Abstract
Sphingolipids are structural lipid components of cell membranes, including membrane of organelles, such as mitochondria or endoplasmic reticulum, playing a role in signal transduction as well as in the transport and intermixing of cell membranes. Sphingolipid microdomains, also called lipid rafts, participate in several metabolic and catabolic cell processes, including apoptosis. However, the defined role of lipid rafts in the autophagic flux is still unknown. In the present study we analyzed the role of gangliosides, a class of sphingolipids, in autolysosome morphogenesis in human and murine primary fibroblasts by means of biochemical and analytical cytology methods. Upon induction of autophagy, by using amino acid deprivation as well as tunicamycin, we found that GD3 ganglioside, considered as a paradigmatic raft constituent, actively contributed to the biogenesis and maturation of autophagic vacuoles. In particular, fluorescence resonance energy transfer (FRET) and coimmunoprecipitation analyses revealed that this ganglioside interacts with phosphatidylinositol 3-phosphate and can be detected in immature autophagosomes in association with LC3-II as well as in autolysosomes associated with LAMP1. Hence, it appears as a structural component of autophagic flux. Accordingly, we found that autophagy was significantly impaired by knocking down ST8SIA1/GD3 synthase (ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase 1) or by altering sphingolipid metabolism with fumonisin B1. Interestingly, exogenous administration of GD3 ganglioside was capable of reactivating the autophagic process inhibited by fumonisin B1. Altogether, these results suggest that gangliosides, via their molecular interaction with autophagy-associated molecules, could be recruited to autophagosome and contribute to morphogenic remodeling, e.g., to changes of membrane curvature and fluidity, finally leading to mature autolysosome formation.
Keywords: autophagy, autophagosome, rafts, gangliosides, mitochondria
Introduction
There are 3 different forms of autophagy that are commonly described, which include macroautophagy, microautophagy, and chaperone-mediated autophagy.1 Macroautophagy (hereafter autophagy) has been extensively investigated and described as an essential homeostatic process by which cells break down their own components. During autophagy, parts of the cytoplasm and organelles are encapsulated in double-membraned vacuoles called autophagosomes, which finally fuse with lysosomes to degrade the incorporated material by acidic hydrolases.2 The autophagy machinery is thought to have evolved as a stress response that allows unicellular eukaryotic organisms to survive during harsh conditions, probably by regulating energy homeostasis and/or by protein quality control. This is primarily performed by the ubiquitin-proteasome system, but autophagy plays a key role too. In fact, terminally misfolded proteins that are not timely removed tend to form aggregates and their clearance requires autophagy. In addition, autophagy also serves in intracellular quality control by selectively segregating defective organelles, including mitochondria, and targeting them for degradation by the lysosomes. Autophagy interfaces with most cellular stress-response pathways, including those involved in controlling several human morbidity states.3 It is thus nowadays considered either a pathogenetic determinant or a therapeutic target in a plethora of human diseases.4
Much progress has been made in the field of autophagy in terms of identification of the molecular components involved in the process. The overall mechanics of the autophagy-lysosome pathway are now completely understood: more than 30 autophagy-related proteins (ATG proteins) have been identified, 18 of which are essential for autophagosome formation.5 However, how these vary in different cell types needs to be established. Generation of the phagophore requires the BECN1/Beclin 1-class III phosphatidylinositol (PtdIns) 3-kinase (PtdIns3K) complex, as well as generation and insertion of LC3 (microtubule-associated protein 1 light chain 3)-II into the autophagosomal membrane.6 This is a consistent key step in autophagosomal formation and its level reflects the relative amount of autophagosomes in the cell. At the final step, fusion of autophagosomes to lysosomes may be mediated by the RAB7A small GTPase and the lysosomal-associated membrane protein family LAMP.7,8
While the ATG proteins functioning in autophagy have extensively been analyzed, little is known about the lipid molecules that form autophagosomes. In particular, gangliosides are molecules composed of a glycosphingolipid (ceramide and oligosaccharide) with one or more sialic acids linked on the sugar chain. They are abundant lipid components of eukaryotic plasma membranes, which can play a role in several biological processes, including proliferation, apoptosis, membrane transport, signal trasduction, and cell adhesion.9-11 Emerging evidence indicates that although autophagy and apoptosis utilize distinct machinery, these processes are highly interconnected by many key regulators, including different sphingolipids. For instance, current literature supports the notion that ceramide-induced autophagy functions to promote cell death, either through the induction of autophagic cell death,11,12 or by “switching” off autophagy and inducing apoptosis through the CAPN/calpain-mediated cleavage of ATG5,13 and/or DISC formation.14 Interestingly, acid sphingomyelinase (SMPD1), a hydrolase enzyme that is involved in sphingolipid metabolism reactions and is responsible for breaking sphingomyelin down into phosphocholine and ceramide, appeared to be responsible for the induction of autophagy during amino acid depletion, the prototypical preautophagic stimulus.15
Recently, Hwang et al. have demonstrated that gangliosides, sialic acid-containing glycosphingolipids, induce autophagic cell death of astrocytes by activation of the IKBKB/IKK-NFKB/NF-kB pathway.16 Furthermore, ganglioside-induced autophagic cell death has been shown to be dependent on the generation of ROS, inhibition of AKT-MTOR, activation of MAPK1 (mitogen-activated protein kinase 1)-MAPK3/ERK2-ERK1, and “lipid raft” integrity.17 A recent study demonstrates that the inhibition of sphingolipid synthesis in Saccharomyces cerevisiae suppressed autophagosome biogenesis through a mechanism that was both independent of the ATG12–ATG5 and ATG8–phosphatidylethanolamine conjugates as well as the formation of phagophores.18 However, a defined role for lipid rafts or individual gangliosides in regulating autophagy induction as well as autophagosome biogenesis and/or maturation is unclear. In this study, we analyzed whether gangliosides play a role in autophagosome morphogenic remodeling leading to autophagosome formation. To do this, we decided to use well established but untransformed cell models (primary human fibroblasts and mouse embryo fibroblasts), where GM3, GD3 and GD1a gangliosides are constitutively expressed.19
Results
Autophagy induction in primary fibroblasts
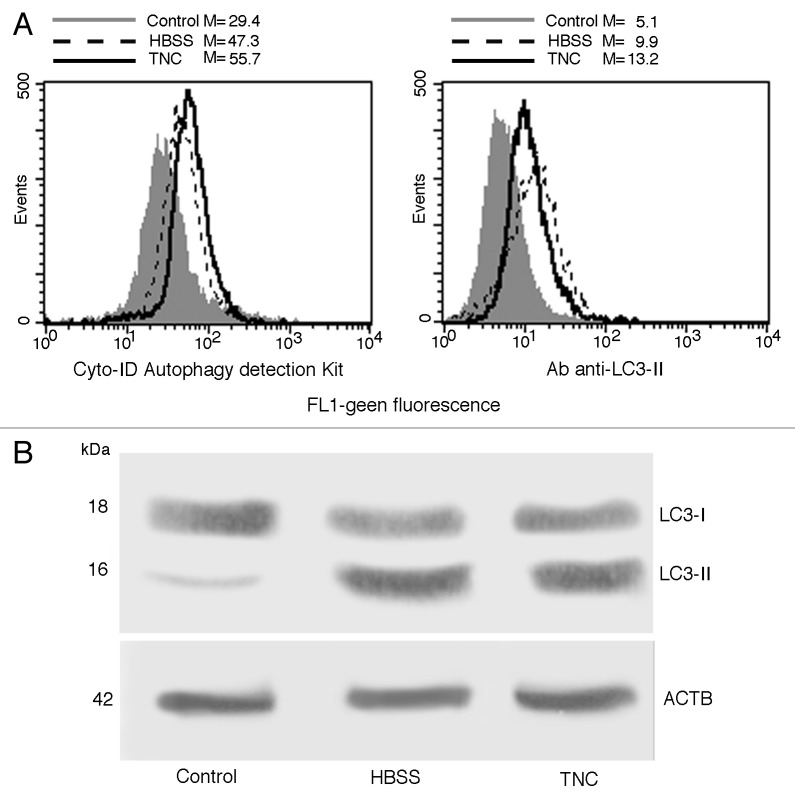
We preliminarily analyzed autophagy triggering. Common cellular stresses known to induce autophagy include amino acid starvation, glucose withdrawal, and ER stress. To trigger autophagic machinery in our experimental model we used a classical autophagic stimulus, such as amino acid starvation (HBSS) and a pharmacological stimulus able to induce ER stress, tunicamycin (TNC). As shown in Figure 1A (left panel), flow cytometry analysis of cells stained with Cyto-ID Autophagy detection kit showed a significant (P < 0.01) increase of green fluorescence emission in cells starved in HBSS or treated with TNC in comparison with untreated control cells. These data were also confirmed by using a specific antibody able to recognize cleaved form of LC3 (Fig. 1A, right panel). Flow cytometry results were confirmed by western blot analysis, which revealed a band corresponding to LC3-II either after cell starvation or in cells treated with TNC (Fig. 1B).
Figure 1. Autophagy induction in primary fibroblasts. (A) Semiquantitative flow cytometry analysis of autophagy performed in primary human fibroblasts untreated (full gray curves), starved 18 h with HBSS medium (dashed lines) or treated 18 h with TNC (empty curves) performed by Cyto-ID Autophagy detection kit (left panel) or by LC3-II cell staining (right panel). Numbers represent the median fluorescence intensity. A representative experiment among 3 is shown. (B) Primary fibroblasts, untreated or treated with HBSS or TNC (5 µM for 18 h at 37 °C), were lysed in lysis buffer, subjected to 10% SDS-PAGE and analyzed by western blot using anti-LC3 polyclonal antibody. Loading control was evaluated using anti-ACTB Mab.
Engagement of sphingolipids in the autophagic process
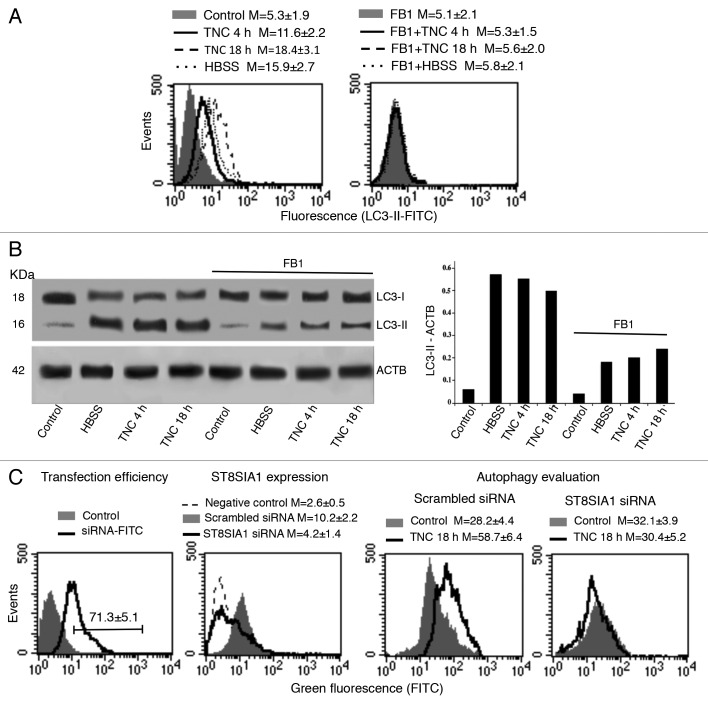
Starting from the observation that sphingolipids have already been hypothesized to be related to autophagy in cancer cells,20 we first investigated their possible involvement in autophagic progression by impairing their biosynthesis. For this purpose, we pretreated primary human fibroblasts with fumonisin B1 (FB1), an inhibitor of sphingosine (sphinganine) N-acyltransferase and de novo sphingolipid biosynthesis, before the autophagic stimulus. Flow cytometry analysis after cell staining with anti-LC3-II antibodies clearly showed that cell pretreatment with FB1 significantly inhibited autophagy either induced by HBSS or by TNC after 4 or 18 h (Fig. 2A). We therefore analyzed LC3 expression by western blot after autophagy triggering, in the presence or in the absence of FB1, under the same experimental conditions (Fig. 2B). Our results confirmed that in the presence of FB1 the conversion of soluble LC3-I to lipid-membrane-bound LC3-II, considered as the prototypical marker of autophagy, was significantly lower (Fig. 2B, see densitometric analysis). Similar results were obtained in experiments performed with mouse fibroblasts (MEFs, see Fig. S1).
Figure 2. Engagement of sphingolipids in the autophagic process. (A) Semiquantitative flow cytometry analysis of autophagy performed after human fibroblast staining with anti-LC3-II antibody. The numbers represent the mean ± SD of the median fluorescence intensity obtained in 3 independent measurements. A representative experiment is shown. (B) Human fibroblasts, untreated or treated with TNC (5 µM for 4 h or 18 h at 37 °C) or HBSS, in the presence or in the absence of pretreatment with FB1, were analyzed by western blot using anti-LC3 polyclonal antibody. Loading control was evaluated using anti-ACTB Mab (left panel). Densitometric LC3-II-ACTB ratios are shown (right panel). (C) In the first panel cytofluorimetric analysis of fluorescence emission in fibroblasts transfected with FITC-siRNA is shown. The percentage of FITC-positive cells was considered indicative of the transfection efficiency. The number represents the mean percentage ± SD of FITC-positive cells (corresponding to transfected cells) obtained in 3 independent measurements. In the second panel cytofluorimetric evaluation of ST8SIA1 expression level 48 h after siRNA transfection is shown. The numbers represent the mean ± SD of the median fluorescence intensity obtained in 3 independent experiments. A representative experiment is shown. In the third and fourth panels a semiquantitative flow cytometry analysis performed by Cyto-ID Autophagy detection kit of autophagy induced by TNC in cells knocked down for ST8SIA1 (fourth panel) or in cells transfected with scrambled siRNA (third panel) is shown. The numbers represent the mean ± SD of the median fluorescence intensity obtained in 3 independent measurements. A representative experiment is shown.
In order to clarify the role of GD3 during autophagic process, a small interfering RNA (siRNA) was employed to knock down ST8SIA1 gene expression. In our experimental conditions, we found that 48 h after siRNA addition: i) about 70% of cells (Fig. 2C, first panel) were transfected (positive control siGLO laminin A/C siRNA-FITC) and that ii) in cells transfected with ST8SIA1 siRNA a significant reduction (more than 70%) of ST8SIA1, with respect to scrambled siRNA transfected cells, was detectable (Fig. 2C, second panel). Flow cytometry analyses of siRNA-treated cells (Fig. 2C, third panel), incubated with TNC as above, revealed that autophagy was significantly reduced as compared with scrambled siRNA transfected cells (Fig. 2C, fourth panel).
Implication of endocytosis in autophagy induction
Since a relationship of the classical endosomal pathway with autophagic process has recently been reported,21 we evaluated either the possible involvement of GD3 in endocytosis or the contribution of endocytosis to the autophagy induced by TNC. For the first, we quantified the endocytosis in fibroblasts, untreated or treated with sphingolipid biosynthesis inhibitor FB1. As expected, we found a very low basal endocytic activity in these cells. However, FB1 that significantly impaired autophagy downmodulating GD3 synthesis did not influence endocytosis (Fig. S2A). In addition, we also observed that D609, an inhibitor of endocytosis that selectively inhibits phosphatidylcholine-specific phospholipase C, did not alter TNC-induced autophagy (Fig. S2B). Finally, immunofluorescence analyses, performed in order to verify whether GD3 was present in late endosomes, did not show any colocalization between RAB7A, a marker of late endosomes, and GD3 neither in control untreated cells nor in TNC-treated cells (Fig. S2C). Altogether, these data seem to indicate that in our experimental setting ganglioside GD3 plays a minor role in the endocytic pathway, which, in turn, seems to play an ancillary role in TNC-induced autophagy.
Involvement of GD3 in autophagic machinery
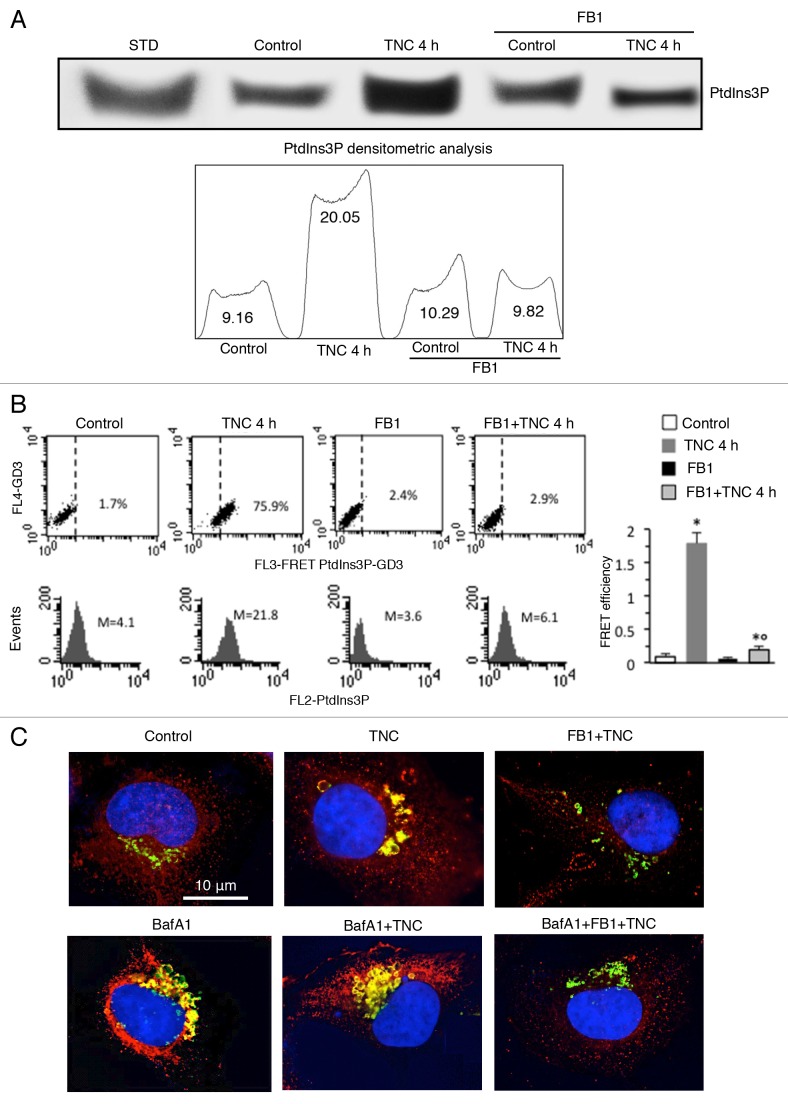
Phosphatidylinositol 3-phosphate (PtdIns3P) has been suggested to create a membrane platform to concentrate and spatially coordinate specific effectors necessary for signal transduction and progression of autophagy.22 With the aim to evaluate which step of autophagosome formation may act to block sphingolipid biogenesis, we analyzed the effects of disturbed sphingolipid metabolism on PtdIns3P levels. Cells, either untreated or treated with TNC (5 μM for 4 h at 37 °C), in the presence or in the absence of pretreatment with FB1, were subjected to phosphoinositide extraction. The samples were separated by HPTLC and the plates were immunostained with anti-PtdIns3P monoclonal antibody (Mab). The results showed that TNC administration increased PtdIns3P synthesis, which was significantly reduced by cell pretreatment with FB1 (Fig. 3A).
Figure 3. Involvement of GD3 in the autophagic machinery. (A) Human fibroblasts, untreated or treated with TNC (5 µM for 4 h at 37 °C), in the presence or in the absence of pretreatment with FB1, were subjected to phosphoinositides extraction. The extracts were run on HPTLC aluminum-backed silica gel and were analyzed for the presence of PtdIns3P, using an anti-PtdIns3P Mab. Densitometric scanning analysis of PtdIns3P levels was performed by MAC OS X using NIH Image 1.62 software. (B) Flow cytometry analysis of GD3-PtdIns3P association by FRET technique in: control cells (first panel), cells treated with FB1 (second panel) or TNC 4 h in the absence (third panel) or in the presence (fourth panel) of FB1. Numbers indicate the percentage of FL3-positive events obtained in one experiment representative of 4. In the bottom row is shown PtdIns3P intracellular amount in the corresponding sample and was quantitatively expressed by the median fluorescence intensity. Bar graph shows the evaluation of FE, according to the Riemann algorithm, of GD3-PtdIns3P association. Results represent the mean ± SD from 3 independent experiments. Statistical analyses indicated that: i) GD3-PtdIns3P association was significantly higher in cells treated 4 h with TNC than in control cells and that ii) cell pretreatment with FB1 significantly prevents TNC-induced associations. *P < 0.01 vs. control; °P < 0.01 vs. TNC 4 h. (C) IVM analysis after cell staining for GD3 and LC3-II in control cells (first picture) and in cells treated with TNC in the absence (second picture) or presence (third picture) of FB1 and challenged (bottom row) or not (upper row) with BafA1. Yellow fluorescence observed in the merge picture indicates GD3-LC3-II colocalization.
We thus investigated the possible physical interaction of GD3 with PtdIns3P by FRET methodology (Fig. 3B). Quantitative analyses were performed by pooling together data obtained from 3 independent experiments (Fig. 3B, bar graph) by calculating the FRET efficiency (FE) by using the following algorithm: FE = [FL3DA − FL2DA/a − FL4DA/b]/FL3DA, where A is the acceptor and D the donor and where a = FL2D/FL3D and b = FL4A/FL3A. This quantitative analysis indicated a strict molecular interaction between GD3 and PtdIns3P 4 h after TNC addition (Fig. 3B, in the second panel a representative experiment is shown), which was significantly prevented by pretreatment of cells with FB1 (Fig. 3B, in the fourth panel a representative experiment is shown). In accordance with the results reported in Figure 3A, the increased amount of PtdIns3P induced by TNC treatment was significantly prevented by FB1 (Fig. 3B, see histograms in the second row).
To better understand the role of GD3 in the autophagic flux, specific experiments were performed by using bafilomycin A1 (BafA1). This is an inhibitor of the late phase of autophagy, since it prevents maturation of autophagosomes, by hindering fusion between autophagosomes and lysosomes by inhibiting vacuolar H+ ATPase (V-ATPase).23 Human fibroblasts pretreated with BafA1 and then added with TNC, alone or in combination with FB1, were subjected to IVM analysis after triple staining with anti-LC3-II antibodies (green), anti-GD3 antibodies (red) and Hoechst dye (blue). Our results showed that BafA1 induced an appreciable increase of LC3-II staining, due to the autophagosomes accumulation, either in control cells or, to a greater extent, in TNC-treated cells, which substantially colocalized with GD3 (Fig. 3C, left and central panels). By contrast, BafA1 was ineffective in inducing autophagosome accumulation in fibroblasts treated with FB1 before TNC administration (Fig. 3C, right panels).
GD3 intracellular localization during autophagic process
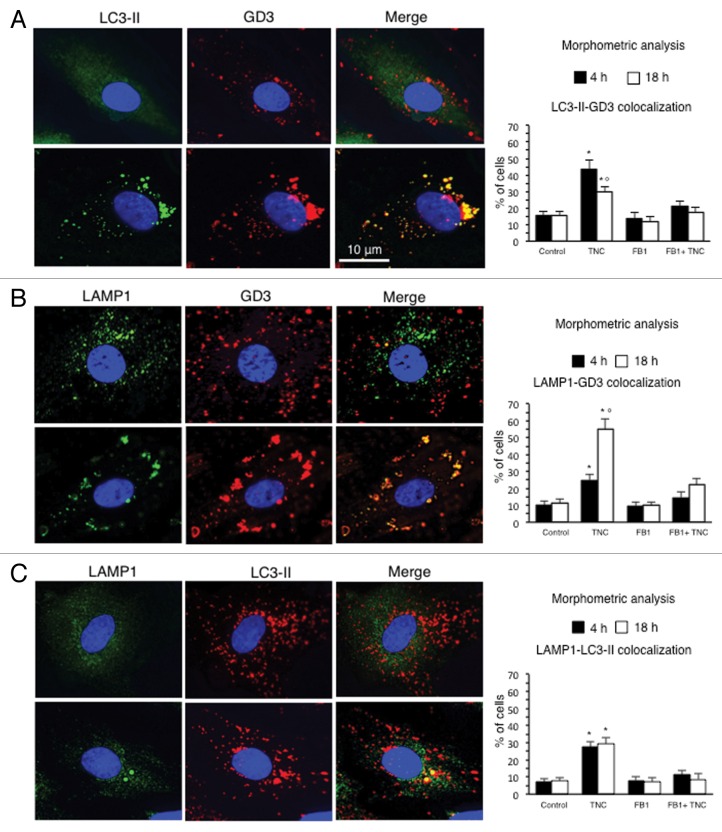
In light of these results, we analyzed the behavior of ganglioside GD3, which can be considered as a paradigmatic raft component,24 performing a time-course analysis by immunofluorescence, after cell double staining with anti-GD3 antibodies (red) and anti-LC3 (green) or anti-LAMP1 (green) antibodies in TNC-treated cells (Fig. 4). As expected, in control untreated cells either LC3 or LAMP1 appeared diffused throughout the cell cytoplasm and GD3-LC3 (Fig. 4A, upper panel) as well as GD3-LAMP1 (Fig. 4B, upper panel) colocalization areas were very barely detectable (yellow spots in merge pictures). By contrast, in TNC-treated cells, colocalization of GD3 with LC3 (Fig. 4A, bottom line, yellow fluorescence in merge micrograph), as well as of GD3 with LAMP1 (Fig. 4B, bottom line) were well evident, as revealed by a number of punctate yellow spots in merge pictures. Importantly, this colocalization was detectable soon after TNC treatment (4 h), but was also observable at later time points, i.e., up to 18 h. It has been reported that LC3 colocalizes with LAMP1 during autophagy.25 According to this observation, we also found that autolysosomes stained with anti-LC3 antibodies (Fig. 4C, second micrograph) were also positive to LAMP1 (Fig. 4C, first micrograph), as revealed by yellow fluorescence in the merge picture (Fig. 4C, third micrograph).
Figure 4. GD3 intracellular localization during the autophagic process. IVM analysis after cell staining for (A) GD3 and LC3-II, (B) GD3 and LAMP1 and (C) LC3-II and LAMP1 in control cells (upper row) and in cells treated with TNC (bottom row). Yellow fluorescence observed in the merge picture in TNC-treated cells indicates GD3-LC3-II, GD3-LAMP1 or LC3-II-LAMP1 colocalization. Bar graphs, showing morphometric analysis, evaluate GD3-LC3-II, GD3-LAMP1 and LC3-II-LAMP1 colocalization. In ordinate the percentage of cells in which we observed yellow fluorescence is shown. Data are reported as mean ± SD of the results obtained in 3 independent experiments. *P < 0.01 vs. control; °P < 0.01 vs. TNC 4 h.
Morphometric analysis, performed as stated in Materials and Methods, revealed a different kinetics in GD3-LC3 and GD3-LAMP1 association (bar graphs). In particular, GD3-LC3 colocalization, measured considering yellow spots, was higher 4 h after TNC addition (Fig. 4A, bar graph), whereas GD3-LAMP1 colocalization peaked 18 h after TNC treatment (Fig. 4B, bar graph). As far as LC3-LAMP1 association was concerned, we did not observe any significant difference between 4 h and 18 h of TNC treatment (Fig. 4C, bar graph). Importantly, FB1 significantly prevented TNC-induced colocalization of both LC3 and LAMP1 with GD3 as well as of LC3 with LAMP1. These experiments seem to suggest that ganglioside GD3 was present on autophagic vacuoles during their maturation stages.
Ultrastructural evidence of GD3 on the autolysosomal membrane
Immunogold electron microscopy was also performed in order to better assess the presence of ganglioside GD3 on the autophagic vacuoles in our samples. After treatment with TNC, several autolysosomes were detectable (Fig. 5A) and gold particles labeling GD3 ganglioside were observed on putative autolysosomes containing digested materials (Fig. 5B, arrows),
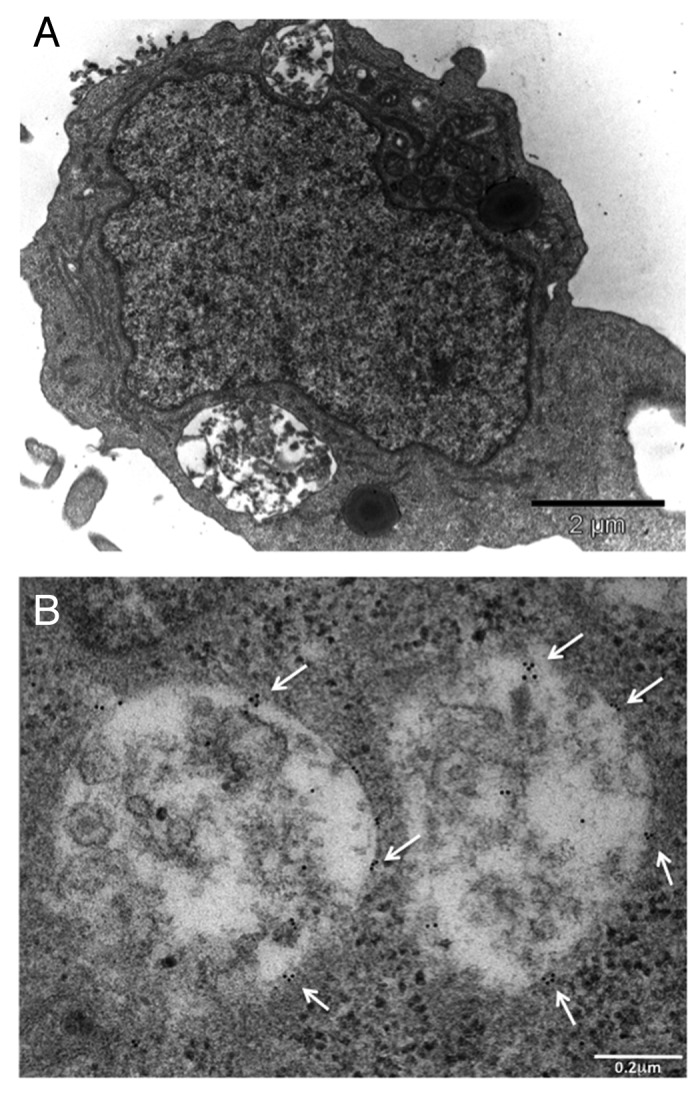
Figure 5. Ultrastructural evidence of GD3 on the autolysosomal membrane. (A) Transmission and (B) immunogold electron microscopy micrographs showing the presence of putative autolysosomes containing semidigested cell materials after treatment with TNC (A and B). Several gold particles indicating the presence of ganglioside GD3 are visible (B, arrows). Note that the gold particles do not appear randomly distributed but as small clusters (arrows).
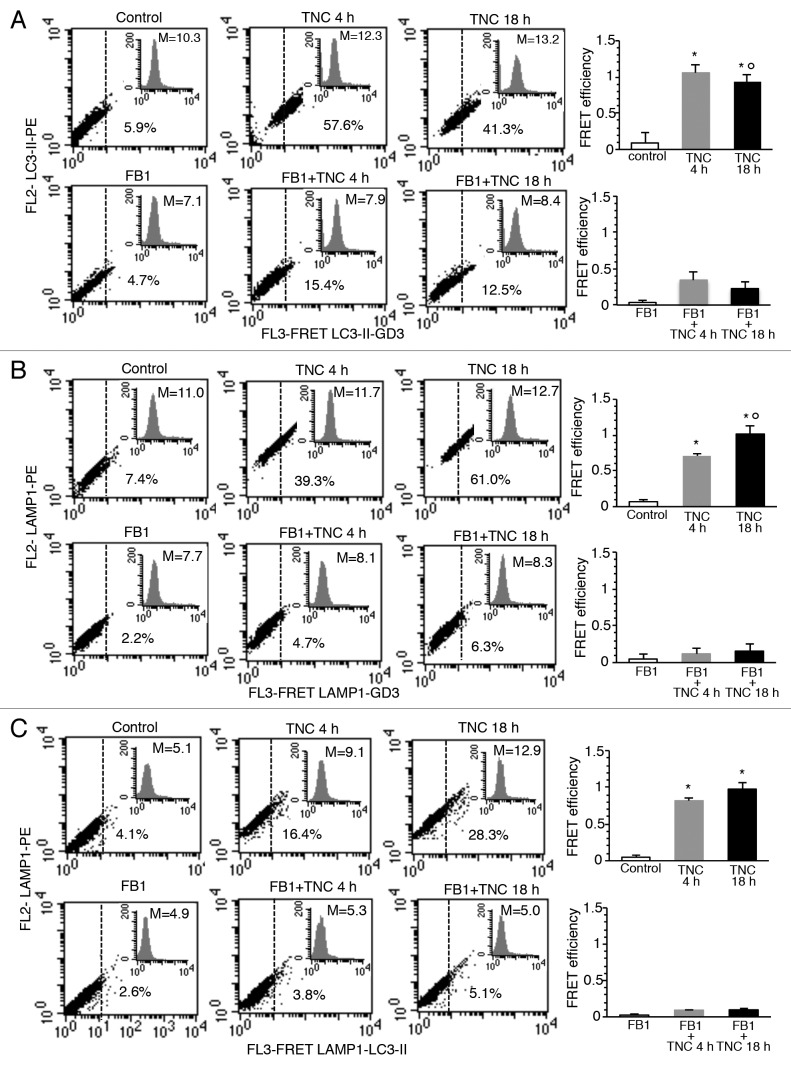
GD3 association with LC3 and LAMP1 during the autophagic process
To investigate the intracytoplasmic redistribution of GD3 in autophagy-triggered cells, possible molecular association of GD3 with LC3 or LAMP1 was analyzed by FRET methodology (Fig. 6). Statistical analyses were then performed by pooling together data obtained from four independent experiments (Fig. 6, right panels) and by calculating the FRET efficiency (FE). We used the following algorithm: FE = [FL3DA − FL2DA/a − FL4DA/b]/FL3DA, where A is the acceptor and D the donor and where a = FL2D/FL3D and b = FL4A/FL3A (Fig. S3–S5). According to morphometric analysis reported above, this quantitative analysis indicated a strict molecular interaction of GD3 either with LC3 or LAMP1, although with different kinetics. In fact, GD3/LC3 interaction peaked 4h after TNC addition, while GD3-LAMP1 association reached the higher FE only later (18 h), when the molecular association between GD3 and LC3 instead decreased. Cell treatment with FB1, given before TNC administration, significantly prevented the molecular association of GD3 either with LC3 or with LAMP1. In accordance with the known mechanism of action of FB1 that blocks sphingolipid biosynthesis, this drug also decreased the intracellular levels of GD3 (Fig. 6, see insets). FRET analysis also revealed a significant association of LC3 with LAMP1 in cells treated with TNC for 4 h or 18 h, but not in control cells. According to the above results, pretreatment of cells with FB1 significantly prevented this association (Fig. 6).
Figure 6. GD3 association with LC3 and LAMP1 during the autophagic process. Flow cytometry analysis of (A) GD3-LC3-II, (B) GD3-LAMP1 and (C) LC3-II-LAMP1 association by FRET technique in: control cells (first panel), cells treated with TNC 4 h (second panel) or 18 h (third panel), in the absence (upper row) or in the presence (bottom row) of FB1. Numbers indicate the percentage of FL3-positive events obtained in one experiment representative of 4. Inserts represent GD3 intracellular amount in the corresponding sample and was quantitatively expressed by the median fluorescence intensity. Bar graphs in (A–C) show evaluation of FE, according to the Riemann algorithm, of GD3-LC3-II, GD3-LAMP1 and LC3-II-LAMP1 association. Results represent the mean ± SD from four independent experiments. Statistical analyses indicated that: i) GD3-LC3-II association was significant either 4 h or 18 h after TNC administration, but was higher in cells treated with TNC for 4 h; ii) GD3-LAMP1 association was significant either 4 h or 18 h after TNC administration but was higher in cells treated with TNC for 18 h and that iii) LC3-II-LAMP1 association was significant either 4 h or 18 h after TNC administration. In addition, evaluation of FE also indicates that cell pretreatment with FB1 significantly prevents all the above associations. *P < 0.01 vs. control; °P < 0.01 vs. TNC 4 h.
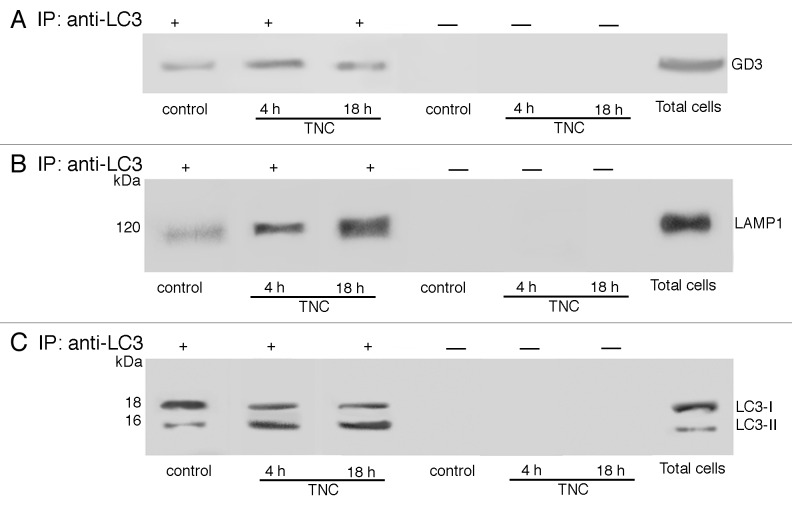
To verify by a sanctioned biochemical method whether GD3 may interact with LC3, cell lysates, obtained from TNC-treated (4 and 18 h) or starved in HBSS (not shown) and untreated cells were immunoprecipitated with the anti-LC3 Ab, followed by protein G-acrylic beads. Results of TLC immunostaining analysis, presented in Figure 7A, showed that in control unstimulated cells GD3 was slightly associated with LC3. On the contrary, after triggering with TNC, a significant proportion of GD3 became associated with LC3, which was more evident in cells stimulated with TNC for 4 h. No bands were detected after TNC stimulation in control immunoprecipitation experiments with an IgG having irrelevant specificity.
Figure 7. GD3 associates with LC3 and LAMP1. Coimmunoprecipitation of LC3 with GD3 (A) and LAMP1 (B) in control cells and in cells treated with TNC 4 h or 18 h. Primary fibroblasts, untreated or treated with TNC, were lysed in lysis buffer, followed by immunoprecipitation with anti-LC3 polyclonal antibody. A rabbit IgG isotypic control was employed. The immunoprecipitates were subjected to ganglioside extraction. The extracts were run on HPTLC aluminum-backed silica gel and were analyzed for the presence of GD3, using an anti-GD3 Mab (GMR19) (A). Alternatively, the immunoprecipitates were analyzed for the presence of LAMP-1 by western blot analysis, using anti-LAMP1 Mab (B). As a control, the immunoprecipitates were assessed by immunoblot with anti-LC3 polyclonal antibody (C).
In the same immunoprecipitates we also evaluated the association of LAMP1 with LC3 and observed a positive band of coimmunoprecipitation (Fig. 7B), which was more evident in cells stimulated with TNC for 18 h. No bands were detected after TNC stimulation in control immunoprecipitation experiments with an IgG having irrelevant specificity. LC3 immunoprecipitation was verified by western blot (Fig. 7C). Altogether, FRET and coimmunoprecipitation analyses clearly suggest that the induction of autophagy could trigger a molecular interaction between ganglioside GD3 and 2 key molecules involved in autolysosome formation and maturation, such as LC3 and LAMP1.
Effect of exogenous GD3 supplementation on the autophagic susceptibility of ganglioside-depleted cells
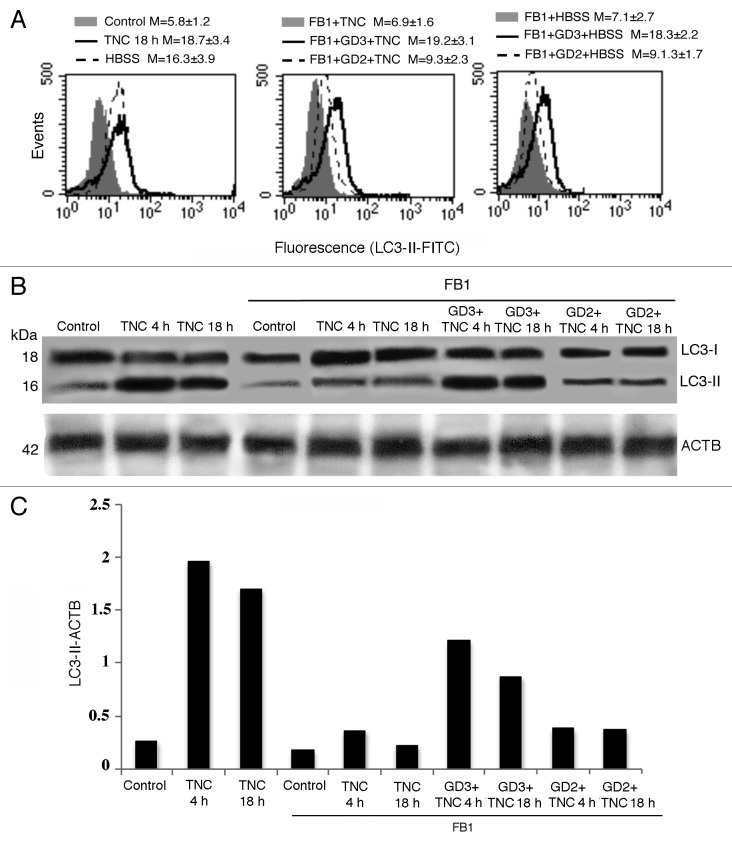
To verify whether GD3 has a primary role in autophagic processes, we pretreated human primary fibroblasts with FB1 and then we treated them with exogenous GD3 before the autophagic triggering. Flow cytometry analysis after cell staining with anti-LC3-II antibodies clearly showed that, while cell pretreatment with FB1 significantly inhibited TNC-induced autophagy as mentioned above, the addition of exogenous GD3 was able to “reactivate” the autophagic process (Fig. 8A). As a further control, we also performed the same experiments by using a different ganglioside of relevance in cell physiology, i.e., GD2 disialoganglioside. We found that the effectiveness of this ganglioside in reactivating authophagic process in cells treated with FB1 was very low. Of note, statistical analysis indicated that GD3 was significantly more efficient in reactivating autophagic process than GD2 (FB1+GD3+TNC and FB1+GD3+HBSS vs. FB1+GD2+TNC and FB1+GD2+HBSS, P < 0.01), suggesting that the action of GD3 could be quite specific. We therefore analyzed LC3 expression by western blot after autophagy triggering, in the presence or in the absence of FB1 plus exogenous GD3 or GD2, under the same experimental conditions (Fig. 8B). Again, our results confirmed that, in the presence of FB1, only treatment with GD3 restored the conversion of soluble LC3-I to lipid-membrane-bound LC3-II induced by TNC (Fig. 8C, see densitometric LC3-II/ACTB ratios), whereas GD2 was virtually ineffective. Similar results were obtained by using the starvation procedure (not shown). Altogether, these data confirm the above results suggesting that ganglioside GD3 could actively participate in the autophagic process but, also, that a histotype-dependent role of different gangliosides cannot be ruled out.
Figure 8. Effect of exogenous GD3 supplementation on the autophagic susceptibility of ganglioside-depleted cells. (A) Semiquantitative flow cytometry analysis of autophagy performed in primary human fibroblasts performed by LC3-II cell staining. The numbers represent the mean ± SD of the median fluorescence intensity obtained in 3 independent measurements. A representative experiment is shown. (B) western blot analysis (using anti-LC3 polyclonal antibody) of human primary fibroblasts untreated or treated with TNC (5 µM for 18 h at 37 °C) in the presence or in the absence of pretreatment with FB1, either incubated or not with GD3 or GD2 (50 µg/ml). Loading control was evaluated using anti-ACTB Mab. (C) Densitometric LC3-II-ACTB ratios are shown.
Discussion
Lipid microdomains are normally detected at the cell plasma membrane, where they exert a key catalytic function facilitating molecular interactions.26 Under preapoptotic stimulation, the presence of raft-like microdomains at mitochondrial level has also been demonstrated.27,28 It has been hypothesized that they could contribute to mitochondrial remodeling, e.g., fission processes and changes of membrane curvature, associated with apoptosis.29 Furthermore, it has also been suggested that glycosphingolipids could contribute to trafficking and scrambling of membranes among organelles in cell physiopathology.26 In the present study we demonstrated for the first time a functional role for gangliosides in the autophagic process, suggesting that they may play a role in autophagosome morphogenesis, leading to mature autophagosome formation.
As a general rule, the adaptive mechanism of autophagy process, in parallel with proteasome activity, allows the recycling of unfolded proteins and, more importantly, of cytoplasmic components, including organelles, into basic components. For instance, selective autophagy of mitochondria, known as mitophagy, is an important mechanism able to hijack specific mitochondrial components to the lysosomal pathway. In this case, autophagy serves as a mechanism to sequester selected mitochondrial cargos and then to deliver those mitochondrial components to the lysosome for degradation.30 In the present paper, we observed that early after autophagic ignition (4 h) the main ganglioside of fibroblasts, GD3, is associated with PtdIns3P and LC3. These molecular associations are compatible with a role for GD3 in the initiation phase of autophagy and with autophagosome biogenesis. Interestingly, at later time points (18 h) a GD3-LAMP1 association was also detected, suggesting that GD3 could also play a role in the maturation of the autophagosome into autolysosome.
Although gangliosides, in particular ceramides, have previously been associated with autophagy induction,11,31 the contribution of constitutive glycosphingolipids to cellular autophagic processes is still unknown. Since ceramides are suppressors of AKT,32 it can be argued that inactivation of MTOR, a well-known preautophagic signaling molecule that acts downstream of AKT, may represent a key mechanism of ceramide-induced autophagy. In fact, it has been demonstrated that accumulation of ceramide suppresses AKT activity to stimulate autophagy both in human colon cancer and breast cancer cell lines.31 Accordingly, it has been suggested that amino acid starvation could activate the “sphingolipid rheostat” signaling, inactivating MTOR, activating the MAPK1-MAPK3 pathway and inducing autophagy.15,17 Moreover, in astrocytes, gangliosides have also been hypothesized to promote the autophagic process by activating NFKB,16 probably via generation of reactive oxygen species,17 known activators of this transcription factor. Finally, a recent work also suggested that sphingolipids could act as regulators of the crosstalk between apoptosis and autophagy.33
In the present work, in agreement with these observations,15-18 and with those underlying the presence of GD3 not only on the plasma membrane, but also in the cell cytoplasm,34,35 including the ER-mitochondria associated membranes,36 we found that the inhibitor of ceramide synthase FB1 significantly prevented autophagy induced either by amino acid starvation or by tunicamycin. Furthermore, we point out a specific role for ganglioside GD3, since we found that the knocking down of GD3 synthase significantly hindered autophagy. This finding was strongly supported by the observation that treatment with exogenous GD3 significantly counteracted the effects of FB1, restoring autophagic susceptibility to TNC or HBSS. Conversely, at least in our cell system, i.e., human and murine fibroblasts, the role of other gangliosides, such as GD2, known to be abundant in other cell types, e.g., neuronal cells, appeared as negligible. We can thus hypothesize that different gangliosides could act differently depending on the cell histotype.
The importance of the sphingolipid-protein molecular interaction has already been suggested. In particular, it has been hypothesized that ganglioside-protein interactions, which can be evaluated by different techniques, e.g., FRET analysis and mass spectroscopy,37,38 could act as regulators of membrane organization and may be implicated in the pathogenesis of human diseases.39-41 Accordingly, by using FRET and coimmunoprecipitation analyses, we provided proof of a direct molecular association between GD3 ganglioside and autophagy determinants, i.e., GD3-LC3 and GD3-LAMP1. Importantly, these results came from analyses performed in freshly isolated nontransformed fibroblasts, where the genomic and metabolic alterations due to the number of subseedings are minimized.
On these bases, we hypothesize that the biophysical and biochemical properties of ganglioside GD3 could be fundamental in the autophagic process, contributing to autophagosome biogenesis and/or maturation. At which step of the autophagic flux gangliosides could play a critical role still remains to be clarified. As a general rule our data seem to suggest that the block of sphingolipid biogenesis may affect autophagosome formation. However, our results also showed that i) the contribution of the endocytic process appears negligible, at least in the present experimental model, and that ii) the block of sphingolipid biogenesis may affect PtdIns3P synthesis. It has been reported that during autophagosome formation, the balance between the production of PtdIns3P and its turnover is critical in regulating both the size and the rate of production of autophagosomes.42 In particular, the synthesized PtdIns3P takes part in the creation of a platform that helps in the recruitment of specific effectors for membrane-trafficking events, including autophagosome biogenesis.43 Thus, our results seem to suggest that ganglioside GD3 may act at an early step of the autophagic flux, i.e., during autophagosome biogenesis, but also carry on its action throughout the autophagosome maturation steps. In fact, other analyses reported here, e.g., GD3-LAMP1 association observed by FRET, seem to indicate that the presence of ganglioside GD3 could be pivotal in all the steps leading to the morphogenesis of mature autolysosomes. Conversely, experiments performed with the inhibitor of vacuolar H+ ATPase BafA1 seem to indicate that the role of ganglioside in the late events, such as acidification, could be negligible.
A further important point to be considered here is derived from the fact that tunicamycin, the preautophagic drug used in this work, is also considered as a paradigmatic ER stress inducer.44 ER stress is known to activate a complex intracellular signaling transduction pathway, called the unfolded protein response, essentially devoted to reestablish ER homeostasis. However, it is also well established that ER stress, i.e., a physiopathological condition in which protein folding is perturbed, represents an essential step of the autophagic process. During autophagy the membrane trafficking and intermixing leads to the formation of autophagosomes, but the origin of autophagosomal membranes appears still unclear. In fact, at present, there is much controversy about the organelle from which the membranes originate: the endoplasmic reticulum (ER), the mitochondria, or the plasma membrane.2 Very recently, it has been suggested that autophagosomes form at the ER-mitochondria contact site, at least in mammalian cells.45 Here, we provide evidence that gangliosides could participate to this intermixing activity thanks to their molecular interaction with key molecules involved in autophagosome biogenesis and maturation, e.g., PtdIns3P, LC3, and LAMP1.
Considering that sphingolipids are known to strongly influence the biophysical properties of the membranes, as well as their curvature,29,46 we can speculate that ganglioside GD3 may play a pivotal role in morphogenetic remodeling of autophagosomes during the autophagic process.
Although sphingolipids are involved in many aspects of cell life and death and although sphingolipid metabolism has been proposed as a possible target for the generation of novel chemotherapeutics and sphingomimetics in different fields of investigation, including cancer,33 the study of the “sphingolipidome,” due to its complexity, is still at the beginning. Better knowledge of the role of gangliosides in the autophagic pathway may thus provide useful insights in basic as well as in translational medicine.
Materials and Methods
Cells and autophagy induction
Primary human fibroblast cultures obtained from skin biopsy and mouse embryo fibroblasts (MEFs), prepared as previously described,47 were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma, D5796), containing 10% fetal calf serum (FCS) plus 100 units/ml penicillin, 10 mg/ml streptomycin, at 37 °C in humified CO2 atmosphere. For autophagy induction, cells were stimulated under condition of nutrient deprivation with Hank’s Balanced Salt Solution (HBSS) (Sigma, H9269) or treated with tunicamycin (from Streptomyces lysosuperficus; TNC, Calbiochem, 654380), 5 µM, for 4 h or 18 h at 37 °C. The optimal incubation time with HBSS and tunicamycin was selected on the basis of preliminary experiments. To inhibit autophagy, cells were pretreated with bafilomycin A1 (BafA1, 1 μM 1 h before autophagy induction) (Sigma-Aldrich, B1793). After treatments, cells were collected and prepared for experimental procedures described below. Results obtained with MEFs were substantially superimposable to that obtained with human fibroblasts (reported in Fig. S1).
Analysis of autophagy
Flow cytometry analyses
Detection of autophagy was performed by using Cyto-ID Autophagy Detection Kit (Enzo Life Sciences, ENZ-51031-K200). The kit was optimized for detection of autophagy in live cells by flow cytometry. This assay provides a rapid, specific, and quantitative approach for monitoring autophagic activity at the cellular level by using a 488 nm-excitable probe that becomes fluorescent in vesicles produced during autophagy.48 Alternatively, to validate data obtained by using Cyto-ID Autophagy Detection Kit we quantified autophagy after cell staining with rabbit polyclonal anti-LC3-II antibody (Abgent, AP1805A). Briefly, cells (untreated or treated with HBSS or TNC, 5 µM for 18 h at 37 °C), fixed with 4% paraformaldehyde and permeabilized by 0.5% (v/v) Triton X-100 were incubated with rabbit polyclonal anti-LC3-II antibody for 1 h at 4 °C, followed by AlexaFluor 488-conjugated anti-rabbit (Molecular Probes, A-11008) for additional 30 min. After washing, cells were analyzed by flow cytometer. As showed in Figure 1A, these 2 alternative methods to quantify autophagy revealed completely overlapping results.
Western blot analysis
Cells, untreated or treated with HBSS or TNC (5 µM for 18 h at 37 °C), were lysed in lysis buffer containing 1% Triton X-100, 10 mM TRIS-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4 and 75 U of aprotinin and allowed to stand for 20 min at 4 °C. The cell suspension was mechanically disrupted by Dounce homogenization (10 strokes). The lysate was centrifuged for 5 min at 1300 × g to remove nuclei and large cellular debris. After evaluation of the protein concentration by Bradford Dye Reagent assay (Bio-Rad, 500-0006) the lysate was subjected to 10% sodium-dodecyl sulfate polyacrilamide gel electrophoresis (SDS-PAGE). The proteins were electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad, 162-0177). Membranes were blocked with 5% defatted dried milk in TBS, containing 0.05% Tween 20 and probed with rabbit polyclonal anti-LC3 antibody (MBL Int Corporation, PD014) or with anti-ACTB/β-actin Mab (Sigma, A5316). Bound antibodies were visualized with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Sigma-Aldrich, A1949) or anti-mouse IgG (Sigma-Aldrich, A9044) and immunoreactivity assessed by chemiluminescence reaction, using the ECL western detection system (Amersham, RPN2106). Densitometric scanning analysis was performed by Mac OS X (Apple Computer International), using NIH Image 1.62 software. The density of each band in the same gel was analyzed, values were totaled, and then the percent distribution across the gel was detected.
Analysis of the effects of fumonisin B1 on autophagy induction
Primary fibroblasts, untreated or treated with HBSS or TNC (5 µM for 4 h or 18 h at 37 °C), were incubated with 30 µM fumonisin B1 (FB1, Sigma, F1147) (inhibitor of ceramide synthases) for 24 h at 37 °C to evaluate the effects of sphingolipid depletion on autophagy induction.
In parallel experiments, cells, incubated with 30 µM fumonisin B1 for 24 h at 37 °C were treated with GD3 (Alexis Biochemicals, 302-010), 50 µg/ml, or GD2 (Sigma, G0776), 50 µg/ml, for 30 min.
Briefly, after evaluation of the protein concentration by Bradford Dye Reagent assay the lysates were analyzed by western blot using the anti-LC3 polyclonal antibody, as reported above.
Assays of endocytosis
Control and treated cells were washed 3 times, resuspended in DMEM and then incubated for additional 30 min with dextran-FITC (1 mg/ml, Sigma-Aldrich, 53379) at 37 °C. After 3 washings, cells were resuspended in PBS and immediately analyzed by a cytometer. To distinguish the ingested dextran particles from those attached on the cell surface we used fluorescence quenching with trypan blue as previously described.49 To inhibit endocytosis, cells were pretreated 30 min with 20 μM tricyclodecan-9-yl-xanthogenate (D609, Millipore, 251400), an inhibitor of endocytosis that selectively inhibits phosphatidylcholine-specific phospholipase C,50 before autophagic induction.
ST8SIA1 (GD3-synthase) siRNA
Primary fibroblasts, were cultured in an antibiotic-free medium containing 10% FCS and transfected with Dharma FECT 1 reagent (Dharmacon, T-2001-03), according to the manufacturer's instructions, using 25 nM Smart pool siRNA ST8SIA1 (GD3 synthase) (Dharmacon, M-011775-02) and its related scrambled siRNA as control (Dharmacon, D-001810-10-05). The transfection efficiency was confirmed by using a Dharmacon’s positive silencing control, siGLO laminin A/C siRNA (D-001620-02). Forty eight h after transfection cells were treated with TNC for 4 h, as described above.
After 48 h, the effect of transfection on ST8SIA1 (GD3-synthase) expression level was verified by flow cytometry by using an anti-ST8SIA1 (K-18) polyclonal antibody (Santa Cruz Biotechnology, sc-44587).
PtdIns3P analysis
Primary fibroblasts, either untreated or treated with TNC (5 μM for 4 h at 37 °C), in the presence or in the absence of pretreatment with FB1, were subjected to phosphoinositide extraction, as previously described.51 Samples were normalized for protein content, quantified as reported above. Briefly, chloroform/methanol/HCL extraction was followed by lipid drying under N2. Samples were separated by high-performance thin layer chromatography (HPTLC) using aluminum-backed Silica gel 60 (20 × 20) plates (Merck, 1.05547.0001) saturated with 1% potassium oxalate in 50% methanol. Chromatography was performed in chloroform:acetone:methanol:acetic acid:water (40:15:13:12:8) (v:v:v:v:v). The plates were immunostained for 1 h at room temperature with anti-PtdIns3P Mab (Echelon Biosciences Inc., Z-P003) and then with HRP-conjugated anti-mouse IgG. Immunoreactivity was assessed by chemiluminescence reaction, using the ECL western detection system. PtdIns3P was quantified from TLC plates by densitometric scanning analysis, using NIH Image 1.62 software.
Immunofluorescence analysis
For triple fluorescence analysis, control and treated cells were fixed with 4% paraformaldehyde and then permeabilized by 0.5% (v/v) Triton X-100. After washing, cells were incubated with rabbit polyclonal anti-LC3-II antibody or anti-LAMP1 Mab (Santa Cruz Biotechnology, sc18821) or anti-PtdIns3P Mab (Echelon Biosciences Inc., Z-P003) or with goat polyclonal antibody to RAB7 (Santa Cruz Biotechnology, sc-6563) for 1 h at 4 °C. After washing cells were incubated with AlexaFluor 488-conjugated anti-rabbit (Molecular Probes, A-11034) anti-mouse IgG (Molecular Probes, A-11001) or anti-goat (Molecular Probes, A-11055) for additional 30 min. After washing, cells were incubated for 1 h at 4 °C with IgM anti-GD3 (Seikagaku Corporation, 370635) or anti-GD2 Mab (Abcam, ab68456) followed by AlexaFluor 594-conjugated anti-mouse IgM (Molecular Probes, 4-21044). Finally, after washing, all samples were counterstained with Hoechst 33258 (Sigma, 94403), 1 mg/ml in PBS, and then mounted in glycerol/PBS (ratio 1:1, pH 7.4). The images were acquired by intensified video microscopy (IVM) with an Olympus fluorescence microscope (Olympus Corporation of the Americas, Center Valley, Pa.), equipped with a Zeiss charge-coupled device (CCD) camera (Carl Zeiss).
Morphometric analysis
Quantitative evaluation of cells in which GD3 colocalized with LC3 or with LAMP1 or cells in which LAMP1 colocalized with LC3-II was performed by analyzing fluorescence images (magnification 500 × ) by counting at least 200 cells for each experimental point. Only those cells in which GD3-LC3-II, GD3-LAMP1 or LAMP1-LC3-II overlapped (yellow fluorescence) were considered in our analysis.
Transmission electron microscopy (TEM) studies
For TEM examination, cells were fixed in 2.5% cacodylate-buffered (0.1 M, pH 7.2) glutaraldehyde (TAAB, G004) for 20 min at room temperature and post-fixed in 1% OsO4 (Electron Microscopy Sciences, 19100) in cacodylate buffer for 1 h at room temperature. Fixed specimens were dehydrated through a graded series of ethanol solutions and embedded in Agar 100 (Electron Microscopy Sciences, 10200). Ultrathin sections were collected on 200-mesh grids and counterstained with uranyl acetate (Electron Microscopy Sciences, 22400) and lead citrate.
Immunogold electron microscopy
Immunoelectron microscopy was also performed as previously described.28,37 Ganglioside GD3 antigenicity was in fact demonstrated as resistant to fixation procedures preserving cell ultrastructural features.
Briefly, thin sections were treated with PBS containing 1% (w/v) gelatin, 1% BSA, 5% FCS and 0.05% Tween 20 and then incubated with anti-GD3 abs diluted 1:30 in the same buffer w/o gelatin overnight at 4 °C. After washing for 1 h at room temperature (RT), sections were labeled with anti-mouse IgG-10 nm gold conjugate (1:10) for 1 h at RT and washed again. Negative controls were incubated with the gold conjugate alone and then counterstained with uranyl acetate and lead citrate. No non-specific gold labeling was detectable in these conditions.
Both TEM and IEM samples were observed with a Philips 208 electron microscope (Royal Philips, Amstelplein; Amsterdam) at 80 kV.
Fluorescence resonance energy transfer (FRET analysis)
We applied fluorescence resonance energy transfer (FRET) analysis by flow cytometry, in order to study the molecular association of GD3-PtdIns3P, GD3-LC3-II, GD3-LAMP1, and LC3-II-LAMP1.52 Briefly, cells were fixed and permeabilized as above, washed twice in cold PBS and then labeled with antibodies tagged with donor (Phycoerythrin, PE) or acceptor (Cy5) dyes. PtdIns3P and LAMP1 staining were performed using mouse unlabeled antibodies and LC3-II staining was performed using unlabeled rabbit antibodies and saturating amount of PE-labeled anti-mouse (for PtdIns3P and LAMP1, Sigma, P9670) or anti-rabbit (for LC3-II, Sigma, P9537). GD3 was detected by using a mouse IgM anti-GD3, followed by biotinylated anti-mouse IgM (BD Biosciences PharMingen, 553519) and then by saturating concentrations of streptavidin-Cy5 (BD Biosciences PharMingen, 554062). After staining, cells were washed twice, resuspended in PBS and analyzed with a dual-laser FACScalibur flow cytometer (BD Biosciences Franklin Lakes, New Jersey). For determination of FRET efficiency (FE), changes in fluorescence intensities of donor plus acceptor labeled cells were compared with the emission signal from cells labeled with donor-only and acceptor-only fluorophores. As a further control, the cross-reactivity among all the different primary and secondary antibodies was also assessed. All data were corrected for background by subtracting the binding of the isotype controls. Efficient energy transfer resulted in an increased acceptor emission on cells stained with both donor and acceptor dyes. The FE was calculated according to Riemann (see details in Fig. S3–S5).53
LC3 immunoprecipitates
Primary fibroblasts, untreated or treated with HBSS and TNC (5 µM for 18 h at 37 °C), were lysed in lysis buffer (10 nM TRIS-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mg/ml leupeptin). Cell-free lysates were mixed with protein G-acrylic beads (Sigma-Aldrich, P3296) and stirred by a rotary shaker for 2 h at 4 °C to preclear nonspecific binding. After centrifugation (500 × g for 1 min), the supernatant was immunoprecipitated with anti-LC3 polyclonal (MBL Int Corporation, PD015) plus protein G-acrylic beads.
A rabbit IgG isotypic control (Sigma, I5006) was used. The immunoprecipitates were split into 2 aliquots. The first one was subjected to ganglioside extraction, according to the method of Svennerholm and Fredman.54 The second one was checked by western blot analysis, using anti-LAMP1 Mab.
Ganglioside analysis in the immunoprecipitates of LC3
Briefly, immunoprecipitates were extracted twice in chloroform/methanol/water (4:8:3) (v/v/v) and subjected to Folch partition by the addition of water resulting in a final chloroform/methanol/water ratio of 1:2:1.4. The upper phase, containing polar glycosphingolipids, was purified of salts and low molecular weight contaminants using Bond elut C18 columns (Superchrom, 07807T, Restek Corporation, Bellefonte, PA), according to the method of Williams and McCluer.55 The eluted glycosphingolipids were dried down and separated by HPTLC, using aluminum-backed silica gel 60 (20 × 20) plates (Merck). Chromatography was performed in chloroform/methanol/ aqueous KCl (0.25%) (5:4:1) (v/v/v). The dried chromatograms were soaked for 90 s in a 0.5% (w/v) solution of poly(iso)butylmethacrylate (Sigma-Aldrich) dissolved in hexane. The plates were immunostained for 1 h at room temperature with GMR19 anti-GD3 Mab and then with HRP-conjugated anti-mouse IgM (Sigma, A8786). Immunoreactivity was assessed by chemiluminescence reaction, using the ECL western detection system.
Immunoblotting analysis of LC3 immunoprecipitates
The immunoprecipitates, obtained as reported above, were subjected to SDS-PAGE. The proteins were electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes. Membrane were blocked with 5% defatted dried milk in TBS, containing 0.05% Tween 20 and probed with anti-LAMP1 Mab or anti-LC3 polyclonal antibody. Bound antibodies were visualized with HRP-conjugated anti-mouse IgG or anti-rabbit IgG and immunoreactivity assessed by chemiluminescence reaction, using the ECL western detection system.
Data analysis and statistics
For flow cytometry studies all samples were analyzed by a dual-laser FACScalibur cytometer equipped with a 488 argon laser and with a 635 red diode laser. At least 20,000 events/sample were acquired. Data were recorded and statistically analyzed by a Macintosh computer using CellQuestPro Software. Collected data analysis was performed by using ANOVA 2-way test for repeated samples by using Graphpad software. All data reported in this paper were verified in at least 3 different experiments and reported as mean ± standard deviation (SD). Only P values of less than 0.01 were considered as significant.
Supplementary Material
Glossary
Abbreviations:
- ACTB
actin, beta
- ATG
autophagy-related
- BafA1
bafilomycin A1
- BECN1
Beclin 1, autophagy-related
- CCD camera
charge-coupled device
- DISC
death-inducing signaling complex
- D609
tricyclodecan-9-yl-xanthogenate
- ER
endoplasmic reticulum
- FB1
fumonisin B1
- FE
FRET efficiency
- FRET
fluorescence resonance energy transfer
- GD3
aNeu5Ac(2-8)aNeu5Ac(2-3)bDGalp(1-4)bDGlcp(1-1)ceramide
- HBSS
Hank's Balanced Salt Solution
- HRP
horseradish peroxidase
- IKBKB
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
- IVM
intensified video microscopy
- LAMP1
lysosomal-associated membrane protein 1
- Mab
monoclonal antibodies
- MAP1LC3 (LC3)
microtubule-associated protein 1 light chain 3
- MAPK
mitogen-activated protein kinase
- MTOR
mechanistic target of rapamycin
- NFKB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- PtdIns3K
phosphatidylinositol 3-kinase
- PtdIns3P
phosphatidylinositol 3-phosphate
- RAB7A
RAB7A, member RAS oncogene family
- ROS
reactive oxygen species
- SD
standard deviation
- SMPD1
sphingomyelin phosphodiesterase 1, acid lysosomal
- ST8SIA1/GD3 Synthase
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
- TNC
tunicamycin
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported in part by grants from the Ministry of Education, University and Research (Rome, Italy), the Ministry of Health (Ricerca 2010 to WM), the Italian Association for Cancer Research (MCO-9998 and 11505) (WM); Arcobaleno Onlus (WM); Peretti Foundation (WM); PRIN project 2009 (MS); PRIN project 2011 (RM) and Sapienza University project 2012 (MS).
References
- 1.Yang Z, Klionsky DJ. . Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124 - 31; http://dx.doi.org/ 10.1016/j.ceb.2009.11.014; PMID: 20034776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tooze SA, Yoshimori T. . The origin of the autophagosomal membrane. Nat Cell Biol 2010; 12:831 - 5; http://dx.doi.org/ 10.1038/ncb0910-831; PMID: 20811355 [DOI] [PubMed] [Google Scholar]
- 3.Choi AM, Ryter SW, Levine B. . Autophagy in human health and disease. N Engl J Med 2013; 368:651 - 62; http://dx.doi.org/ 10.1056/NEJMra1205406; PMID: 23406030 [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein DC, Codogno P, Levine B. . Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709 - 30; http://dx.doi.org/ 10.1038/nrd3802; PMID: 22935804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Yoshimori T, Ohsumi Y. . The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107 - 32; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005; PMID: 21801009 [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Klionsky DJ. . The regulation of autophagy - unanswered questions. J Cell Sci 2011; 124:161 - 70; http://dx.doi.org/ 10.1242/jcs.064576; PMID: 21187343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. . Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004; 117:4837 - 48; http://dx.doi.org/ 10.1242/jcs.01370; PMID: 15340014 [DOI] [PubMed] [Google Scholar]
- 8.Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. . The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 2005; 1:23 - 36; http://dx.doi.org/ 10.4161/auto.1.1.1495; PMID: 16874023 [DOI] [PubMed] [Google Scholar]
- 9.Hakomori S, Handa K, Iwabuchi K, Yamamura S, Prinetti A. . New insights in glycosphingolipid function: “glycosignaling domain,” a cell surface assembly of glycosphingolipids with signal transducer molecules,involved in cell adhesion coupled with signaling. Glycobiology 1998; 8:xi - xix; http://dx.doi.org/ 10.1093/oxfordjournals.glycob.a018822; PMID: 9840984 [DOI] [PubMed] [Google Scholar]
- 10.Lingwood D, Simons K. . Lipid rafts as a membrane-organizing principle. Science 2010; 327:46 - 50; http://dx.doi.org/ 10.1126/science.1174621; PMID: 20044567 [DOI] [PubMed] [Google Scholar]
- 11.Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. . Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res 2004; 64:4286 - 93; http://dx.doi.org/ 10.1158/0008-5472.CAN-03-3084; PMID: 15205343 [DOI] [PubMed] [Google Scholar]
- 12.Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. . Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci U S A 2008; 105:17402 - 7; http://dx.doi.org/ 10.1073/pnas.0802781105; PMID: 18981422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lépine S, Allegood JC, Edmonds Y, Milstien S, Spiegel S. . Autophagy induced by deficiency of sphingosine-1-phosphate phosphohydrolase 1 is switched to apoptosis by calpain-mediated autophagy-related gene 5 (Atg5) cleavage. J Biol Chem 2011; 286:44380 - 90; http://dx.doi.org/ 10.1074/jbc.M111.257519; PMID: 22052905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park MA, Zhang G, Norris J, Hylemon PB, Fisher PB, Grant S, Dent P. . Regulation of autophagy by ceramide-CD95-PERK signaling. Autophagy 2008; 4:929 - 31; PMID: 18719356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi M, Kitatani K, Kondo T, Hashimoto-Nishimura M, Asano S, Hayashi A, Mitsutake S, Igarashi Y, Umehara H, Takeya H, et al. . Regulation of autophagy and its associated cell death by “sphingolipid rheostat”: reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. J Biol Chem 2012; 287:39898 - 910; http://dx.doi.org/ 10.1074/jbc.M112.416552; PMID: 23035115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang J, Lee HJ, Lee WH, Suk K. . NF-κB as a common signaling pathway in ganglioside-induced autophagic cell death and activation of astrocytes. J Neuroimmunol 2010; 226:66 - 72; http://dx.doi.org/ 10.1016/j.jneuroim.2010.05.037; PMID: 20554329 [DOI] [PubMed] [Google Scholar]
- 17.Hwang J, Lee S, Lee JT, Kwon TK, Kim DR, Kim H, Park HC, Suk K. . Gangliosides induce autophagic cell death in astrocytes. Br J Pharmacol 2010; 159:586 - 603; http://dx.doi.org/ 10.1111/j.1476-5381.2009.00563.x; PMID: 20067473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagata M, Obara K, Kihara A. . Sphingolipid synthesis is involved in autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun 2011; 410:786 - 91; http://dx.doi.org/ 10.1016/j.bbrc.2011.06.061; PMID: 21703229 [DOI] [PubMed] [Google Scholar]
- 19.Sciannamblo M, Chigorno V, Passi A, Valaperta R, Zucchi I, Sonnino S. . Changes of the ganglioside pattern and content in human fibroblasts by high density cell population subculture progression. Glycoconj J 2002; 19:181 - 6; http://dx.doi.org/ 10.1023/A:1024249707516; PMID: 12815229 [DOI] [PubMed] [Google Scholar]
- 20.Ghidoni R, Houri JJ, Giuliani A, Ogier-Denis E, Parolari E, Botti S, Bauvy C, Codogno P. . The metabolism of sphingo(glyco)lipids is correlated with the differentiation-dependent autophagic pathway in HT-29 cells. Eur J Biochem 1996; 237:454 - 9; http://dx.doi.org/ 10.1111/j.1432-1033.1996.0454k.x; PMID: 8647085 [DOI] [PubMed] [Google Scholar]
- 21.Lamb CA, Dooley HC, Tooze SA. . Endocytosis and autophagy: Shared machinery for degradation. Bioessays 2013; 35:34 - 45; http://dx.doi.org/ 10.1002/bies.201200130; PMID: 23147242 [DOI] [PubMed] [Google Scholar]
- 22.Simonsen A, Tooze SA. . Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 2009; 186:773 - 82; http://dx.doi.org/ 10.1083/jcb.200907014; PMID: 19797076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. . Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 1998; 23:33 - 42; http://dx.doi.org/ 10.1247/csf.23.33; PMID: 9639028 [DOI] [PubMed] [Google Scholar]
- 24.Malorni W, Giammarioli AM, Garofalo T, Sorice M. . Dynamics of lipid raft components during lymphocyte apoptosis: the paradigmatic role of GD3. Apoptosis 2007; 12:941 - 9; http://dx.doi.org/ 10.1007/s10495-007-0757-1; PMID: 17453161 [DOI] [PubMed] [Google Scholar]
- 25.Furuta N, Yoshimori T, Amano A. . Mediatory molecules that fuse autophagosomes and lysosomes. Autophagy 2010; 6:417 - 8; http://dx.doi.org/ 10.4161/auto.6.3.11418; PMID: 20400858 [DOI] [PubMed] [Google Scholar]
- 26.Sorice M, Garofalo T, Misasi R, Manganelli V, Vona R, Malorni W. . Ganglioside GD3 as a raft component in cell death regulation. Anticancer Agents Med Chem 2012; 12:376 - 82; http://dx.doi.org/ 10.2174/187152012800228670; PMID: 21554197 [DOI] [PubMed] [Google Scholar]
- 27.Garofalo T, Giammarioli AM, Misasi R, Tinari A, Manganelli V, Gambardella L, Pavan A, Malorni W, Sorice M. . Lipid microdomains contribute to apoptosis-associated modifications of mitochondria in T cells. Cell Death Differ 2005; 12:1378 - 89; http://dx.doi.org/ 10.1038/sj.cdd.4401672; PMID: 15947792 [DOI] [PubMed] [Google Scholar]
- 28.Sorice M, Manganelli V, Matarrese P, Tinari A, Misasi R, Malorni W, Garofalo T. . Cardiolipin-enriched raft-like microdomains are essential activating platforms for apoptotic signals on mitochondria. FEBS Lett 2009; 583:2447 - 50; http://dx.doi.org/ 10.1016/j.febslet.2009.07.018; PMID: 19616549 [DOI] [PubMed] [Google Scholar]
- 29.Ciarlo L, Manganelli V, Garofalo T, Matarrese P, Tinari A, Misasi R, Malorni W, Sorice M. . Association of fission proteins with mitochondrial raft-like domains. Cell Death Differ 2010; 17:1047 - 58; http://dx.doi.org/ 10.1038/cdd.2009.208; PMID: 20075943 [DOI] [PubMed] [Google Scholar]
- 30.Ashrafi G, Schwarz TL. . The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ 2013; 20:31 - 42; http://dx.doi.org/ 10.1038/cdd.2012.81; PMID: 22743996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, Ghidoni R, Codogno P. . Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem 2004; 279:18384 - 91; http://dx.doi.org/ 10.1074/jbc.M313561200; PMID: 14970205 [DOI] [PubMed] [Google Scholar]
- 32.Zhou H, Summers SA, Birnbaum MJ, Pittman RN. . Inhibition of Akt kinase by cell-permeable ceramide and its implications for ceramide-induced apoptosis. J Biol Chem 1998; 273:16568 - 75; http://dx.doi.org/ 10.1074/jbc.273.26.16568; PMID: 9632728 [DOI] [PubMed] [Google Scholar]
- 33.Young MM, Kester M, Wang HG. . Sphingolipids: regulators of crosstalk between apoptosis and autophagy. J Lipid Res 2013; 54:5 - 19; http://dx.doi.org/ 10.1194/jlr.R031278; PMID: 23152582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorice M, Matarrese P, Manganelli V, Tinari A, Giammarioli AM, Mattei V, Misasi R, Garofalo T, Malorni W. . Role of GD3-CLIPR-59 association in lymphoblastoid T cell apoptosis triggered by CD95/Fas. PLoS One 2010; 5:e8567; http://dx.doi.org/ 10.1371/journal.pone.0008567; PMID: 20052288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorice M, Matarrese P, Tinari A, Giammarioli AM, Garofalo T, Manganelli V, Ciarlo L, Gambardella L, Maccari G, Botta M, et al. . Raft component GD3 associates with tubulin following CD95/Fas ligation. FASEB J 2009; 23:3298 - 308; http://dx.doi.org/ 10.1096/fj.08-128140; PMID: 19509307 [DOI] [PubMed] [Google Scholar]
- 36.Mattei V, Matarrese P, Garofalo T, Tinari A, Gambardella L, Ciarlo L, Manganelli V, Tasciotti V, Misasi R, Malorni W, et al. . Recruitment of cellular prion protein to mitochondrial raft-like microdomains contributes to apoptosis execution. Mol Biol Cell 2011; 22:4842 - 53; http://dx.doi.org/ 10.1091/mbc.E11-04-0348; PMID: 22031292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malorni W, Garofalo T, Tinari A, Manganelli V, Misasi R, Sorice M. . Analyzing lipid raft dynamics during cell apoptosis. Methods Enzymol 2008; 442:125 - 40; http://dx.doi.org/ 10.1016/S0076-6879(08)01406-7; PMID: 18662567 [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Liu L, Daneshfar R, Kitova EN, Li C, Jia F, Cairo CW, Klassen JS. . Protein-glycosphingolipid interactions revealed using catch-and-release mass spectrometry. Anal Chem 2012; 84:7618 - 21; http://dx.doi.org/ 10.1021/ac3023857; PMID: 22920193 [DOI] [PubMed] [Google Scholar]
- 39.Sonnino S, Mauri L, Ciampa MG, Prinetti A. . Gangliosides as regulators of cell signaling: ganglioside-protein interactions or ganglioside-driven membrane organization?. J Neurochem 2013; 124:432 - 5; http://dx.doi.org/ 10.1111/jnc.12088; PMID: 23351079 [DOI] [PubMed] [Google Scholar]
- 40.Hicks DA, Nalivaeva NN, Turner AJ. . Lipid rafts and Alzheimer’s disease: protein-lipid interactions and perturbation of signaling. Front Physiol 2012; 3:189; http://dx.doi.org/ 10.3389/fphys.2012.00189; PMID: 22737128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciarlo L, Manganelli V, Matarrese P, Garofalo T, Tinari A, Gambardella L, Marconi M, Grasso M, Misasi R, Sorice M, et al. . Raft-like microdomains play a key role in mitochondrial impairment in lymphoid cells from patients with Huntington’s disease. J Lipid Res 2012; 53:2057 - 68; http://dx.doi.org/ 10.1194/jlr.M026062; PMID: 22773688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vergne I, Deretic V. . The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett 2010; 584:1313 - 8; http://dx.doi.org/ 10.1016/j.febslet.2010.02.054; PMID: 20188094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dall’Armi C, Devereaux KA, Di Paolo G. . The role of lipids in the control of autophagy. Curr Biol 2013; 23:R33 - 45; http://dx.doi.org/ 10.1016/j.cub.2012.10.041; PMID: 23305670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakaki K, Wu J, Kaufman RJ. . Protein kinase Ctheta is required for autophagy in response to stress in the endoplasmic reticulum. J Biol Chem 2008; 283:15370 - 80; http://dx.doi.org/ 10.1074/jbc.M710209200; PMID: 18356160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. . Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389 - 93; http://dx.doi.org/ 10.1038/nature11910; PMID: 23455425 [DOI] [PubMed] [Google Scholar]
- 46.Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, et al. . Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta 2006; 1758:1864 - 84; http://dx.doi.org/ 10.1016/j.bbamem.2006.08.009; PMID: 17052686 [DOI] [PubMed] [Google Scholar]
- 47.D’Eletto M, Farrace MG, Falasca L, Reali V, Oliverio S, Melino G, Griffin M, Fimia GM, Piacentini M. . Transglutaminase 2 is involved in autophagosome maturation. Autophagy 2009; 5:1145 - 54; http://dx.doi.org/ 10.4161/auto.5.8.10040; PMID: 19955852 [DOI] [PubMed] [Google Scholar]
- 48.Mizushima N, Yoshimori T, Levine B. . Methods in mammalian autophagy research. Cell 2010; 140:313 - 26; http://dx.doi.org/ 10.1016/j.cell.2010.01.028; PMID: 20144757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matarrese P, Manganelli V, Garofalo T, Tinari A, Gambardella L, Ndebele K, Khosravi-Far R, Sorice M, Esposti MD, Malorni W. . Endosomal compartment contributes to the propagation of CD95/Fas-mediated signals in type II cells. Biochem J 2008; 413:467 - 78; http://dx.doi.org/ 10.1042/BJ20071704; PMID: 18442358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandra F, Degli Esposti M, Ndebele K, Gona P, Knight D, Rosenquist M, Khosravi-Far R. . Tumor necrosis factor-related apoptosis-inducing ligand alters mitochondrial membrane lipids. Cancer Res 2005; 65:8286 - 97; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-1913; PMID: 16166305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galandrini R, Tassi I, Mattia G, Lenti L, Piccoli M, Frati L, Santoni A. . SH2-containing inositol phosphatase (SHIP-1) transiently translocates to raft domains and modulates CD16-mediated cytotoxicity in human NK cells. Blood 2002; 100:4581 - 9; http://dx.doi.org/ 10.1182/blood-2002-04-1058; PMID: 12393695 [DOI] [PubMed] [Google Scholar]
- 52.Stryer L. . Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem 1978; 47:819 - 46; http://dx.doi.org/ 10.1146/annurev.bi.47.070178.004131; PMID: 354506 [DOI] [PubMed] [Google Scholar]
- 53.Riemann D, Tcherkes A, Hansen GH, Wulfaenger J, Blosz T, Danielsen EM. . Functional co-localization of monocytic aminopeptidase N/CD13 with the Fc gamma receptors CD32 and CD64. Biochem Biophys Res Commun 2005; 331:1408 - 12; http://dx.doi.org/ 10.1016/j.bbrc.2005.04.061; PMID: 15883031 [DOI] [PubMed] [Google Scholar]
- 54.Svennerholm L, Fredman P. . A procedure for the quantitative isolation of brain gangliosides. Biochim Biophys Acta 1980; 617:97 - 109; http://dx.doi.org/ 10.1016/0005-2760(80)90227-1; PMID: 7353026 [DOI] [PubMed] [Google Scholar]
- 55.Williams MA, McCluer RH. . The use of Sep-Pak C18 cartridges during the isolation of gangliosides. J Neurochem 1980; 35:266 - 9; http://dx.doi.org/ 10.1111/j.1471-4159.1980.tb12515.x; PMID: 7452256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.