Abstract
Wastewater discharged from clinical isolation and general wards at two hospitals in Singapore was examined to determine the emerging trends of antibiotic resistance (AR). We quantified the concentrations of 12 antibiotic compounds by analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS), antibiotic-resistant bacteria (ARB), the class 1 integrase gene (intI1), and 16 antibiotic resistance genes (ARGs) that confer resistance to 10 different clinically relevant antibiotics. A subset of 119 antibiotic-resistant isolates were phylogenetically classified and tested for the presence of ARGs encoding resistance to β-lactam antibiotics (blaNDM, blaKPC, blaSHV, blaCTX-M), amikacin [aac(6′)-Ib], co-trimoxazole (sul1, sul2, dfrA), ciprofloxacin (qnrA, qnrB), and the intI1 gene. Among these resistant isolates, 80.7% were detected with intI1 and 66.4% were found to carry at least 1 of the tested ARGs. Among 3 sampled locations, the clinical isolation ward had the highest concentrations of ARB and the highest levels of ARGs linked to resistance to β-lactam (blaKPC), co-trimoxazole (sul1, sul2, dfrA), amikacin [aac(6′)-Ib], ciprofloxacin (qnrA), and intI1. We found strong positive correlations (P < 0.05) between concentrations of bacteria resistant to meropenem, ceftazidime, amikacin, co-trimoxazole, and ciprofloxacin and abundances of blaKPC, aac(6′)-Ib, sul1, sul2, dfrA, qnrA, and intI1 genes.
INTRODUCTION
The emergence of antibiotic resistance (AR) has led to heightened global concern due to the reduced efficacy of currently available antibiotics in the treatment of bacterial infections (1, 2). Gram-negative pathogens such as Escherichia coli and Klebsiella pneumoniae have shown increased resistance to a variety of antibiotics typically used in the treatment of infections and diseases caused by these bacteria (3). Hospital wastewater is considered a hot spot for AR as a consequence of receiving a cocktail of antibiotic compounds, disinfectants, and inputs of bacterial sheddings and metabolized drugs from patient excrement, which potentially contain multidrug-resistant (MDR) pathogens (4–6). As such, hospital wastewaters provide an environment for the exchange of antibiotic resistance genes (ARGs) between clinical pathogens and other environmental bacteria in recipient sewers, which could result in broader epidemiological consequences extending beyond the hospital setting.
Integrons enable the recombination and expression of mobile gene cassette arrays which contain ARGs and are used as indicators to gather information on trends of AR development and dissemination in bacterial communities (7, 8). Gram-negative bacteria bearing multiple bla genes (e.g., blaNDM, blaKPC, blaCTX-M, and blaSHV) are a problem and are increasingly found in hospital wastewaters (4, 9), together with other common and related drug-resistant ARGs such as qnr (fluoroquinolone), erm (macrolide), sul (sulfonamides), and tet (tetracycline). High densities of MDR bacteria in hospital wastewaters could facilitate the propagation and dissemination of ARGs by horizontal gene transfer via plasmids, transposons, and integrons (8, 10–12). In a preventive effort aimed at reducing the spread of antimicrobial resistance and formulating intervention strategies, clinical institutions have taken steps to conduct surveillance studies on antibiotic prescriptions in hospitals through longitudinal assessments of antibiotic prescription patterns (13). More specifically, linking information from monitoring studies of the fate of antibiotics to data on the occurrence of the type and abundances of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) will provide a direct and concise measure of dissemination patterns of antibiotic resistance.
The objective of this study was to quantify antibiotic residues (AB) using liquid chromatography-tandem mass spectrometry (LC-MS/MS), antibiotic-resistant bacteria (ARB) using a culture-based method, and antibiotic resistance genes (ARGs) using quantitative PCR (qPCR) in hospital wastewaters in Singapore. This improves our current knowledge of the potential impacts of hospital wastewater on antibiotic resistance in the environment under tropical conditions. The correlation between these antibiotic resistance factors and the class 1 integron was analyzed to evaluate the potential dissemination of multidrug resistance across the microbial community. The results provide important baseline data and highlight the more prevalent ARB and ARGs found in local hospitals in Asia, as well as levels of antibiotics present.
MATERIALS AND METHODS
Collection of hospital wastewater samples.
Eight clinical wastewater samples were collected from two hospitals in Singapore. Two hospital blocks (i.e., block a of hospital 1 [H1a] and block b of hospital 1 [H1b]) (1,597 beds) were sampled once a week over a period of 2 weeks from a manhole receiving direct sewage from each block. These two blocks were differentiated based on their ward types, with block A (i.e., H1a) consisting of clinical isolation wards and block B (i.e., H1b) consisting of more-general wards. For hospital 2 (i.e., H2; 1,500 beds), samples were collected once a week over a period of 1 month from the main manhole discharging mixed wastewater from the entire hospital. All samples were collected on weekdays in the morning between 9:30 a.m. and 11:30 a.m. during September 2014 at ambient temperatures ranging from 24°C to 31°C. A volume of 1 liter of wastewater was collected from each site using sterile plastic bottles, preserved on ice, and transported to the laboratory (within 2 h) for chemical and microbiological analyses. The 1-liter sample was divided into two 0.5-liter samples which were reserved for DNA extraction and chemical analysis.
DNA extraction.
A volume of 0.5 liter of hospital wastewater was filtered through a 0.45-μm-pore-size cellulose nitrate membrane (Sartorius Stedim, Gottingen, Germany). Total DNA was extracted from the biomass using a PowerWater DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. DNA concentration and purity were measured using a NanoDrop spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific, Wilmington, DE, USA).
Quantification of antibiotics.
The 0.5 liter of hospital wastewater samples was processed immediately upon arrival in the laboratory by filtration through a 0.1-μm-pore-size Millipore glass fiber filter (Millipore, Bedford, MA, USA) and adjusted to a pH of 2.4. Extraction of antibiotics was performed using two solid-phase extraction (SPE) cartridges in series: first with a 500-mg LC-NH2 (Supelco, Sigma-Aldrich, MO, USA) SPE anionic-exchange cartridge to remove part of the impurities and then with a polymeric 500-mg Oasis HLB (Waters, Milford, CT) to retain target compounds. Final compounds were eluted with 9 ml of methanol in amber glass vials, dried under a stream of nitrogen, and reconstituted with 0.9 ml methanol. An LC-electrospray ionization (ESI)-MS/MS analysis performed on an Agilent 1290 Infinity LC coupled to a 6490 Triple Quad MS/MS system (Agilent Technologies Inc. Santa Clara, CA) was used to detect concentrations of 12 antibiotics in the hospital wastewater samples: meropenem (MEM), lincomycin, sulfamethazine, trimethoprim, tetracycline, ciprofloxacin (CIP), sulfamethoxazole, chloramphenicol (CHL), azithromycin, clindamycin (CLI), erythromycin (ERI), and clarithromycin. To identify these antibiotic targets, the retention times and the area ratios of the qualitative and quantitative ions in each sample were compared with those of the calibration standards. The results were accepted when (i) the retention time was within ±15 s of the average retention time in the calibration standards; (ii) the uncertainty of two product ions was within ±20% of that in the standards; and (iii) the signal/noise (S/N) ratio of the quantitative ions was higher than 3. The quantitation process followed USEPA method 1694 and was performed in duplicate.
The SPE recoveries (see Table S1 in the supplemental material) were tested by spiking four replicates of reagent water with antibiotics and processing the spiked waters through the entire analytical method. The overall recoveries, taking into consideration both the SPE and matrix recoveries, were tested by spiking four different sewage samples with antibiotics and analyzing the spiked samples through the entire analytical method. Antibiotics (in methanol) were spiked at 1 μg/liter. The procedural recoveries and overall recoveries and their relative standard deviations are listed in Table S1. Samples were analyzed in duplicate; one procedural blank consisting of reagent water was processed together with each batch of sample to demonstrate that the analytical system and glassware were free of contamination.
Quantification of ARB.
To determine the concentration of bacteria resistant to each of the 10 antibiotics tested in the study, hospital wastewater samples were serially diluted and cultured on LB agar (BD Diagnostics, Sparks, MD, USA) supplemented with each of the 10 antibiotics. Antibiotics (Sigma-Aldrich, USA) were used in the screening process at the following concentrations: amikacin (AMK) at 17 μg/ml; meropenem (MEM) at 4 μg/ml; ceftazidime (CAZ) at 8 μg/ml; clindamycin (CLI) at 0.5 μg/ml; erythromycin (ERY) at 0.5 μg/ml; ciprofloxacin (CIP) at 2 μg/ml; co-trimoxazole (SXT; trimethoprim at 3 μg/ml and sulfamethoxazole at 57 μg/ml); tetracycline (TET) at 4 μg/ml; vancomycin (VAN) at 4 μg/ml; and chloramphenicol (CHL) at 8 μg/ml. All of the antibiotic concentrations used for screening were higher than the susceptible breakpoints published in the 2013 Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing report, except for the concentrations of erythromycin and clindamycin, which were used at their breakpoints. Raw water samples were serially diluted in 1× phosphate-buffered saline (PBS) (Vivantis Technologies, Malaysia), and 100-μl volumes of samples were spread-plated in triplicate and incubated at 35°C for 24 h and subsequently at room temperature the next day for an additional 24 h. Dilution-containing colonies within the range of 30 to 100 cells were counted in order to minimize the error caused by the too-numerous-to-count problem.
Quantification of ARGs.
Specific primers for real-time PCR were selected from the literature and designed in this study for quantification of the 16S rRNA gene (14, 15), intI1 (16), and 16 other ARGs, including blaKPC (17), blaNDM (this study), blaSHV (18), blaCTX-M (19), sul1 (this study), sul2 (20), dfrA (21), aac(6′)-Ib (this study), tet(M) (22), tet(O) (23), erm(B) (24), mef(A/E) (25), cfr (26), vanA (this study), qnrA (27), and qnrB (27). These genes confer resistance to the 10 antibiotics used in our phenotypic resistance screening (see Table S2 in the supplemental material). Total genomic DNA directly extracted from hospital wastewaters (see “DNA extraction” section) was used for qPCR assays, and analysis of each sample was performed in triplicate within the same run together with a calibration curve and a no-template control (NTC). The calibration curves were built using a 10-fold dilution series of ARG standards. Each qPCR mixture contained 10 μl SYBR Select master mix (Applied Biosystems, CA, USA), 0.5 μl each of 10 μM concentrations of forward primers and reverse primers, and 20 to 50 ng of DNA template and was made up to a volume of 20 μl with PCR-grade water. The qPCR assays were conducted in a StepOnePlus real-time PCR system (Applied Biosystems, CA, USA) under the following conditions: holding at 50°C for 2 min and 95°C for 3 min; 40 cycles of 95°C for 15 s and annealing temperature for 1 min; and, finally, a melting curve analysis performed at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Only amplicons with a single peak in the melting curve analysis were considered specific amplified products of targeted genes. All qPCR assays were performed according to the manufacturer's specifications. Calculation of the absolute gene copy numbers followed a previously described procedure (28, 29). Relative gene abundances were calculated by normalizing the absolute number of ARG copies to that of 16S rRNA gene copies.
Identification and ARG detection of ARB isolates.
A total of 236 colonies were randomly picked from plates containing 1 of the 10 antibiotics and were subcultured to ensure that pure isolates were obtained. Single colonies were inoculated into liquid LB broth and allowed to grow overnight at 37°C, and 10 μl of culture suspension was subsequently subjected to 16S rRNA gene PCR amplification using the 27F (5′-AGA GTT TGA TYM TGG CTC AG-3′) and 1492R (5′-GGY TAC CTT GTT ACG ACT T-3′) primers. The PCR products were purified using an Expin cleanup kit (GeneAll Biotechnology, Seoul, South Korea) and sent for capillary sequencing at AITbiotech (Singapore). Sequences were manually edited using Bioedit software and queried against the National Center for Biotechnology Information (NCBI) 16S rRNA gene database for archaea and bacteria. The bacterial gene sequences were deposited in GenBank (see below for accession numbers).
In addition, bacterial isolates showing phenotypic resistance to the following antibiotics were screened for the presence of associated ARGs (in parentheses): β-lactam (blaNDM, blaKPC, blaCTX-M, blaSHV), amikacin [aac(6′)-Ib], co-trimoxazole (sul1, sul2, dfrA), ciprofloxacin (qnrA, qnrB), and integrase 1 (intI1). Conventional PCR assay was used, and the working conditions were as follows: 95°C for 5 min; 35 repeats of a cycle of 95°C for 30 s, an annealing temperature for 30 s, and 72°C for 30 s; and, finally, 72°C for 10 min. The optimal annealing temperature for each primer was tested with a temperature gradient from 55°C to 65°C, as shown in Table S2 in the supplemental material. The PCR products were sequenced and queried against the NCBI nonredundant database using BLAST.
Statistical analysis.
Statistical analysis was performed using a software package (Primer version 6 and Origin version 9.0). For every analysis of AB, ARB, and ARGs, three replicates were performed, and the averages of the results were calculated. Differences in average antibiotic, ARB, and ARG concentrations among samples were statistically verified using analysis of variance (ANOVA) and/or Student's t test. Correlations between ARB concentrations, ARG abundances, and antibiotic concentrations were analyzed using Spearman and Pearson tests. A principal component analysis (PCA) was employed to assess relationships between measured concentrations of antibiotic residues and cultivated ARB and ARGs for each sampled location.
Accession number(s).
The bacterial gene sequences determined in this work were deposited in GenBank with accession numbers KU312692 to KU312927.
RESULTS
Occurrence of antibiotic residues in hospital wastewater.
Trimethoprim, sulfamethoxazole, erythromycin, azithromycin, and clarithromycin were detected in all samples at various concentrations fluctuating between 0.78 and 71.8 μg/liter, 0.94 and 28.36 μg/liter, 0.30 and 17.63 μg/liter, 0.11 and 1.51 μg/liter, and 0.76 and 72.87 μg/liter, respectively, with azithromycin having the lowest concentration among the five antibiotics (Table 1). Ciprofloxacin was detected at higher levels in location H1b, and tetracycline was detected only in location H2 at a low level whereas meropenem, lincomycin, sulfamethazine, chloramphenicol, and clindamycin levels were relatively low or below the method detection limit (MDL). As a general trend, the concentrations of antibiotics in the location H1b samples were slightly higher than those in the wastewater samples from the two other locations.
TABLE 1.
Concentration range of antibiotics detected for hospital wastewater samplesa
| Antibiotic | Concentration(s) (μg/liter) |
Method detection limit (μg/liter) | ||
|---|---|---|---|---|
| Location H1a | Location H1b | Location H2 | ||
| Azithromycin | 0.18–1.511 | 0.13–1.16 | 0.11–0.72 | 0.0036 |
| Chloramphenicol | <MDL–0.65 | <MDL | <MDL | 0.0378 |
| Ciprofloxacin | <MDL | 8.74–76.44 | 1.72–4.34 | 0.1405 |
| Clarithromycin | 1.29–20.49 | 0.76–72.87 | 0.76–5.11 | 0.0185 |
| Clindamycin | <MDL | <MDL | <MDL–1.43 | 0.0559 |
| Erythromycin | 1.64–8.09 | 0.49–13.71 | 0.3–17.63 | 0.0332 |
| Lincomycin | <MDL | <MDL | <MDL–0.015 | 0.0030 |
| Meropenem | <MDL–0.19 | <MDL–1.01 | <MDL–1.07 | 0.0303 |
| Sulfamethazine | <MDL | <MDL | <MDL | 0.0160 |
| Sulfamethoxazole | 19.90–24.43 | 7.21–28.36 | 0.94–23.92 | 0.0445 |
| Tetracycline | <MDL | <MDL | <MDL–0.82 | 0.1017 |
| Trimethoprim | 8.63–13.83 | 6.61–71.8 | 0.78–11.87 | 0.0155 |
H1a, hospital 1, at block A; H1b, hospital 1, at block B; H2, hospital 2, at the mixed manhole (i.e., the main manhole discharging mixed wastewater from the entire hospital); MDL, method detection limit.
Occurrence of ARB in hospital wastewater.
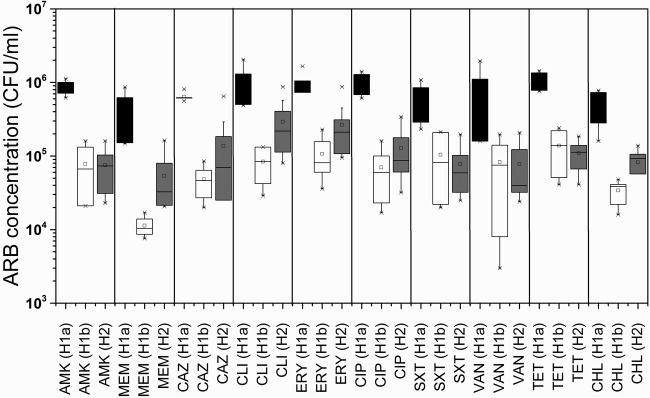
The average concentration of cultivable ARB in the H1a samples (7.64 × 106 CFU/ml) was 6-fold higher than that in the H2 samples (1.30 × 106 CFU/ml) and 10-fold higher than that in the H1b samples (7.57 × 105 CFU/ml). The concentrations of ARB exhibiting resistance to specific classes differed across different hospitals and ward types; however, across all samples, the average concentrations of ARB resistant to amikacin (1.06 × 106 CFU/ml), clindamycin (1.37 × 106 CFU/ml), erythromycin (1.24 × 106 CFU/ml), ciprofloxacin (1.14 × 106 CFU/ml), and tetracycline (1.30 × 106 CFU/ml) were at least 1 order of magnitude higher than those of ARB resistant to meropenem (4.79 × 105 CFU/ml), ceftazidime (8.22 × 105 CFU/ml), vancomycin (9.19 × 105 CFU/ml), chloramphenicol (6.08 × 105 CFU/ml), and co-trimoxazole (2.54 ×105 CFU/ml) (Fig. 1). The concentrations of ARB for each of the 10 antibiotics in sample H1a were consistently higher than those determined in the other samples (ANOVA and Tukey test, P < 0.05).
FIG 1.
Concentration (in CFU per milliliter) of antibiotic-resistant bacteria in hospital wastewaters for the locations of clinical isolation ward H1a (black), general ward H1b (white), and mixed wastewater at hospital H2 (gray).
Occurrence of ARGs in hospital wastewater.
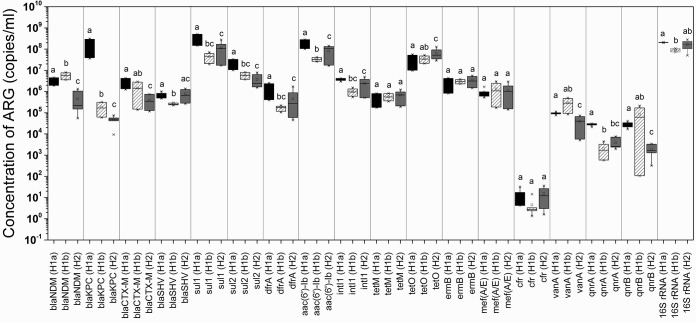
The abundances of all 16 ARGs corresponding to 10 antibiotic classes in the hospital wastewaters were quantified using real-time PCR assay. The average concentrations of ARGs ranged between 104 and 109 gene copies/ml, with the exception of the cfr gene, which was the least prevalent, with an average concentration of 11.0 gene copies/ml (Fig. 2; see also Fig. S1 in the supplemental material). The 16S rRNA gene was the most abundant across all samples, with an average concentration of 1.58 × 108 gene copies/ml (Fig. 2; see also Fig. S1). The integrase intI1 gene, which is used as a proxy for the horizontal gene transfer (HGT) of ARGs, was detected at an average concentration of 2.31 × 106 gene copies/ml.
FIG 2.
Concentration (in numbers of copies per milliliter) of ARGs at H1a (black), H1b (diagonal lines), and H2 (gray). Significant differences (P value < 0.05 [Student's t test]) are indicated by different letters (i.e., a and b, a and bc) at the top of each bar. The absence of significant differences is indicated by identical letters (i.e., a and a, a and ab).
The four ARG targets for β-lactam resistance, including blaNDM, blaKPC, blaCTX-M, and blaSHV, were detected at average concentrations of 2.29 × 106, 4.08 × 107, 1.25 × 106, and 6.19 × 105 gene copies/ml, respectively (Fig. 2). blaNDM and blaKPC were slightly more prevalent than blaCTX-M and blaSHV in the hospital wastewaters. The relative abundances of ARGs were calculated by normalizing the gene copy numbers of each ARG to the 16S rRNA gene copy numbers in the same samples (see Fig. S1 in the supplemental material). Among the four bla genes, blaKPC was the most prevalent gene in H1a, while blaNDM was the most prevalent in H1b, and no significant differences in blaCTX-M and blaSHV levels were found between the isolation ward (H1a) and general ward (H1b) locations.
Identification and ARG detection of ARB isolates.
Colony PCR assays of meropenem- and ceftazidime-resistant isolates (n = 48) indicated the percentages of the total isolates carrying the blaNDM, blaKPC, blaCTX-M, and blaSHV genes (see Table S3 and S4 in the supplemental material). Most isolates carrying blaKPC were of the Enterobacteriaceae family (i.e., Enterobacter spp., Pantoea spp., K. pneumoniae, and Kluyvera georgiana), while those harboring blaNDM belonged to both Enterobacteriaceae and non-Enterobacteriaceae species (i.e., Pseudomonas spp., Acinetobacter spp., and Comamonas testosteroni).
Among the co-trimoxazole-resistant isolates (n = 24), which were tested for sul1, sul2, and drfA ARGs, sul1 was the most predominant gene detected (at 83.3%), while the other two ARGs, sul2 and dfrA, were not found in any isolates. This pattern of detection in isolates reflects the gene prevalence in the hospital wastewater determined by qPCR, in which sul1 was the most dominant co-trimoxazole resistance gene (see Table S5 in the supplemental material). Most of the amikacin-resistant isolates (n = 22) were positive for the aac(6′)-Ib (95.5%) and intI1 (86.4%; see Table S6) genes, while most of the ciprofloxacin-resistant isolates (n = 23) were negative for the qnrA and qnrB genes, suggesting that another mechanism may be implicated in ciprofloxacin resistance for these particular strains (see Table S7).
Correlations between antibiotic residues, ARB, and ARGs.
Both Pearson and Spearman correlation analyses were employed to examine correlations between the concentrations of antibiotic residues, ARB (in CFU per milliliter), and the associated ARGs (in numbers of gene copies per milliliter) for meropenem, ceftazidime, co-trimoxazole, amikacin, and ciprofloxacin antibiotics. We found positive correlations between the concentrations of (i) MEM (r = 0.984, P = 1.06 × 10−5) and CAZ (r = 0.709, P = 4.91 × 10−2) and ARB and blaKPC genes; (ii) SXT ARB and sul1 (r = 0.937, P = 6.07 × 10−4), sul2 (r = 0.994, P = 5.03 × 10−7), and dfrA (r = 0.910, P = 1.68 × 10−3) genes; (iii) AMK ARB and the aac(6′)-Ib (r = 0.872, P = 4.8 × 10−3) gene; and (iv) CIP ARB and the qnrA (r = 0.977, P = 3.06 × 10−5) gene (Table 2, Table 3, and Table 4). Among the antibiotics measured in the hospital wastewaters, significant correlations were found only between ciprofloxacin concentrations and qnrB genes (r = 0.994). The intI1 gene, a potential ARG-transferring vector, showed positive correlations with the blaSHV, sul1, dfrA, and aac(6′)-Ib genes. A PCA plot (see Fig. S2 in the supplemental material) indicated that H1a was one of locations with the highest sources of antibiotic resistance in terms of concentrations of ARB, intI1, and 7 of the 16 ARGs tested [the blaKPC, blaCTX-M, sul1, sul2, dfrA, aac(6′)-Ib, and qnrA genes]. Wastewaters from this location (H1a), however, did not have the highest measured levels of ciprofloxacin, sulfamethoxazole, and trimethoprim antibiotics, which in this case were higher at H1b (Table 1). Other ARGs such as blaSHV, blaNDM, and qnrB were more abundant at other locations (see Fig. S1 in the supplemental material).
TABLE 2.
Correlation between blaNDM, blaKPC, blaCTX-M, and blaSHV genes, meropenem and ceftazidime ARB, and meropenem residuesa
| Parameter | Analysis method | Correlation coefficient |
|||
|---|---|---|---|---|---|
| blaNDM (GCN/ml) | blaKPC (GCN/ml) | blaCTX-M (GCN/ml) | blaSHV (GCN/ml) | ||
| intI1 (GCN/ml) | Pearson | −0.063 | 0.323 | 0.478 | 0.795 |
| Spearman | 0.167 | −0.071 | 0.667 | 0.762 | |
| MEM (CFU/ml) | Pearson | 0.273 | 0.984 | 0.177 | 0.057 |
| Spearman | 0.024 | 0.262 | 0.548 | 0.571 | |
| CAZ (CFU/ml) | Pearson | 0.159 | 0.709 | 0.551 | 0.444 |
| Spearman | 0.286 | 0.238 | 0.690 | 0.619 | |
| Meropenem (ppb) | Pearson | −0.219 | −0.991 | −0.050 | 0.250 |
| Spearman | −0.500 | −1.000 | −0.500 | 0.500 | |
The Pearson and Spearman correlation coefficients which were statistically significant at a P value of <0.05 are bold and underlined in the table. GCN, gene copy numbers; MEM, meropenem; CAZ, ceftazidime.
TABLE 3.
Correlation between sul1, sul2, and dfrA genes and the combination of trimethoprim-and-sulfamethoxazole-resistant ARB and trimethoprim and sulfamethoxazole residuesa
| Parameter | Analysis method | Correlation coefficient |
||
|---|---|---|---|---|
| sul1 (GCN/ml) | sul2 (GCN/ml) | dfrA (GCN/ml) | ||
| intI1 (GCN/ml) | Pearson | 0.640 | 0.436 | 0.574 |
| Spearman | 0.929 | 0.571 | 0.905 | |
| SXT (CFU/ml) | Pearson | 0.937 | 0.994 | 0.910 |
| Spearman | 0.786 | 0.833 | 0.857 | |
| Trimethoprim (ppb) | Pearson | −0.126 | 0.079 | −0.136 |
| Spearman | 0.119 | 0.571 | 0.262 | |
| Sulfamethoxazole (ppb) | Pearson | 0.293 | 0.395 | 0.189 |
| Spearman | 0.262 | 0.476 | 0.405 | |
The Pearson and Spearman correlation coefficients which were statistically significant at a P value of <0.05 are bold and underlined in the table. GCN, gene copy numbers; SXT, trimethoprim and sulfamethoxazole (co-trimoxazole).
TABLE 4.
Correlation between aac(6′)-Ib, qnrA, and qnrB genes, amikacin and ciprofloxacin ARB, and ciprofloxacin residuesa
| Parameter | Analysis method | Correlation coefficient |
||
|---|---|---|---|---|
| aac(6′)-Ib (GCN/ml) | qnrA (GCN/ml) | qnrB (GCN/ml) | ||
| intI1 (GCN/ml) | Pearson | 0.678 | 0.459 | −0.112 |
| Spearman | 0.881 | 1.000 | 0.143 | |
| AMK (CFU/ml) | Pearson | 0.872 | 0.984 | 0.165 |
| Spearman | 0.714 | 0.690 | 0.595 | |
| CIP (CFU/ml) | Pearson | 0.903 | 0.977 | 0.093 |
| Spearman | 0.810 | 0.810 | 0.548 | |
| Ciprofloxacin (ppb) | Pearson | −0.279 | 0.074 | 0.994 |
| Spearman | −0.314 | −0.314 | −0.200 | |
The Pearson and Spearman correlation coefficients which were statistically significant at a P value of <0.05 are bold and underlined in the table. GCN, gene copy numbers; AMK, amikacin; CIP, ciprofloxacin.
DISCUSSION
Hospital wastewater, which receives high loads of antimicrobial agents and human pathogens, is considered a reservoir for antibiotic resistance and other genetic factors which promote the potential spread of AMR to the environment (30, 31). In attempt to control this critical point, this surveillance study aimed at obtaining an in-depth characterization of AMR in hospital wastewater in Singapore by quantifying the presence of antibiotic resistance factors, including antibiotic residues, resistant bacteria, and genetic determinants (i.e., ARGs, integron) in hospital discharge. Local levels of ciprofloxacin, azithromycin, clarithromycin, sulfamethoxazole, and trimethoprim were much lower (∼10-fold) than those reported in another study (12). Sulfamethoxazole and trimethoprim are used in combination as a therapeutic agent (known as co-trimoxazole), and the trend of high concentrations detected at all locations is consistent with most other studies of hospital wastewaters (12). Although hospitals are known to be a source of carbapenems, the levels of meropenem measured in our study were low. The macrolide clarithromycin was detected at higher levels in both H1 samples, which suggests that most inpatients in these two wards had respiratory or skin infections, which are commonly treated with this antibiotic (32). In Singapore, clarithromycin is the standard therapy of community-acquired pneumonia, which is a common diagnosis.
Screening for phenotypic resistance showed that concentrations of ARB differed between locations and ranged between 105 and 106 CFU/ml, which fell within the range determined in another study of hospital wastewater in Iran (33). In our study, we found that average concentrations of ARB across all hospitals and wards indicated that resistance to amikacin, clindamycin, erythromycin, ciprofloxacin, and tetracycline appears to be more prevalent than resistance to meropenem, ceftazidime, vancomycin, chloramphenicol, and co-trimoxazole. Antibiotic prescription trends in local hospitals from 2005 to 2010 reported that a high average daily dose prescription of ciprofloxacin for inpatient users which may result in higher concentrations of ARB resistant to ciprofloxacin (34). The effects of cross-resistance between amikacin and ciprofloxacin have been observed in other studies (35, 36), which offers an explanation for the ARB concentrations observed in both cases.
We found significantly higher concentrations of ARB in location H1a than in the H1b and H2 locations. This implies that although concentrations of specific antibiotics may be high in wastewaters, this does not necessarily translate to high concentrations of bacteria resistant to the antibiotic. The overall abundance of ARB in hospital wastewaters is likely driven by patient-derived shedding and patient demographics in various wards. Wastewater discharge from clinical isolation wards housing patients recovering from severe infections tends to harbor higher levels of ARB detection.
In this study, the average level of intI1 genes detected was at least 2 orders of magnitude lower than that detected in hospital wastewaters in China, with tet and sul genes detected in a lower range as well (37). Class 1 integrons are responsible for the spread and carriage of antibiotic resistance gene cassettes in Gram-negative pathogens (38, 39). The high concentrations of class 1 integrons detected in raw wastewater and the occurrence in ARB isolates were consistent with recent literature reports, e.g., 97% for E. coli isolates in Iran (40), 76% for E. coli and 43% for Salmonella isolates in Netherlands (41), 67% for E. coli isolates in Spain (42), and 63% for E. coli isolates in Thailand (43). This suggests that hospital wastewater is a potential source of class 1 integrons associated with antibiotic resistance. Our results also indicated that the tetracycline [i.e., tet(M) and tet(O)]- and MLSB [i.e., erm(B) and mef(A/E)]-related ARGs did not display much variation across all sampling locations. The low abundances of qnrA and qnrB in raw wastewater measured by qPCR analysis and in resistance isolates detected by conventional PCR in this study were consistent with other literature reports (44, 45). A survey of hospital wastewaters in Spain indicated levels of blaTEM, erm(B), qnrS, sul1, and tet(W) of between 105 and 106 gene copies/ml, which is slightly lower than the measured concentrations of ARGs in our study (12). In comparison to the domestic wastewaters in China, the levels of the 4 β-lactamase ARGs in our study (i.e., blaNDM, blaKPC, blaSHV, and blaCTX-M) were 1 order of magnitude higher than those of the carbapenemase ARGs (i.e., blaNDM, blaKPC, blaOXA, and blaIMP) detected in China (46, 47). However, the concentrations of total 16S rRNA genes in our hospital wastewaters were slightly lower than those in the domestic wastewaters, which suggests a higher prevalence of these ARGs in hospital wastewaters.
Further investigation of bacterial isolates exhibiting phenotypic resistance to meropenem and ceftazidime confirmed that isolates that were resistant to meropenem were likely carrying the blaNDM and blaKPC genes whereas ceftazidime-resistant bacteria were found to be more closely associated with blaCTX-M and blaSHV. The blaNDM gene was more frequently detected in meropenem-resistant bacteria isolated from H1b, including bacterial species such as Acinetobacter junii, Comamonas testosterone, Enterobacter sp., and Pseudomonas sp. In contrast, blaKPC genes were found to be more prevalent in meropenem-resistant isolates from H1a, which consisted of bacteria belonging to Enterobacteriaceae. This result is consistent with other reports where metallo-β-lactamases was found across various bacterial species and the presence of serine-based carbapenemase was mainly restricted to Enterobacteriaceae (48, 49). The detection of the blaNDM gene in a variety of bacterial species in hospital wastewaters demonstrates the ability of the blaNDM-bearing plasmid to successfully establish itself in other strains, providing other possible routes of transmission and dissemination to other bacteria within the hospital wastewater community.
The observed correlations among the detected levels of AB residues, ARB, and ARGs in different sampled locations suggest that some targeted ARGs such as blaNDM, blaKPC, sul1, aac(6′)-Ib, and intI1 are well correlated with the resistance mechanism of the relevant antibiotics and that the H1a clinical isolation ward on average had higher concentrations of ARB, ARGs, and intI1 genes. On the basis of the results determined for the cultured ARB, the clinical isolation ward was less diverse and more enriched in members of the Enterobacteriaceae group, which could explain the high concentrations of blaKPC genes detected in H1a wastewaters and the prevalence of the blaKPC gene in meropenem-resistant isolates. Bacteria in this group are known to be the main culprits responsible for hospital-associated infections and for the spread of the blaKPC gene in tertiary care hospitals (50). This work provided an in-depth study of hospital wastewaters as a source of antibiotic resistance in a tropical country by applying both quantitative and qualitative methods to determine resistance factors (i.e., AB, ARB, and ARGs). Although ARB were detected in the wastewater effluents of both hospitals of different ward types, our results indicate that effluents from clinical isolation wards consistently displayed higher loads of ARB and could play a major role in the emergence and spread of antibiotic resistance if not properly treated in the downstream environment. We detected the 16 relevant ARG targets in all hospital wastewater samples where sul and aac(6′)-Ib were among the most abundant genes and cfr was the least. We detected the blaKPC carbapenem resistance gene in Enterobacteriaceae isolates, while the blaNDM gene was detected in various species of Enterobacteriaceae, Pseudomonas, and Acinetobacter. This study showed the widespread occurrence of ARB, ARGs, and genetic elements which potentially aid the transfer of antibiotic resistance gene cassette arrays in hospital wastewaters. This underscores the need to ensure that hospital wastewaters are sufficiently treated in wastewater treatment plants to reduce the risk of spread of antimicrobial resistance vectors.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Singapore National Research Foundation under its Environmental & Water Technologies Strategic Research Programme (Reference: 1102-IRIS-12-02) and administered by the Environment & Water Industry Programme Office (EWI) of the PUB. Laboratory administration is supported by the National University of Singapore Environmental Institute (NERI).
Funding Statement
The funders had no contributions in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01556-16.
REFERENCES
- 1.Rolain JM, Canton R, Cornaglia G. 2012. Emergence of antibiotic resistance: need for a new paradigm. Clin Microbiol Infect 18:615–616. doi: 10.1111/j.1469-0691.2012.03902.x. [DOI] [PubMed] [Google Scholar]
- 2.Cantón R, Morosini MI. 2011. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev 35:977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 3.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chagas TP, Seki LM, Cury JC, Oliveira JA, Davila AM, Silva DM, Asensi MD. 2011. Multiresistance, beta-lactamase-encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J Appl Microbiol 111:572–581. doi: 10.1111/j.1365-2672.2011.05072.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang X, Meyer MT, Liu X, Zhao Q, Chen H, Chen J-a, Qiu Z, Yang L, Cao J, Shu W. 2010. Determination of antibiotics in sewage from hospitals, nursery and slaughter house, wastewater treatment plant and source water in Chongqing region of Three Gorge Reservoir in China. Environ Pollut 158:1444–1450. doi: 10.1016/j.envpol.2009.12.034. [DOI] [PubMed] [Google Scholar]
- 6.Galvin S, Boyle F, Hickey P, Vellinga A, Morris D, Cormican M. 2010. Enumeration and characterization of antimicrobial-resistant Escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl Environ Microbiol 76:4772–4779. doi: 10.1128/AEM.02898-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bengtsson-Palme J, Larsson DGJ. 2015. Antibiotic resistance genes in the environment: prioritizing risks. Nat Rev Microbiol 13:396. doi: 10.1038/nrmicro3399-c1. [DOI] [PubMed] [Google Scholar]
- 8.Stalder T, Barraud O, Jove T, Casellas M, Gaschet M, Dagot C, Ploy MC. 2014. Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. ISME J 8:768–777. doi: 10.1038/ismej.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Lu X, Zong Z. 2012. Enterobacteriaceae producing the KPC-2 carbapenemase from hospital sewage. Diagn Microbiol Infect Dis 73:204–206. doi: 10.1016/j.diagmicrobio.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Kelly BG, Vespermann A, Bolton DJ. 2009. Gene transfer events and their occurrence in selected environments. Food Chem Toxicol 47:978–983. doi: 10.1016/j.fct.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Knapp CW, Zhang W, Sturm BSM, Graham DW. 2010. Differential fate of erythromycin and beta-lactam resistance genes from swine lagoon waste under different aquatic conditions. Environ Pollut 158:1506–1512. doi: 10.1016/j.envpol.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Mozaz S, Chamorro S, Marti E, Huerta B, Gros M, Sanchez-Melsio A, Borrego CM, Barcelo D, Balcazar JL. 2015. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res 69:234–242. doi: 10.1016/j.watres.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 13.WHO. 2001. WHO global strategy for containment of antimicrobial resistance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 14.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barraud O, Baclet MC, Denis F, Ploy MC. 2010. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J Antimicrob Chemother 65:1642–1645. doi: 10.1093/jac/dkq167. [DOI] [PubMed] [Google Scholar]
- 17.Hindiyeh M, Smollen G, Grossman Z, Ram D, Davidson Y, Mileguir F, Vax M, Ben David D, Tal I, Rahav G, Shamiss A, Mendelson E, Keller N. 2008. Rapid Detection of blaKPC carbapenemase genes by real-time PCR. J Clin Microbiol 46:2879–2883. doi: 10.1128/JCM.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xi C, Zhang Y, Marrs CF, Ye W, Simon C, Foxman B, Nriagu J. 2009. Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol 75:5714–5718. doi: 10.1128/AEM.00382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birkett CI, Ludlam HA, Woodford N, Brown DFJ, Brown NM, Roberts MTM, Milner N, Curran MD. 2007. Real-time TaqMan PCR for rapid detection and typing of genes encoding CTX-M extended-spectrum beta-lactamases. J Med Microbiol 56:52–55. doi: 10.1099/jmm.0.46909-0. [DOI] [PubMed] [Google Scholar]
- 20.Dahmen S, Mansour W, Boujaafar N, Arlet G, Bouallègue O. 2010. Distribution of cotrimoxazole resistance genes associated with class 1 integrons in clinical isolates of Enterobacteriaceae in a university hospital in Tunisia. Microb Drug Resist 16:43–47. doi: 10.1089/mdr.2009.0091. [DOI] [PubMed] [Google Scholar]
- 21.Lee JC, Oh JY, Cho JW, Park JC, Kim JM, Seo SY, Cho DT. 2001. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichi coli in Korea. J Antimicrob Chemother 47:599–604. doi: 10.1093/jac/47.suppl_2.6. [DOI] [PubMed] [Google Scholar]
- 22.Dorsch MR. 2007. Rapid detection of bacterial antibiotic resistance: preliminary evaluation of PCR assays targeting tetracycline resistance genes. Human Protection and Performance Division, DSTO Defence Science and Technology Organisation, Edinburgh, Australia. [Google Scholar]
- 23.Smith MS, Yang RK, Knapp CW, Niu Y, Peak N, Hanfelt MM, Galland JC, Graham DW. 2004. Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl Environ Microbiol 70:7372–7377. doi: 10.1128/AEM.70.12.7372-7377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koike S, Aminov RI, Yannarell AC, Gans HD, Krapac IG, Chee-Sanford JC, Mackie RI. 2010. Molecular ecology of macrolide-lincosamide-streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microb Ecol 59:487–498. doi: 10.1007/s00248-009-9610-0. [DOI] [PubMed] [Google Scholar]
- 25.Gygax SE, Schuyler JA, Kimmel LE, Trama JP, Mordechai E, Adelson ME. 2006. Erythromycin and clindamycin resistance in group B streptococcal clinical isolates. Antimicrob Agents Chemother 50:1875–1877. doi: 10.1128/AAC.50.5.1875-1877.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marti E, Balcazar JL. 2013. Real-time PCR assays for quantification of qnr genes in environmental water samples and chicken feces. Appl Environ Microbiol 79:1743–1745. doi: 10.1128/AEM.03409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (ed). 1992. Short protocols in molecular biology. John Wiley, New York, NY. [Google Scholar]
- 29.Le TH, Sivachidambaram V, Yi X, Li X, Zhou Z. 2014. Quantification of polyketide synthase genes in tropical urban soils using real-time PCR. J Microbiol Methods 106:135–142. doi: 10.1016/j.mimet.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Hocquet D, Muller A, Bertrand X. 2016. What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect 93:395–402. doi: 10.1016/j.jhin.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, Burgmann H, Sorum H, Norstrom M, Pons MN, Kreuzinger N, Huovinen P, Stefani S, Schwartz T, Kisand V, Baquero F, Martinez JL. 2015. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol 13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 32.Piscitelli SC, Danziger LH, Rodvold KA. 1992. Clarithromycin and azithromycin: new macrolide antibiotics. Clin Pharm 11:137–152. [PubMed] [Google Scholar]
- 33.Aali R, Nikaeen M, Khanahmad H, Hassanzadeh A. 2014. Monitoring and comparison of antibiotic resistant bacteria and their resistance genes in municipal and hospital wastewaters. Int J Prev Med 5:887–894. [PMC free article] [PubMed] [Google Scholar]
- 34.Liew YX, Krishnan P, Yeo CL, Tan TY, Lee SY, Lim WP, Lee W, Hsu LY; Network for Antimicrobial Resistance Surveillance Singapore. 2011. Surveillance of broad-spectrum antibiotic prescription in Singaporean hospitals: a 5-year longitudinal study. PLoS One 6:e28751. doi: 10.1371/journal.pone.0028751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simberkoff MS, Rahal JJ Jr. 1986. Bactericidal activity of ciprofloxacin against amikacin- and cefotaxime-resistant gram-negative bacilli and methicillin-resistant staphylococci. Antimicrob Agents Chemother 29:1098–1100. doi: 10.1128/AAC.29.6.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shalit I, Dan M, Gutman R, Gorea A, Berger SA. 1990. Cross resistance to ciprofloxacin and other antimicrobial agents among clinical isolates of Acinetobacter calcoaceticus biovar anitratus. Antimicrob Agents Chemother 34:494–495. doi: 10.1128/AAC.34.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Cheng W, Xu L, Strong PJ, Chen H. 2015. Antibiotic-resistant genes and antibiotic-resistant bacteria in the effluent of urban residential areas, hospitals, and a municipal wastewater treatment plant system. Environ Sci Pollut Res Int 22:4587–4596. doi: 10.1007/s11356-014-3665-2. [DOI] [PubMed] [Google Scholar]
- 38.Roy Chowdhury P, Ingold A, Vanegas N, Martinez E, Merlino J, Merkier AK, Castro M, Gonzalez Rocha G, Borthagaray G, Centron D, Bello Toledo H, Marquez CM, Stokes HW. 2011. Dissemination of multiple drug resistance genes by class 1 integrons in Klebsiella pneumoniae isolates from four countries: a comparative study. Antimicrob Agents Chemother 55:3140–3149. doi: 10.1128/AAC.01529-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betteridge T, Partridge SR, Iredell JR, Stokes HW. 2011. Genetic context and structural diversity of class 1 integrons from human commensal bacteria in a hospital intensive care unit. Antimicrob Agents Chemother 55:3939–3943. doi: 10.1128/AAC.01831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehdipour Moghaddam MJ, Mirbagheri AA, Salehi Z, Habibzade SM. 2015. Prevalence of class 1 integrons and extended spectrum beta lactamases among multi-drug resistant Escherichia coli isolates from north of Iran. Iran Biomed J 19:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Essen-Zandbergen A, Smith H, Veldman K, Mevius D. 2007. Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in the Netherlands. J Antimicrob Chemother 59:746–750. doi: 10.1093/jac/dkl549. [DOI] [PubMed] [Google Scholar]
- 42.Machado E, Cantón R, Baquero F, Galán J-C, Rollán A, Peixe L, Coque TM. 2005. Integron content of extended-spectrum-β-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob Agents Chemother 49:1823–1829. doi: 10.1128/AAC.49.5.1823-1829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phongpaichit S, Wuttananupan K, Samasanti W. 2008. Class 1 integrons and multidrug resistance among Escherichia coli isolates from human stools. Southeast Asian J Trop Med Public Health 39:279–287. [PubMed] [Google Scholar]
- 44.Akter F, Amin MR, Osman KT, Anwar MN, Karim MM, Hossain MA. 2012. Ciprofloxacin-resistant Escherichia coli in hospital wastewater of Bangladesh and prediction of its mechanism of resistance. World J Microbiol Biotechnol 28:827–834. doi: 10.1007/s11274-011-0875-3. [DOI] [PubMed] [Google Scholar]
- 45.Park CH, Robicsek A, Jacoby GA, Sahm D, Hooper DC. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob Agents Chemother 50:3953–3955. doi: 10.1128/AAC.00915-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang F, Mao D, Zhou H, Luo Y. 2016. Prevalence and fate of carbapenemase genes in a wastewater treatment plant in northern China. PLoS One 11:e0156383. doi: 10.1371/journal.pone.0156383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo Y, Yang F, Mathieu J, Mao D, Wang Q, Alvarez PJJ. 2014. Proliferation of multidrug-resistant New Delhi metallo-β-lactamase genes in municipal wastewater treatment plants in northern China. Environ Sci Technol Lett 1:26–30. doi: 10.1021/ez400152e. [DOI] [Google Scholar]
- 48.Bonomo RA. 2011. New Delhi metallo-beta-lactamase and multidrug resistance: a global SOS? Clin Infect Dis 52:485–487. doi: 10.1093/cid/ciq179. [DOI] [PubMed] [Google Scholar]
- 49.Rasmussen BA, Bush K. 1997. Carbapenem-hydrolyzing beta-lactamases. Antimicrob Agents Chemother 41:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Molton JS, Tambyah PA, Ang BS, Ling ML, Fisher DA. 2013. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin Infect Dis 56:1310–1318. doi: 10.1093/cid/cit020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.