Abstract
Seven nonredundant ertapenem-resistant Klebsiella pneumoniae isolates were collected between May 2014 and 19 January 2015 in the nephrology and hematology units of Batna University Hospital in Algeria. All strains coproduced the blaOXA-48, blaCTX-M-15, blaSHV-1, and blaTEM-1D genes. Six of these isolates belonged to the pandemic clone sequence type 101 (ST101). The blaOXA-48 gene was located on a conjugative IncL/M-type plasmid. This is the first known outbreak of OXA-48-producing K. pneumoniae isolates involving an ST101 clone in Batna University Hospital.
TEXT
Carbapenems are a group of β-lactam drugs that constitute the last therapeutic option available for treating infections caused by multidrug-resistant Enterobacteriaceae (1). However, due to the emergence of carbapenem resistance, the antimicrobial activity of these drugs is no longer effective (2). Production of carbapenemases is one of the main mechanisms for carbapenem resistance (3). These carbapenemases belong to different Ambler classes (A, B, and D) and have been reported worldwide among Enterobacteriaceae isolates (4). The OXA-48 class D β-lactamase is called the “phantom menace” due to its weak but significant carbapenemase activity. This enzyme hydrolyzes penicillins at a high level and carbapenems at a low level but spares expanded-spectrum cephalosporins (5). OXA-48 was initially described in a clinical Klebsiella pneumoniae isolate from Istanbul, Turkey, in 2001 (6), and since then, several sporadic cases and outbreaks have been reported, especially in the Mediterranean countries (4).
Here, we describe a nosocomial outbreak in Batna University Hospital, Algeria, of ertapenem-resistant K. pneumoniae clinical isolates expressing OXA-48 associated with CTXM-15, SHV-1, and TEM-1D β-lactamases involving a sequence type 101 (ST101) clone.
Between May 2014 and 19 January 2015, seven patients hospitalized in the nephrology and hematology units at Batna University Hospital, Algeria, were infected with ertapenem-resistant K. pneumoniae. During the period of this study, the first OXA-48-positive K. pneumoniae strain was isolated in the hematology unit in May 2014 from a patient with myelosuppression and Fanconi anemia. This strain (Kp1) was assigned to sequence type 985 (ST985); however, it represented the only case until the outbreak. The index isolate Kp2 assigned to the pandemic ST101 was retrieved in August 2014 in the nephrology unit from pus on a catheter sample of a patient with chronic renal failure. During the outbreak, five patients in the hematology unit were infected with an ertapenem-resistant K. pneumoniae isolate of an ST101 clone, suggesting transmission via the hospital staff because the nephrology unit is in front of the hematology unit (Table 1).
TABLE 1.
Clinical features of the K. pneumoniae isolate outbreak
| Patient no. | Isolate | Site of isolation | Date of positive isolate | Unit | Reason for hospitalization | Treatment(s) | Outcome |
|---|---|---|---|---|---|---|---|
| 2 | Kp2 | Pus | August 2014 | Nephrology | Chronic renal failure | Last step: colistin | Improved |
| 3 | Kp3 | Blood | October 2014 | Hematology | Acute myeloid leukemia, stage 2 | 1st step: cefotaxime, gentamicin, and metronidazole | Died |
| 2nd step: amikacin and piperacillin | |||||||
| 3rd step: imipenem, vancomycin, and ofloxacin | |||||||
| 4 | Kp4 | Mouth ulcers | December 2014 | Hematology | Acute myeloid leukemia, stage 5 | 1st step: cefotaxime, gentamicin, and metronidazole | Died |
| 2nd step: amikacin and piperacillin | |||||||
| 3rd step: imipenem, vancomycin, and ofloxacin | |||||||
| 5 | Kp5 | Blood | December 2014 | Hematology | Acute myeloid leukemia, stage 1 | 1st step: cefotaxime, gentamicin, and metronidazole | Died |
| 2nd step: amikacin and piperacillin | |||||||
| 3rd step: imipenem, vancomycin, and ofloxacin | |||||||
| 6 | Kp6 | Blood | December 2014 | Hematology | Acute myeloid leukemia, stage 4 | 1st step: cefotaxime, gentamicin, and metronidazole | Died |
| 2nd step: amikacin and piperacillin | |||||||
| 3rd step: imipenem, vancomycin, and ofloxacin | |||||||
| 7 | Kp7 | Blood | January 2015 | Hematology | Severe myelosuppression | 1st step: cefotaxime, gentamicin, and metronidazole | Died |
| 2nd step: amikacin and piperacillin | |||||||
| 3rd step: imipenem, vancomycin, and ofloxacin |
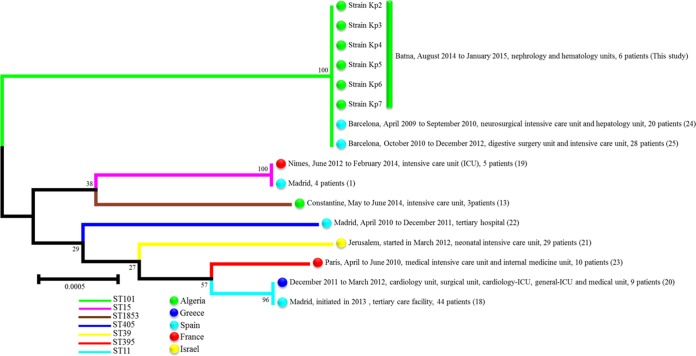
Antimicrobial drug susceptibility was determined by the standard disc diffusion method recommended by the Antibiogram Committee of the French Society for Microbiology (http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM_EUCAST_V1_2015.pdf). In addition, the MICs of imipenem and colistin were determined using Etest strips (bioMérieux). All isolates were resistant to amoxicillin, cefoxitin, cefotaxime, ceftazidime, cefepime, aztreonam, amoxicillin-clavulanic acid, ertapenem, tobramycin, gentamicin, ciprofloxacin, and trimethoprim-sulfamethoxazole, but they were sensitive to colistin and displayed MIC values within the intermediate range for imipenem (3 to 4 μg/ml), except for imipenem-resistant strain Kp1 (MIC, >32 μg/ml). All the strains were positive for phenotypic carbapenemase production as tested by the modified Carba NP (MCNP) test (7). Real-time and standard PCR and sequencing for β-lactamase-encoding genes, which were performed using specific primers for the blaCTX-M, blaSHV, blaTEM, blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48 genes (8, 9), indicated the presence of the blaCTX-M-15, blaSHV-1, blaTEM-1D, and blaOXA-48 genes in all isolates. Multilocus sequence typing (MLST) was performed according to the Institute Pasteur scheme (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html) and showed that all strains belonged to sequence type 101 (ST101) except for the Kp1 strain, which was assigned to ST985. An MLST concatenated gene sequence-based phylogenetic tree of our K. pneumoniae ST101 isolates with those responsible for most nosocomial OXA-48 outbreaks occurring in the Mediterranean countries was constructed to illustrate their phylogenetic position and clustering. MLST allele sequences of the strains not included in this work were retrieved from the K. pneumoniae MLST database through its website (http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html). The phylogenetic tree was built using Mega 6 software (10–12) (Fig. 1). Transferability of the blaOXA-48 gene was tested using conjugation experiments among the K. pneumoniae isolates Kp2 and Kp4 and Escherichia coli strain J53, which is resistant to sodium azide. The obtained transconjugants were MCNP positive and resistant to only ertapenem and amoxicillin-clavulanic acid. PCR for β-lactamase determinants in transconjugants identified only the blaOXA-48 gene. The incompatibility group of the plasmid carrying the blaOXA-48 gene was determined by PCR using previously published primers (9) to be IncL/M.
FIG 1.
MLST-based phylogenetic tree of clinical K. pneumoniae isolates in this study and those responsible for most OXA-48 outbreaks in Mediterranean countries. The evolutionary history was inferred using the neighbor-joining method (11). The evolutionary distances were computed using the Kimura 2-parameter method (12). The numbers at the nodes are the levels of bootstraps from 1,000 replicates.
With this study, we describe the first reported outbreak caused by OXA-48-producing K. pneumoniae isolates in Batna University Hospital in Algeria. This represents the second reported outbreak of OXA-48-producing K. pneumoniae in Algeria, after the recent study by Cuzon et al. (13), who characterized an outbreak of OXA-48-producing K. pneumoniae ST1853 in Constantine University Hospital in Algeria. In 2012, Poirel et al. (5) suggested that the OXA-48 enzyme may be endemic in Algeria, since its dissemination has been described worldwide (4), particularly in the Mediterranean area, including Egypt, Lebanon, Morocco, and Tunisia (14–17). This suggestion is supported by the isolation of OXA-48-producing Enterobacteriaceae strains from patients hospitalized in French hospitals who had recently travelled to or were hospitalized in Algeria (13). Previous studies reported the isolation of clinical K. pneumoniae strains coproducing extended-spectrum β-lactamase (ESBL) and the blaOXA-48 gene (1, 13, 18). In this study, we identified the association of blaOXA-48 with three different β-lactamase genes (blaCTX-M-15, blaSHV-1, and blaTEM-1D) that had not been previously described in Algeria; the same association was observed in Tunisia, except for the TEM variant (17). In the Mediterranean area, outbreaks involving OXA-48-producing isolates have implicated diverse sequence types, such as ST1853, ST15, ST11, ST39, ST405, ST395, and ST101 (Fig. 1) (13, 19–25). However, in this investigation, the outbreak was due to the pandemic ST101, which was detected for the first time in Algeria. The introduction of this ST101 clone in our hospital environment was attributed to index patient 2, who was probably infected during a prior hospitalization. OXA-48-producing K. pneumoniae isolates belonging to ST101 have already been implicated in two nosocomial outbreaks in Spain (Fig. 1) (24, 25). In addition to its involvement in the nosocomial outbreaks, there have been some sporadic cases reported in Tunisia, Switzerland, Bulgaria, and Germany (17, 26–28).
In conclusion, this study identified the first OXA-48-positive K. pneumoniae coproducing CTX-M-15, SHV-1, and TEM-1D β-lactamases in Batna University Hospital. These data, along with those of earlier studies and the more recent description of OXA-48-producing Enterobacteriaceae from companion animals and hospital cockroaches, confirm the widespread presence of OXA-48-producing Enterobacteriaceae in Algeria (29, 30).
ACKNOWLEDGMENTS
We are very grateful to Linda Hadjadj for technical assistance. We also thank TradOnline for English language corrections.
This work was partly funded by the Centre National de la Recherche Scientifique (CNRS7278) and the IHU Méditérranée Infection.
The authors report no conflicts of interest relevant to this article.
REFERENCES
- 1.Arana DM, Saez D, Garcia-Hierro P, Bautista V, Fernandez-Romero S, Angel de la Cal M, Alos JI, Oteo J. 2015. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin Microbiol Infect 21:148e1–148e4. doi: 10.1016/j.cmi.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Beyrouthy R, Robin F, Delmas J, Gibold L, Dalmasso G, Dabboussi F, Hamze M, Bonnet R. 2014. IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of bla OXA-48 into the Escherichia coli chromosome. Antimicrob Agents Chemother 58:3785–3790. doi: 10.1128/AAC.02669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Dortet L, Poirel L. 2012. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Djahmi N, Dunyach-Remy C, Pantel A, Dekhil M, Sotto A, Lavigne JP. 2014. Epidemiology of carbapenemase-producing Enterobacteriaceae and Acinetobacter baumannii in Mediterranean countries. Biomed Res Int 2014:305784. doi: 10.1155/2014/305784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 6.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakour S, Garcia V, Loucif L, Brunel JM, Gharout-Sait A, Touati A, Rolain JM. 2015. Rapid identification of carbapenemase-producing Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii using a modified Carba NP test. New Microbes New Infect 7:89–93. doi: 10.1016/j.nmni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mellouk FZ, Bakour S, Meradji S, Al-Bayssari C, Bentakouk MC, Zouyed F, Djahoudi A, Boutefnouchet N, Rolain JM. 17 June 2016. First detection of VIM-4-producing Pseudomonas aeruginosa and OXA-48-producing Klebsiella pneumoniae in northeastern (Annaba, Skikda) Algeria. Microb Drug Resist doi: 10.1089/mdr.2016.0032. [DOI] [PubMed] [Google Scholar]
- 9.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 13.Cuzon G, Bentchouala C, Vogel A, Hery M, Lezzar A, Smati F, Dortet L, Naas T. 2015. First outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Constantine, Algeria. Int J Antimicrob Agents 46:725–727. doi: 10.1016/j.ijantimicag.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Poirel L, Abdelaziz MO, Bernabeu S, Nordmann P. 2013. Occurrence of OXA-48 and VIM-1 carbapenemase-producing Enterobacteriaceae in Egypt. Int J Antimicrob Agents 41:90–91. doi: 10.1016/j.ijantimicag.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Beyrouthy R, Robin F, Dabboussi F, Mallat H, Hamze M, Bonnet R. 2014. Carbapenemase and virulence factors of Enterobacteriaceae in north Lebanon between 2008 and 2012: evolution via endemic spread of OXA-48. J Antimicrob Chemother 69:2699–2705. doi: 10.1093/jac/dku181. [DOI] [PubMed] [Google Scholar]
- 16.Hays C, Benouda A, Poirel L, Elouennass M, Nordmann P. 2012. Nosocomial occurrence of OXA-48-producing enterobacterial isolates in a Moroccan hospital. Int J Antimicrob Agents 39:545–547. doi: 10.1016/j.ijantimicag.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Lahlaoui H, Poirel L, Barguellil F, Moussa MB, Nordmann P. 2012. Carbapenem hydrolyzing class D beta-lactamase OXA-48 in Klebsiella pneumoniae isolates from Tunisia. Eur J Clin Microbiol Infect Dis 31:937–939. doi: 10.1007/s10096-011-1389-5. [DOI] [PubMed] [Google Scholar]
- 18.Branas P, Villa J, Viedma E, Mingorance J, Orellana MA, Chaves F. 2015. Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital in Madrid: successful establishment of an OXA-48 ST11 clone. Int J Antimicrob Agents 46:111–116. doi: 10.1016/j.ijantimicag.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Pantel A, Richaud-Morel B, Cazaban M, Bouziges N, Sotto A, Lavigne JP. 2016. Environmental persistence of OXA-48-producing Klebsiella pneumoniae in a French intensive care unit. Am J Infect Control 44:366–368. doi: 10.1016/j.ajic.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 20.Voulgari E, Zarkotou O, Ranellou K, Karageorgopoulos DE, Vrioni G, Mamali V, Themeli-Digalaki K, Tsakris A. 2013. Outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in Greece involving an ST11 clone. J Antimicrob Chemother 68:84–88. doi: 10.1093/jac/dks356. [DOI] [PubMed] [Google Scholar]
- 21.Adler A, Solter E, Masarwa S, Miller-Roll T, Abu-Libdeh B, Khammash H, Najem K, Dekadek S, Stein-Zamir C, Nubani N, Kunbar A, Assous MV, Carmeli Y, Schwaber MJ. 2013. Epidemiological and microbiological characteristics of an outbreak caused by OXA-48-producing Enterobacteriaceae in a neonatal intensive care unit in Jerusalem, Israel. J Clin Microbiol 51:2926–2930. doi: 10.1128/JCM.01049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pano-Pardo JR, Ruiz-Carrascoso G, Navarro-San FC, Gomez-Gil R, Mora-Rillo M, Romero-Gomez MP, Fernandez-Romero N, Garcia-Rodriguez J, Perez-Blanco V, Moreno-Ramos F, Mingorance J. 2013. Infections caused by OXA-48-producing Klebsiella pneumoniae in a tertiary hospital in Spain in the setting of a prolonged, hospital-wide outbreak. J Antimicrob Chemother 68:89–96. doi: 10.1093/jac/dks364. [DOI] [PubMed] [Google Scholar]
- 23.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. 2011. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitart C, Sole M, Roca L, Fabrega A, Vila J, Marco F. 2011. First outbreak of a plasmid-mediated carbapenem-hydrolyzing OXA-48 beta-lactamase in Klebsiella pneumoniae in Spain. Antimicrob Agents Chemother 55:4398–4401. doi: 10.1128/AAC.00329-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cubero M, Cuervo G, Dominguez MA, Tubau F, Marti S, Sevillano E, Gallego L, Ayats J, Pena C, Pujol M, Linares J, Ardanuy C. 2015. Carbapenem-resistant and carbapenem-susceptible isogenic isolates of Klebsiella pneumoniae ST101 causing infection in a tertiary hospital. BMC Microbiol 15:177. doi: 10.1186/s12866-015-0510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seiffert SN, Marschall J, Perreten V, Carattoli A, Furrer H, Endimiani A. 2014. Emergence of Klebsiella pneumoniae co-producing NDM-1, OXA-48, CTX-M-15, CMY-16, QnrA and ArmA in Switzerland. Int J Antimicrob Agents 44:260–262. doi: 10.1016/j.ijantimicag.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Markovska R, Stoeva T, Schneider L, Boyanova L, Popova V, Dacheva D, Kaneva R, Bauernfeind A, Mitev V, Mitov L. 2015. Clonal dissemination of multilocus sequence type ST15 KPC-2-producing Klebsiella pneumoniae in Bulgaria. APMIS 123:887–894. doi: 10.1111/apm.12433. [DOI] [PubMed] [Google Scholar]
- 28.Kaase M, Schimanski S, Schiller R, Beyreiss B, Thurmer A, Steinmann J, Kempf VA, Hess C, Sobottka L, Fenner L, Ziesing S, Burckhardt L, Von ML, Hamprecht A, Tammer I, Wantia N, Becker K, Holzmann T, Furitsch M, Volmer G, Gatermann SG. 2016. Multicentre investigation of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in German hospitals. Int J Med Microbiol 306:415–420. doi: 10.1016/j.ijmm.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Yousfi M, Touati A, Mairi A, Brasme L, Gharout-Sait A, Guillard T, De Champs C. 2016. Emergence of carbapenemase-producing Escherichia coli isolated from companion animals in Algeria. Microb Drug Resist 22:342–346. doi: 10.1089/mdr.2015.0196. [DOI] [PubMed] [Google Scholar]
- 30.Loucif L, Gacemi-Kirane D, Cherak Z, Chamlal N, Grainat N, Rolain JM. 2016. First report of German cockroaches (Blattella germanica) as reservoirs of CTX-M-15 extended-spectrum-β-lactamase- and OXA-48 carbapenemase-producing Enterobacteriaceae in Batna University Hospital, Algeria. Antimicrob Agents Chemother 60:6377–6380. doi: 10.1128/AAC.00871-16. [DOI] [PMC free article] [PubMed] [Google Scholar]