Abstract
The distinct epidemiology of original hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) and early community-associated MRSA (CA-MRSA) is largely unexplained. S. aureus carries either five or six rRNA operon copies. Evidence is provided for a scenario in which MRSA has adapted to the hospital environment by rRNA operon loss (six to five copies) due to antibiotic pressure. Early CA-MRSA, in contrast, results from wild-type methicillin-susceptible S. aureus (MSSA) that acquired mecA without loss of an rRNA operon. Of the HA-MRSA isolates (n = 77), 67.5% had five rRNA operon copies, compared to 23.2% of the CA-MRSA isolates (n = 69) and 7.7% of MSSA isolates (n = 195) (P < 0.001). In addition, 105 MSSA isolates from cystic fibrosis patients were tested, because these patients are repeatedly treated with antibiotics; 32.4% of these isolates had five rRNA operon copies. For all subsets, a correlation between resistance profile and rRNA copy number was found. Furthermore, we showed that in vitro antibiotic pressure may result in rRNA operon copy loss. We also showed that without antibiotic pressure, S. aureus isolates containing six rRNA copies are more fit than isolates with five copies. We conclude that HA-MRSA and cystic fibrosis isolates most likely have adapted to an environment with high antibiotic pressure by the loss of an rRNA operon copy. This loss has facilitated resistance development, which promoted survival in these niches. However, strain fitness decreased, which explains their lack of success in the community. In contrast, CA-MRSA isolates retained six rRNA operon copies, rendering them fitter and thereby able to survive and spread in the community.
INTRODUCTION
An important fraction of Staphylococcus aureus isolates from humans are methicillin resistant (MRSA) (1). The emergence of MRSA is the result of the acquisition of SCCmec by methicillin-susceptible S. aureus (MSSA) (2). The epidemiology and genetic characteristics of hospital-associated MRSA (HA-MRSA) and community-associated-MRSA (CA-MRSA), especially in the first decade after its emergence, show considerable differences (3–5). A prime example is USA300 in the United States (6, 7). First, original HA-MRSA consisted of only a limited number of lineages, in contrast to early CA-MRSA, which was much more diverse (8, 9). Second, before the emergence of CA-MRSA, HA-MRSA carried SCCmec type I, II, or III, whereas early CA-MRSA carried the relatively small SCCmec type IV, V, or VI (10). Third, early CA-MRSA isolates were less often multidrug resistant than the HA-MRSA isolates (10), although nowadays these differences in resistance are less clear (11). Fourth, HA-MRSA carrying SCCmec type I, II, or III seems unable to spread to the community, whereas CA-MRSA is transmitted in the community, although CA-MRSA has also increasingly been found in the hospital setting during the last decade (12).
The difference in epidemiology between the original HA-MRSA and CA-MRSA is as yet largely unexplained (13). Some studies suggest that the relatively small SCCmec cassettes present in CA-MRSA impose a lower genetic burden on the bacterium, thereby retaining bacterial fitness required for competitive growth and spread in the community. HA-MRSA, with its bulky SCCmec cassette, needs the selective pressure of antibiotic use in a hospital setting to be able to compete (14–17). However, S. aureus carries either five or six copies of the rRNA operon, which might also contribute to the observed epidemiological differences. In other bacterial species, a correlation between the number of rRNA operon copies present in the bacterial cell and fitness, via its effect on protein synthesis rates, has been quantified in vitro by growth rate differences and less efficient adaptation to a changing environment (18–22). There might even be a link with antibiotic pressure, as loss of a rRNA operon in S. aureus has been described for follow-up isolates from a patient after prolonged treatment with linezolid (23). It has been suggested that loss of one wild-type (WT) operon helps to increase the number of mutated and therefore functional ribosomes. Variation in rRNA operon copy number, bacterial fitness, and antibiotic resistance may thus be linked and thereby explain at least part of the epidemiological behavior. A relation between antibiotic resistance, fitness characteristics, and the different epidemiologies of original HA- and CA-MRSA has been suggested before (24, 25). However, it has never been examined whether variation in rRNA operon copy number is related to S. aureus epidemiology. The aim of the current study was to assess whether variation in rRNA operon copy number may be a novel factor that contributes to the epidemiology, differences in fitness, and antibiotic resistance profiles of CA-MRSA and HA-MRSA.
MATERIALS AND METHODS
Ethics statement.
The isolates cannot be linked to any patients, as patient data were anonymized. The Medical Ethical Committee of the University Medical Center Utrecht was consulted if informed consent was needed according to Dutch law and the policies of the University Medical Center Utrecht, which was not required for this study. The cystic fibrosis (CF) isolates were collected from our routine diagnostic laboratory over a 7-month period (May to November 2009) when CF patients came in for routine outpatient checkups, as recommended by the Dutch guideline. Also, for these isolates we had no access to any patient data.
Isolates.
This study examined a collection of 195 MSSA and 146 MRSA isolates originating from 15 countries. In addition, 105 MSSA isolates from cystic fibrosis patients from the Netherlands were collected from our diagnostic laboratory between May and November 2009. The position of these isolates in the S. aureus population is shown in a multilocus sequence typing (MLST)-based minimal spanning tree in Fig. S1 in the supplemental material.
CA-MRSA isolates were defined as MRSA isolates carrying SCCmec IV, V, or VI. In contrast, HA-MRSA isolates were defined as MRSA isolates carrying SCCmec I, II, or III (4, 6, 11). Strains COL, MW2, MSSA476, MRSA252, Mu50, N315, RF122, USA300-FPR3757, USA300-TCH1516, and S0385 were used as controls for PCRs and Southern blotting for determination of the rRNA operon copy number.
DNA extraction.
Bacteria were cultured overnight on blood agar plates (Becton Dickinson, Erembodegem-Aalst, Belgium). Chromosomal DNA was isolated using the Nucleospin tissue kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's protocol, except for the lysis step. Lysis was performed in 20 mM Tris-HCl (pH 8.0), 2 mM EDTA, 1% Triton X-100, 0.2 mg/ml of lysostaphin, achromopeptidase, and RNase.
MLST.
MLST was performed according to the protocol of Enright et al. (26). New sequences were submitted to the MLST database (http://www.mlst.net).
rRNA copy number-specific PCR.
A seminested PCR was used to determine the presence or absence of the third rRNA operon (Fig. 1; see also Table S1 in the supplemental material). The first PCR (PCR-1) amplified a fragment spanning the 5Sp-16S intergenic region from the second rRNA operon and/or the 5S-16S intergenic region between the second and third rRNA operons (5S-F, ATAGCAAGGAGGTCACAC, and 16S-R, AGGCCCGGGAACGTATTCAC) in a 50-μl reaction mixture containing 10 pmol of each primer, 0.4 mM deoxynucleoside triphosphates (dNTPs; Invitrogen, Carlsbad, CA), 2 mM MgCl2, 0.3 U of AmpliTaq (Applied Biosystems, Carlsbad, CA), and genomic DNA. Touchdown amplification was performed with an initial step of 95°C for 5 min, followed by 10 cycles of 95°C for 30 s, annealing starting at 65°C for 30 s (decreasing by 0.8°C/cycle), and extension for 2.5 min at 72°C. This step was followed by 20 cycles of 95°C for 30 s, 57°C for 30 s, 72°C for 2.5 min, and a final extension at 72°C for 7 min.
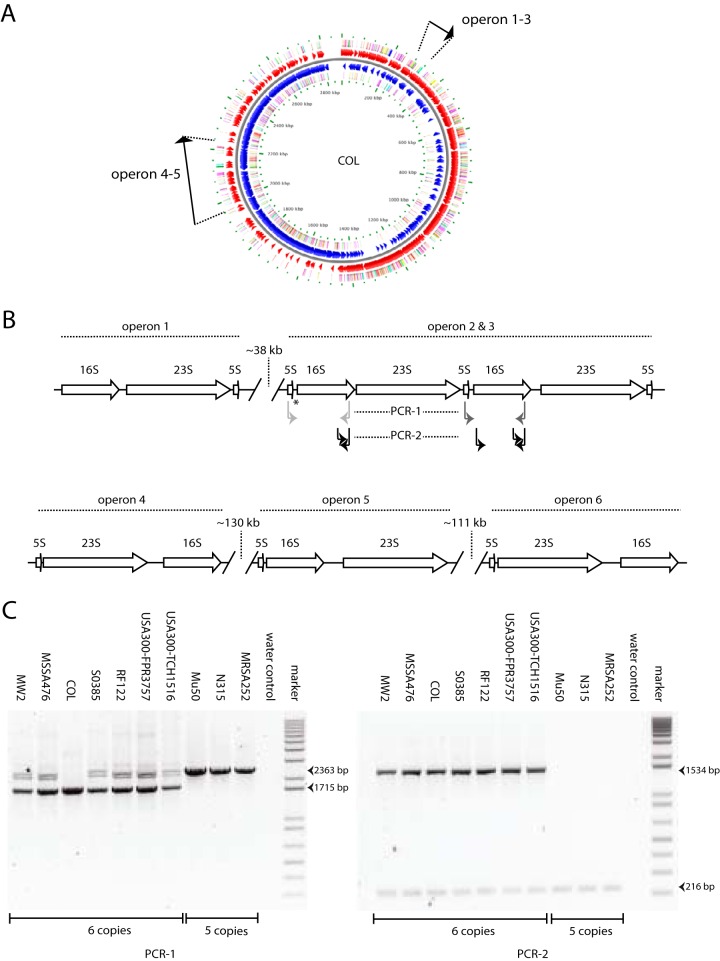
FIG 1.
Schematic representation of rRNA operons in S. aureus and PCR strategy. (A) Locations of the six rRNA operons on the genome of strain COL. (B) Organization of the rRNA operons. Operon 3 is lacking in strains with five operon copies. Two PCRs were performed: PCR-1 is indicated with light and dark gray arrows. In isolates with five copies of the rRNA operon in PCR-1, only a product of 2,363 bp (light gray arrows) will be amplified, whereas in isolates with six copies, both the 2,363-bp product and a 1,715-bp fragment (dark gray arrows) will be yielded (COL is an exception due to a smaller intergenic region between the 5S and 16S genes, where the amplification products will be of similar sizes; indicated by an asterisk). The black arrows indicate the products formed by the seminested PCR (PCR-2), consisting of a product of 1,534 bp when the third operon is present and a 16S internal control (216 bp). (C) Amplification products formed for strains with either six or five rRNA operon copies.
A seminested PCR (PCR-2) was performed on this product, which amplified the unique 5S-16S intergenic region between the second and third rRNA operons in a 50-μl reaction mixture containing 10 pmol of op2-3Int-F (GTATAATTAATTCTTGTCGGTA), 5 pmol of 16S-R (AGGCCCGGGAACGTATTCAC), 2.5 pmol of 16S-F (GAGGAAGGTGGGGATGACGT) as an internal control, 0.4 mM dNTPs, 2 mM MgCl2, 0.3 U of AmpliTaq, and 1/50-diluted template from the first PCR. The first cycle consisted of 5 min of denaturation at 95°C, followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 2 min. In the final cycle, the extension time was increased to 7 min at 72°C. All products were analyzed on a 0.8% agarose gel, stained with ethidium bromide, and visualized under UV light.
Southern blotting.
EcoRI- or ClaI-digested chromosomal DNA was separated overnight on 0.7% SeaKem Gold agarose and transferred to a Hybond N+ membrane (GE Healthcare, Piscataway, NJ) using a Vacu-blot system (Biometra, Göttingen, Germany). A 23S rRNA gene (forward [F], CGCGACAGGACGGAAAGACC, and reverse [R], CAGCCCCAGGATGCGATGAG) or 16S rRNA gene (F, GAGGAAGGTGGGGATGACGT, and R, AGGCCCGGGAACGTATTCAC) digoxigenin-labeled probe was used for hybridization according to the manufacturer's protocol (GE Healthcare).
Whole-genome mapping.
Whole-genome mapping of S. aureus genomes was performed as described before (27). Briefly, high-molecular-weight DNA is applied to Mapcards (OpGen, Gaithersburg, MD) and digested with AflII restriction endonuclease. This process retains the order of the restriction fragments. The restriction fragments are seized and assembled into a whole-genome map. The maps were analyzed for the presence of the restriction fragment carrying the third operon copy.
Antibiotic resistance.
Susceptibility testing was performed on a Phoenix automated test system (BD Diagnostic Systems, Sparks, MD). The multiple antibiotic resistance (MAR) index (28) was calculated for clindamycin, erythromycin, gentamicin, fusidic acid, linezolid, mupirocin, quinupristin-dalfopristin, and tetracycline, as these act on protein synthesis.
Induction of operon deletion by linezolid.
Six isolates containing six rRNA operons and susceptible to linezolid were cultured for 21 days in Mueller-Hinton (MH) broth with a dilution series of linezolid (Pfizer, New York, NY) ranging from 0 to 32 μg/ml. MICs were monitored, and daily, 10-μl samples with 80% growth, compared with growth in MH broth only, were used to inoculate a new dilution series (1 ml). From the isolates that gained resistance, genomic DNA was isolated and checked by PCR for loss of the third rRNA operon. Multiple colonies of each daily sample were analyzed.
In vitro growth rates.
Bacteria were grown overnight on blood agar plates. A single colony was used to inoculate 5 ml of Luria broth (LB) and grown overnight at 28°C to mimic temperatures in the nose. Cultures were diluted to a final concentration of 2.5 × 105 CFU/ml, confirmed by plating. Duplicates of each sample with a volume of 200 μl were run in the Bioscreen C system (Oy Growth Curves Ab Ltd., Helsinki, Finland) at 28°C for 48 h with continuous, moderate shaking, and the optical density at 420 to 580 nm (OD420–580) was measured every 20 min. Growth data (ln[OD/ODo]) (ODo, OD at time zero/start of the experiment) were fitted to the Gompertz model (29), from which three biological parameters were calculated: lag time (λ), maximum specific growth rate (μmax), and maximum population density (A). Experiments were performed independently three times.
Statistical analysis.
Statistical significance of the rRNA operon distributions was determined by Fisher's exact test. The Wilcoxon rank sum test with continuity correction was used to examine the association between rRNA operon number and the MAR index. For the growth data, a Student t test was used for the comparison of the maximum specific growth rates of the tested isolates; for the lag time, a Mann-Whitney test was used. A P value of <0.05 was considered to be statistically significant.
RESULTS
rRNA operon copy number in S. aureus.
Analysis of 10 published genomes showed that S. aureus carries either five or six copies of the rRNA operon. This difference in copy number is invariably due to the presence or absence of the third rRNA operon downstream from OriC (Fig. 1).
Results of the two PCRs distinguishing between isolates lacking the third rRNA operon (five copies) and isolates carrying this operon (six copies) agreed completely (Fig. 1). Sequence analysis of the rRNA operons showed that 120 bp in front of rRNA copy 1 are repeated in front of rRNA copy 3; however, a distinction can be made because 80 unique additional base pairs are present. This was confirmed by sequencing of PCR products. Finally, Southern blotting also was in agreement with the presence of six rRNA copies (data not shown). However, the results for USA300-TCH1516, USA300-FPR3757, and RF122 did not match the whole-genome data; according to PCR, partial sequencing, and Southern blotting, these isolates contained six rRNA operon copies instead of five copies in the published genome sequences.
The loss of an rRNA operon has been suggested as a mechanism in the development of resistance in S. aureus (23). To generate a set of controls, a susceptible S. aureus isolate carrying six rRNA operon copies was exposed to sub-MICs of linezolid. Twelve days of culturing yielded cells which differed in rRNA operon copy number, including cultures with both five and six rRNA copies. Bacterial strains containing five rRNA operons had higher MICs than strains with six rRNA operons. Already after 15 days of culturing, all bacterial colonies tested carried five rRNA operons, as demonstrated by third-operon-specific PCRs. Sequencing of the flanking regions of the third rRNA operon confirmed the absence of the operon. The isolates were indistinguishable from the original wild-type isolate by pulsed-field gel electrophoresis (PFGE), MLST, spa typing, and partial sequencing (data not shown).
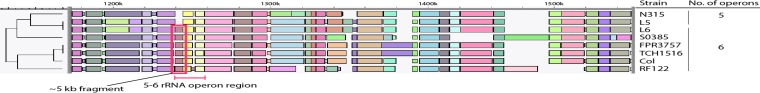
To further confirm the findings, whole-genome mapping was performed. With whole-genome mapping a complete ordered restriction map of a genome is created. Isolates of USA300-TCH1516, USA300-FPR3757, and RF122, together with S0385 (six rRNA copies), COL (six rRNA copies), and N315 (five rRNA copies) as controls, were subjected to whole-genome mapping. In addition, linezolid-susceptible and -resistant isolates (L5 and L6, with five and six rRNA operon copies, respectively) were selected. The results showed the lack of a restriction fragment for both L5 and N315 in the region of the third rRNA copy (Fig. 2). This further confirms the presence of six rRNA copies in the USA300 and RF122 strains in contrast to the published sequence.
FIG 2.
Detail of the whole-genome maps showing the alignment of the restriction fragments in the third rRNA operon region. The restriction fragments and their different sizes are represented by different colors. Fragments with the same size have the same color. Isolates from N315 and L5 lack a restriction fragment of approximately 5 kb, whereas this fragment is present in the other isolates (red box).
This combined evidence showed that indeed USA300-TCH1516, USA300-FPR3757, and RF122 clearly contained six instead of five rRNA operon copies. Additionally, we also analyzed 15 USA300 isolates from different patients that could not be linked and 11 ST-matched bovine mastitis isolates (30) from our collection by PCR, partial sequencing, and Southern blotting. The results showed the presence of six rRNA operon copies.
MSSA and HA- and CA-MRSA differ in rRNA operon copy number.
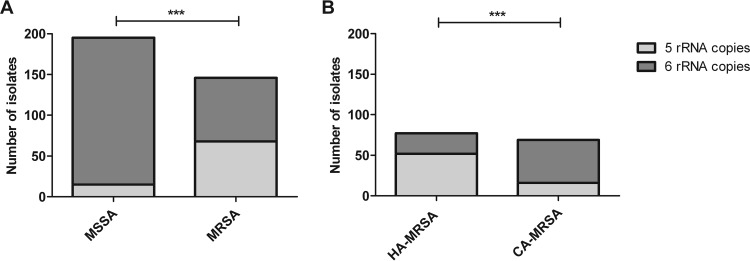
We analyzed a large collection of clinical isolates of S. aureus for the number of rRNA copies. Fifteen out of 195 MSSA isolates (7.7%) contained only five rRNA copies, in comparison to 68 out of 146 MRSA isolates (46.6%) (Fisher's exact test, P < 0.0001; odds ratio [OR], 0.09559; 95% confidence interval [CI], 0.05146 to 0.1775) (Fig. 3A). Within the subgroup of MRSA isolates, 52 out of 77 HA-MRSA isolates (67.5%), in comparison to 16 out of 69 CA-MRSA isolates (23.2%), contained five copies of the rRNA operon (Fisher's exact test, P < 0.0001; OR, 6.890; 95% CI, 3.303 to 14.37) (Fig. 3B).
FIG 3.
rRNA operon analysis of MSSA and MRSA. (A) Among MSSA isolates, 180 of the 195 carried six rRNA operons (92.3%). Among MRSA isolates, 78 of the 146 carried six rRNA operons (53.4%). ***, P < 0.0001 (Fisher's exact test). (B) MRSA isolates were subdivided into HA-MRSA and CA-MRSA according to the criteria described in Materials and Methods. Of the 78 HA-MRSA isolates, 52 (66.7%) had five rRNA operon copies; 16 (23.2%) of the 69 CA-MRSA isolates had five operon copies.
In most of the MLST-based clonal complexes (CCs), both isolates with five and isolates with six rRNA operons were found. Thus, the difference in distribution of rRNA operon copies between MRSA and MSSA isolates does not simply reflect a difference in genetic background (see Fig. S1 in the supplemental material).
rRNA operon copy number relates to antibiotic resistance.
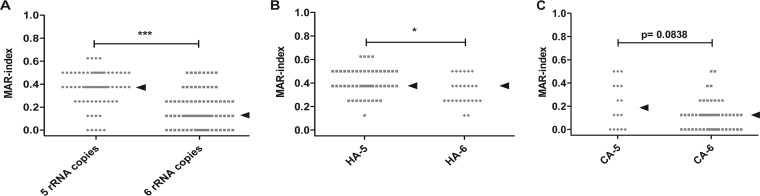
The loss of an rRNA operon under linezolid pressure, which coincided with the gain of resistance in a clinical isolate (23), prompted us to investigate the relationship between rRNA operon copy number and resistance to antibiotics acting on the ribosome. First, the clinical observed phenomenon could be replicated in vitro as described above. Next, a multiple antibiotic resistance (MAR) index (28) was calculated for eight antibiotics. A strong association between isolates carrying five rRNA operons and resistance was found in MRSA (Fig. 4A). For the subset of HA-MRSA isolates, the differences in antibiotic resistance between isolates carrying five and six rRNA operons remained significant. For CA-MRSA, a trend was observed in the association between resistance and carriage of five rRNA operon copies (Fig. 4B and C). Due to the limited number of MSSA isolates with five rRNA copies, no relation between resistance and rRNA copy number could be determined.
FIG 4.
Multiple antibiotic resistance index in MRSA. The MAR index is defined as a/b, where a represents the number of antibiotics to which the isolate was resistant and b represents the number of antibiotics for which the isolate was tested. The MAR index is shown in relation to rRNA operon copy number in MRSA (A); isolates with five rRNA copies are more resistant. ***, P < 0.0001 (Wilcoxon rank sum test with continuity correction). Also in the subset of hospital-associated MRSA (B), isolates with five operon copies show more resistance. *, P = 0.01504 (Wilcoxon rank sum test with continuity correction). In community-associated MRSA (C), a clear trend is seen toward more resistance with five rRNA operons. The P value was obtained by Wilcoxon rank sum test with continuity correction. Arrowheads represent medians.
rRNA operon copy number and antibiotic resistance in CF isolates.
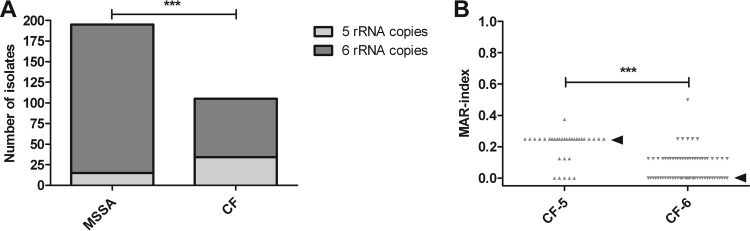
The role of antibiotics and rRNA operon copy number was further investigated in MSSA isolates from cystic fibrosis (CF) patients, who repeatedly receive (prophylactic) antibiotics. MSSA from CF patients and non-CF patients exhibit the same degree of heterogeneity. CF isolates were compared to the total group of non-CF MSSA isolates used in this study. Thirty-four out of the 105 (32.4%) CF isolates had five instead of six copies of the rRNA operon, whereas only 15 (7.7%) of the 195 non-CF MSSA isolates had five rRNA operon copies (Fisher's exact test, P < 0.0001; OR, 0.1740; 95% CI, 0.08932 to 0.3390) (Fig. 5A). MAR indices of the CF isolates showed that strains containing five rRNA operon copies are more resistant than isolates with six copies (Fig. 5B).
FIG 5.
rRNA operon copy number and MAR analysis of cystic fibrosis isolates. (A) Distribution of rRNA operon copy numbers in MSSA isolates from CF patients compared to non-CF isolates. Of CF isolates, 32.4% carried five rRNA operons; in non-CF isolates, 7.7% carried five rRNA operon copies. ***, P < 0.0001 (Fisher's exact test). (B) MAR index in relation to rRNA operon copy numbers for these isolates. Isolates with five rRNA operon copies are more resistant. ***, P < 0.0001 (Wilcoxon rank sum test with continuity correction). Arrowheads represent medians.
Five-rRNA-operon-copy isolates are less fit.
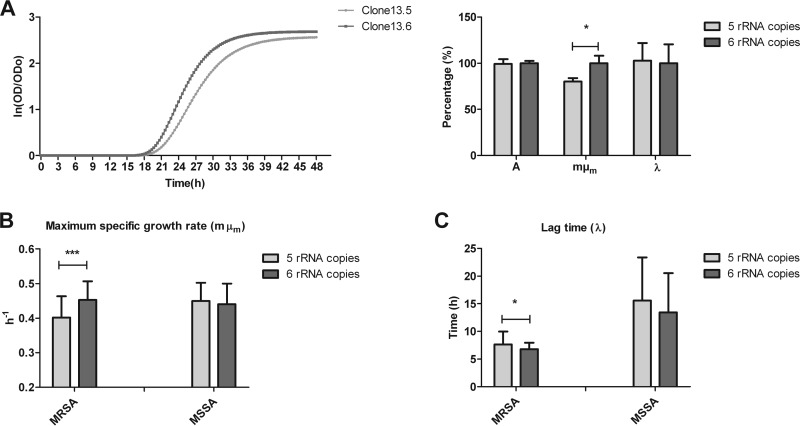
To test the effects of rRNA copy number on fitness, the in vitro growth rates were compared between a five-operon isolate that had lost an rRNA copy due to linezolid treatment and an isolate with six copies from the same experiment and after the same period of linezolid exposure. Remarkably, the growth characteristics showed a difference in maximum specific growth rate (μmax), while maximum ODs and lag times were not different (Fig. 6A). This difference in maximum specific growth rate was observed by both OD measurement and CFU counts (data not shown).
FIG 6.
In vitro growth analysis. (A) The growth curves for the isolate from the linezolid induction experiment are shown. Colonies isolated on the same day which differ in rRNA operon copy number were compared; growth parameters (A, μmax, and λ) were analyzed. Results are expressed as a percentage of the isolate with six rRNA operon copies (clone 13.6), set at 100%. The isolates with only five rRNA copies showed a decreased maximum specific growth rate. *, P = 0.0193 (Student t test). For a larger set of MRSA and MSSA isolates, μmax (B) and λ (C) were analyzed. In MRSA, isolates with five rRNA operon copies have a decreased maximum specific growth rate and a longer lag time. ***, P < 0.0001 (Student t test); *, P = 0.0135 (Mann-Whitney test).
Next, growth characteristics were tested for 51 MRSA isolates which were matched for ST and SCCmec type (six different ST types, including three SCCmec types) but carrying either five or six rRNA operon copies. We also investigated MSSA, but due to the relatively small number of MSSA isolates with five rRNA operon copies, only 31 MSSA isolates were available, and these could be matched only at the level of clonal complex (CC) instead of sequence type (six different CCs), leading to a genetically less homogeneous set of isolates within a group.
Under suboptimal conditions (growth at 28°C [to mimic temperatures in the nose, a common site for carriage] in LB medium), a clear difference in growth rates was observed. MRSA isolates carrying five rRNA operon copies showed a decreased maximum specific growth rate (Student t test, P < 0.0001) and increased lag time (Mann-Whitney test, P = 0.0135) compared to those of isolates carrying six rRNA operon copies (Fig. 6B and C). When results were broken down by MLST, five-copy-number isolates had decreased maximum specific growth rate and increased lag time in most STs. Also in MSSA isolates with five rRNA operon copies an increased lag time was observed in most CCs. Interestingly, a decreased maximum specific growth rate for five-copy-isolates was observed in only four of the seven CCs tested (see Fig. S2 in the supplemental material).
DISCUSSION
We identified variation in rRNA operon copy number as a novel factor that correlates with S. aureus epidemiology, antibiotic resistance, and in vitro fitness. This factor might be important in the epidemiology of S. aureus and could provide an explanation for the emergence and distinct epidemiology of early CA-MRSA and HA-MRSA carrying SCCmec type I, II, or III, although final proof will be nearly impossible, because we cannot replicate the emergence of HA-MRSA. Since the vast majority of MSSA strains have six rRNA operons (92.3%), we consider the presence of six rRNA operons to represent the wild type. Most MLST clusters contain isolates with both five and six rRNA operons. This suggests that variation in rRNA operon copy number is part of an adaptation process, rather than the result of the spread of a few ancestor strains. However, the distribution of strains with five or six copies is not random, as exemplified by our findings with MSSA, CA-MRSA, HA-MRSA, and CF isolates. Whereas the vast majority of MSSA and CA-MRSA carry six rRNA operon copies, the isolates frequently exposed to antibiotics (HA-MRSA and CF-MSSA) are dominated by strains that carry five rRNA operons. Moreover, CF-MSSA and MRSA isolates were more resistant to antibiotics targeting the ribosome than were isolates carrying six operons. In addition, we could show in vitro that prolonged exposure to linezolid resulted in deletion of an rRNA operon accompanied by a significant increase in MIC against linezolid, as was also described by Meka et al. (23) for sequential patient isolates. These data suggest that ribosome-targeting antibiotics can lead to deletion of the third rRNA operon, although we could not show this effect for gentamicin or streptomycin, probably because we could not mimic the in vivo conditions. Loss of the third rRNA operon could lead to an increment of the proportion of mutant (less susceptible) ribosomes and may be the effect of restoring the balance in ribosome assembly after antibiotic treatment, as was demonstrated for E. coli. Treatment with chloramphenicol increased rRNA production in E. coli and increasingly inhibited protein synthesis, resulting in dissociation between the synthesis rates of ribosomal proteins and those of rRNA. This would result in decreased numbers of complete ribosomes (31, 32).
Only the third operon is deleted and the mechanism of excision is unknown, but whatever the mechanism may be, deletion of an rRNA operon comes at a fitness cost. Studies with E. coli showed that deletion of rRNA operons resulted in increased lag times and decreased growth rates, thereby leading to a slower ability to adapt to novel environments (20, 22).
In S. aureus isolates, a clear overall decrease in maximum specific growth rate (μmax) and an increased lag time (λ) were observed in isolates carrying five instead of six rRNA operon copies. To exclude the influence of the genetic background, isolates were matched for ST and, if present, SCCmec type. Both in vitro parameters (μmax and λ) may correlate to in vivo colonization efficiency. The increased lag time may reflect a slower ability to adapt to a novel niche (e.g., during adhesion and initial colonization phase), whereas lower growth rates may result in outgrowth by competing colonizing strains after the initial colonization phase. A link between rRNA copy number and the ability to adapt to changing environments like temperature or nutrient shifts at 37°C was not observed (data not shown).
To prove the causality between rRNA operon copy number and growth rates, we attempted to genetically manipulate S. aureus. However, deletion of an rRNA operon failed due to the large homology between the sequences flanking the rRNA operons, and introduction of wild-type rRNA operons led to a substantial number of mutations in 16S, 23S, and 5S rRNAs. This was observed in several strains. This might be due to the fact that S. aureus may be restricted in the number of rRNA operons it can carry (five or six). In agreement with this rigidity is the fact that apparently only the third rRNA operon is dispensable.
Based on our results we present a completely novel hypothesis for a major driving factor behind S. aureus epidemiology. Due to antibiotic pressure in hospitals, the major original HA-MRSA pandemic clones have successfully adapted to the hospital environment by the loss of an rRNA operon copy, which facilitated resistance development. At the same time, strain fitness decreased, which explains lack of success in the community where no antibiotic pressure is present. However, the very successful community-associated USA300 clone and the bovine mastitis isolate RF122 would not fit in this picture, as in the published genomes five rRNA operons are present (33–35). Therefore, we analyzed these isolates together with 15 USA300 isolates from unrelated patients and 11 ST-matched bovine mastitis isolates (30) from our collection. In contrast to the published genome sequences, we found the completely sequenced USA300 isolates, RF122, 15 independent USA300 strains, and 11 bovine mastitis isolates to harbor six rRNA operon copies, as confirmed by PCR, Southern blotting, and genome mapping. The discrepancies between the whole-genome sequences and the other methods can easily be explained by the fact that due to (inverted) repeats in rRNA operons, whole-genome sequencing leads to partial rRNA genes at the end of a contig or separate contigs for each rRNA operon. Depending on, among other things, the chosen backbone, rRNA operons can be missed while assembling contigs. De novo assembly of contigs will even have a greater risk of missing an rRNA operon. We also tried to sequence the region of the third rRNA operon in RF122 and USA300 strains TCH1516 and FPR3757, but this was unsuccessful, probably for reasons mentioned above. Therefore, these isolates were subjected to whole-genome mapping to further confirm the presence of six rRNA operons in these isolates. The results agree with the presence of six rRNA copies in the USA300 and RF122 strains.
In conclusion, CA-MRSA strains in general carry six rRNA copies, which most likely contributes to their relative fitness and spread in the community, whereas the original HA-MRSA clones had lost a copy, increasing their fitness in environments with high antibiotic pressure at the cost of their ability to spread in the community. Variation in rRNA operon copy number thus provides a novel mechanism that explains the distinct epidemiology of early CA-MRSA and HA-MRSA.
Supplementary Material
ACKNOWLEDGMENT
We thank Marcel Zwietering for his advice and expertise on growth curves.
Funding Statement
This work, including the efforts of A. C. Fluit, was funded in part by European Commission (EC) (CONCORD-HEALTH-F3-2008-222718).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01613-16.
REFERENCES
- 1.Palavecino E. 2007. Clinical, epidemiological, and laboratory aspects of methicillin-resistant Staphylococcus aureus (MRSA) infections. Methods Mol Biol 391:1–19. doi: 10.1007/978-1-59745-468-1_1. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa M, de Lencastre H. 2004. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol Med Microbiol 40:101–111. doi: 10.1016/S0928-8244(03)00370-5. [DOI] [PubMed] [Google Scholar]
- 3.Bradley SF. 2005. Staphylococcus aureus pneumonia: emergence of MRSA in the community. Semin Respir Crit Care Med 26:643–649. doi: 10.1055/s-2005-925528. [DOI] [PubMed] [Google Scholar]
- 4.Gosbell IB. 2005. Epidemiology, clinical features and management of infections due to community methicillin-resistant Staphylococcus aureus (cMRSA) Intern Med J 35(Suppl 2):S120–S135. [DOI] [PubMed] [Google Scholar]
- 5.Hinrichs JH. 2006. CA-MRSA: an old foe develops new fangs. Mol Med 103:129–132. [PubMed] [Google Scholar]
- 6.Otto M. 2013. Community-associated MRSA: what makes them special? Int J Med Microbiol 303:324–330. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated methicillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groom AV, Wolsey DH, Naimi TS, Smith K, Johnson S, Boxrud D, Moore KA, Cheek JE. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 286:1201–1205. doi: 10.1001/jama.286.10.1201. [DOI] [PubMed] [Google Scholar]
- 9.Naimi TS, LeDell KH, Boxrud DJ, Groom AV, Steward CD, Johnson SK, Besser JM, O'Boyle C, Danila RN, Cheek JE, Osterholm MT, Moore KA, Smith KE. 2001. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996–1998. Clin Infect Dis 33:990–996. doi: 10.1086/322693. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Kuwahara K, Hiramatsu K. 2007. Staphylococcal cassette chromosome mec (SCCmec) analysis of MRSA. Methods Mol Biol 391:87–102. doi: 10.1007/978-1-59745-468-1_7. [DOI] [PubMed] [Google Scholar]
- 11.Otter JA, French GL. 2012. Community-associated meticillin-resistant Staphylococcus aureus: the case for a genotypic definition. J Hosp Infect 81:143–148. doi: 10.1016/j.jhin.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Patel M, Waites KB, Hoesley CJ, Stamm AM, Canupp KC, Moser SA. 2008. Emergence of USA300 MRSA in a tertiary medical centre: implications for epidemiological studies. J Hosp Infect 68:208–213. doi: 10.1016/j.jhin.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Chen CJ, Su LH, Hu S, Yu J, Chiu CH. 2008. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol Rev 32:23–37. doi: 10.1111/j.1574-6976.2007.00086.x. [DOI] [PubMed] [Google Scholar]
- 14.Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, DeGirolami PC, Eliopoulos GM, Moellering RC Jr, Gold HS. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol 40:4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lencastre H, Oliveira D, Tomasz A. 2007. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol 10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ender M, McCallum N, Adhikari R, Berger-Bächi B. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob Agents Chemother 48:2295–2297. doi: 10.1128/AAC.48.6.2295-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bächi B, Cook GM. 2007. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother 51:1497–1499. doi: 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monshupanee T, Fa-aroonsawat S, Chungjatupornchai W. 2006. A cyanobacterial strain with all chromosomal rRNA operons inactivated: a single nucleotide mutation of 23S rRNA confers temperature-sensitive phenotypes. Microbiology 152:1417–1425. doi: 10.1099/mic.0.28691-0. [DOI] [PubMed] [Google Scholar]
- 19.Shrestha PM, Noll M, Liesack W. 2007. Phylogenetic identity, growth-response time and rRNA operon copy number of soil bacteria indicate different stages of community succession. Environ Microbiol 9:2464–2474. doi: 10.1111/j.1462-2920.2007.01364.x. [DOI] [PubMed] [Google Scholar]
- 20.Stevenson BS, Schmidt TM. 2004. Life history implications of rRNA gene copy number in Escherichia coli. Appl Environ Microbiol 70:6670–6677. doi: 10.1128/AEM.70.11.6670-6677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klappenbach JA, Dunbar JM, Schmidt TM. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol 66:1328–1333. doi: 10.1128/AEM.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Condon C, Liveris D, Squires C, Schwartz I, Squires CL. 1995. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J Bacteriol 177:4152–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meka VG, Pillai SK, Sakoulas G, Wennersten C, Venkataraman L, DeGirolami PC, Eliopoulos GM, Moellering RC Jr, Gold HS. 2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J Infect Dis 190:311–317. doi: 10.1086/421471. [DOI] [PubMed] [Google Scholar]
- 24.Laurent F, Lelièvre H, Cornu M, Vandenesch F, Carret G, Etienne J, Flandrois JP. 2001. Fitness and competitive growth advantage of new gentamicin-susceptible MRSA clones spreading in French hospitals. J Antimicrob Chemother 47:277–283. doi: 10.1093/jac/47.3.277. [DOI] [PubMed] [Google Scholar]
- 25.Noto MJ, Fox PM, Archer GL. 2008. Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 52:1221–1229. doi: 10.1128/AAC.01164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bosch T, Verkade E, van Luit M, Pot B, Vauterin P, Burggrave R, Savelkoul P, Kluytmans J, Schouls L. 2013. High resolution typing by whole-genome mapping enables discrimination of LA-MRSA (CC398) strains and identification of transmission events. PLoS One 8:e66493. doi: 10.1371/journal.pone.0066493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krumperman PH. 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwietering MH, Jongenburger I, Rombouts FM, van ‘t Riet K. 1990. Modeling of the bacterial growth curve. Appl Environ Microbiol 56:1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikawaty R, Brouwer EC, Jansen MD, van Duijkeren E, Mevius D, Verhoef J, Fluit AC. 2009. Characterization of Dutch Staphylococcus aureus from bovine mastitis using a multiple locus variable number tandem repeat analysis. Vet Microbiol 136:277–284. doi: 10.1016/j.vetmic.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 31.Dodd J, Kolb JM, Nomura M. 1991. Lack of complete cooperativity of ribosome assembly in vitro and its possible relevance to in vivo ribosome assembly and the regulation of ribosomal gene expression. Biochimie 73:757–767. doi: 10.1016/0300-9084(91)90055-6. [DOI] [PubMed] [Google Scholar]
- 32.Nomura-M. 1999. Regulation of ribosome biosynthesis in Echerichia coli and Saccharomyces cerevisiae: diversity and common principles. J Bacteriol 181:6857–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 34.Highlander SK, Hulten KG, Qin X, Jian H, Yerrapragada S, Mason EO Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. 2007. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. 2007. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One 2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.