Abstract
Chikungunya virus (CHIKV; genus Alphavirus) is the causative agent of chikungunya fever. CHIKV replication can be inhibited by some broad-spectrum antiviral compounds; in contrast, there is very little information about compounds specifically inhibiting the enzymatic activities of CHIKV replication proteins. These proteins are translated in the form of a nonstructural (ns) P1234 polyprotein precursor from the CHIKV positive-strand RNA genome. Active forms of replicase enzymes are generated using the autoproteolytic activity of nsP2. The available three-dimensional (3D) structure of nsP2 protease has made it a target for in silico drug design; however, there is thus far little evidence that the designed compounds indeed inhibit the protease activity of nsP2 and/or suppress CHIKV replication. In this study, a set of 12 compounds, predicted to interact with the active center of nsP2 protease, was designed using target-based modeling. The majority of these compounds were shown to inhibit the ability of nsP2 to process recombinant protein and synthetic peptide substrates. Furthermore, all compounds found to be active in these cell-free assays also suppressed CHIKV replication in cell culture, the 50% effective concentration (EC50) of the most potent inhibitor being ∼1.5 μM. Analysis of stereoisomers of one compound revealed that inhibition of both the nsP2 protease activity and CHIKV replication depended on the conformation of the inhibitor. Combining the data obtained from different assays also indicates that some of the analyzed compounds may suppress CHIKV replication using more than one mechanism.
INTRODUCTION
Chikungunya virus (CHIKV; genus Alphavirus, family Togaviridae) is the causative agent of chikungunya fever, a disease that has affected millions in the last decade. It is characterized by high fever, arthralgia, myalgia, headache, and rash. The disease is usually self-limiting; however long-lasting chronic symptoms are observed in nearly 50% of CHIKV-infected patients (1). As there is no approved vaccine or specific licensed antiviral compounds (2), the treatment of CHIKV infection is largely based on relief of symptoms.
CHIKV infection can be inhibited by targeting host factors essential for virus infection. Targeting of several metabolic pathways has revealed anti-CHIKV effects of several licensed drugs (3). Compounds most likely targeting virus entry or host cell-specific components required for virus infection have been described (4–8). Several nucleosides or nucleotides, acting as pseudosubstrates for CHIKV RNA-dependent RNA polymerase and/or using another, more general mechanism of action, have shown anti-CHIKV activity (9). In addition, anti-CHIKV effects have been described for several groups of novel synthetic compounds (10–12).
Computer-aided design based on molecular docking and molecular dynamics simulations and pharmacophore approach allows identifying potential in silico hits as active inhibitors for different CHIKV replicase proteins. This approach, however, requires the three-dimensional (3D) structures of targeted proteins, advanced knowledge of the functions of the viral replicase, and availability of robust assays. CHIKV replicase proteins, called nonstructural (ns) proteins 1 to 4 (nsP1 to -4), are translated as P1234 polyprotein precursors directly from the 11.8-kb genomic RNA of the virus (13). nsP1 is a cap methyl- and guanylyltransferase and serves as the membrane anchor of replicase complexes (14, 15). nsP2 has protease, NTPase, RNA triphosphatase, and RNA helicase activities (16–18). The N-terminal domain of nsP3 has ADP-ribose protein hydrolase and relatively poor ADP-ribose 1″-phosphohydrolase activities (19, 20), while nsP4 is the RNA-dependent RNA polymerase and, most likely, also a terminal adenylyltransferase (13). The activities of CHIKV nsP2 are relatively easy to analyze using purified recombinant proteins, and very recently, an assay for inhibitors of CHIKV nsP1 was developed (21). A combination of cell-based and cell-free assays was successfully used to identify inhibitors targeting nsP1 of CHIKV (22). In addition, the activities of ns proteins can indirectly be analyzed using recently developed CHIKV trans-replication system or an assay based on the use of extracts derived from infected cells (23, 24).
Thus far, only the structures of the C-terminal region of nsP2 (PDB code 3TRK) and the N-terminal macro domain of nsP3 (25), together comprising only ∼20% of the ns region of CHIKV, have been resolved. A combination of virtual screening and molecular dynamic simulations has allowed finding three different target sites in the macro domain of CHIKV nsP3. Altogether, 1,541 compounds were subsequently screened by docking them to these nsP3 sites. The best binding free energies (ΔG) of lead compounds that presumably target CHIKV nsP3 ranged between −8.3 and 11.1 kcal/mol (26). Notably, the structures of these compounds were very different, and their ability to inhibit nsP3 or the actual virus has not been demonstrated.
Alphavirus nsP2 protease consists of two different domains (27). This region is folded similarly in nsP2s of different alphaviruses and thus represents a potential target both for specific and pan-alphavirus protease inhibitors. Virtual screening in the ZINC database using the Glide module by Schrödinger LLC has been carried out (28–30). Molecular docking was carried out using the hypothetical CHIKV nsP2 protease active site in the area of the catalytic amino acid residues Cys1013, His1083, and Trp1084 (here and below, the amino acid residues are numbered from the N terminus of P1234). For the top 10 hits, the best binding free energies were found to be in the range of −9.145 to 9.609 kcal/mol. The compounds with the highest predicted activity, 5-fluoro-N-[2-(2-imidazol-1-yletoxy)phenyl]-1H-indol-2-carboxamide and N-[2-(imidazole-1-ylmethyl)phenyl]-2-methylbenzamide, have rather different molecular structures, although in both cases the benzamide bridge is involved as a linker. Similarly, several potential inhibitors were identified using a combination of molecular docking, virtual screening, and molecular dynamics simulations on the crystal structure of CHIKV nsP2 protease (31). The best compounds were characterized by binding free energies up to−10.6 kcal/mol. However, in these studies the ability of the predicted compounds to inhibit the protease activity of CHIKV nsP2 was not demonstrated.
Venezuelan equine encephalitis virus (VEEV) nsP2 protease structure (PDB code 2HWK [27]) was initially used for modeling the 3D structure of CHIKV protease (32), and using molecular docking, several potential virus inhibitors were predicted. Although no quantitative correlation was established between docking efficiency and experimentally measured virus inhibition, some inhibitors were shown to be active in cell-based assays with 50% effective concentrations (EC50s) between 3.2 and 6.4 μM. Similarly, different arylalkylidene derivatives of 1,3-thiazolidin-4-one, predicted to interact with the protease part of nsP2, were found to inhibit CHIKV replication in Vero cells at concentrations between 0.42 and 40.1 μM (30). However, again, neither of these studies provided experimental proof that the protease activity of nsP2 was indeed targeted by the analyzed compounds. Other inhibitors targeting nsP2 include peptidomimetic compounds designed using quantum mechanics-based ligand descriptors. These compounds were verified using cell culture assays, in which the most active of them had EC90s of 8.76 to 9.57 μg/ml (33). For the protease of VEEV, a crystal structure of the enzyme with such an inhibitor has been recently resolved, clearly confirming the targeting of nsP2 and demonstrating the mode of binding of peptidomimetic inhibitors (34).
As highlighted above, thus far the common drawback of CHIKV inhibitors originating from computer-aided design is the lack of experimental verification of their antiviral activity and/or target specificity. The protease activity of nsP2 is robust and can be easily analyzed (35, 36). Such analysis represents the only option to provide direct proof regarding the true molecular target of the designed compounds, information regarding the exact mechanism of inhibition, and valuable input data for subsequent optimization of inhibitor structure. In this study, for the first time, we combined the power of computer-aided inhibitor design with careful verification of the effects of selected compounds both on the protease activity of CHIKV nsP2 and on the replication of CHIKV in cell culture. From our rationally designed and tested compounds, the most promising compound shows an EC50 of ∼1.5 μM, with a ΔG of −8.61 kcal/mol. Overall, the results represent, to our knowledge, the first set of compounds proven to inhibit the protease activity of CHIKV nsP2 and also demonstrated to directly inhibit CHIKV replication.
MATERIALS AND METHODS
Molecular design.
The crystal structure of CHIKV nsP2 protease was obtained from the Protein Data Bank (PDB code 3RTK). The hydrophobic hydrogen atoms were added to the structure for further modeling (37), and docking was performed essentially as previously described (30). In docking simulations, the nsP2 protein was kept as a rigid molecule. The ligands were optimized before molecular docking using the semiempirical quantum-chemical RM1 method within the program Maestro 9.5 (37). In all simulations the active site was first surrounded with a grid box at 70 by 70 by 70 Å. The AutoDock 4.2 specific force-field (37) was used for calculating interactions between CHIKV nsP2 protease and the predicted inhibitor molecules. All compounds, except those synthesized in-house, were obtained from MolPort.
Synthesis of compounds 1a to 1d and 1aL to 1dL.
Diastereomeric compounds 1a to 1d were synthesized starting from commercially available 3,4-dimethylbenzaldehyde as shown below in Fig. 6. The latter was first converted into 3,4-dimethylstyrene by Wittig olefination, and the obtained olefin was cyclopropanated with ethyl diazoacetate to afford a mixture of cis- and trans-ethyl esters of 2-arylcyclopropanecarboxylic acid (cis/trans ratio= 35:65). After selective alkaline hydrolysis, the pure trans-isomer of 2-arylcyclopropanecarboxylic acid was isolated. Unreacted cis-ethyl ester was further converted into the corresponding cis-2-arylcyclopropanecarboxylic acid by treatment with excess alkali solution at an elevated temperature (see the supplemental material for detailed experimental procedures). The obtained cis- and trans-acids were transformed into the corresponding acid chlorides prior to the acylation of d-(−)-phenylglycinamide to afford target compounds 1a-1b and 1c-1d as equimolecular mixtures of diastereomers in 63% and 79% combined yields, respectively. Small amounts (∼50 mg) of the obtained mixtures were separated by column chromatography on a silica gel using a gradient of toluene in ethyl acetate as an eluent. The configuration of the stereocenters in the cyclopropane ring was assigned with the aid of nuclear magnetic resonance (NMR) spectroscopy and assisted conformational analysis. Enantiomeric counterparts 1aL to 1dL were prepared in the same manner using l-(+)-phenylglycinamide instead of its d-(−)-enantiomer in the last step. Details of the synthesis, purification, and NMR spectra of the compounds and determination of the relative conformations of compounds 1a to 1d are provided in the supplemental material.
FIG 6.
Schema for the synthesis of isomers 1a to 1d of compound 1. Reagents and conditions: (a) Ph3PMe+Br−, t-BuOK, tetrahydrofuran (THF); (b) N2CHCO2Et, Rh2(OAc)4 (0.5 mol%), dichloromethane (DCM); (c) 2 M aqueous NaOH (0.65 eq), THF-EtOH, room temperature, and then extraction and chromatography; (d) 2 M aqueous NaOH (5 eq), THF-EtOH, 65°C; (e) (COCl)2, THF; (f) d-(−)-phenylglycinamide, N-ethylmorpholine, 4-(dimethylamino)pyridine (DMAP), THF.
Cell lines and viruses.
BHK-21 cells (ATCC CCL-10) were grown at 37°C in a 5% CO2 atmosphere in Glasgow's minimal essential medium (GMEM; Gibco) containing 10% fetal bovine serum, 2% tryptose phosphate broth, 200 mM HEPES, 100 U/ml of penicillin, and 0.1 mg/ml of streptomycin.
In virus-based experiments, wild-type (wt) CHIKV isolate LR-2006-OPY1 (East/Central/South African genotype) derived from an infectious cDNA clone (5) was used. For EC50 determination, a derivative of this clone harboring a sequence encoding a Renilla luciferase (Rluc) marker in the nsP3 coding region, referred to as CHIKV-Rluc (5), was used.
Production of protease and substrates.
Full-length recombinant CHIKV nsP2 was used as the protease in all cell-free assays. Recombinant protein substrate contained the nsP2 cleavage site (P10 to P′5) from the nsP1/nsP2 junction, placed between enhanced green fluorescent protein (EGFP) and thioredoxin. The recombinant proteins were expressed and purified as described in detail earlier (16, 36). Briefly, CHIKV nsP2 was expressed in Escherichia coli and the thioredoxin tag was removed by autocatalytic cleavage. Recombinant nsP2 was purified using metal affinity, cation-exchange, and size exclusion chromatography. The recombinant protease substrate was expressed and purified using the same procedures except that the clarified bacterial lysate was first passed through preswollen DE52 anion-exchange resin. Protein concentrations were measured using a NanoDrop spectrophotometer (Thermo Scientific, USA), and purified proteins were flash frozen and stored at −80°C.
Cell-free protease inhibition assays.
Compounds obtained from commercial sources were given specific serial numbers 1 to 12, while isomers of compound 1 synthesized in-house were named 1a to 1d and 1aL to 1dL. Stocks were prepared by dissolving compounds in sterile dimethyl sulfoxide (DMSO; Sigma, USA) at 10 mM, aliquoted, and stored at −20°C until further use.
The maximal tolerated DMSO concentration was determined by varying the DMSO concentration from 2 to 30% in protease assay buffer A (20 mM HEPES [pH 7.2], 2 mM dithiothreitol [DTT]). CHIKV nsP2 (final concentration, 348 nM) was added, the mixture was incubated for 10 min at 22°C, after which recombinant protein substrate was added to a final concentration of 5.9 μM. The reaction was carried out at 30°C for 1 h in a 10-μl volume. A protease inhibition assay using a recombinant protease substrate was carried out as described above except that the compounds were added to the reaction mixture at a final concentration of 1 mM; 10% DMSO was used as a solvent control. Products of protease reaction were resolved by 10% SDS-PAGE and detected using Coomassie blue staining as described earlier (35, 36).
A protease inhibition assay with a peptide substrate was carried out using purified CHIKV nsP2 and a substrate representing the nsP3/nsP4 cleavage site of CHIKV P1234 polyprotein. The peptide had the sequence DELRLDRAGG↓YIFSS (arrow indicates the scissile bond), a quencher (4-{[4-(dimethylamino)phenyl]-azo} benzoic acid; DABCYL) at the N terminus, and a fluorescent molecule {5-[(2′-aminoethyl)-amino] naphthalenesulfonic acid; EDANS} at the C terminus (35). Compounds were diluted in buffer A to a final concentration of 200 μM (10 to 200 μM in some experiments); DMSO was used as a solvent control. nsP2 was added to a final concentration of 78 nM, and the mixture was incubated at 22°C for 10 min, after which the protease substrate was added to a final concentration of 15 μM. Continuous fluorescence resonance energy transfer (FRET) measurement was carried out with an excitation wavelength of 340 nm and emission wavelength of 490 nm at 30°C using a Synergy M microplate reader (BioTek, USA). The data were normalized and processed using MS Excel and expressed as relative fluorescence units (RFU).
Antiviral activity and toxicity measurements in cell culture.
Antiviral activity and toxicity measurements in cell culture were performed as previously described (7). Briefly, BHK-21 cells seeded on 96-well white-bottom culture plates (PerkinElmer) were infected with CHIKV-Rluc at a multiplicity of infection (MOI) of 0.01 PFU/cell in infection medium containing minimal essential medium, 0.2% bovine serum albumin, 20 mM HEPES buffer, and 1 mM l-glutamine. Protease inhibitors at final concentrations ranging from 0.02 μM to 200 μM (100 μM for compound 1 and its isomers) or 0.1% DMSO (solvent control) were added along with the virus inoculum and were present throughout the course of the infection. The assay was carried out in triplicate wells. At 16 h postinfection the medium was discarded, cells were lysed, and Rluc activity was measured using a Renilla luciferase assay system (Promega) according to the manufacturer's instructions. Percent inhibition was calculated by comparing values obtained from compound-treated wells with those from infected wells treated with 0.1% DMSO. In parallel, BHK-21 cells were mock infected and treated with inhibitors at final concentrations up to 200 μM. At 16 h posttreatment, cells were lysed and cell viability was measured using a Cell Titer Glo cytotoxicity assay (Promega) according to the manufacturer's instructions. Percent inhibition was calculated based on the readout from mock-infected cells containing 0.1% DMSO. EC50 and 50% cell cytotoxicity (CC50) were calculated by generating dose-response curves using OriginPro software.
Northern blot analysis.
Ninety percent confluent BHK-21 cell cultures (∼3 × 106 cell/plate) were infected with wt CHIKV at an MOI of 10 in the presence of compounds 1c, 3, and 11 (each at 12.5 μM, 25 μM, 50 μM, or 100 μM) or 8 (0.75 μM, 1.5 μM, 3 μM, 6 μM, 12.5 μM, 25 μM, 50 μM, or 100 μM) in GMEM containing 2% bovine serum albumin (BSA). Control cells were infected in 1% DMSO used as a solvent control. After 1 h of incubation at 37°C, the inoculum was removed and replaced with growth medium containing the same concentration of compounds or DMSO. Cells were incubated at 37°C for 6 h, collected, and lysed, and total RNA was purified using TRIzol reagent (Thermo Scientific). Northern blotting was carried out as previously described (36). Briefly, equal amounts of total RNA samples (5 μg) were denatured, separated by electrophoresis in a 1% agarose and 6.6% formaldehyde-containing denaturing gel, and transferred to a Hybond-N+ membrane (GE Healthcare). Membranes were probed with a digoxigenin (DIG)-labeled RNA detection probe complementary to the 3′ untranslated region of the CHIKV genome, washed, and incubated with detection antibody (anti-digoxigenin-alkaline phosphatase [AP] Fab fragments; Roche); CDP-Star (ready to use; Roche) was used to detect the hybridized signals. Finally, the membrane was exposed to X-ray film (medical X-ray films, blue; AGFA).
Time-of-addition assays with compounds 1c, 3, 8, and 11.
For the time-of-addition assay, BHK-21 cell cultures (∼3 × 106 cell/plate) were infected with CHIKV at an MOI of 10; DMSO was used as a solvent control. To analyze the effects of compounds on viral RNA synthesis, cells were pretreated for 2 h with 100 μM concentrations of compounds that were discarded prior to infection with CHIKV. Alternatively, the compounds were added together with virus or at 2 h postinfection and were present until cells were harvested at 6 h postinfection. Total RNA was purified and analyzed as described above. For the analysis of infectious virus release, cells were treated as described above except that in one setup the compounds were added at 4 h postinfection and the virus-containing media were collected at 8 h postinfection. The amounts of released virions were determined using plaque titration in BHK-21 cells.
RESULTS
Molecular design.
Currently there are almost no data on the specific inhibition of the protease activity of CHIKV nsP2, either at the level of individual enzyme or in the context of CHIKV infection. We started from the hypothesis that compounds recently predicted by Bassetto and coauthors using molecular docking modeling and then tested in cell culture experiments (32) indeed act as nsP2 protease inhibitors. In order to find new CHIKV nsP2 inhibitors, compound B1 (here prefix B is used to designate compounds originating from the study by Bassetto et al. [32]) was selected as the lead structure for hit generation.
The B1 molecule was divided into fragments, and a pharmacophore approach was applied. The design of new inhibitors started from the definition of the central (scaffold) structure of the compound that has two fixed planar angles (Fig. 1). The first of them is determined by either cis- or trans-substitution at the cyclopropane ring group, whereas the second rigid angle is fixed by the cis- or trans-substitution at the CH=N double bond. The phenyl group bound to the cyclopropane ring was also included into the central structure, as it can be responsible for potential hydrophobic interactions with the enzyme. The heteroatoms (O and N) that are located in the main structure of B1 are potential hydrogen bond acceptors. The —CH=N— link is attached to comparatively hydrophobic 3,4-diethoxyphenyl group that also contains oxygen atoms as potential hydrogen bond acceptors. In construction of the new compounds, different hydrophobic groups and hydrogen bond acceptors were used as pharmacophore replacements near the cyclopropane ring and —CH=N— link. A modification of the central structure, where the cyclopropane ring was replaced by the —C=C— double bond linker was also examined.
FIG 1.
Pharmacophoric characterization of the starting compound, B1. The compound has two terminal aromatic/hydrophobic groups and hydrogen bonding donor and acceptor centers in the bridge structure.
By displacing substituents with different pharmacophoric analogues, a set of 60 new potential inhibitor structures was generated. The fragments having the biggest effect on the binding energy were selected for further virtual screening of the MolPort and ZINC (29) databases. Using the substructure search, 70 compounds with suitable pharmacophoric groups were found from these databases. Their affinity was again estimated by docking to the CHIKV nsP2 active site. The used docking site was surrounded with amino acid residues Tyr1079, Asn1082, Ser1048, Cys1013, Ala1046, Tyr1047, Trp1084, Leu1205, and Asp1246; among these, Cys1013 (from the catalytic dyad) was the most important residue. This docking site is similar to that used in several previous studies. Thus, in modeling the thiazolidone derivates as CHIKV nsP2 protease inhibitors, the most important binding amino acid residue was found to be Tyr1047 (30), while in another study by the same group, residues Tyr1047, Trp1084, and Asp1246 were determined as the most important ones (38). Similarly, Bassetto and coworkers (32) described the docking pose of inhibitors with the enzyme residues His1083, Cys1013, Asn1082, and Trp1084.
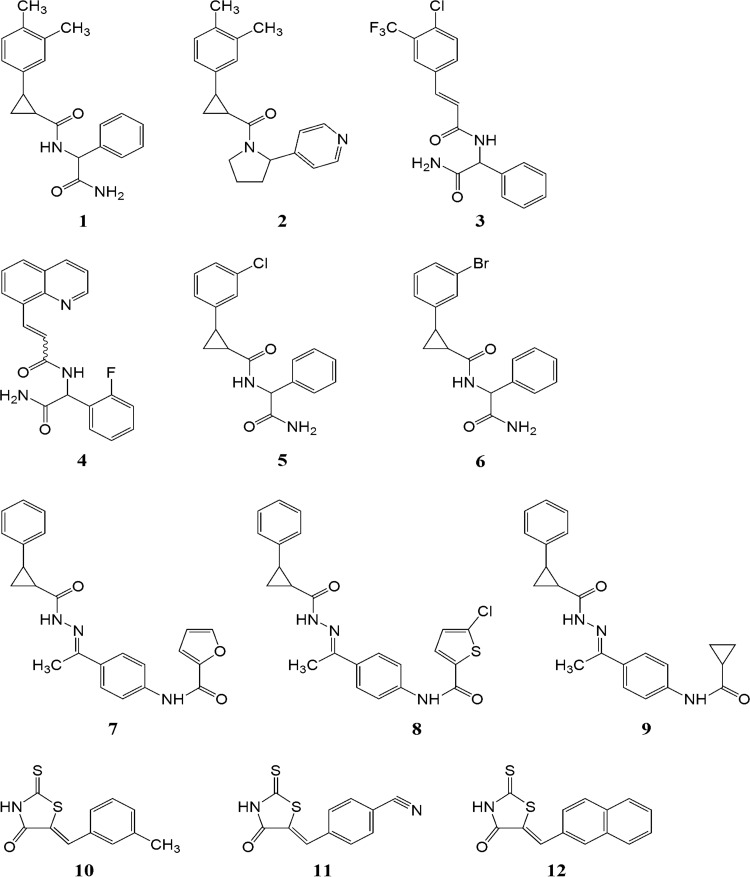
Next the affinities of the generated compounds to the CHIKV nsP2 protease active site were calculated. The molecular docking data revealed that the largest effect on the binding free energy (ΔG) and ligand efficiency (LE) was caused by the variation of hydrophobic groups. The generated compounds also possess the cis-, trans-isomerism at the cyclopropane ring or at the double bond and possible chirality at the terminal acetamidophenyl groups. For final calculations, isomers and conformations with the highest binding energy were used. In total 25 compounds were obtained; of these, 13 compounds were excluded from inhibition assays due to poor solubility and/or cytotoxicity. Thus, the final list of analyzed inhibitors consisted of 12 compounds, designated 1 to 12 (Fig. 2); their docking free energies and LE values are summarized in Table 1.
FIG 2.
Structures of compounds 1 to 12.
TABLE 1.
Docking free energies and ligand efficiencies of analyzed inhibitors
| Compound no. | Molecular mass (Da) | ΔG (kcal/mol) | Ligand efficiencya | EC50 (μM) | CC50 (μM) | SIb |
|---|---|---|---|---|---|---|
| 1 | 322.4 | NAc | NA | 27 | >200 | >7.4 |
| 1a | 322.4 | −8.02 | −0.334 | 82 | >200 | >2.4 |
| 1b | 322.4 | −6.65 | −0.277 | >100 | >200 | NA |
| 1c | 322.4 | −7.34 | −0.306 | 50 | >200 | >4 |
| 1d | 322.4 | −6.97 | −0.290 | 57 | >200 | >3.5 |
| 2 | 320.43 | −7.49 | −0.300 | 73 | >200 | >2.7 |
| 3 | 382.764 | −6.53 | −0.312 | 33 | >200 | >6 |
| 4 | 349.358 | −7.3 | −0.292 | 92 | >200 | >2.1 |
| 5 | 328.793 | −7.04 | −0.320 | 73 | >200 | >2.7 |
| 6 | 373.244 | −7.5 | −0.341 | 71 | >200 | >2.8 |
| 7 | 387.4311 | −8.16 | −0.291 | 11 | >200 | >18.1 |
| 8 | 437.942 | −8.61 | −0.297 | 1.5 | >200 | >133.3 |
| 9 | 361.437 | −8.03 | −0.297 | 22 | >200 | >9 |
| 10 | 235.32 | −6.8 | −0.453 | 69 | >200 | >2.8 |
| 11 | 246.3 | −6.3 | −0.394 | 25 | >200 | >8 |
| 12 | 271.35 | −6.77 | −0.376 | 47 | >200 | >4.2 |
ΔG/number of heavy atoms.
SI, selectivity index (CC50/EC50).
NA, not applicable.
Determination of inhibitory activities using cell-free protease assays.
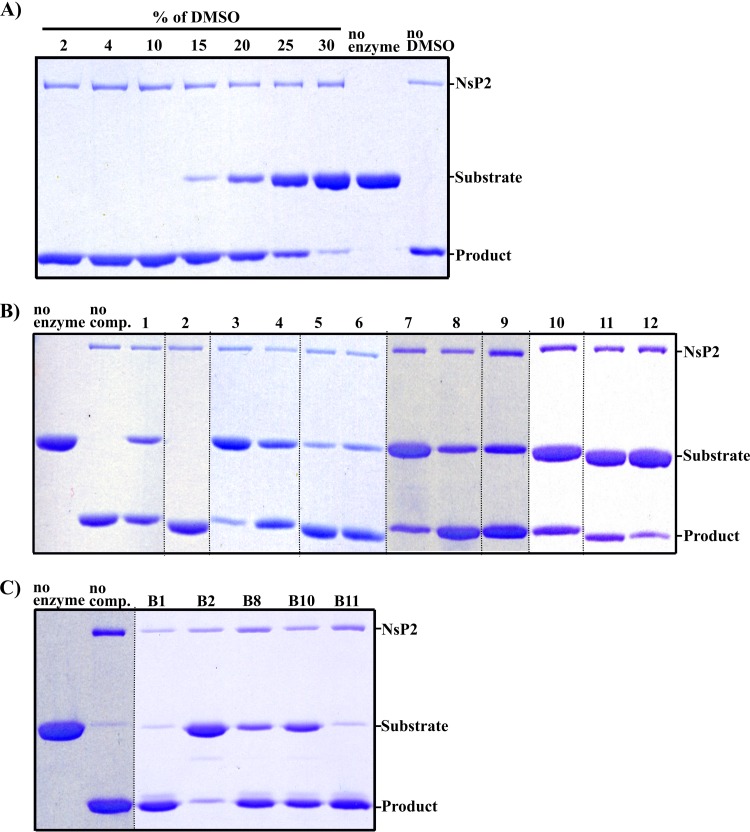
In contrast to previous studies of potential inhibitors of CHIKV nsP2, we first systematically analyzed the ability of the selected compounds to inhibit the protease activity of the purified enzyme in cell-free assays. For the assay, based on the use of recombinant protein substrate, we first determined the maximally allowed concentration of DMSO (used as a solvent). It was found that CHIKV nsP2 protease was fully active in the presence of 10% DMSO (Fig. 3A). Thus, as all stock solutions of compounds were 10 mM, they could be applied at concentrations up to 1 mM. At this concentration all compounds, except compound 2, showed at least some inhibitory activity; the most prominent inhibition was observed for compounds 1, 3, 4, and 7 to 12 (Fig. 3B). It should be, however, noted that under these reaction conditions, compounds 7, 8, and 9 formed a visible precipitate in the reaction mixture; thus, the results with these three compounds in this assay may not be reliable. Furthermore, attempts to use this type of assay to reveal concentration dependence of inhibition failed to produce consistent results (data not shown). As a result, we were unable to make direct comparison of their inhibitory activities, though they clearly affected the cleavage of recombinant protein substrate to different extents (Fig. 3B). We also assayed compound B1 along with compounds B2, B8, B10, and B11 (32). Interestingly, B1 and B11 failed to show notable inhibition (Fig. 3C). Compounds B2, B8, and B10 inhibited the cleavage (Fig. 3C), but they also caused precipitation of components of the reaction mixture. Thus, the experimentally revealed properties of B1 did not corroborate our initial hypothesis. Nevertheless, if somewhat surprisingly, we were able to select 11 new CHIKV nsP2 protease inhibitors, including several rather potent ones.
FIG 3.
Rationally selected compounds inhibit the ability of CHIKV nsP2 to process a recombinant protein substrate. The ratio of enzyme to substrate was ∼1:17. Reactions were carried out at 30°C for 60 min; products were separated by SDS-PAGE and visualized using Coomassie blue staining. (A) Determination of maximally tolerated solvent (DMSO) concentration. (B) Effects of 1 mM compounds 1 to 12 (indicated at the top) on the protease activity of CHIKV nsP2. The image combines three gels that have been rearranged to support the final numbering of compounds; the dotted lines show where different lanes have been merged. (C) Effects of 1 mM compounds B1, B2, B8, B10, and B11 (indicated at the top) on the protease activity of CHIKV nsP2. The dotted lines show where different lanes have been merged. The experiments for each panel were repeated at least three times, with highly similar results.
To compare the efficiencies of different inhibitors, a fluorescence resonance energy transfer (FRET)-based assay, originally described for HIV protease (39), was developed and used. This continuous assay allows gathering of valuable information concerning the initial period of the reaction and can be easily used in a high-throughput format. The FRET-based assay can also use recombinant protein based substrates, such as a pair of fluorescent protein joined by a cleavable linker (40). However, as CHIKV nsP2 cleaves recombinant protein and properly designed peptide substrates with similar efficiencies (K. Rausalu, A. Utt, T. Quirin, F. S. Varghese, E. Žusinaite, P. K. Das, T. Ahola, and A. Merits, submitted for publication), in this study, the simpler synthetic peptide substrate was used.
Continuous monitoring of the reaction progress can be used to identify different modes of inhibition. For this type of assay, it is recommended that the substrate be used at a concentration similar to the Km (41). The Km of CHIKV nsP2 for our peptide substrate is approximately 3 μM (Rausalu et al., submitted); however, 3 μM substrate was found to be insufficient for measuring confident FRET signal intensity (data not shown). Therefore, the peptide substrate was used at 15 μM, which allowed us to balance between a confident signal window and substrate binding modality (41). Initially, this assay was carried out in the presence of a 200 μM concentration of each inhibitor. The obtained results were highly similar to those obtained using recombinant protein substrates. Again, compound 2 failed to inhibit CHIKV nsP2 protease; the same was the case for compounds 5 and 6 (Fig. 4A), which were also the least potent inhibitors in the previous assay (Fig. 3B). All the remaining compounds acted as inhibitors; especially prominent inhibition of the protease activity was observed with the lower-molecular-mass compounds (Table 1) 10 to 12 (Fig. 4A and B). Although we did not specifically intend to test the mode of inhibition (in part because not all of the analyzed compounds were promising inhibitors in virus-based assays; see subsequent sections), it was still evident that compounds 1, 7, 8, and 9 appear to be slowly binding inhibitors (Fig. 4A), while compounds 10 to 12 (and possibly compounds 3 and 4 as well) seem to act as tight binding inhibitors (Fig. 4B) (42).
FIG 4.
Compounds 1 to 12 inhibit the ability of CHIKV nsP2 to process a peptide substrate. In all assays the enzyme concentration was 78 nM, while the concentration of the substrate was 15 μM. Reactions were carried out at 30°C for 60 min. Vertical axes show EDANS fluorescence in relative fluorescence units (RFU). Horizontal axes show reaction time. (A) Effects of compounds 1, 2, 5, 6, 7, 8, and 9 at 200 μM. (B) Effects of compounds 3, 4, 10, 11, and 12 at 200 μM. (C to F) Concentration-dependent inhibition of protease activity by compounds 1 (C), 3 (D), 8 (E), and 11 (F) is shown. Each graph represents averages from at least two independent experiments.
Finally, compounds 1, 3, 8, and 11, which have EC50s between 1.5 to 34 μM in virus inhibition assays (Table 1; see subsequent sections for details), were also shown to inhibit the cleavage of peptide substrate in a concentration-dependent manner (Fig. 4C to F). In this cell-free assay, compound 1 was the least efficient, with an estimated 50% inhibitory concentration (IC50) of ∼200 μM (Fig. 4C); in contrast, compounds 3 and 11 have IC50s of ∼50 μM (Fig. 4D and F). Compound 8, when used at 20 to 100 μM, inhibited the protease activity of CHIKV nsP2 to a roughly similar extent (Fig. 4E), indicating that its IC50 was also about 50 μM.
Synthesis of isomers of compound 1 and analysis of their inhibitory properties.
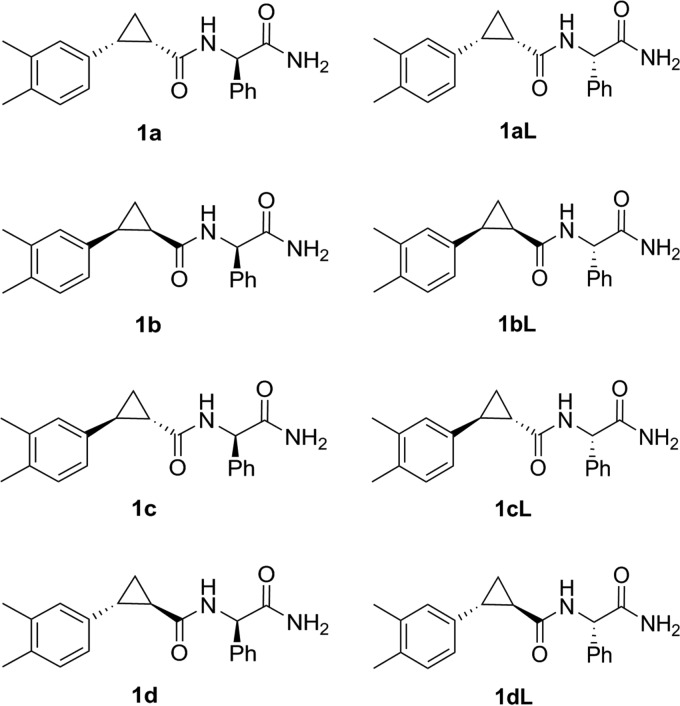
Analysis of the inhibitory properties of different isomers of the same compound can provide valuable information regarding the binding mechanism of the inhibitor to its target (43). Therefore, this aspect was analyzed with compound 1, which was shown to be active in both types of cell-free assays (Fig. 3B and 4A and C). Compound 1 (2-{[2-(3,4-dimethylphepyl)cyclopropyl]formamido}-2-phenylacetamide) has three asymmetric atoms; hence, the material used in previous assays represents a mixture of 8 different isomers (Fig. 5). In order to examine which of these isomers are capable of inhibiting the protease activity of CHIKV nsP2, all of them (designated 1a to 1d and 1aL to 1dL [Fig. 5]) were synthesized (Fig. 6), purified, and analyzed using the above-described assays.
FIG 5.
Structures of isomers of compound 1.
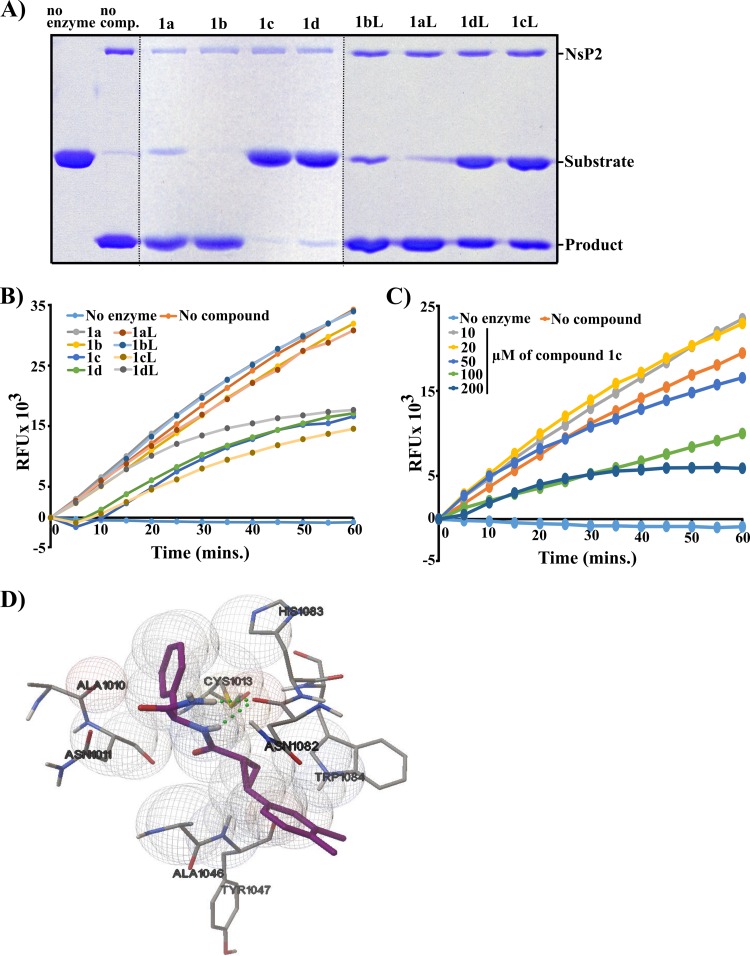
It was found that trans-isomers of compound 1 (1c, 1d, 1cL, and 1dL) acted as potent inhibitors of protease activity (Fig. 7A and B). In contrast, at 200 μM cis-isomers 1a, 1b, 1aL, and 1bL had no detectable inhibitory activity in the FRET-based assay (Fig. 7B). At 1 mM, 1a, 1aL, and 1bL were able to cause moderate inhibition of cleavage of the recombinant protein substrate, while compound 1b displayed no inhibitory activity. Thus, the inhibitory activity of compound 1 clearly depends on cis- or trans-isomerism at the cyclopropane ring (Fig. 5). The fact that compounds 1aL to 1dL had properties very similar to those of their counterparts (1a to 1d) indicates that the orientation of the phenyl group (Fig. 5) has little or no effect on the inhibitory properties of the compound 1 isomers (Fig. 7A and B); based on these data, the isomers 1aL to 1dL were excluded from further analysis.
FIG 7.
Inhibitory properties of different stereoisomers of compound 1. (A) Effects of compounds 1a to 1d and 1aL to 1dL on the ability of CHIKV nsP2 to cleave a recombinant protein substrate. The assay was performed and data are presented as described for Fig. 3. (B) Effects of compounds 1a to 1d and 1aL to 1dL on the ability of CHIKV nsP2 to cleave a peptide substrate. The assay was performed and data are presented as described for Fig. 4A. (C) Concentration-dependent inhibition of protease activity by compound 1c. (D) Molecular docking of compound 1c to the active site of CHIKV nsP2 protease. Hydrogen bonding between either of the amide N-H hydrogens of the inhibitor molecule and the oxygen atom of Asn1082 is shown with a dotted line.
The concentration dependence of inhibition by compound 1c (the most active isomer in the recombinant-protein-based assay [Fig. 7A]) was analyzed using the FRET-based assay. As expected, clear concentration-dependent inhibition was observed, allowing rough estimation of the IC50 of compound 1c, which was ∼100 μM (Fig. 7C).
Anti-CHIKV effects of nsP2 inhibitors in infected cells.
Replication of alphaviruses tolerates considerable variation in P1234 processing efficiencies. Thus, viruses lacking processing at 1/2 and/or 2/3 cleavage sites are viable and can replicate to high titers (44–46). The cleavage of 3/4 sites is absolutely required for infectivity, but even in this case a huge reduction of processing efficiency had little to no effect on virus infection (47). Thus, an efficient inhibitor of nsP2 protease activity does not necessarily cause significant reduction of virus replication. Therefore, the effects of selected compounds on CHIKV multiplication were analyzed next.
In order to minimize the impact of indirect effects of reduced protease activity on virus propagation, the assay was carried out in interferon-negative BHK-21 cells (48). Dose-response curves for EC50 determination (provided in the supplemental material) were generated for all 12 compounds that were tested in the cell-free protease assay. Cytotoxicity assays with these compounds were also performed in the same cells in parallel. None of the compounds caused any cytotoxicity up to 200 μM, the highest concentration that was tested. It was observed that compound 1 and its trans-isomers (1c and 1d) clearly inhibited CHIKV replication, though the EC50s (∼27 to 58 μM) were rather modest. Consistent with data from cell-free assays (Fig. 7A and B), cis-isomers of compound 1 showed little (1a) to no (1b) activity in the antiviral assay (Table 1). Compounds 2, 5, and 6, none of which inhibited cleavage of peptide substrate in the cell-free assay (Fig. 4A), also inhibited CHIKV replication with low efficiency (EC50, >70 μM). Compounds 3, 11, and 12, which prominently inhibited the protease activity in the previous assays, also had lower EC50s, between 25 and 47 μM. In contrast, compounds 4 and 10, which in the protease assay behaved similarly to compounds 3 and 11, respectively (Fig. 4B), did not show corresponding antiviral activity (Table 1). Perhaps most surprisingly, compounds 7, 8, and 9, which were slow-binding inhibitors and showed only moderate efficiency in the cell-free assay (Fig. 4A), were the most effective, with EC50s of 11, 1.5, and 22 μM, respectively (Table 1). This suggests that at least in part their antiviral activity may be due to effects other than inhibition of protease activity of nsP2.
Compounds 1c, 3, 8, and 11 suppress CHIKV RNA synthesis and infectious virus release.
Next, the effects of selected inhibitors on the synthesis of viral positive-strand RNAs were analyzed. For this assay, compounds 1c, 3, 8, and 11, all capable of inhibiting protease activity in a concentration-dependent manner (Fig. 4D to F and 7C) and having EC50s between 1.5 and 50 μM in a virus inhibition assay (Table 1), were selected. Again, the experiment was carried out in BHK-21 cells, but in order to synchronize the infection, a high MOI, 10, was used. The compounds in concentrations up to 100 μM were added to the cells together with the virus and were present in growth media for the entire duration of the assay.
Northern blot analysis revealed that compound 1c had only a modest effect on the synthesis of CHIKV positive-strand RNAs (Fig. 8A). Compound 3 displayed classical concentration-dependent inhibition that was detectable already at 50 μM and became more prominent at 100 μM (Fig. 8A). Compound 8 was unable to suppress RNA replication at concentrations around its EC50 and showed relatively weak inhibition only at the two highest concentrations (>34-fold above its EC50 for virus propagation). Compound 11 had a moderate effect on RNA synthesis at concentrations of ≤50 μM but caused almost a complete block of RNA synthesis at 100 μM (∼4-fold above its EC50). Thus, it was confirmed that compounds used in this assay reduced the accumulation of CHIKV positive-strand RNAs.
FIG 8.
Compounds 3, 8, and 11 inhibit CHIKV positive-strand RNA synthesis and release of infectious virions. (A) Demonstration of concentration-dependent inhibition. Compounds (names and concentrations are shown above the panels) were added to the cells together with virus and present throughout experiment. Control cells were treated with an appropriate amount of a solvent control (DMSO). Total RNAs were extracted and separated; CHIKV positive-stand RNAs were detected using a probe complementary to the 3′ untranslated region of the virus genome. Arrows point out the positions of CHIKV genomic and subgenomic RNAs. (B) Time-of-addition assay (inhibition of positive-strand RNA synthesis). The illustration at the top represents the schema of the experiment. All compounds were used at 100 μM; samples were analyzed and results are presented as described for panel A. (C) Time-of-addition assay (inhibition of infectious virus release). The illustration at the top represents the schema of the experiment. All compounds were used at 100 μM; amounts of released virions were determined using plaque titration. Results were normalized to those from DMSO-treated control cells and expressed as log10 fold reduction of infectious virus titers. Each column represents an average from two experiments performed in triplicate; error bars represent standard deviations.
To obtain more information about the effects of the compounds on CHIKV RNA synthesis, a time-of-addition experiment was carried out (Fig. 8B). In this assay, all compounds were present at higher (100 μM) concentrations. Only compound 8 reduced virus RNA synthesis in pretreated cells (Fig. 8B). Consistent with the results of the previous experiment (Fig. 8A), all four compounds were active when added together with the virus, the effect caused by compound 11 being the most prominent (Fig. 8B). When compounds were added at 2 h postinfection, their effect on CHIKV RNA synthesis was, with the exception of compound 11, minimal (Fig. 8B).
We also analyzed the effects of the compounds on the release of infectious virus progeny. The experiment was performed as described above, except that virus-containing media were harvested 2 h later (at 8 h postinfection) to account for the time needed for infectious virion formation and release (Fig. 8C). The results were consistent with those of the RNA synthesis assay. Again, only compound 8 caused clear reduction of infectious virus production in pretreated cells (Fig. 8B). In contrast, when present throughout the assay, all compounds caused clear reduction of virus yield. Compound 11 was the most efficient inhibitor, resulting in nearly a 1,000-fold reduction of infectious virus release, followed by compounds 8 and 1c (Fig. 8C); exactly the same order of relative potencies was observed in the previous assay (compare Fig. 8B and C). Finally, with the possible exception of compound 1c, the efficiencies of all the inhibitors were reduced when they were added 4 h postinfection; again, compound 11 stood out as the most efficient inhibitor (Fig. 8C). The striking similarity of the results from Northern blot and titration experiments clearly indicates that the reduction of RNA synthesis, caused by the presence of the inhibitors, is the main contributor to the reduced release of infectious virions.
DISCUSSION
CHIKV outbreaks in the Indian Ocean region and currently in the Caribbean/Americas have affected the lives of millions of people. Therefore, the WHO recognizes CHIKV as one of three viral agents causing neglected tropical diseases (http://www.who.int). Continued spread of Aedes mosquitoes, capable of transmitting CHIKV, creates possibilities of new outbreaks in territories with immunologically naive populations. Thus, development of efficient compounds, capable of inhibiting CHIKV infection, is essential. Furthermore, chemical inhibitors also serve as important tools for studies of the molecular biology of a virus and its interactions with the host.
The application of target-based modeling resulted in selection of 12 compounds that were predicted to interact with the active site of CHIKV nsP2 protease. This represents the first set of CHIKV nsP2 protease inhibitors for which, in addition to the in silico predictions, the abilities to inhibit the enzymatic activity of nsP2 protease and virus replication in cell culture have been analyzed. At 200 μM, 75% of these compounds inhibited the protease activity of nsP2 (Fig. 4A and B); the exceptions were compounds 2, 5, and 6. Importantly, with the exception of compounds 4 and 10, all compounds that inhibited the protease activity of nsP2 were also capable of inhibiting CHIKV replication with EC50s of <50 μM. Thus, the efficiency of the applied in silico approaches was appreciably high, even taking into account that 13 predicted potential inhibitors were excluded from detailed analysis due to their toxicity and/or poor solubility. At a glance, this outcome is somewhat unexpected, as our predictions were based on the structure of compound B1 (32), which was subsequently demonstrated to be a very inefficient inhibitor of nsP2 protease activity (Fig. 3C). It should be mentioned, however, that in their study, Bassetto and colleagues did not demonstrate that compound B1 inhibits the enzymatic activity of nsP2 protease; instead, it was shown that it inhibits virus replication with an EC50 of ∼5 μM (32). The fact that in this study the majority of compounds, designed based on B1, were inhibitors of nsP2 (Fig. 3 and 4) is consistent with the possibility that compound B1 binds, as predicted by Bassetto and colleagues, to the active-site region of nsP2 but this binding does not result in the inhibition of protease activity. Thus, the observed inhibition of CHIKV infection may result from compromising some other activity of nsP2 instead. This possibility is indirectly supported by observation that several compounds (most notably compound 8) analyzed in this study also inhibit CHIKV replication with much lower EC50s than could be deduced from their ability to inhibit the protease activity of nsP2 (Fig. 4E and Table 1).
The synthesis and subsequent analysis of stereoisomers of compound 1 clearly demonstrated that the ability of this compound to inhibit nsP2 protease activity and also its antiviral efficiency depend on the conformation of the inhibitor (Fig. 7 and Table 1). The cis-trans-isomerism of the cyclopropane ring was found to be the critical determinant. The docking configuration for the active isomer 1c in the active site of CHIKV nsP2 protease reveals that the inhibitor molecule is surrounded by amino acid residues Ala1010, Asn1011, Ala1046, Tyr1047, Trp1084, Asn1082, and His1083 (Fig. 7D). The aromatic groups of the inhibitor are favorably located close to the hydrophobic area surrounded by Cys1013, Ala1010, Ala1046, and Trp1084. There is also important hydrogen bonding between either of the amide N—H hydrogens of the inhibitor molecule and the oxygen atom of the Asn1082 residue (Fig. 7D). At the same time, the phenyl group of compound 1 is turned away from the enzyme binding pocket, and thus, the relative configuration of it should have little influence on the compound's activity, an assumption that was clearly supported by experimental data (Fig. 7A and B). These findings once again highlight excellent correlation between predicted and experimentally revealed properties of the compounds used in this study. Furthermore, conformation-specific inhibition of protease activity demonstrates the high specificity of the inhibitor and also the high quality of the recombinant protein, opening the possibility to cocrystallize the enzyme with its inhibitor, as was recently done using a peptidomimetic inhibitor of VEEV protease (34). Such structures may further facilitate development of CHIKV nsP2 protease inhibitors as well as provide valuable information about the properties and functions of nsP2.
In this study, the activities of the compounds were not only predicted in silico but also experimentally measured using two different cell-free protease assays (a recombinant protein and a FRET-based peptide cleavage assay) and cell-culture virus inhibition assay; in addition, the effects of four compounds on the synthesis of positive-stand RNAs and on the release of progeny virions were revealed. It was anticipated that the use of different approaches allows confirmation of the findings and, in some cases, reveals additional important properties of the compounds. Indeed, it was observed that compounds that affect the cleavage of recombinant proteins in a similar manner may have different effects on the cleavage of a peptide substrate. Our data suggest that there are at least two different types of inhibitors among the analyzed compounds. Compound 3 displayed a clear concentration-dependent mode of inhibition (Fig. 4D). In contrast, several compounds, including compound 1 (Fig. 4C) and its active isomers (Fig. 7B and C), had little to no effect on the initial speed of cleavage of peptide substrate, and the inhibition became apparent only at later time points; this behavior is consistent with properties of slow-binding inhibitors (49). The existence of different mechanisms of inhibition should be taken into account for analysis of structure-function relationships, as it may have significant impact for additional in silico searches for more active compounds.
In most cases the different assays, from in silico predictions to the analysis of inhibition of virus replication and viral positive-strand RNA synthesis, produced remarkably consistent results. Thus, compounds inactive in protease assays were always poor inhibitors of virus replication; from isomers of compound 1, compound 1b, which had the smallest absolute LE value (Table 1), was also the only one that also lacked any inhibitory properties (Fig. 7A and B). The existence of such correlations serves as an important parameter for the reliability of the results. On this background, however, compounds that produced seemingly conflicting results deserve specific attention. Compounds 4, 10, and, to a lesser extent, 12 were all among the most effective inhibitors of the protease activity of nsP2 (Fig. 3 and 4) but had a low capacity to inhibit CHIKV replication in cell culture. Likely reasons for this include poor ability of these compounds to enter cells and/or their low stability in cell culture assay. It is, however, also possible that these compounds could only bind to and inactivate free nsP2 but were unable to do the same in infected cells, where nsP2 exists as part of a multienzyme complex and its ability to process its cleavage sites is altered by presentation of these sequences to the enzyme (45).
The ability of compounds 1c, 3, 8, and 11 to inhibit the production of progeny virions clearly correlates with their ability to suppress viral positive-strand RNA synthesis (Fig. 8B and C). The inhibitory effects were always more prominent when compounds were present throughout the infection. However, remarkably, when added as late as 4 h postinfection, all these compounds were still able to suppress infectious virus production. Thus, these compounds inhibit some process still ongoing at this stage of infection, nsP2-mediated cleavage of CHIKV P1234 polyprotein being an obvious candidate for this role. This does not, however, exclude the possibility of other targets for these inhibitors. As discussed below, compound 8 almost certainly has another mode(s) of action. Interestingly also, at 100 μM compound 11 is a very efficient inhibitor of viral RNA synthesis, and this inhibition is clearly translated into prominent suppression of progeny virus release; below 100 μM, however, this compound sharply loses its activity (Fig. 8A). As the inhibition profile of virus positive-strand RNA synthesis (Fig. 8A) is clearly different from that of peptide substrate cleavage (Fig. 4F), compound 11 may also have other targets in the virus-infected cells. Additional experiments (such as isolation and characterization of drug-resistant CHIKV mutants) are needed to characterize the true modes of actions of the different inhibitors.
The most interesting behavior was observed for compound 8 and, to a lesser extent, for compound 7. Both of these compounds were clearly more potent inhibitors of virus replication (EC50s, 1.5 μM and 11 μM, respectively) than could be expected based on their potency to inhibit the protease activity of nsP2. Thus, the IC50 of compound 8 in the FRET assay was ∼50 μM, >25-fold higher than its EC50 in the virus inhibition assay (Table 1). This resembles the properties of compound B1, which was reported to have an EC50 of ∼5 μM (32), yet it was virtually unable to inhibit the protease activity of CHIKV nsP2 (Fig. 3C), suggesting the existence of an additional mechanism(s) of action. This possibility was further emphasized by the finding that at concentrations of <50 μM, compound 8 is a rather inefficient inhibitor of CHIKV positive-strand RNA synthesis (Fig. 8A). Among compounds analyzed in this study, compound 8 has the biggest calculated absolute ΔG value (−8.61 kcal/mol [Table 1]), indicating efficient binding to nsP2. It should also be noted that compound 8 has also the highest molecular mass among the analyzed compounds (Table 1). It is possible that binding of compound 8 has only a moderate effect on the protease activity of nsP2 and, consequently, also on the synthesis of viral RNAs. Instead, it could be speculated that binding of compound 8 to its target affects more prominently some other function(s) of nsP2; the same may be true for other compounds, such as 7 and B1. As the pronounced inhibitory effect of compound 8 was detected in experiments carried out using low MOI (virus inhibition) but not using a high MOI (positive-strand RNA synthesis), it can be speculated that compound 8 may also inhibit virus spread in cell culture; this property is crucial under low-MOI, but not under high-MOI, conditions. Previous studies carried out using VEEV have established a functional link between nsP2 and alphavirus particle formation. Thus, it has been observed that replication of a VEEV mutant in which the viral subgenomic promoter was replaced by an encephalomyocarditis virus internal ribosomal entry site was strongly enhanced by adaptive mutations located in the helicase region nsP2 (50). Recently, it was also shown that both titers and cytotoxic effects of a VEEV mutant harboring multiple mutations in the capsid protein were increased by adaptive mutations mapped to the protease domain of nsP2 or to the N-terminal part of nsP2 (51), which is known to act as a cofactor of alphavirus nsP2 protease (52). Therefore, it could be put forth that indirect inhibition efficiencies of some compounds may originate from effects of these compounds on the formation and release of virus particles; this topic is under investigation in our laboratories. In addition, pretreatment of cells with compound 8 inhibits both viral positive-strand RNA synthesis and infectious virus production (Fig. 8B and C). This also indicates an existence of yet another mechanism of action which is unlikely to be mediated by the suppression of nsP2's activities. Therefore, the mechanism may be cell mediated rather than based on the direct inhibition of CHIKV replication; its analysis also represents a topic for further studies.
To our knowledge, this study represents the first example of a rational search for inhibitors against CHIKV nsP2 protease in which in silico predictions were combined with the use of cell-free protease assays and cell-based virus inhibition assays. Several novel inhibitors were identified, providing new information for development of further antiviral compounds as well as tools to study CHIKV infection. Furthermore, taking into account that the known 3D structures of alphaviral nsP2 proteases are similar to each other, it is also reasonable to expect that at least some of the inhibitors analyzed in this study are also active against other alphaviruses which, similar to CHIKV, have the potential to become serious human pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Kai Rausalu for her help in performing cell-free protease assays and Tõnis Pehk for relative conformation assignment of stereoisomers of compound 1. We acknowledge the Drug Discovery and Chemical Biology Network funded by Biocenter Finland, for providing access to screening instrumentation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01421-16.
REFERENCES
- 1.Weaver SC, Lecuit M. 2015. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med 372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 2.Ahola T, Couderc T, Courderc T, Ng LFP, Hallengärd D, Powers A, Lecuit M, Esteban M, Merits A, Roques P, Liljeström P. 2015. Therapeutics and vaccines against chikungunya virus. Vector Borne Zoonotic Dis 15:250–257. doi: 10.1089/vbz.2014.1681. [DOI] [PubMed] [Google Scholar]
- 3.Karlas A, Berre S, Couderc T, Varjak M, Braun P, Meyer M, Gangneux N, Karo-Astover L, Weege F, Raftery M, Schönrich G, Klemm U, Wurzlbauer A, Bracher F, Merits A, Meyer TF, Lecuit M. 2016. A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat Commun 7:11320. doi: 10.1038/ncomms11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lani R, Hassandarvish P, Chiam CW, Moghaddam E, Chu JJH, Rausalu K, Merits A, Higgs S, Vanlandingham D, Abu Bakar S, Zandi K. 2015. Antiviral activity of silymarin against chikungunya virus. Sci Rep 5:11421. doi: 10.1038/srep11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pohjala L, Utt A, Varjak M, Lulla A, Merits A, Ahola T, Tammela P. 2011. Inhibitors of alphavirus entry and replication identified with a stable Chikungunya replicon cell line and virus-based assays. PLoS One 6:e28923. doi: 10.1371/journal.pone.0028923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rathore APS, Haystead T, Das PK, Merits A, Ng M-L, Vasudevan SG. 2013. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antiviral Res 103C:7–16. [DOI] [PubMed] [Google Scholar]
- 7.Varghese FS, Kaukinen P, Gläsker S, Bespalov M, Hanski L, Wennerberg K, Kümmerer BM, Ahola T. 2016. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral Res 126:117–124. doi: 10.1016/j.antiviral.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Varghese FS, Thaa B, Amrun SN, Simarmata D, Rausalu K, Nyman TA, Merits A, McInerney GM, Ng LFP, Ahola T. 17 August 2016. The antiviral alkaloid berberine reduces chikungunya virus-induced mitogen-activated protein kinase (MAPK) signaling. J Virol doi: 10.1128/JVI.01382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pohjala L, Barai V, Azhayev A, Lapinjoki S, Ahola T. 2008. A luciferase-based screening method for inhibitors of alphavirus replication applied to nucleoside analogues. Antiviral Res 78:215–222. doi: 10.1016/j.antiviral.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Ching K-C, Kam Y-W, Merits A, Ng LFP, Chai CLL. 2015. Trisubstituted thieno[3,2-b]pyrrole 5-carboxamides as potent inhibitors of alphaviruses. J Med Chem 58:9196–9213. doi: 10.1021/acs.jmedchem.5b01047. [DOI] [PubMed] [Google Scholar]
- 11.Gigante A, Canela M-D, Delang L, Priego E-M, Camarasa M-J, Querat G, Neyts J, Leyssen P, Pérez-Pérez M-J. 2014. Identification of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones as novel inhibitors of Chikungunya virus replication. J Med Chem 57:4000–4008. doi: 10.1021/jm401844c. [DOI] [PubMed] [Google Scholar]
- 12.Staveness D, Abdelnabi R, Near KE, Nakagawa Y, Neyts J, Delang L, Leyssen P, Wender PA. 2016. Inhibition of Chikungunya virus-induced cell death by salicylate-derived bryostatin analogues provides additional evidence for a PKC-independent pathway. J Nat Prod 79:680–684. doi: 10.1021/acs.jnatprod.5b01017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and nonstructural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahola T, Kääriäinen L. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci U S A 92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahola T, Lampio A, Auvinen P, Kääriäinen L. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J 18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das PK, Merits A, Lulla A. 2014. Functional crosstalk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity. J Biol Chem 289:5635–5653. doi: 10.1074/jbc.M113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karpe YA, Aher PP, Lole KS. 2011. NTPase and 5′-RNA triphosphatase activities of chikungunya virus nsP2 protein. PLoS One 6:e22336. doi: 10.1371/journal.pone.0022336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastorino BAM, Peyrefitte CN, Almeras L, Grandadam M, Rolland D, Tolou HJ, Bessaud M. 2008. Expression and biochemical characterization of nsP2 cysteine protease of Chikungunya virus. Virus Res 131:293–298. doi: 10.1016/j.virusres.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egloff M-P, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, Ahola T, Canard B. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Debing Y, Jankevicius G, Neyts J, Ahel I, Coutard B, Canard B. 2016. Viral macro domains reverse protein ADP-ribosylation. J Virol 90:8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bullard-Feibelman KM, Fuller BP, Geiss BJ. 2016. A sensitive and robust high-throughput screening assay for inhibitors of the chikungunya virus nsP1 capping enzyme. PLoS One 11:e0158923. doi: 10.1371/journal.pone.0158923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delang L, Li C, Tas A, Quérat G, Albulescu IC, De Burghgraeve T, Guerrero NAS, Gigante A, Piorkowski G, Decroly E, Jochmans D, Canard B, Snijder EJ, Pérez-Pérez MJ, van Hemert MJ, Coutard B, Leyssen P, Neyts J. 2016. The viral capping enzyme nsP1: a novel target for the inhibition of chikungunya virus infection. Sci Rep 6:31819. doi: 10.1038/srep31819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albulescu IC, Tas A, Scholte FEM, Snijder EJ, van Hemert MJ. 2014. An in vitro assay to study chikungunya virus RNA synthesis and the mode of action of inhibitors. J Gen Virol 95(Part 12):2683–2692. doi: 10.1099/vir.0.069690-0. [DOI] [PubMed] [Google Scholar]
- 24.Utt A, Quirin T, Saul S, Hellström K, Ahola T, Merits A. 2016. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One 11:e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malet H, Coutard B, Jamal S, Dutartre H, Papageorgiou N, Neuvonen M, Ahola T, Forrester N, Gould EA, Lafitte D, Ferron F, Lescar J, Gorbalenya AE, de Lamballerie X, Canard B. 2009. The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol 83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen PTV, Yu H, Keller PA. 2014. Discovery of in silico hits targeting the nsP3 macro domain of chikungunya virus. J Mol Model 20:2216. doi: 10.1007/s00894-014-2216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo AT, White MA, Watowich SJ. 2006. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 14:1449–1458. [DOI] [PubMed] [Google Scholar]
- 28.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. 2006. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 29.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. 2012. ZINC: a free tool to discover chemistry for biology. J Chem Infect Model 52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jadav SS, Sinha BN, Hilgenfeld R, Pastorino B, de Lamballerie X, Jayaprakash V. 2015. Thiazolidone derivatives as inhibitors of chikungunya virus. Eur J Med Chem 89:172–178. doi: 10.1016/j.ejmech.2014.10.042. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen PTV, Yu H, Keller PA. 2015. Identification of chikungunya virus nsP2 protease inhibitors using structure-base approaches. J Mol Graph Model 57:1–8. doi: 10.1016/j.jmgm.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Bassetto M, De Burghgraeve T, Delang L, Massarotti A, Coluccia A, Zonta N, Gatti V, Colombano G, Sorba G, Silvestri R, Tron GC, Neyts J, Leyssen P, Brancale A. 2013. Computer-aided identification, design and synthesis of a novel series of compounds with selective antiviral activity against chikungunya virus. Antiviral Res 98:12–18. doi: 10.1016/j.antiviral.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 33.El-labbad EM, Ismail MAH, Abou Ei Ella DA, Ahmed M, Wang F, Barakat KH, Abouzid KAM. 2015. Discovery of novel peptidomimetics as irreversible CHIKV NsP2 protease inhibitors using quantum mechanical-based ligand descriptors. Chem Biol Drug Des 86:1518–1527. doi: 10.1111/cbdd.12621. [DOI] [PubMed] [Google Scholar]
- 34.Hu X, Compton JR, Leary DH, Olson MA, Lee MS, Cheung J, Ye W, Ferrer M, Southall N, Jadhav A, Morazzani EM, Glass PJ, Marugan J, Legler PM. 19 May 2016. Kinetic, mutational, and structural studies of the Venezuelan equine encephalitis virus nonstructural protein 2 cysteine protease. Biochemistry doi: 10.1021/acs.biochem.5b00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stapleford KA, Rozen-Gagnon K, Das PK, Saul S, Poirier EZ, Blanc H, Vidalain P-O, Merits A, Vignuzzi M. 2015. Viral polymerase-helicase complexes regulate replication fidelity to overcome intracellular nucleotide depletion. J Virol 89:11233–11244. doi: 10.1128/JVI.01553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Utt A, Das PK, Varjak M, Lulla V, Lulla A, Merits A. 2015. Mutations conferring a noncytotoxic phenotype on chikungunya virus replicons compromise enzymatic properties of nonstructural protein 2. J Virol 89:3145–3162. doi: 10.1128/JVI.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha GB, Freire RO, Simas AM, Stewart JJP. 2006. RM1: a reparameterization of AM1 for H, C, N, O, P, S, F, Cl, Br, and I. J Comput Chem 27:1101–1111. doi: 10.1002/jcc.20425. [DOI] [PubMed] [Google Scholar]
- 38.Jadav SS, Jayaprakash V, Basu A, Sinha BN. 2012. Chikungunya protease domain–high throughput virtual screening. Int Scholarly Sci Res Innovation 6:718. [Google Scholar]
- 39.Matayoshi ED, Wang GT, Krafft GA, Erickson J. 1990. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 40.Emmott E, Sweeney TR, Goodfellow I. 2015. A cell-based fluorescence resonance energy transfer (FRET) sensor reveals inter- and intragenogroup variations in norovirus protease activity and polyprotein cleavage. J Biol Chem 290:27841–27853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acker MG, Auld DS. 2014. Considerations for the design and reporting of enzyme assays in high-throughput screening applications. Perspect Sci 1:56–73. doi: 10.1016/j.pisc.2013.12.001. [DOI] [Google Scholar]
- 42.Strelow J, Dewe W, Iversen PW, Brooks HB, Radding JA, McGee J, Weidner J. 2004. Mechanism of action assays for enzymes, p 60–86. In Sittampalam GS, Coussens NP, Nelson H, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Iversen PW, Li Z, McGee J, McManus O, Minor L, Napper A, Peltier JM, Riss T, Trask OJ, Weidner J (ed), Assay guidance manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD. [PubMed] [Google Scholar]
- 43.Desai MC, Meanwell NA (ed). 2013. Successful strategies for the discovery of antiviral drugs. Royal Society of Chemistry, Cambridge, UK. [Google Scholar]
- 44.Gorchakov R, Frolova E, Sawicki S, Atasheva S, Sawicki D, Frolov I. 2008. A new role for ns polyprotein cleavage in Sindbis virus replication. J Virol 82:6218–6231. doi: 10.1128/JVI.02624-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lulla V, Karo-Astover L, Rausalu K, Merits A, Lulla A. 2013. Presentation overrides specificity: probing the plasticity of alphaviral proteolytic activity through mutational analysis. J Virol 87:10207–10220. doi: 10.1128/JVI.01485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mai J, Sawicki SG, Sawicki DL. 2009. Fate of minus-strand templates and replication complexes produced by a p23-cleavage-defective mutant of Sindbis virus. J Virol 83:8553–8564. doi: 10.1128/JVI.00056-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lulla A, Lulla V, Tints K, Ahola T, Merits A. 2006. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J Virol 80:5413–5422. doi: 10.1128/JVI.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breakwell L, Dosenovic P, Karlsson Hedestam GB, D'Amato M, Liljeström P, Fazakerley J, McInerney GM. 2007. Semliki Forest virus nonstructural protein 2 is involved in suppression of the type I interferon response. J Virol 81:8677–8684. doi: 10.1128/JVI.02411-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waley SG. 1993. The kinetics of slow-binding and slow, tight-binding inhibition: the effects of substrate depletion. Biochem J 294:195–200. doi: 10.1042/bj2940195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volkova E, Frolova E, Darwin JR, Forrester NL, Weaver SC, Frolov I. 2008. IRES-dependent replication of Venezuelan equine encephalitis virus makes it highly attenuated and incapable of replicating in mosquito cells. Virology 377:160–169. doi: 10.1016/j.virol.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim DY, Atasheva S, Frolova EI, Frolov I. 2013. Venezuelan equine encephalitis virus nsP2 protein regulates packaging of viral genome into infectious virions. J Virol 87:4202–4213. doi: 10.1128/JVI.03142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lulla A, Lulla V, Merits A. 2012. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J Virol 86:553–565. doi: 10.1128/JVI.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.