated to reduce the acquisition of antibiotic-resistant microorganisms and the incidence of hospital-acquired infections (4–6). While decolonization has traditionally been employed in health care settings, the emergence of methicillin-resistant S. aureus (MRSA) in the community and the resultant epidemic of skin and soft tissue infections (SSTIs) have led to the extrapolation of decolonization to outpatients to prevent recurrent SSTIs (7–10). As these broad-spectrum therapies are not pathogen specific (e.g., for MRSA or Enterobacteriaceae), these agents may suppress or eliminate other organisms on the skin and nasal mucosa, thereby potentially disrupting the balance of the microbiota, an important component of host defense against pathogenic organisms (11, 12). Indeed, this dysbiosis has been demonstrated for the intestinal microbiota following administration of oral antibiotics (13, 14).
Culture-independent molecular methods, including 16S rRNA gene sequencing and metagenomic sequence analysis, have become common approaches in studies evaluating the microbiome (15–18). While microbial culture has traditionally been regarded as insensitive for detecting a majority of the microbiota, molecular methods have informed augmented culture approaches, yielding enhanced recovery of these microorganisms (19–26). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a rapid and cost-effective technique that utilizes proteomic profiling for organism identification (27). MALDI-TOF MS facilitates the field of “culturomics,” which employs a variety of conditions for microbial cultivation, followed by identification by mass spectrometry (24). MALDI-TOF MS provides excellent species-level resolution, even for closely related Gram-positive bacteria that are major contributors to the skin microbiota. This process supplants traditional limitations of 16S rRNA gene sequencing for closely related species.
Using a culturomic approach, our objective was to discern the identity and diversity of microorganisms recovered from multiple anatomic niches in individuals at risk for infection with MRSA. As a myriad of factors influence the skin and nasal microbiota, many of which differ between outpatients and hospitalized patients, we enrolled two distinct cohorts (community-dwelling individuals and hospitalized individuals in an ICU) for this investigation. Additionally, the decolonization regimens prescribed can vary. Thus, within cohorts, we aimed to compare the richness and diversity of microorganisms within individuals prior to and following the use of topical antimicrobial decolonization measures.
MATERIALS AND METHODS
Study populations.
Two participant cohorts comprised of individuals at risk for MRSA infection were enrolled. The community-dwelling cohort consisted of 8 adults living with a child (aged 6 months to 13 years) recently treated for a MRSA SSTI (these individuals represent a subset of participants in a study of MRSA transmission dynamics among households of children with MRSA infections [28]). Community-dwelling individuals completed a 5-day decolonization protocol with twice-daily application of 2% mupirocin (Teva Pharmaceuticals USA) to the anterior nares and daily bathing in dilute bleach water (1/4 cup of bleach [Clorox; The Clorox Company] to 1/4 bathtub-full of water), with soaking for ≥15 min (8, 29). The hospitalized cohort consisted of 7 adults in the 15-bed cardiac care ICU at Barnes-Jewish Hospital (BJH) in St. Louis, MO. As a component of routine clinical care, BJH ICU patients receive daily body washes from the neck down with chlorhexidine-based soap (Exidine 4%; Cardinal Health). Specifically, one 4-ounce bottle of 4% chlorhexidine-based soap is added to 4 quarts of water in a bath basin (to a final concentration of 0.125%, which has been proven to be effective at preventing MRSA transmission and infection in ICUs [30]); this solution is applied to the patient with a wash cloth and allowed to air dry (30, 31). These chlorhexidine baths are discontinued upon transfer to the general medicine unit. Although application of nasal mupirocin is not routinely prescribed in the BJH ICU, two patients received one dose each of preoperative mupirocin prophylaxis. A standardized case report form was used to collect participant demographics, health history, antibiotic use, and hygiene factors. Study procedures were approved by the Washington University Institutional Review Board, and participants provided written informed consent.
Specimen collection and processing.
A standardized culturing protocol (BD Eswab; Becton Dickinson) was employed by study personnel to obtain samples from five anatomic sites, including the anterior nares and the subject's dominant-side axilla, inguinal fold, dorsal surface of the forearm, and ventral surface of the lower leg. For all sites except the anterior nares, a 2-in. by 2-in. (5.1 × 5.1 cm) area was swabbed using a collection template, with application of consistent pressure. Community-dwelling participants were sampled at three time points: day 0 (prior to initiating the 5-day decolonization protocol described above), day 5 (immediately after performing the decolonization protocol), and day 30 (1 month after completing the decolonization protocol). Hospitalized patients were sampled up to five times (in the ICU or on the general medicine unit), depending on their length of hospitalization, as follows: day 0 (upon ICU admission, prior to the first chlorhexidine bath), day 3, day 7, upon discharge from the ICU, and upon discharge from the hospital.
Microorganism identification and characterization.
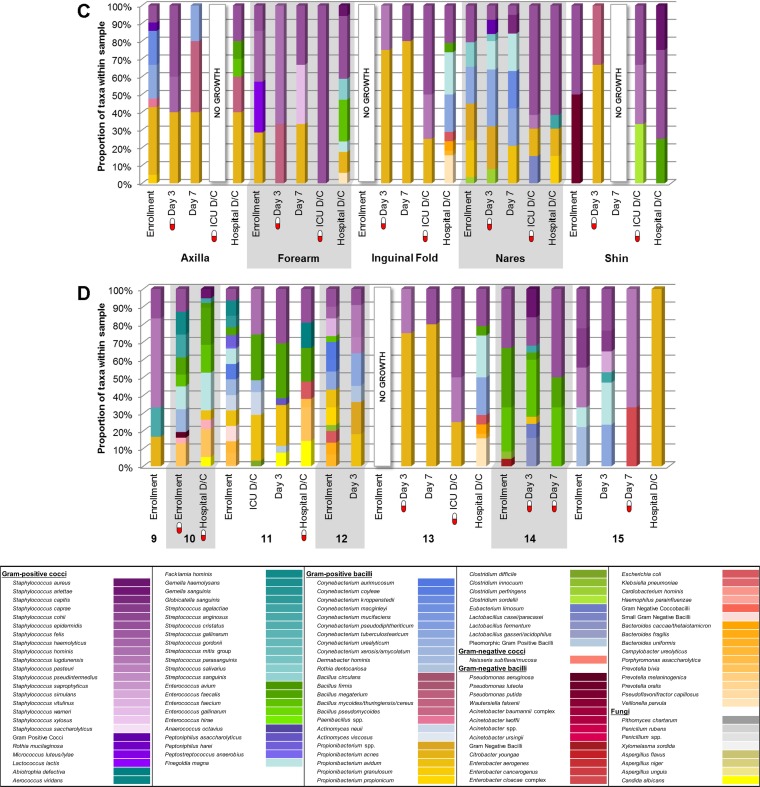
After vortexing, each Eswab eluate was inoculated to 9 selective and differential media and incubation environments, selected to support the growth of organisms anticipated to be recovered from the nares and skin (see Table S1 in the supplemental material) (11, 24, 32). A four-quadrant streaking method was employed. The relative abundance of each morphotype recovered in culture was approximated as follows: 0.5+, growth in broth only; 1+, rare; 2+, few; 3+, moderate; and 4+, abundant. Distinct morphotypes were selected for MALDI-TOF MS species identification as previously described (33–37). In brief, yeast and bacteria were routinely identified using the Vitek MS IVD v2.0 Knowledge Base according to the manufacturer's protocols. Any isolates not identified using this method were analyzed using the Vitek MS SARAMIS database. For a very small number of isolates, this approach was not successful and the isolate was analyzed using the Bruker Biotyper RUO database. Any isolate that remained unidentified was assigned a descriptive identity based on the Gram stain reaction and growth conditions. Filamentous fungi were identified using the Vitek MS v3.0 Knowledge Base following extraction and inactivation; any isolate that was unidentified using this method was identified using sequence-based analysis.
Disk diffusion testing was performed on all S. aureus isolates to detect resistance to cefoxitin (as an indicator of methicillin resistance) and mupirocin (38). Multiplex PCR was performed to detect mupA and qacA/B, conferring high-level mupirocin resistance and chlorhexidine tolerance, respectively, as described elsewhere (39).
Statistical analysis.
Richness is defined as the number of different microbial species identified at a given collection site and time point (19, 32). Diversity is defined as a measure of community composition accounting for the relative abundances of species as determined by Shannon's diversity index (which considers species richness and evenness) (32, 40). Shannon's diversity index was calculated based on the relative abundance of each morphotype recovered in culture.
To evaluate trends in richness over time and across body sites, with accounting for correlation within subjects, repeated-measures GEE models assuming a Poisson distribution were used; similarly, for Shannon's diversity index, repeated-measures mixed models assuming a normal distribution were deployed. To assess the correlation between S. aureus and other relevant bacteria, with accounting for within-subject correlation, repeated-measures log-linear models were utilized, with adjustments for body site and time. In log-linear models, the presence of the two taxa is assessed simultaneously, with checks for the effect of the presence of one on the presence of the other. To compare skin microbial compositions at different time points, the numbers of body sites colonized with various organisms per subject over time were analyzed using related-samples Friedman's two-way analysis of variance by ranks. Analyses were performed with R statistical software, version 3.2, and SPSS 22 for Windows (IBM SPSS, Chicago, IL).
RESULTS
Participants and microbiology.
The overall study population was comprised of 15 participants with a median age of 43 years (range, 26 to 66 years). Ten (67%) participants were female, seven (47%) were Caucasian, and six (40%) had experienced an SSTI in the previous year.
Overall, 174 unique organisms representing 65 genera and 158 species were identified: 137 organisms from the community-dwelling cohort and 99 from the hospitalized cohort. Sixty-two (36%) of the organisms were shared between the two cohorts; 75 (43%) were unique to the community-dwelling cohort (notably Klebsiella pneumoniae and Staphylococcus pseudintermedius), and 37 (21%) were unique to the hospitalized cohort (notably Clostridium difficile and Pseudomonas aeruginosa). In addition to the Gram-positive organisms we expected to find (e.g., Staphylococcus spp., Streptococcus spp., Corynebacterium spp., and Propionibacterium spp.), many participants were also colonized with enteric Gram-negative bacteria and fungi (e.g., Aspergillus spp. and dematiaceous fungi) (Table 1). The most commonly recovered taxa were Staphylococcus epidermidis, Staphylococcus aureus, Propionibacterium acnes, Corynebacterium tuberculostearicum, Propionibacterium avidum, Enterococcus faecalis, Staphylococcus hominis, Bacillus spp., Finegoldia magna, and Staphylococcus lugdunensis. Twenty species of Staphylococcus, 4 species of Propionibacterium, and 14 species of Corynebacterium were identified. Anaerobic bacteria were recovered from all 15 subjects and 168 of 225 (75%) sites sampled.
TABLE 1.
Comparison of skin microbial compositions at different time pointsa
| Organism | Median no. (IQR) [range] of body sites colonized per subject |
|||||
|---|---|---|---|---|---|---|
| Community-dwelling cohort |
Hospitalized cohort |
|||||
| Enrollment (n = 8) | Day 5 (n = 8) | Day 30 (n = 8) | Enrollment (n = 7) | ICU (n = 5) | General medicine unit (final) (n = 5) | |
| Fungi | 0 (0–1.75) [0–3] | 0 (0–1.5) [0–3] | 0 (0–0) [0–3] | 0 (0–2) [0–2] | 0 (0–1) [0–2] | 0 (0–1) [0–1] |
| Aspergilli | 0 (0–0.75) [0–1] | 0 (0–0.75) [0–1] | 0 (0–0) [0–1] | 0 (0–0) [0–2] | 0 (0–0) [0–0] | 0 (0–0) [0–0] |
| Black mold | 0 (0–0) [0–3] | 0 (0–0) [0–1] | 0 (0–0) [0–1] | 0 (0–0) [0–1] | 0 (0–0) [0–0] | 0 (0–0) [0–0] |
| Yeast | 0 (0–0) [0–0] | 0 (0–0) [0–0] | 0 (0–0) [0–0] | 0 (0–0) [0–1] | 0 (0–1) [0–2] | 0 (0–1) [0–1] |
| Bacteria | 5 (5–5) [4–5] | 5 (5–5) [4–5] | 5 (5–5) [5–5] | 5 (5–5) [4–5] | 5 (4–5) [3–5] | 5 (5–5) [5–5] |
| Gram-positive rods | 5 (4–5) [3–5] | 5 (3.5–5) [2–5] | 4.5 (4–5) [3–5] | 4 (2–5) [2–5] | 3 (2.5–5) [2–5] | 4 (1.5–4.5) [1–5] |
| Gram-positive cocci | 5 (5–5) [4–5] | 5 (4.25–5) [4–5] | 5 (5–5) [4–5] | 5 (5–5) [4–5] | 5 (3.5–5) [3–5] | 5 (4–5) [4–5] |
| Aerobes | 5 (5–5) [4–5] | 5 (4.25–5) [4–5] | 5 (5–5) [5–5] | 5 (5–5) [4–5] | 5 (4–5) [3–5] | 5 (4–5) [4–5] |
| Gram-negative aerobes | 1.5 (0–2) [0–5] | 1 (0–1.75) [0–5] | 1 (0–1) [0–2] | 1 (0–4) [0–5] | 0 (0–2.5) [0–3] | 1 (0.5–1.5) [0–2] |
| Enterobacteriaceae | 0 (0–1.75) [0–3] | 0.5 (0–1) [0–2] | 0 (0–0.75) [0–1] | 0 (0–1) [0–2] | 0 (0–1.5) [0–2] | 1 (0.5–1.5) [0–2] |
| Nonfermenting bacteria | 0 (0–0) [0–0] | 0 (0–0) [0–1] | 0 (0–0.75) [0–2] | 0 (0–1) [0–5] | 0 (0–0.5) [0–1] | 0 (0–0) [0–0] |
| Fastidious bacteria | 0.5 (0–1) [0–2] | 0 (0–0) [0–1] | 0 (0–0) [0–1] | 0 (0–1) [0–1] | 0 (0–0) [0–0] | 0 (0–0.5) [0–1] |
| Gram-positive aerobes | 5 (5–5) [4–5] | 5 (4.25–5) [4–5] | 5 (5–5) [5–5] | 5 (5–5) [4–5] | 5 (4–5) [3–5] | 5 (4–5) [4–5] |
| Cocci | 5 (5–5) [4–5] | 5 (4.25–5) [4–5] | 5 (5–5) [4–5] | 5 (5–5) [4–5] | 5 (3.5–5) [3–5] | 5 (4–5) [4–5] |
| S. aureus | 3 (2.25–4) [1–4] | 1.5 (1–3.5) [1–5] | 0.5 (0–1.75)b [0–3] | 0 (0–2) [0–2] | 2 (0–4.5) [0–5] | 2 (0–3) [0–3] |
| Other Staphylococcus spp. | 5 (4.25–5) [4–5] | 5 (4.25–5) [4–5] | 5 (4.25–5) [4–5] | 5 (5–5) [4–5] | 5 (3.5–5) [3–5] | 5 (4–5) [4–5] |
| Enterococcus spp. | 0 (0–0.75) [0–3] | 0 (0–2.5) [0–3] | 1.5 (0–2) [0–4] | 2 (0–3) [0–4] | 4 (0–4) [0–4] | 3 (0–4.5) [0–5] |
| Streptococcus-like bacteria | 1 (1–1.75) [0–3] | 1 (0.25–2.75) [0–4] | 2 (1–2) [0–3] | 1 (0–3) [0–3] | 1 (0–2) [0–3] | 1 (0–1.5) [0–2] |
| Rods | 2.5 (2–4) [1–5] | 3.5 (2–4.75) [1–5] | 3.5 (2.25–4.75) [2–5] | 3 (2–4) [0–5] | 2 (2–4) [2–5] | 2 (0.5–2) [0–2] |
| Corynebacterium spp. | 1.5 (0.25–4) [0–5] | 2 (1–4) [0–4] | 2.5 (1.25–4) [1–4] | 3 (2–4) [0–5] | 1 (0.5–3) [0–4] | 1 (0.5–1.5) [0–2] |
| Spore-forming bacteria | 1 (1–2) [0–5] | 2 (0.25–3.75) [0–5] | 2.5 (1–4.5) [0–5] | 0 (0–1) [0–1] | 1 (0.5–2) [0–2] | 0 (0–1) [0–1] |
| Bacillus spp. | 1 (1–2) [0–5] | 2 (0.25–3.75) [0–5] | 2.5 (1–4.5) [0–5] | 0 (0–0) [0–1] | 1 (0.5–2) [0–2] | 0 (0–1) [0–1] |
| Other spore-forming bacteria | 0 (0–0) [0–1] | 0 (0–0) [0–1] | 0 (0–0) [0–0] | 0 (0–1) [0–1] | 0 (0–0.5) [0–1] | 0 (0–0) [0–0] |
| Anaerobes | 4 (3.25–4.75) [2–5] | 5 (3.25–5) [2–5] | 3.5 (3–5) [1–5] | 4 (3–4) [2–5] | 4 (2–4.5) [1–5] | 4 (2.5–4.5) [1–5] |
| Clostridium/spore-forming bacteria | 0 (0–0) [0–0] | 0 (0–0) [0–1] | 0 (0–0) [0–2] | 0 (0–1) [0–1] | 0 (0–1) [0–1] | 0 (0–0.5) [0–1] |
| Propionibacterium spp. | 4 (2.25–4) [2–5] | 3.5 (2–4.75) [1–5] | 3 (1.5–3.75) [1–4] | 2 (1–4) [0–5] | 1 (1–3.5) [1–4] | 3 (0.5–4.5) [0–5] |
| Other non-spore-forming bacteria | 2 (0.25–2.75) [0–3] | 2 (0.25–4.75) [0–5] | 2 (0.5–3) [0–3] | 1 (0–4) [0–4] | 3 (0.5–4) [0–5] | 1 (0–2.5) [0–3] |
Abbreviations: IQR, interquartile range (25th to 75th percentiles); ICU, intensive care unit.
P = 0.004 across samplings within community-dwelling participants by related-samples Friedman's two-way analysis of variance by ranks; for the remaining organisms, there was not a statistically significant change in the number of body sites colonized over time.
Characteristics and microbiota of community-dwelling participants.
The community-dwelling cohort (n = 8) had a median age of 33 years (range, 26 to 43 years). Five (63%) participants were female, five (63%) were Caucasian, and five (63%) had experienced an SSTI in the previous year. Two (25%) community-dwelling participants had taken systemic antibiotics (ciprofloxacin and trimethoprim-sulfamethoxazole) in the month prior to study enrollment; two participants were taking systemic antibiotics at the time of sample collection (ciprofloxacin and trimethoprim-sulfamethoxazole) (indicated by a capsule graphic in the figures).
For community-dwelling participants, the median number of distinct taxa (i.e., richness) across all body sites combined was 21 (interquartile range [IQR], 13.75 to 26.5) at enrollment, 17.5 (IQR, 15.25 to 26.75) at day 5, and 20 (IQR, 14.25 to 26) at day 30 (Table 2). Richness did not differ significantly between sample collections across time points (P = 0.48) (see Fig. S1 in the supplemental material). Richness did differ across body sites (P < 0.01), with the largest number of taxa recovered from the inguinal fold and the lowest from the axilla. Specifically, the inguinal fold richness was significantly greater than that of the axilla, anterior nares, and leg (P < 0.01 for each comparison).
TABLE 2.
Sample richness at each body site over timea
| Site | Median no. of species (IQR) [range] |
|||||
|---|---|---|---|---|---|---|
| Community-dwelling cohort |
Hospitalized cohort |
|||||
| Enrollment (n = 8) | Day 5 (n = 8) | Day 30 (n = 8) | Enrollment (n = 7) | ICU (n = 5) | General medicine unit (final) (n = 5) | |
| Axilla | 5 (3.25–5) [1–11] | 4 (2–5.75) [0–8] | 3.5 (1.25–5.75) [1–7] | 5 (3–7) [2–10] | 5 (3–6.5) [3–7] | 4 (2–6.5) [1–8] |
| Forearm | 8 (6.25–9) [6–10] | 7.5 (6–10.25) [4–13] | 6.5 (5.25–9.75) [4–15] | 6 (5–7) [4–10] | 5 (1–6.5) [0–7] | 4 (3–6.5) [2–7] |
| Inguinal fold | 10 (4.75–13) [4–15] | 8 (5.25–11) [4–13] | 7 (6.25–10.75) [6–14] | 5 (4–13) [0–15] | 6 (2–9) [2–10] | 7 (2–8) [1–9] |
| Nares | 5.5 (4.25–8) [1–10] | 7 (3.75–9.25) [3–10] | 7 (4.5–9.5) [4–10] | 6 (5–7) [1–7] | 7 (4.5–7) [4–7] | 4 (2.5–7) [1–8] |
| Shin | 7 (5.25–8) [3–10] | 6.5 (2.25–10.25) [2–11] | 6 (4.25–6.75) [1–9] | 6 (4–6) [2–9] | 3 (1–6.5) [0–8] | 3 (1.5–3) [1–3] |
| All sites | 21 (13.75–26.5) [11–27] | 17.5 (15.25–26.75) [14–34] | 20 (14.25–26) [11–28] | 16 (14–24) [10–29] | 12 (10–17.5) [9–20] | 15 (7.5–15.5) [5–16] |
Abbreviations: IQR, interquartile range (25th to 75th percentiles); ICU, intensive care unit. Note that within the community-dwelling cohort, richness did not differ significantly between collections across time points in a repeated-measures model (P = 0.48). Within the hospitalized cohort, richness decreased over time compared to that for the enrollment sampling, with a trend for lower richness at the second ICU sampling (after initiation of chlorhexidine bathing; P = 0.08) and significantly lower richness at the third sampling (after discharge from the ICU to the general medicine unit; P = 0.01).
For community-dwelling participants, the mean (± standard deviation) Shannon's diversity index across all body sites combined was 1.71 (±0.55) at enrollment, 1.65 (±0.52) at day 5, and 1.61 (±0.61) at day 30 (Table 3). Diversity did not differ significantly between samplings across time points (P = 0.71) (see Fig. S2 in the supplemental material). The inguinal fold had the most diversity, which was significantly greater than that of the axilla and anterior nares (P < 0.01 and P = 0.05, respectively); there was a trend for greater diversity in the inguinal folds than on the shin (P = 0.08). The axilla had the least diversity.
TABLE 3.
Sample diversity at each body site over timea
| Site | Mean Shannon's diversity index ± SD |
|||||
|---|---|---|---|---|---|---|
| Community-dwelling cohort |
Hospitalized cohort |
|||||
| Enrollment (n = 8) | Day 5 (n = 8) | Day 30 (n = 8) | Enrollment (n = 7) | ICU (n = 5) | General medicine unit (final) (n = 5) | |
| Axilla | 1.31 ± 0.63 | 1.26 ± 0.46 | 0.98 ± 0.70 | 1.43 ± 0.46 | 1.45 ± 0.43 | 1.17 ± 0.74 |
| Forearm | 1.99 ± 0.20 | 1.90 ± 0.38 | 1.88 ± 0.41 | 1.74 ± 0.26 | 1.43 ± 0.54 | 1.33 ± 0.42 |
| Inguinal fold | 2.03 ± 0.49 | 1.87 ± 0.39 | 2.00 ± 0.33 | 1.92 ± 0.60 | 1.34 ± 0.69 | 1.34 ± 0.84 |
| Nares | 1.44 ± 0.63 | 1.62 ± 0.45 | 1.69 ± 0.38 | 1.45 ± 0.64 | 1.60 ± 0.31 | 1.21 ± 0.75 |
| Shin | 1.79 ± 0.37 | 1.56 ± 0.69 | 1.50 ± 0.64 | 1.58 ± 0.45 | 1.31 ± 0.57 | 0.76 ± 0.46 |
| All sites | 1.71 ± 0.55 | 1.65 ± 0.52 | 1.61 ± 0.61 | 1.61 ± 0.50 | 1.43 ± 0.48 | 1.16 ± 0.64 |
Abbreviations: SD, standard deviation; ICU, intensive care unit. Note that within the community-dwelling cohort, diversity did not differ significantly between samplings across time points (P = 0.71) in a repeated-measures model. Within the hospitalized cohort, diversity decreased over time, with a trend for less diversity at the second ICU collection (after initiation of chlorhexidine bathing; P = 0.09) and significantly less diversity at the third collection (after ICU discharge to the general medicine unit; P = 0.01).
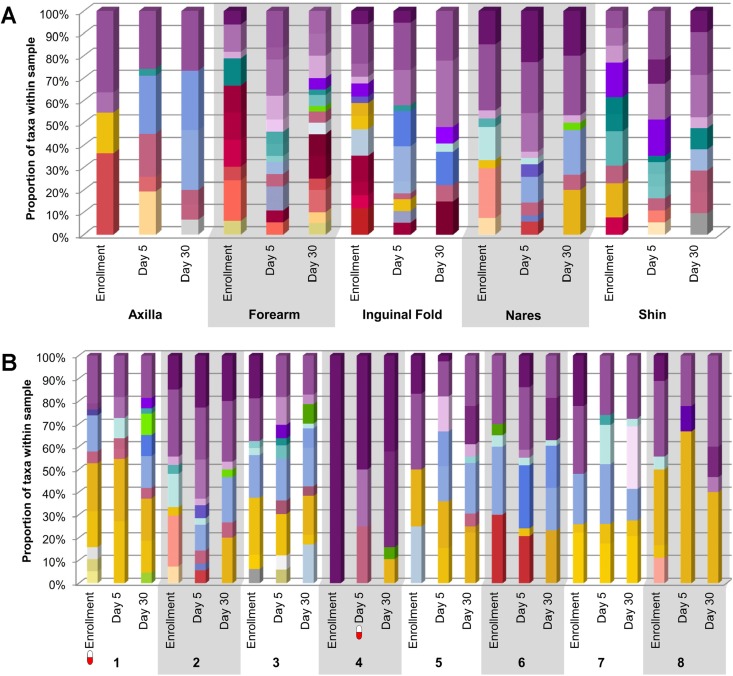
The composition of skin flora for each body site over time is displayed in Fig. 1A for one representative community-dwelling participant. The core Gram-positive flora (e.g., coagulase-negative staphylococci and Corynebacterium spp.) were relatively consistent over time and across body sites, while the accessory bacteria (e.g., Enterobacteriaceae, enterococci, and anaerobes) were more variable. The composition of nasal flora for all community-dwelling participants over time is displayed in Fig. 1B. While the core Gram-positive organisms within the nares were well represented across all individuals, the accessory bacteria were more similar within individuals over time than between individuals.
FIG 1.
Skin microbiota. The height of each color within each bar represents the relative abundance of the corresponding taxon within each sample. A red and white “capsule” symbol represents a participant who was taking systemic antibiotics at the time of sampling. No growth, no organisms were cultivated from the sample. D/C, discharge. (A) Community-dwelling participant (ID 2). The data show the skin microbiota at each body site and time point for one community-dwelling participant. (B) Community-dwelling cohort anterior nares. The data show the nasal microbiota for all community-dwelling participants at each time point. (C) Hospitalized patient (ID 13). The data show the skin microbiota at each body site and time point for one hospitalized patient. (D) Hospitalized cohort inguinal folds. The data show the inguinal fold microbiota for all hospitalized patients at each time point.
Characteristics and microbiota of hospitalized participants.
The hospitalized cohort (n = 7) had a median age of 56 years (range, 47 to 66 years). Five (71%) patients were female, two (29%) were Caucasian, and one (14%) had experienced an SSTI in the previous year. Four (57%) hospitalized patients had taken systemic antibiotics in the month prior to study enrollment; five patients were receiving at least one systemic antibiotic (up to four antibiotics per sampling) at ≥1 sampling time points (indicated by a capsule graphic in the figures).
For the hospitalized cohort, the median richness over all body sites combined was 16 (IQR, 14 to 24) at enrollment, 12 (IQR, 10 to 17.5) at the second ICU sampling, and 15 (IQR, 7.5 to 15.5) after discharge to the general medicine unit (Table 2). Within the hospitalized cohort, richness decreased over time compared to that of the enrollment sampling, with a trend for lower richness at the second ICU sampling (after initiation of chlorhexidine bathing; P = 0.08) and significantly lower richness at the third sampling (after discharge from the ICU to the general medicine unit; P = 0.01) (see Fig. S3 in the supplemental material). There was a trend for differing richness levels across body sites overall (P = 0.06); the inguinal fold again had the highest richness, and the shin had the lowest richness.
For the hospitalized cohort, the mean (± standard deviation) Shannon's diversity index over all body sites combined was 1.61 (±0.50) at enrollment, 1.43 (±0.48) at the second ICU sampling, and 1.16 (±0.64) after discharge to the general medicine unit (Table 3). Diversity decreased over time, with a trend for less diversity at the second ICU collection (after initiation of chlorhexidine bathing; P = 0.09) and significantly less diversity at the third collection (after ICU discharge to the general medicine unit; P = 0.01) (see Fig. S4 in the supplemental material). The inguinal fold had the most diversity and the shin the least diversity, though these differences were not statistically significant.
The composition of skin flora for each body site over time is displayed in Fig. 1C for one representative hospitalized patient; while the overall Staphylococcus spp. were maintained across anatomic sites and time, the accessory microbiota were less variable over time than those in the community-dwelling participant (Fig. 1A). Note that Propionibacterium spp. comprised a larger portion of the microbiota in the hospitalized patient than in the community participant (Fig. 1C and A, respectively). The composition of inguinal fold flora for all hospitalized participants over time is displayed in Fig. 1D.
Relationships between S. aureus and other microbes.
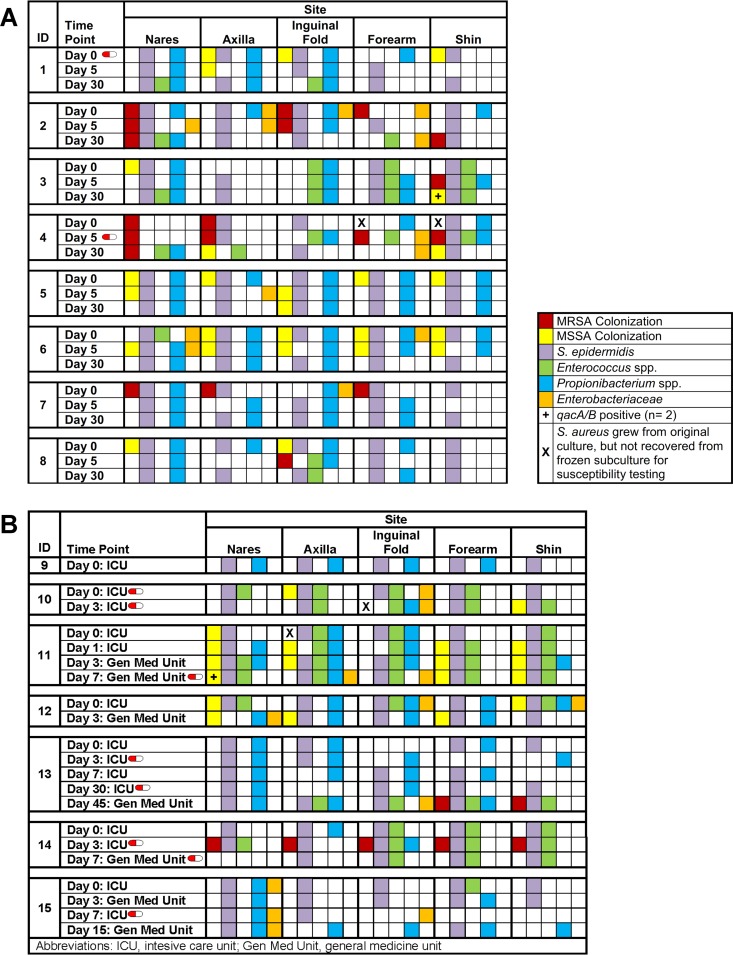
S. aureus was recovered from 13 participants (all eight community-dwelling and five hospitalized subjects) at ≥1 time points over the study period. Of the 71 S. aureus isolates available for susceptibility testing, 25 (35%) isolates (from seven participants) were MRSA. None of the isolates exhibited mupirocin resistance; two isolates, both of which were methicillin-susceptible S. aureus (MSSA) (one from a community-dwelling and one from a hospitalized participant), possessed genes conferring chlorhexidine tolerance (qacA/B) (Fig. 2). Of six community-dwelling participants colonized with S. aureus in the nares at enrollment, two (colonized with MRSA) remained colonized at all three samplings despite application of mupirocin, while four participants (one with MRSA and three with MSSA) were decolonized (Fig. 2A).
FIG 2.
Presence of S. aureus, S. epidermidis, Enterococcus spp., Propionibacterium spp., and Enterobacteriaceae at each body site over time. A red and white “capsule” symbol represents a participant who was taking systemic antibiotics at the time of sampling. (A) Community-dwelling cohort. (B) Hospitalized cohort.
Microbial richness and diversity within each cohort over time and across body sites (detailed above) were also evaluated in the context of S. aureus; adjusting for the presence of S. aureus did not affect the richness or diversity models. The relationships between S. aureus and other relevant organisms (i.e., inhibition of or coexistence with S. aureus by other components of the microbiota) were evaluated, with adjustments for multiple body sites within an individual across all time points. In both community-dwelling and hospitalized populations, the presence of S. aureus was independent of colonization with S. epidermidis, Propionibacterium spp., Enterococcus spp., or Enterobacteriaceae. The presence of these organisms for each participant at each body site over time is depicted in Fig. 2. In both cohorts, colonization with these organisms was relatively consistent over time, despite decolonization efforts and, for some participants, administration of systemic antibiotics. Note that in the community-dwelling cohort, the number of body sites colonized with S. aureus significantly decreased over time (P = 0.004) (Table 1 and Fig. 2A).
DISCUSSION
Given the effectiveness of decolonization in infection prevention across broad study populations, the practice of topical antimicrobial application has become common in health care and community settings (4, 5, 8, 9). While the outcomes of these decolonization studies have focused on reducing the colonization burden and infection incidence with a specific organism, the overall community composition of the skin microbiota has not been evaluated. These topical antimicrobials may result in a modulation of the normal skin microbiota, accommodating both the presence and prosperity of potential pathogens. Furthermore, a reduction in microbiome diversity has been associated with inflammatory skin disorders (41).
Although many studies evaluating microbial communities have applied a genomic approach, culturomics is emerging as a robust technique for detailed microbial identification to the species level (19–26). As demonstrated herein, this level of detail is especially important for the skin and nasal flora, where closely related Gram-positive species predominate. Sequencing reference databases are less comprehensive for taxa common to these body niches than for taxa comprising the gut microbiota. Furthermore, depending on the specific sequencing technique used and/or the portion of the 16S rRNA gene evaluated, important taxonomic groups, such as Propionibacterium spp., may be underestimated (16). Using our culturomic approach, we were able to recover and identify a wide variety of bacteria with high species-level resolution, including 20 staphylococcal species and 14 species of Corynebacterium. Additionally, we recovered fungi from several participants, which would not have been identified by 16S rRNA gene sequencing. With that said, the genera commonly recovered in this study mirror those frequently encountered using 16S rRNA gene sequencing or metagenomic approaches to interrogation of the skin microbiota (16, 17, 32).
In the present study, unique communities of organisms were recovered from the hospitalized and community participants, which is an important consideration in interpreting similar investigations, as findings from one population may not be generalizable. In our cohort of hospitalized patients who were bathed daily with chlorhexidine, the microbial richness and diversity decreased over time, a finding which may be affected by use of broad-spectrum systemic antibiotics and medical and surgical interventions occurring concomitantly with topical antisepsis. Similarly, in a study of adult ICU patients in France, microbial diversity was lower among patients receiving chlorhexidine body washes than among those bathed with soap and water (19). Among the members of our community-dwelling cohort, although adherence to the prescribed decolonization protocol was high (data not shown), the richness and diversity of the microbiota did not change significantly upon performance of the 5-day decolonization regimen with intranasal mupirocin application and bleach-water baths, although it is important that these indices do not discern the specific organisms present. Additionally, while the burden of S. aureus (i.e., number of body sites colonized) was reduced following decolonization, topical antimicrobials did not eradicate or significantly reduce the burden of other components of the microbiota.
In both the community-dwelling and hospitalized cohorts, microbial richness and diversity differed significantly between anatomic niches. In the community-dwelling population, the axilla yielded the lowest richness and diversity (which may perhaps be attributed to additional exogenous factors, such as deodorant use), while the shin was the site with the lowest richness and diversity in the hospitalized cohort. Among both cohorts, the inguinal fold consistently maintained the highest richness and diversity, which may be reflective of its proximity to the perirectal area as well as the temperature and humidity in this region (32). Note that enteric Gram-negative rods were found not only in the inguinal fold but also at all body sites. Overall, our data suggest that even following decolonization, an individual's skin microbiota is a “personal trait” and is temporally stable, similar to recent findings by Oh and colleagues (17). Thus, over time, the microbiota of the skin of an individual is more similar to “self” than to “others.”
This study has numerous strengths, such as the inclusion of both hospitalized and community-dwelling individuals, with multiple longitudinal samplings, and the robust number of taxa identified to the species level, a degree of granularity that is imperative for assessing the skin microbiota. However, this study is not without limitations, including the limited sample size, the application of different decolonization regimens between the cohorts (which may have differential activity on the microbiota and thus precludes comparisons between cohorts), and the variable number of samplings obtained from hospitalized patients.
As endogenous microbial communities exist in a delicate balance within a niche, many microorganisms produce toxins and bacteriocins or augment host immune responses to ward off potentially pathogenic organisms. We were thus encouraged by the findings of the present study demonstrating that topical antimicrobial therapies do not completely eradicate the commensal microbes, or so-called “good bacteria,” on the skin. Although prior decolonization investigations in hospital and community settings have demonstrated reduced burdens of S. aureus colonization and infection with the administration of topical antimicrobials (4, 5, 8), important considerations of these therapies include the longevity of effectiveness as well as microbial development of resistance to these agents (39, 42–44). Thus, development of novel preventive strategies is essential, and one promising approach is that of “bacterial interference,” or the application of nonpathogenic organisms to impede or outcompete colonization or infection by potential pathogens. Indeed, this practice was implemented in the 1960s to abrogate nursery outbreaks caused by hypervirulent S. aureus strains (phage type 80/81) (45–48).
In conducting the present study, we postulated that the concept of bacterial interference might have clinical utility in the contemporary era as well. While we anticipated identification of one or two key organisms sufficient and necessary to outcompete S. aureus within a niche, this hypothesis was not substantiated. In prior investigations of S. aureus in the context of other constituents of the skin and nasal microbiota, an inverse relationship was observed between the presence of S. aureus and that of a variety of other flora, including S. epidermidis, Corynebacterium spp., Streptococcus spp., and Propionibacterium spp., suggesting competition or, again, bacterial interference between these organisms within the niche (49–53). In the present study, however, S. aureus often coexisted with Propionibacterium spp. and S. epidermidis, and in contrast to studies suggesting that S. aureus predominates in the niche, the presence of S. aureus did not affect microbial richness or diversity (52). While the concept of a single-organism probiotic is an attractive “natural” method for S. aureus eradication and infection prevention, it is likely that a community of organisms is required, similar to the approach of normal flora restoration therapy for Clostridium difficile infection. Restoration of the normal skin and nasal flora as an infection prevention approach is an exciting avenue for future investigation.
In conclusion, we demonstrated that topical antimicrobials reduced the burden of S. aureus on the skin of community-dwelling and hospitalized individuals while preserving other components of the skin and nasal microbiota. Specific taxa permitting or precluding S. aureus colonization were not identified.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Children's Discovery Institute of Washington University and St. Louis Children's Hospital, the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (grant K23-AI091690 to S.A.F.), the NIH/National Center for Advancing Translational Sciences (grant UL1-TR000448 to C.-A.D.B. and S.A.F.), and the Agency for Healthcare Research and Quality (grants R01-HS021736 and R01-HS024269 to S.A.F.).
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Agency for Healthcare Research and Quality.
We thank Sondra Seiler, Mary Boyle, and Carol Muenks for their assistance with participant enrollment and specimen collection, Ryley Thompson for his assistance with creating the REDCap (Research Electronic Data Capture) database, and Ryley Thompson and David Ross for their assistance with figure design.
C.-A.D.B. conceptualized and designed the study, supervised the microbial cultivation, identification, antimicrobial resistance testing, and data analysis, drafted and revised the manuscript, and approved the final manuscript as submitted. P.G.H. conducted the initial analyses, drafted and revised the manuscript, created the figures, and approved the final manuscript as submitted. M.A.W. conducted the microbial cultivation, identification, and antimicrobial resistance testing, critically reviewed the manuscript, and approved the final manuscript as submitted. E.D. conducted statistical analyses, critically reviewed the manuscript, and approved the final manuscript as submitted. W.S. participated in study design, supervised the statistical analyses, critically reviewed the manuscript, and approved the final manuscript as submitted. D.K.W. participated in study design and data collection and analysis, critically reviewed the manuscript, and approved the final manuscript as submitted. S.A.F. conceptualized and designed the study, supervised the human specimen and data collection, drafted and revised the manuscript, and approved the final manuscript as submitted.
Burnham has received research support from bioMérieux, Cepheid, and Accelerate Diagnostics. Warren has served on an advisory board for Becton Dickinson and has received research support from Cepheid. The other authors have no conflicts of interest relevant to this article to disclose.
Funding Statement
These funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data, preparation, review, or approval of the manuscript, or decision to submit the manuscript for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01289-16.
REFERENCES
- 1.Klevens RM, Edwards JR, Richards CL Jr, Horan TC, Gaynes RP, Pollock DA, Cardo DM. 2007. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122:160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 3.von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 4.Climo MW, Yokoe DS, Warren DK, Perl TM, Bolon M, Herwaldt LA, Weinstein RA, Sepkowitz KA, Jernigan JA, Sanogo K, Wong ES. 2013. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med 368:533–542. doi: 10.1056/NEJMoa1113849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R, CDC Prevention Epicenters Program, AHRQ DECIDE Network and Healthcare-Associated Infections Program. 2013. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassir N, Thomas G, Hraiech S, Brunet J, Fournier PE, La Scola B, Papazian L. 2015. Chlorhexidine daily bathing: impact on health care-associated infections caused by gram-negative bacteria. Am J Infect Control 43:640–643. doi: 10.1016/j.ajic.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Creech CB, Beekmann SE, Chen Y, Polgreen PM. 2008. Variability among pediatric infectious diseases specialists in the treatment and prevention of methicillin-resistant Staphylococcus aureus skin and soft tissue infections. Pediatr Infect Dis J 27:270–272. doi: 10.1097/INF.0b013e31815c9068. [DOI] [PubMed] [Google Scholar]
- 8.Fritz SA, Camins BC, Eisenstein KA, Fritz JM, Epplin EK, Burnham CA, Dukes J, Storch GA. 2011. Effectiveness of measures to eradicate Staphylococcus aureus carriage in patients with community-associated skin and soft-tissue infections: a randomized trial. Infect Control Hosp Epidemiol 32:872–880. doi: 10.1086/661285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritz SA, Hogan PG, Hayek G, Eisenstein KA, Rodriguez M, Epplin EK, Garbutt J, Fraser VJ. 2012. Household versus individual approaches to eradication of community-associated Staphylococcus aureus in children: a randomized trial. Clin Infect Dis 54:743–751. doi: 10.1093/cid/cir919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan SL, Forbes A, Hammerman WA, Lamberth L, Hulten KG, Minard CG, Mason EO. 2014. Randomized trial of “bleach baths” plus routine hygienic measures vs. routine hygienic measures alone for prevention of recurrent infections. Clin Infect Dis 58:679–682. doi: 10.1093/cid/cit764. [DOI] [PubMed] [Google Scholar]
- 11.Cogen AL, Nizet V, Gallo RL. 2008. Skin microbiota: a source of disease or defence? Br J Dermatol 158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belkaid Y, Hand TW. 2014. Role of the microbiota in immunity and inflammation. Cell 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dethlefsen L, Relman DA. 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108(Suppl 1):S4554–S4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meisel JS, Hannigan GD, Tyldsley AS, SanMiguel AJ, Hodkinson BP, Zheng Q, Grice EA. 2016. Skin microbiome surveys are strongly influenced by experimental design. J Invest Dermatol 136:947–956. doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh J, Byrd AL, Park M, NISC Comparative Sequencing Program, Kong HH, Segre JA. 2016. Temporal stability of the human skin microbiome. Cell 165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. 2012. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med 4:77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassir N, Papazian L, Fournier PE, Raoult D, La Scola B. 2015. Insights into bacterial colonization of intensive care patients' skin: the effect of chlorhexidine daily bathing. Eur J Clin Microbiol Infect Dis 34:999–1004. doi: 10.1007/s10096-015-2316-y. [DOI] [PubMed] [Google Scholar]
- 20.Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, Raoult D. 2013. The proof of concept that culturomics can be superior to metagenomics to study atypical stool samples. Eur J Clin Microbiol Infect Dis 32:1099. doi: 10.1007/s10096-013-1843-7. [DOI] [PubMed] [Google Scholar]
- 21.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A 108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouba N, Raoult D, Drancourt M. 2014. Eukaryote culturomics of the gut reveals new species. PLoS One 9:e106994. doi: 10.1371/journal.pone.0106994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greub G. 2012. Culturomics: a new approach to study the human microbiome. Clin Microbiol Infect 18:1157–1159. doi: 10.1111/1469-0691.12032. [DOI] [PubMed] [Google Scholar]
- 24.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 25.Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, Raoult D. 2015. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev 28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood NJ. 2011. Microbiome: human gut microbiota can be readily cultured, manipulated and archived. Nat Rev Gastroenterol Hepatol 8:241. doi: 10.1038/nrgastro.2011.60. [DOI] [PubMed] [Google Scholar]
- 27.McElvania Tekippe E, Shuey S, Winkler DW, Butler MA, Burnham CA. 2013. Optimizing identification of clinically relevant Gram-positive organisms by use of the Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry system. J Clin Microbiol 51:1421–1427. doi: 10.1128/JCM.02680-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fritz SA, Hogan PG, Singh LN, Thompson RM, Wallace MA, Whitney K, Al-Zubeidi D, Burnham CA, Fraser VJ. 2014. Contamination of environmental surfaces with Staphylococcus aureus in households with children infected with methicillin-resistant S aureus. JAMA Pediatr 168:1030–1038. doi: 10.1001/jamapediatrics.2014.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak MJ, Talan DA, Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin Infect Dis 52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 30.Viray MA, Morley JC, Coopersmith CM, Kollef MH, Fraser VJ, Warren DK. 2014. Daily bathing with chlorhexidine-based soap and the prevention of Staphylococcus aureus transmission and infection. Infect Control Hosp Epidemiol 35:243–250. doi: 10.1086/675292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren DK, Prager M, Munigala S, Wallace MA, Kennedy CR, Bommarito KM, Mazuski JE, Burnham CA. 2016. Prevalence of qacA/B genes and mupirocin resistance among methicillin-resistant Staphylococcus aureus (MRSA) isolates in the setting of chlorhexidine bathing without mupirocin. Infect Control Hosp Epidemiol 37:590–597. doi: 10.1017/ice.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequence Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manji R, Bythrow M, Branda JA, Burnham CA, Ferraro MJ, Garner OB, Jennemann R, Lewinski MA, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Westblade LF, Ginocchio CC. 2014. Multi-center evaluation of the VITEK® MS system for mass spectrometric identification of non-Enterobacteriaceae Gram-negative bacilli. Eur J Clin Microbiol Infect Dis 33:337–346. doi: 10.1007/s10096-013-1961-2. [DOI] [PubMed] [Google Scholar]
- 34.Garner O, Mochon A, Branda J, Burnham CA, Bythrow M, Ferraro M, Ginocchio C, Jennemann R, Manji R, Procop GW, Richter S, Rychert J, Sercia L, Westblade L, Lewinski M. 2014. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK® MS system. Clin Microbiol Infect 20:335–339. doi: 10.1111/1469-0691.12317. [DOI] [PubMed] [Google Scholar]
- 35.Richter SS, Sercia L, Branda JA, Burnham CA, Bythrow M, Ferraro MJ, Garner OB, Ginocchio CC, Jennemann R, Lewinski MA, Manji R, Mochon AB, Rychert JA, Westblade LF, Procop GW. 2013. Identification of Enterobacteriaceae by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using the VITEK® MS system. Eur J Clin Microbiol Infect Dis 32:1571–1578. doi: 10.1007/s10096-013-1912-y. [DOI] [PubMed] [Google Scholar]
- 36.Westblade LF, Jennemann R, Branda JA, Bythrow M, Ferraro MJ, Garner OB, Ginocchio CC, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Rychert JA, Sercia L, Burnham CA. 2013. Multicenter study evaluating the Vitek MS system for identification of medically important yeasts. J Clin Microbiol 51:2267–2272. doi: 10.1128/JCM.00680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rychert J, Burnham CA, Bythrow M, Garner OB, Ginocchio CC, Jennemann R, Lewinski MA, Manji R, Mochon AB, Procop GW, Richter SS, Sercia L, Westblade LF, Ferraro MJ, Branda JA. 2013. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization–time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J Clin Microbiol 51:2225–2231. doi: 10.1128/JCM.00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute. 2014. Analysis and presentation of cumulative antimicrobial susceptibility test data; 4th ed, M39-A4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 39.Fritz SA, Hogan PG, Camins BC, Ainsworth AJ, Patrick C, Martin MS, Krauss MJ, Rodriguez M, Burnham CA. 2013. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother 57:559–568. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Aberg N, Perkin MR, Tripodi S, Coates AR, Hesselmar B, Saalman R, Molin G, Ahrne S. 2008. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol 121:129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, NISC Comparative Sequence Program, Murray PR, Turner ML, Segre JA. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNeil JC, Hulten KG, Kaplan SL, Mahoney DH, Mason EO. 2013. Staphylococcus aureus infections in pediatric oncology patients: high rates of antimicrobial resistance, antiseptic tolerance and complications. Pediatr Infect Dis J 32:124–128. doi: 10.1097/INF.0b013e318271c4e0. [DOI] [PubMed] [Google Scholar]
- 43.McNeil JC, Hulten KG, Kaplan SL, Mason EO. 2014. Decreased susceptibilities to retapamulin, mupirocin, and chlorhexidine among Staphylococcus aureus isolates causing skin and soft tissue infections in otherwise healthy children. Antimicrob Agents Chemother 58:2878–2883. doi: 10.1128/AAC.02707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNeil JC, Ligon JA, Hulten KG, Dreyer WJ, Heinle JS, Mason EO, Kaplan SL. 2013. Staphylococcus aureus infections in children with congenital heart disease. J Pediatric Infect Dis Soc 2:337–344. doi: 10.1093/jpids/pit037. [DOI] [PubMed] [Google Scholar]
- 45.Shinefield HR, Ribble JC, Boris M, Eichenwald HF. 1963. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. I. Preliminary observations on artificial colonization of newborns. Am J Dis Child 105:646–654. [PubMed] [Google Scholar]
- 46.Light IJ, Sutherland JM, Schott JE. 1965. Control of a staphylococcal outbreak in a nursery, use of bacterial interference. JAMA 193:699–704. doi: 10.1001/jama.1965.03090090005001. [DOI] [PubMed] [Google Scholar]
- 47.Drutz DJ, Van Way MH, Schaffner W, Koenig MG. 1966. Bacterial interference in the therapy of recurrent staphylococcal infections. Multiple abscesses due to the implantation of the 502A strain of Staphylococcus. N Engl J Med 275:1161–1165. [DOI] [PubMed] [Google Scholar]
- 48.Houck PW, Nelson JD, Kay JL. 1972. Fatal septicemia due to Staphylococcus aureus 502A. Report of a case and review of the infectious complications of bacterial interference programs. Am J Dis Child 123:45–48. [PubMed] [Google Scholar]
- 49.Johnson RC, Ellis MW, Lanier JB, Schlett CD, Cui T, Merrell DS. 2015. Correlation between nasal microbiome composition and remote purulent skin and soft tissue infections. Infect Immun 83:802–811. doi: 10.1128/IAI.02664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uehara Y, Nakama H, Agematsu K, Uchida M, Kawakami Y, Abdul Fattah AS, Maruchi N. 2000. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect 44:127–133. doi: 10.1053/jhin.1999.0680. [DOI] [PubMed] [Google Scholar]
- 51.Alvarez AS, Remy L, Allix-Beguec C, Ligier C, Dupont C, Leminor O, Lawrence C, Supply P, Guillemot D, Gaillard JL, Salomon J, Herrmann JL. 2014. Patient nostril microbial flora: individual-dependency and diversity precluding prediction of Staphylococcus aureus acquisition. Clin Microbiol Infect 20:70–78. doi: 10.1111/1469-0691.12208. [DOI] [PubMed] [Google Scholar]
- 52.Bessesen MT, Kotter CV, Wagner BD, Adams JC, Kingery S, Benoit JB, Robertson CE, Janoff EN, Frank DN. 2015. MRSA colonization and the nasal microbiome in adults at high risk of colonization and infection. J Infect 71:649–657. doi: 10.1016/j.jinf.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 53.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. 2010. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.