Abstract
Metabolic stress induces autophagy as an alternative source of energy and metabolites. Insufficient autophagy in nutrient-deprived cancer cells would be beneficial for cancer therapy. Here, we performed a functional screen in search of novel autophagy regulators from natural products. We showed that oblongifolin C (OC), a natural small molecule compound extracted from Garcinia yunnanensis Hu, is a potent autophagic flux inhibitor. Exposure to OC results in an increased number of autophagosomes and impaired degradation of SQSTM1/p62. Costaining of GFP-LC3B with LysoTracker Red or LAMP1 antibody demonstrates that autophagosome-lysosome fusion is blocked by OC treatment. Furthermore, OC inhibits lysosomal proteolytic activity by altering lysosomal acidification and downregulating the expression of lysosomal cathepsins. Importantly, OC can eliminate the tolerance of cancer cells to nutrient starvation. Starvation dramatically increases the susceptibility of cancer cells to OC-induced CASP3-dependent apoptosis in vitro. Subsequent studies in xenograft mouse model showed that OC has anticancer potency as revealed by increased staining of cleaved CASP3, LC3 puncta, and SQSTM1, as well as reduced expression of lysosomal cathepsins. Combined treatment with OC and caloric restriction potentiates anticancer efficacy of OC in vivo. Collectively, these data demonstrated that OC is a novel autophagic flux inhibitor and might be useful in anticancer therapy.
Keywords: oblongifolin C, autophagy, lysosome, apoptosis, cancer, natural product
Introduction
Macroautophagy (hereafter referred to autophagy for simplicity) is an evolutionarily conserved membrane process that results in the transporting of cellular contents to lysosomes for degradation.1 Autophagy involves the formation of double-membrane vesicles, known as autophagosomes, which engulf intracellular contents such as mitochondria, endoplasmic reticulum, and ribosomes and fuse with lysosomes for degradation.2 Autophagic degradation is an important regulator of cellular homeostasis as this process mediates the turnover of defective organelles, misfolded or aggregated proteins, and certain long-lived molecules.3 However, the role of autophagy extends beyond the general homeostatic removal, degradation, and recycling of damaged proteins and organelles to many specific physiological and pathological processes such as tumorigenesis and cell death.4
The involvement of autophagy within the context of cancer is a double-edged sword. On the one hand, autophagy functions as a tumor suppression mechanism by removing damaged organelles and proteins and preventing genomic instability that drives tumorigenesis. On the other hand, autophagy has been shown to promote the survival of tumor cells within the tumor microenvironment.5-8 For instance, several studies used genetic knockout mice to establish a link between autophagy and cancer development. Loss of BECN1/Beclin 1 or SH3GLB1/BIF-1 resulted in tumor susceptibility in mice,9,10 while ectopic expression of BECN1 or UVRAG could repress the growth of human cancer cell xenografts.11,12 Given these dual effects, it is not surprising that autophagy has implications in both genesis and treatment of malignant disease.5 Therefore, therapeutic modulation of autophagy may serve as an important and challenging endeavor for cancer treatment.13,14
Autophagy is mainly regulated via a group of highly conserved autophagy-related (ATG) proteins, many of which were first identified from yeast, with most of them having orthologs in mammalian cells.15 MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3), a homolog of the yeast protein Atg8, serves as a marker protein for the autophagosome. Changes in autophagy in live cells can be monitored by using a GFP-fused LC3 (GFP-LC3) protein. The number of GFP-LC3 puncta is very low under normal conditions, but rapidly increases when autophagy is activated by rapamycin or stress.16 However, the increase of the GFP-LC3 level is not necessarily dependent on autophagy induction; it may be the result of a lysosomal defect and associated with the inhibition of autophagy. To confirm the function of chemicals as either inducers or inhibitors of autophagy, additional assay criteria such as monitoring autophagic flux are required.17,18 The polyubiquitin binding protein SQSTM1/p62 (sequestosome 1) is selectively incorporated into phagophores, the precursor to autophagosomes, through direct binding to LC3 and is efficiently degraded by autophagy; thus, the total cellular expression levels of SQSTM1 correlate with autophagic activity.18-20 Therefore, by using several different concurrent methods to accurately assess the status and function of autophagic activity in any given biological setting, more specific agents will be developed to modulate autophagy and subsequently for use in anticancer therapy. In addition, novel autophagy regulators may also help to unravel the complex mechanisms in autophagy signaling pathways.
Compounds from natural plants or microbes are important resources for drugs against a wide variety of diseases such as cancer, malaria, and infectious diseases. Many traditional Chinese medicines containing toxic compounds from plants exhibit antitumor effects and have been used for the different stages of cancer therapy.21 Garcinia species have been studied for more than 70 years and many bioactive compounds were identified with anticancer potentials. Xanthones, polycyclic polyprenylated acylphloroglucinols (PPAPs), and benzophenones are the main chemicals from Garcinia plants.22 Gambogic acid, a caged xanthone from Garcinia hanburyi, has been tested in vitro and in vivo as a novel anticancer agent that inhibits cell proliferation, angiogenesis, and metastasis.23-25 The PPAPs have been used in antiseptics, antidepressants, and antibiotics for centuries and their mimics also show fascinating biological activities.26,27 For instance, oblongifolin C (OC), a PPAP purified from G. yunnanensis Hu, can activate a mitochondrial apoptotic pathway in human cervical cancer cells.28 Guttiferone K (GK), a PPAP extracted from G. cowa, inhibits colon cancer cell growth through CDKN1A-mediated cell cycle arrest and finally stimulates CASP3-dependent apoptosis.29 These studies all focused on apoptosis because this process is the major mechanism of cancer cell killing. To explore the diverse activities of natural compounds, it will be interesting to use multiple screening platforms to investigate their functions and detailed mechanisms.
Here, we screened novel autophagic regulators from natural compounds extracted from Garcinia species by use of human cervical carcinoma HeLa cells stably expressing GFP-LC3. We report that OC is a novel autophagic flux inhibitor by blocking autophagosome-lysosome fusion and autophagic degradation. OC also inhibits lysosomal activity by altering lysosomal acidification and downregulating the expression of lysosomal cathepsins. Notably, OC efficiently sensitizes nutrient-deprived cancer cells to apoptosis in vitro. The anticancer activity of OC was also observed in a cervical cancer xenograft mouse model. OC-treated nude mice exhibited increased staining of cleaved CASP3, LC3 puncta, SQSTM1, and reduced expression of lysosomal cathepsins. Our results suggest that screening for novel autophagy modulators from plants may be an efficient approach for the identification of lead compounds for anticancer drug discovery.
Results
A functional screen to identify novel autophagy regulators from natural products
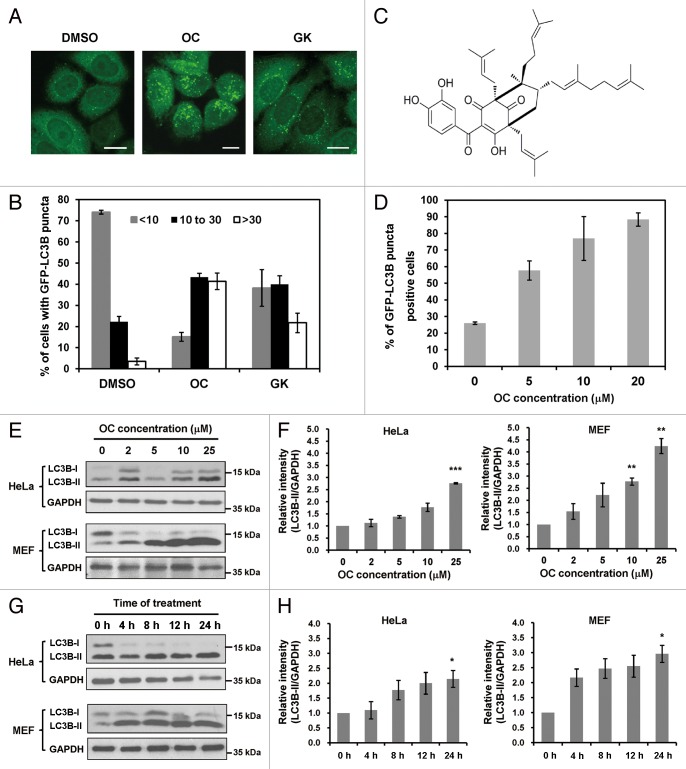
To identify novel autophagy regulators, we performed a functional screen using a cell-based assay. GFP-LC3B, a fluorescent autophagosomal marker, was stably expressed in HeLa cells. Autophagosome accumulation can be detected with a fluorescence microscope. We started the screen with polycyclic polyprenylated acylphoroglucinols (PPAPs) and xanthones extracted from Garcinia species.30-32 Interestingly, several compounds could induce GFP-LC3B puncta accumulation in HeLa cells (For intellectual property protection reasons, the names and structures of these compounds are not shown; Fig. S1). Among all the tested compounds, 2 PPAPs, OC and GK, exhibited preferential activity to induce a GFP-LC3B puncta increase (Fig. 1A). By quantifying the number of GFP-LC3B puncta in the cells, we found that OC is more active than GK to modulate autophagy, and the effect of OC was dose dependent in HeLa cells (Fig. 1B and D). The formula of OC is shown in Figure 1C. In addition, we tested the effect of OC on several human cancer cell lines, including HepG2 (a human liver carcinoma cell line), CNE (a human nasopharyngeal cancer cell line), HCT116 (a human colon carcinoma cell line), MCF7 (a human breast carcinoma cell line), as well as mouse embryonic fibroblasts (MEFs). Consistently, OC treatment resulted in a dramatic increase of GFP-LC3B puncta in these cell lines (Fig. S2A). Therefore, we chose OC for further study to elucidate its mechanisms of action in modulating autophagy.
Figure 1. Screen for novel autophagy regulators from natural compounds extracted from Garcinia species. (A) Accumulation of GFP-LC3B puncta in HeLa cells treated with OC (10 μM), GK (10 μM) or DMSO for 24 h. The distribution of GFP-LC3B was examined by confocal microscopy. Scale bar: 10 μm. (B) Percentage of cells with GFP-LC3B puncta was quantified by analyzing the number of GFP-LC3B dots in the cells. Data are shown as the mean ± SD of 3 independent experiments. (C) The molecular structure of oblongifolin C (OC). (D) Quantification of GFP-LC3B puncta-positive cells at different drug concentrations (5, 10, 20 μM). Each sample was analyzed using a threshold of >10 dots/cell. Data are shown as the mean ± SD of 3 independent experiments. (E) OC increases the amount of LC3B-II protein in a dose-dependent manner. HeLa or MEF cells treated with different concentrations of OC (2, 5, 10, 25 μM) for 24 h were analyzed by western blotting for endogenous LC3B. GAPDH was used as a loading control. (F) ImageJ densitometric analysis of the LC3B-II/GAPDH ratio from LC3B immunoblots (mean ± SD of 3 independent experiments). * indicates a significant difference from the controls. **P < 0.01; ***P < 0.001; Student t test. (G) OC increases the amount of LC3B-II in a time-dependent manner. HeLa or MEF cells were treated with OC at 15 μM concentration over a certain time course; samples were analyzed by western blotting for endogenous LC3B and GAPDH. (H) ImageJ densitometric analysis of the LC3B-II/GAPDH ratio from LC3B immunoblots (mean ± SD of 3 independent experiments). * indicates a significant difference from the controls. *P < 0.05; Student t test.
During autophagy, the cytoplasmic form LC3B-I (18 kD) is processed and recruited to phagophores, where LC3B-II (16 kD) is generated by site-specific proteolysis and lipidation at the C-terminus. Thus the amount of LC3B-II positively correlates with the number of autophagosomes. This characteristic conversion from endogenous LC3B-I to LC3B-II can be used to monitor autophagic activity. We examined the effect of OC on LC3B conversion in both HeLa and MEF cells. Immunoblot analysis showed that OC treatment resulted in dose- and time-dependent accumulation of LC3B-II in both cell lines (Fig. 1E–H). Similarly, the effect of OC on LC3B conversion was confirmed in MDA-MB-231, MCF-7, HCT116, HepG2, and CNE cells (Fig. S2B).
OC inhibits autophagic flux
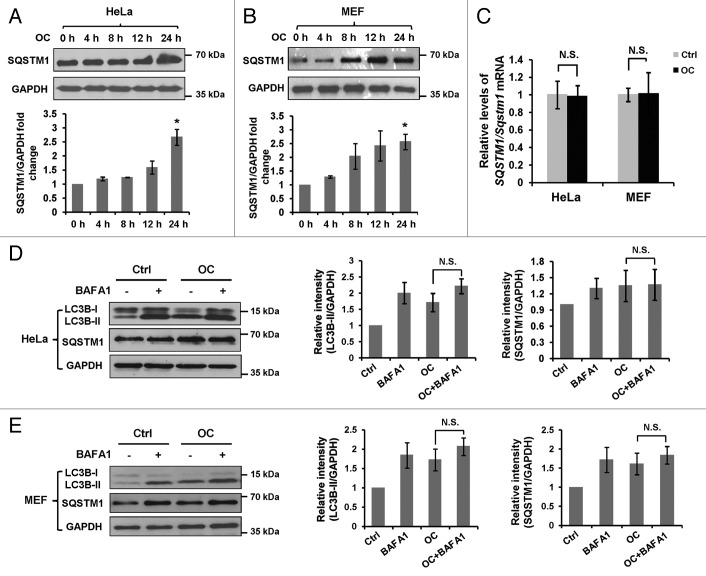
Both induction and suppression of autolysosomal maturation result in increased numbers of autophagosomes. To distinguish whether autophagosome accumulation is due to autophagy induction or rather a block in downstream steps, we performed an autophagic flux assay. SQSTM1 serves as a link between LC3 and ubiquitinated substrates. Inhibition of autophagy correlates with increased levels of SQSTM1 in mammals and Drosophila, suggesting that steady-state levels of this protein reflect the autophagic status.33 We then examined the total cellular amount of SQSTM1 that was delivered to the lysosomes for degradation. Immunoblot analysis showed that a remarkable increase of SQSTM1 was detected at 24 h after OC treatment (Fig. 2A and B), which reflects an inhibition of autophagic degradation. Consistently, increased expression of SQSTM1 protein was observed in MDA-MB-231, MCF-7, HCT116, HepG2 and CNE cells treated with OC (Fig. S2B). To determine whether OC-induced SQSTM1 protein accumulation is due to transcriptional activation, we performed RT-PCR to access SQSTM1 mRNA levels in the presence or absence of OC. As shown in Figure 2C, OC treatment did not alter SQSTM1/Sqstm1 mRNA levels in both HeLa and MEF cells.
Figure 2. OC inhibits autophagic flux. (A) HeLa or (B) MEF cells were treated with OC (15 μM) over a certain time period (4, 8, 12 and 24 h), samples were analyzed by western blotting for endogenous SQSTM1 and GAPDH. ImageJ densitometric analysis of the SQSTM1/GAPDH ratio from immunoblots were shown (mean ± SD of 3 independent experiments, Student t test, *P < 0.05). (C) HeLa or MEF cells were treated with OC (15 μM) for 24 h. Relative SQSTM1/Sqstm1 mRNA levels (compared with GAPDH/Gapdh) was analyzed by quantitative RT-PCR. N.S., not significant. (D to E) HeLa or MEF cells were treated with DMSO or OC (15 μM) for 2 h in the presence or absence of 10 nM BAFA1 as indicated. Western blotting was performed to analyze the status of LC3B, SQSTM1 and GAPDH. ImageJ densitometric analysis of the LC3B-II/GAPDH and SQSTM1/GAPDH ratios from immunoblots is shown (mean ± SD of 3 independent experiments). N.S., not significant, Student t test.
The effect of OC on autophagic flux was evaluated by treating cells with an autophagic inhibitor. Bafilomycin A1 (BAFA1), a vacuolar-type H+-ATPase (V-ATPase) inhibitor, significantly increased the amount of LC3B-II and SQSTM1 in both HeLa and MEF cells. OC-induced accumulation of LC3B-II and SQSTM1 was not significantly enhanced in the presence of BAFA1 (Fig. 2D and E), indicating that OC inhibits degradation of the autophagic contents. We also investigated the effect of OC on starvation-induced autophagy. OC treatment resulted in a significant increase of LC3B-II upon nutrient starvation, which was similar to the result caused by BAFA1, suggesting that OC has the same effect as BAFA1 to inhibit starvation-induced autophagic flux (Fig. S3). Together, these observations indicate that OC is a potent autophagic flux inhibitor; OC-induced autophagosome accumulation is due to impaired autophagic degradation rather than promoting autophagic flux.
OC blocks autophagosome-lysosome fusion
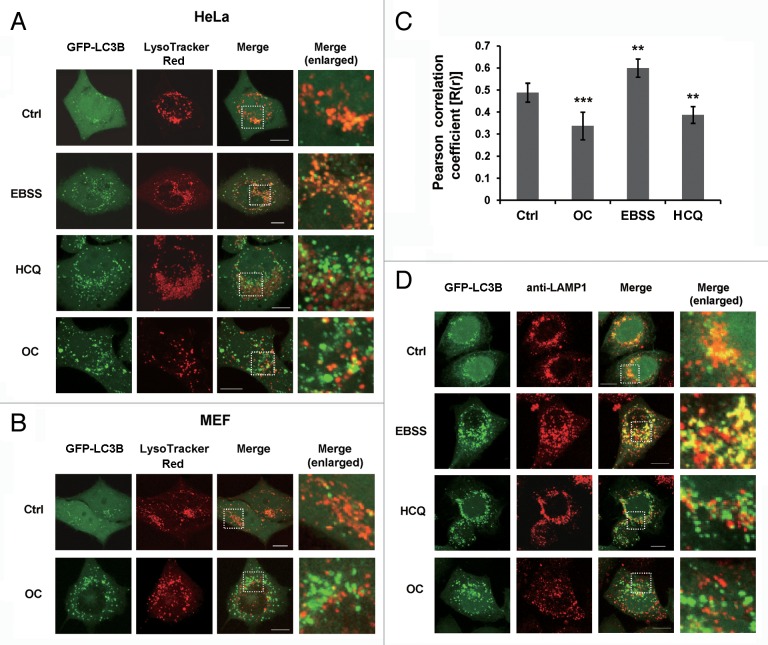
The final step of autophagy is the fusion of autophagosomes with lysosomes; inhibition of this process impairs autophagic degradation. To address whether OC affect autophagosome-lysosome fusion, we examined the colocalization of GFP-LC3B and LysoTracker Red, a specific dye for live cell lysosome labeling. As a positive control, HeLa cells were cultured in EBSS medium to induce nutrient starvation. As shown in Figure 3A, starvation induced a remarkable increase of GFP-LC3B puncta, which were well colocalized with LysoTracker Red, suggesting that autolysosome formation process normally during starvation upon autophagy activation. Hydroxychloroquine (HCQ), a lysosomotropic agent, can prevent endosomal acidification and block autophagosome-lysosome fusion. Notably, we found that OC-treated cells exhibited a significant separation of GFP-LC3B and LysoTracker Red staining (Fig. 3A and B). We used Pearson correlation coefficient (PCC) as a statistic to quantify the extent of colocalization of these 2 signals. PCC values near zero reflect distributions of probes that are uncorrelated with one another. Consistent with confocal microscopy observations, the PCC value of OC or HCQ treated cells was significantly lower than starvation treatment, indicating that both OC and HCQ impair the colocalization of GFP-LC3B and LysoTracker Red (Fig. 3C). The intensity of LysoTracker dye can be changed by pH alteration; therefore we performed immunostaining with anti-LAMP1 antibody, a marker for endosomal and lysosomal membranes. As illustrated in Figure 3D, GFP-LC3B was well colocalized with LAMP1 during starvation, while colocalization was abolished by OC or HCQ treatment.
Figure 3. OC blocks autophagosome-lysosome fusion. (A) HeLa cells stably expressing GFP-LC3B were treated with OC (15 μM), HCQ (50 μM) for 8 h, or cultured in EBSS medium for 2 h. Live cell images of GFP-LC3B and LysoTracker Red were taken using Olympus confocal microscope. Enlarged images are cropped sections from the merge panels (white dash borders) which showing the colocalization of 2 signals. Scale bar: 10 μm. (B) MEFs were transiently transfected with GFP-LC3B and cultured in complete medium with or without OC (15 μM) for 8 h. The colocalization of GFP-LC3B and LysoTracker Red was analyzed by confocal microscopy. Enlarged images show the colocalization of the 2 signals. Scale bar: 10 μm. (C) Quantification of Pearson correlation coefficient as a statistic for quantifying colocalization. More than 30 cells were counted in each condition and data (mean ± SD) are representative of 2 independent experiments (Student t test, **P < 0.01, ***P < 0.001). (D) Immunofluorescence of HeLa GFP-LC3B cells with anti-LAMP1 antibody. Cells were treated with OC (15 μM) or HCQ (50 μM) for 8 h, or cultured in EBSS solution for 2 h. Fusion between autophagosomes (GFP-LC3B) and lysosomes (anti-LAMP1) was evident in EBSS-treated cells (yellow in merged images). A complete separation of green and red signals was observed in HCQ- and OC-treated cells. Scale bar: 10 μm.
OC inhibits lysosomal proteolytic activity
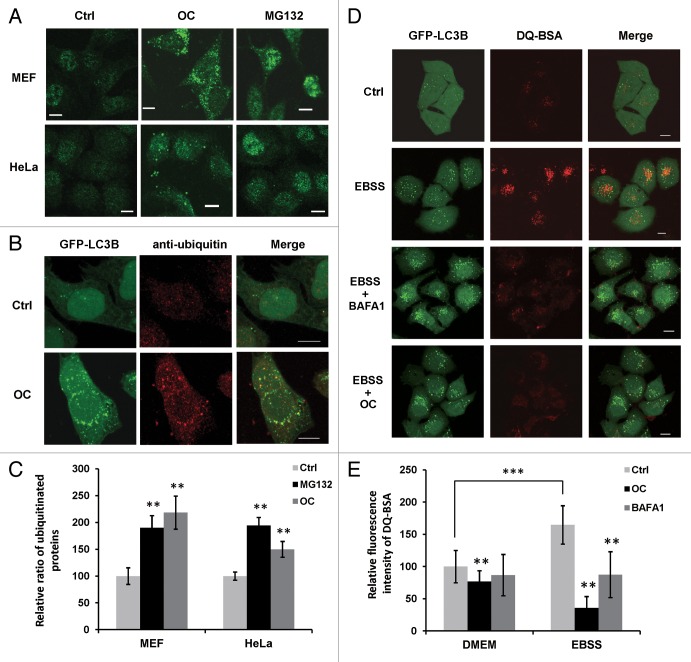
Recent studies reported that lysosomal dysfunction causes increases in ubiquitinated proteins; for example, CQ treatment results in an increased level of ubiquitinated proteins.34,35 Thus, we employed fluorescence microscopy to examine the accumulation of ubiquitinated proteins in OC-treated cells. MG132, a proteasome inhibitor, was used as a positive control. Compared with untreated cells, both OC and MG132 treatment resulted in increased levels of endogenous ubiquitin-staining (Fig. 4A). In addition, confocal images revealed that some GFP-LC3B puncta colocalized with ubiquitinated proteins under OC treatment (Fig. 4B). We quantified the level of ubiquitinated proteins and found that both OC and MG132 significantly induced the accumulation of ubiquitinated proteins in HeLa and MEF cells (Fig. 4C).
Figure 4. OC suppresses lysosomal degradation and lysosomal activity. (A) Immunofluorescence of ubiquitin in HeLa and MEF cells treated with OC (15 μM) for 24 h or MG132 (10 μM) for 8 h. The fluorescence intensity of endogenous ubiquitinated protein in OC- or MG132-treated cells was significantly higher than control. Scale bar: 10 μm. (B) Colocalization of ubiquitin and GFP-LC3B. HeLa cells stable expressing GFP-LC3B were treated with or without OC (15 μM) for 24 h. Cells were fixed and stained with anti-ubiquitin antibody. The images were acquired using a confocal microscope. Scale bar: 10 μm. (C) Quantification of ubiquitinated protein. HeLa or MEF cells treated with control, OC or MG132 were fixed and stained with anti-ubiquitin antibody. Fluorescent images were acquired and the intensity was analyzed with ImageJ software. More than 100 cells were counted in each condition and data are presented as means ± SD from 2 independent experiments, and analyzed using the Student t test (**P < 0.01). (D) HeLa cells stably expressing GFP-LC3B were incubated with DQ-BSA (10 μg/ml) for 12 h, the cells were then washed with PBS before being treated with 15 μM OC or 5 nM BAFA1 for 8 h. Alternatively, cells were starved in EBSS solution for 2 h with or without OC or BAFA1 treatment. The cells were fixed and analyzed for confocal microscopy. Data shown are representative images of control, EBSS-, EBSS/BAF1-, or EBSS/OC-treated samples. Scale bar: 10 μm. (E) The fluorescence intensity of DQ-BSA was quantified with ImageJ software. More than 100 cells were counted in each condition and data are presented as means ± SD from 2 independent experiments, and analyzed using the Student t test (**P < 0.01, ***P < 0.001).
To monitor lysosomal activity during OC treatment, cells were assayed for their ability to process DQ-BSA. This BSA derivative is so heavily labeled that the red fluorophore is self-quenched. Proteolysis of this compound results in dequenching and release of brightly fluorescent fragments. Thus, the use of DQ-BSA is useful for the visualization of intracellular proteolytic activity.36 As a positive control, cells were incubated in starvation medium (EBSS) for 2 h to stimulate lysosomal activity. As shown in Figure 4D, dequenching of DQ-BSA occurred in cells under nutrient starvation, and the red fluorescence was partially colocalized with GFP-LC3B, indicating that starvation stimulated lysosomal activity in autophagic compartments. In contrast, in BAFA1 or OC-treated cells, nearly no dequenching of DQ-BSA was observed. The basal and starvation-induced lysosomal activity was quantified by measuring the red fluorescence intensity of DQ-BSA. Compared with the control, both OC and BAFA1 could reduce basal level and starvation-induced proteolytic activity (Fig. 4E).
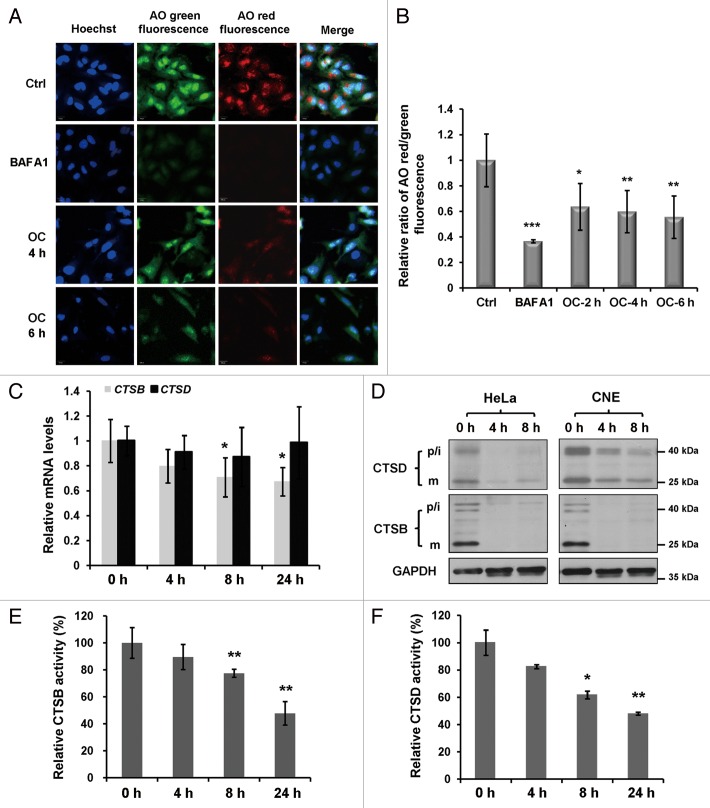
OC raises the pH in acidic compartments
Lysosomal enzymes function optimally over a narrow range of acidic pH values, next we used the acridine orange (AO) to evaluate the lysosomal pH.37 AO is a nucleic acid dye that accumulates in acidic spaces, such as lysosomes. Under low pH conditions, the dye emits red light when excited by blue light. As shown in Figure 5A, unlike the control, the red fluorescence was greatly reduced in both BAFA1- and OC-treated cells. To eliminate the possibility that the reduction of the red fluorescent signal was due to the decrease of AO loading, we calculated the ratio of fluorescent intensity from red and green channels in the cytosolic region. BAFA1 is a selective inhibitor of the V-ATPase, which can raise the pH of acidic compartments. Our measurements indicate that OC has a similar but moderate effect to regulate lysosomal acidification (Fig. 5B).
Figure 5. OC raises lysosomal pH and downregulates lysosomal cathepsins. (A) Acridine orange (AO) staining in control, OC- (15 μM) or BAFA1 (10 nM)-treated HeLa cells. Green fluorescence was acquired at Ex 488 nm, Em 520 to ~560 nm; red fluorescence was acquired with Ex 488 nm, Em 620 nm long pass in Zeiss confocal microscope. (B) Quantification of red/green intensity ratio of (A). The ratio of red and green in the cytosolic region was calculated by Columbus software. The Hoechst staining area (nuclear region) was exclusive (Student t test, *P < 0.05, **P < 0.01; ***P < 0.001). (C) Relative CTSB and CTSD mRNA levels (compared with GAPDH) were analyzed by quantitative RT-PCR. HeLa cells were treated with control or OC (15 μM) for 4 h, 8 h and 24 h, as indicated. Data are presented as means ± SD from 2 independent experiments, and analyzed using the Student t test (*P < 0.05). (D) Western blotting analysis for the processing of endogenous CTSB and CTSD. p/i, precursor/intermediate; m, mature form of CTSB and CTSD. (E and F) Enzymatic activity of CTSB and CTSD in OC-treated HeLa cells. Cells were treated with DMSO or OC (15 μM) at 4 h, 8 h and 24 h as indicated. Enzymatic activity was analyzed by fluorogenic kits. Data are presented as means ± SD from 3 independent experiments and analyzed using the Student t test (*P < 0.05, **P < 0.01).
OC downregulates lysosomal cathepsins
Autophagy is a process involved in the proteolytic degradation of cellular macromolecules in lysosomes, which requires the activity of proteases.38 As the major lysosomal proteases, cathepsins play important roles to maintain cellular homeostasis and differentiation by recycling cellular contents. We next investigated whether OC treatment affects the expression and activation of lysosomal cathepsins. First, we performed an RT-PCR assay to examine mRNA levels of 2 main cathepsins, CTSB and CTSD. OC treatment resulted in a significant reduction of CTSB mRNA expression while CTSD mRNA levels remained the same (Fig. 5C). Cathepsins are synthesized as inactive membrane-associated precursors. The precursors are further cleaved to generate an active form within endosomes or lysosomes.39 Then we evaluated whether OC affects protein expression and the maturation process of CTSB and CTSD. As shown in Figure 5D, OC dramatically downregulated CTSB and CTSD protein levels in HeLa and CNE cells, including both precursors and mature forms. Finally we performed fluorogenic substrate assays to measure the enzymatic activity of CTSB and CTSD. Both CTSB and CTSD activities were reduced in a time-dependent manner upon OC treatment (Fig. 5E and F). Taken together, these results suggest that OC inhibits lysosomal proteolytic activity by altering lysosomal pH and downregulating lysosomal cathepsins.
OC eliminates the tolerance of cancer cells to nutrient starvation
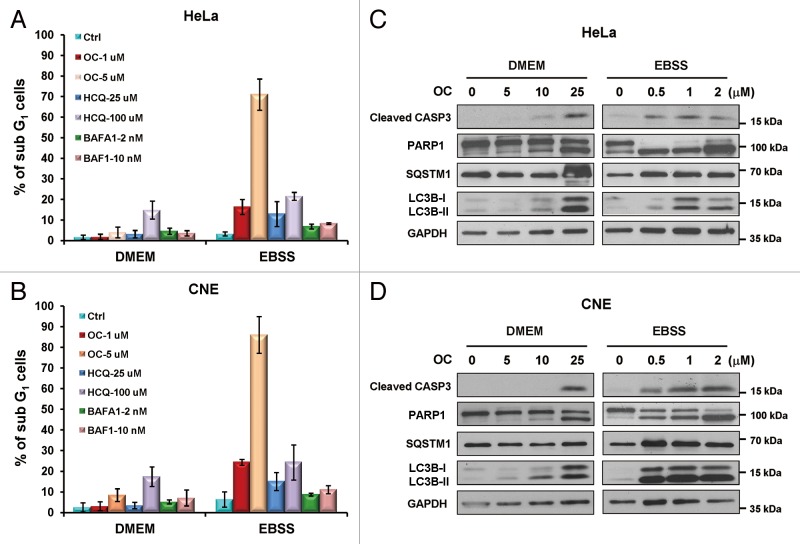
Cancer cells have the ability to tolerate extreme conditions, such as insufficient nutrient/oxygen supply, by modulating their energy metabolism. Metabolic stress induces autophagy as an alternative source of energy and metabolites. Insufficient autophagy in nutrient-deprived cancer cells would be beneficial for tumor therapy. To investigate whether OC-mediated autophagy inhibition and lysosomal dysfunction affect cancer cell survival to nutrient starvation, we performed flow cytometry to quantify the sub-G1 population. Treatment with OC alone (1 μM or 5 μM) in nutrient-rich medium (DMEM) was not toxic to HeLa or CNE cells, whereas in nutrient-deprived medium (EBSS), nearly 20% of the sub-G1 cells were detected from low concentration (1 μM) OC-treated cells, and more than 75% of the cells were killed by a high concentration of OC (5 μM). For comparison, we evaluated the proapoptotic effect of other known autophagy inhibitors, such as HCQ and BAFA1. HCQ at high concentration (100 μM) was able to kill cancer cells even in the absence of nutrient starvation. At lower concentration (25 μM), it decreased the viability of nutrient-deprived cancer cells; the percentage of sub-G1 cells was nearly 15% in HeLa and CNE cells, which was less than seen with the low concentration of OC. BAFA1, a proton pump blocker that potently inhibits lysosomal acidification and subsequent proteolytic digestion, however, caused no significant cytotoxicity at both low (2 nM) and high concentration (10 nM) in either complete medium or nutrient-deprived conditions (Fig. 6A and B). We also tested the susceptibility of noncancer cell lines such as MEFs to OC and starvation treatment. Low concentration OC (1 μM) did not cause an obvious level of cell death upon nutrient starvation, whereas 5 μM OC exhibited the same cytotoxic effect as seen with cancer cells (Fig. S4D).
Figure 6. OC eliminates the tolerance of cancer cells to nutrient starvation. (A) Flow cytometry analysis of the sub-G1 population in OC-treated cells. HeLa or (B) CNE cells were treated with OC (1 μM or 5 μM), HCQ (25 μM or 100 μM) or BAFA1 (2 nM or 5 nM) and cultured in either complete medium (DMEM with serum) or nutrient-deprived medium (EBSS without serum) for 24 h. The cells were fixed and stained with propidium iodide. (C) HeLa or (D) CNE cells cultured in DMEM or EBSS medium were treated with a certain amount of OC for 24 h. Samples were analyzed by western blotting for cleaved CASP3, PARP1, LC3B, and SQSTM1. GAPDH was used as a loading control.
We then examined the OC-induced apoptotic cell death pathway by immunoblot analysis. OC treatment resulted in CASP3 activation, PARP1 cleavage, accumulation of SQSTM1 and LC3B-II in a concentration-dependent manner in both complete and nutrient-deprived medium, indicating that OC-mediated apoptosis is associated with autophagy inhibition (Fig. 6C and D). Starvation dramatically increased the susceptibility of cancer cells to OC-induced apoptosis. Low concentration OC (0.5 μM) could efficiently promote CASP3 activation and PARP1 cleavage under nutrient-starved conditions. However, when cells were cultured in nutrient-rich medium, even 10 μM OC was not able to efficiently activate CASP3 (Fig. 6C and D).
Next, we determined whether silencing the expression of autophagy essential genes or lysosomal cathepsins can phenocopy the proapoptotic effect of OC in response to nutrient deprivation. Flow cytometry analysis showed that ATG5 or ATG7 siRNA did not affect the viability of HeLa cells under starvation treatment. However, depletion of ATG5 or ATG7 resulted in a significant increase of the sub-G1 population (12% and 15%, respectively) in starved CNE cells (Fig. S4A and S4B). Immunoblot results also confirmed that ATG5 or ATG7 siRNA could promote PARP1 cleavage in nutrient-deprived CNE cells (Fig. S4C). In addition, we found that both HeLa and CNE cells transfected with CTSB or CTSD siRNA were not more sensitive to starvation-induced apoptosis than control cells (Fig. S5A and S5B). Collectively, these data indicate that OC can eliminate the tolerance of cancer cells to nutrient starvation; the rapid killing of nutrient-deprived cancer cells by OC is not mediated solely by autophagic flux inhibition and lysosomal dysfunction.
Caloric restriction enhances the anticancer activity of OC in vivo
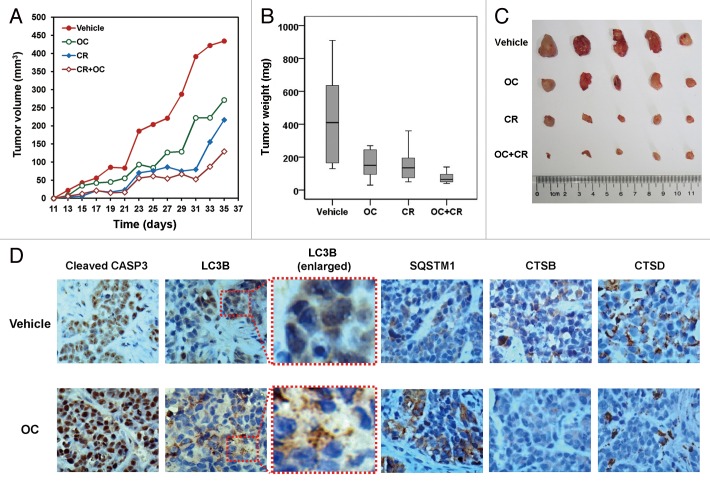
To determine the anticancer activity of OC in vivo, nude mice were injected with human cervical cancer cells and then administrated OC, or vehicle control. Tumor volume and mass increased dramatically in the control group, whereas tumor growth was significantly less prominent in OC-treated mice (Fig. 7A–C). Caloric restriction is the most physiological inducer of autophagy.40,41 Therefore, we investigated the therapeutic potential of combined treatment with OC and caloric restriction in a xenograft mouse model. As reported previously, caloric restriction significantly reduced tumor size and weight.42,43 Interestingly, in animals exposed to a combination of OC and caloric restriction, tumor size, and tumor weight were significantly lower than other groups (Fig. 7A–C). The ability of OC to enhance the anticancer effect of nutrient deprivation is very similar to the autophagy inhibitor chloroquine (CQ), which can rapidly kill serum-starved cancer cells in vitro and inhibit melanoma growth in calorie-restricted mice.42
Figure 7. OC exhibits anticancer activity in cervical cancer xenograft. (A) Four-wk-old nude mice were engrafted with HeLa cells and randomly divided into 4 groups. Tumor-bearing mice were then treated with vehicle, OC (30 mg/kg), caloric restriction (CR, 70% food intake) and OC+caloric restriction (OC+CR, 30 mg/kg OC with 70% food intake) (n = 8) by i.p. once a day for a total of 5 wk. Tumor volumes were calculated by the length and width measured by vernier calipers every 2 d. (B) The weight of tumors from mice from (A). Vehicle (mean: 423.5 mg; median: 410.0 mg; range: 130.0 to 910.9 mg), OC (mean: 212.5 mg; median: 150.2 mg; range: 30.0 to 270.0 mg), CR (mean: 152.5 mg; median: 135.0 mg; range: 50.0 to 360.0 mg) and OC+CR (mean: 75.0 mg; median: 65.0 mg; range: 40.0 to 100.0 mg). Tumors were resected and weighed 1 d after the final injection. (C) Images for representative tumors from (B). (D) Immunohistochemical staining for LC3B, cleaved CASP3, SQSTM1, CSTB, and CSTD in tumor sections treated with vehicle or OC. Four-wk-old nude mice were engrafted with HeLa cells and observed until tumors reached ~100 mm3. Tumor-bearing mice were then treated with vehicle or OC (12 µg) by intratumoral once every 2 d for a total of 7 injections. Mice were killed and tumors were resected 2 d after the final injection. Panels are representative overview images taken at 400× magnification. Panels with dashed borders are cropped sections from the overview panels.
Finally, we performed immunohistochemistry to detect the levels of LC3B, cleaved CASP3, SQSTM1, CTSB, and CTSD. As shown in Figure 7D, LC3B staining was diffusely distributed in the cytoplasm, and no LC3B puncta staining was observed in control mice. However, staining of tumor tissue for LC3B from OC-treated mice revealed that puncta indicative of autophagosomes could clearly be detected. A large number of cells exhibited increased staining of cleaved CASP3, indicating that OC induced CASP3-dependent apoptosis in vivo. Moreover, a significant increase of SQSTM1 and decrease of CTSB/CTSD were observed in further sequential sections from the same tissue block in OC-treated mice (Fig. 7D). In summary, these results indicate that OC-mediated inhibition of tumor growth is associated with autophagy inhibition, lysosome dysfunction, and apoptosis.
Discussion
Cancer cells have the ability to tolerate extreme conditions, and autophagy-related stress tolerance enables cancer cells to survive by maintaining energy production that can lead to tumor growth and therapeutic resistance. As autophagy is clearly a survival pathway utilized by tumor cells to survive metabolic stress, inhibition of autophagy may be therapeutically useful.44-46 Recently, numerous preclinical studies have demonstrated that inhibition of autophagy restores chemosensitivity of a broad array of anticancer agents and enhances tumor cell death.47-50 CQ, HCQ, and lucanthone are the only clinically relevant autophagic inhibitors being used for cancer therapy.14 Therefore, novel inhibitors of autophagy with a lower toxicity and a better therapeutic index are needed.
Compounds from natural plants or microbes are important resources for drugs against a wide variety of diseases including cancer. Many traditional Chinese medicines containing toxic compounds from plants exhibit antitumor effects and have been used for treating different stages of cancer. In this report, we systematically investigated the inhibitory effect of OC on autophagy and its consequence on the crosstalk with apoptosis. Using several different approaches, we show that OC inhibits autophagic flux by blocking autophagosome-lysosome fusion and inhibiting the lysosomal proteolytic activity. Notably, we demonstrate that OC can efficiently sensitize nutrient-deprived cancer cells to apoptosis both in vitro and in vivo. Based on the above, our research indicates that screening novel autophagy modulators from natural products might be an efficient approach for the identification of novel autophagic flux inhibitors and lead compounds for cancer therapy.
To fully understand the possible mechanism of OC on autophagy inhibition, we examined the different actions of OC on autolysosome formation and lysosomal proteolytic activity. LysoTracker Red and GFP-LC3B costaining show that the fusion of autophagosomes and lysosomes is severely impaired by the action of OC (Fig. 3). It has been reported previously that the process of autophagosome-lysosome fusion depends on the pH in acidic compartments.37 Results from AO staining indicate that OC-mediated blockage of autophagosome-lysosome fusion might be due to a rise of lysosomal pH (Fig. 5A and B). However, recent studies suggest that lysosomal positioning proteins, TFEB (transcription factor EB) and RAB7 can affect autophagosome-lysosome fusion.51-53 Thus, other factors may also contribute to the inhibitory effect of OC on autophagosome-lysosome fusion. Autophagy is executed inside the lysosomal compartment, the efficiency of lysosomal degradation determines autophagic flux.38 DQ-BSA is a specific marker that can be used to localize lysosomal degradation activity. Our data report that OC inhibits both basal level and starvation-induced lysosomal proteolytic activity (Fig. 4D and E). CTSB and CTSD are the most abundant lysosomal proteases and participate directly in the execution of autophagy.54 Results from both RT-PCR and western blotting demonstrate that OC downregulates the expression of CTSB and CTSD. Moreover, in vitro cathepsin substrate assays show that OC dramatically reduces the activity of CTSB and CTSD, thus leading to the inhibition of lysosomal activity (Fig. 5C–F). These findings highlight the fact that OC exerts different actions as an autophagic flux inhibitor.
Apart from macroautophagy, both chaperone-mediated autophagy (CMA) and microautophagy also involve the proteolytic lysosomal pathway.38 These 3 types of autophagy act together in cells to deliver cytosolic components into lysosomes. It might be possible that OC inhibits lysosomal degradation activity via CMA or microautophagy. We performed DQ-BSA and in vitro cathepsin substrate assays in both wild-type and autophagy-deficient atg7−/− MEF cells. The results indicate that OC inhibits lysosomal activity to the same extent in wild-type and atg7−/− cells (Fig. S6). Thus, it remains possible that OC works through general inhibition of lysosomal function, like HCQ or BAFA1. While acting as an inhibitor of macroautophagy, OC would also likely affect CMA and microautophagy.
Apoptosis is the best described form of programmed cell death and involves the activation of a family of cysteine proteases. Based on the appearance of cleaved CASP3 from both culture conditions and tumor tissues from nude mice, we demonstrated that OC efficiently sensitizes nutrient-starved cancer cells to apoptosis both in vitro and in vivo. Similarly, a known lysosomotropic autophagy inhibitor, HCQ, exhibits a similar but moderate proapoptotic effect in serum-starved HeLa and CNE cells. Moreover, silencing the expression of ATG5 or ATG7 can promote apoptosis in nutrient-starved CNE cells (Fig. S4). These findings suggest that OC-mediated inhibition of autophagic flux may help to sensitize nutrient-deprived cancer cells to apoptosis in some cell types. However, apoptosis via autophagy inhibition and lysosomal dysfunction is not solely responsible for OC-induced apoptosis. Specific silencing of ATG genes does not significantly contribute to starvation-mediated apoptosis in HeLa cells. In addition, knocking down the expression of CTSB or CTSD does not sensitize HeLa and CNE cells to apoptosis upon nutrient starvation (Fig. S5). Notably, BAFA1, a specific inhibitor of the V-ATPase, which has been shown to pharmacologically inhibit autophagy and lysosomal functions, has a very weak effect on apoptosis in serum-starved HeLa and CNE cells. Furthermore, we observed that OC induced CASP3-dependent apoptosis in autophagy deficient atg7−/− MEFs (unpublished data). A recent study also reports that the autophagy inhibitor chloroquine (CQ) can sensitize breast cancer cells to chemotherapy independent of autophagy.55 Based on these facts, it could be that sensitization to apoptosis and inhibition of lysosomal proteolytic activity/autophagic flux are 2 different mechanisms of OC action.
CTSB and CTSD are of significant importance for cancer therapy as they are involved in various pathologies and oncogenic processes in cancer.56,57 In cancer patients, elevated CTSB activity correlates with poor therapy outcome. Therefore, it is necessary to develop CTSB inhibitors, and some of them (cyctatins, CA-074, etc.) are able to reduce cancer cell motility and invasiveness.58,59 Similarly, CTSD can act as a mitogen on both cancer and stromal cells to stimulate their proinvasive and prometastatic properties.57 CTSD inhibitors not only show promising effects for cancer therapy but also some other diseases such as hypertension and atherosclerosis. Our results suggest OC severely inhibits the expression and activation of CTSB and CTSD, which might be related to cancer cell killing. Further investigation is needed to elucidate the detailed mechanism of OC acting on lysosomal cathepsins.
Materials and Methods
Cell lines and cell culture
HeLa, HCT116, CNE, MCF7, MDA-MB-231, HepG2, and MEF cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco/Invitrogen, 12800-017) supplemented with 10% fetal bovine serum (PAA, A15-101), 10 U/ml penicillin-streptomycin (Gibco/Invitrogen, 15140-122) at 37 °C in a humidified 5% CO2 incubator. For nutrient starvation, HeLa or CNE cells cultured in DMEM were washed 3 times with PBS (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), then cultured in Earle’s Balanced Salt Solution (EBSS, Sigma, E6132) for the indicated time points.
GFP-LC3B translocation and quantitative analyses
MEF, HeLa, CNE, HCT116, MCF7, and HepG2 cells were transfected with pEGFP-LC3B plasmid using lipofectamine 2000 (Invitrogen, 11668-019). One day after transfection, cells were treated with 10 μM OC for 24 h prior to fixation. Image acquisition was done using an Olympus FV1000 (Olympus) confocal microscope. The number of GFP-LC3B dots was counted from at least 150 cells from randomly placed positions within each sample.
Live cell imaging and colocalization analysis
For LysoTracker Red staining, HeLa or MEF cells grown on coverslips were stained with 50 nM LysoTracker Red DND-99 (Molecular Probes/Invitrogen, L7528) in prewarmed medium for 20 min at 37 °C. All of the samples were examined under an Olympus FV1000 confocal microscope equipped with a 63 × oil immersion objective. The confocal images were acquired using FV10-ASW 2.0 software. Calculation of Pearson correlation coefficient was applied to quantify colocalization. PCC was calculated by FV10-ASW 2.0 software between the stack of images from 2 channels.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde in PBS for 15 min followed by permeabilization with 0.25% Triton X-100 in PBS for 10 min at room temperature. Fixed preparations were blocked with 3% BSA in PBS for 1 h, then incubated with primary antibodies against LAMP1 (Cell Signaling Technology, 9091) or ubiquitin (Cell Signaling Technology, 3936) for 1 h. The stained cells were washed and incubated with Alexa Fluor-conjugated secondary antibodies (Alexa Fluor 488 goat anti-rabbit or Alexa Fluor 555 goat anti-mouse, Invitrogen, A11008 and A21422) for 1 h. 4′,6-diamidino-2-phenylindole (DAPI) was used to stain nuclei. All steps were performed at room temperature. Images were captured using an Olympus FV1000 confocal microscope.
Flow cytometry
Cells were treated with OC, hydroxychloroquine sulfate (Sigma, H0915), or bafilomycin A1 (Sigma, B1793) and then cultured in complete (DMEM with serum) or nutrient-deprived medium (EBSS) for 24 h. The cells were fixed in 70% ethanol in PBS overnight. For cell cycle distribution, cells were counterstained with propidium iodide (Sigma, P4170) and analyzed for DNA content by use of a BD Influx™ (BD Biosciences) flow cytometer.
Western blotting
Cells were lysed in ice-cold whole cell extract buffer (50 mM TRIS-HCl, pH 8.0, 4 M urea and 1% Triton X-100), supplemented with complete protease inhibitor mixture (Roche Diagnostics, 04693132001). Cell extracts were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. After blocking with 5% nonfat milk in Tris-buffered saline (50 mM TRIS-HCl,pH 7.5, 150 mM NaCl) containing 0.2% Tween 20, the membranes were probed with the following antibodies: LC3B (sigma, L7543), SQSTM1/p62 (MBL, PM045), GAPDH (Proteintech, 10494-1-AP), CASP3 (Cell Signaling Technology, 9962). Following incubation with horseradish peroxidase coupled secondary anti-mouse (KPL, 074-1806) or anti-rabbit antibodies (KPL, 474-1506), protein bands were visualized using ECL blotting detection reagents (KPL, 54-61-00).
Acridine orange staining
Cell staining with acridine orange (AO; Molecular Probes/Invitrogen, A1301) was performed according to the protocol from the manufacturer, adding a final concentration of 5 μg/ml for a period of 10 min (37 °C, 5% CO2). After washing with PBS 3 times, photographs were obtained using a confocal microscope (Zeiss Observer Z1, Carl Zeiss) equipped with an argon laser (excitation wavelength 488 nm), and a 40× objective lens. AO produces red fluorescence (emission filter 620 nm long pass) in lysosomal compartments, and green fluorescence (emission between 520 and 560 nm) in the cytosolic and nuclear compartments. The red and green intensity ratio in exclusive nuclear region was analyzed by Columbus software (Version 2.3.1).
DQ-BSA staining
Cells were incubated with DQ-BSA (DQTM Red BSA, Molecular Probes/Invitrogen, D-12051) at a concentration of 10 μg/ml for 12 h (37 °C, 5% CO2). The cells were then washed 3 times with PBS before being treated with 15 μM OC or 5 nM BAFA1 for 8 h or starved in EBSS for 2 h. The cells were fixed and photographs were obtained using an Olympus FV1000 confocal microscope with excitation at 542 nm and emission at 600 nm long pass.
Cathepsin activity assay
The catalytic activities of cathespins were determined by CTSB and CTSD activity fluorometric assay kits (BioVision, K140-100, K143-100,). Briefly, 2 × 106 cells were collected by centrifugation and lysed in 200 µl of chilled cell lysis buffer. Then 50 µl of cell lysate was transferred into 96-well plates, mixed with reaction buffer and substrate, and incubated at 37 °C for 2 h. The samples were read in a fluorometer with 400 nm excitation and 505 nm emission filters. The activity was normalized with the samples’ protein concentration.
RNA isolation and quantitative RT-PCR
Total RNA isolation was performed using Trizol reagent (Beyotime, R0016) following the manufacturer’s protocol. Reverse transcription PCR was done using PrimeScript RT reagent kit (TaKaRa, DRR037A). qPCR analysis was performed in a Verti Thermal Cycler (Applied Biosystem, Life Technologies) using SYBR Green Real Time PCR kit (TOYOBO, QPK-201). Data collection was performed using a StepOne Plus Real-Time PCR System Thermal Cycling Block (Applied Biosystems, Life Technologies). Primers for qPCR reactions were as follows:
SQSTM1 (human): 5′-GAACTCCAGT CCCTACAGAT GCC-3′, 5′-CGGGAGATGT GGGTACAAGG-3′; Sqstm1 (mouse): 5′-AAGAGTAACA CTCAGCCAAG CAGC-3′, 5′-ATCTGTTCCT CTGGCTGTCC C-3′; CTSB (human): 5′-AACACGTCAC CGGAGAGATG A-3′, 5′-CCCAGTCAGT GTTCCAGGAG TT-3′; CTSD (human): 5′-GGCTCTGTGG AGGACCTGAT TG-3′, 5′-CGATGCCAAT CTCCCCGTAG TA-3′; GAPDH (human): 5′-TGTTGCCATC AATGACCCCT T-3′, 5′-CTCCACGACG TACTCAGCG-3′; Gapdh (mouse): 5′-ACAACTTTGG CATTGTGGAA-3′, 5′-GATGCAGGGA TGATGTTCTG-3′.
Tumorigenesis in nude mice
Four-wk-old BALB/c nude mice ages were purchased from the Experimental Animal Center of the Chinese Academy of Science (Shanghai, China) and kept in a pathogen-free environment at the Experimental Animal Center in the Shanghai University of Traditional Chinese Medicine. Approximately 2 × 106 HeLa cells were injected into the left and right sides of the animals. Five days after tumor cell implantation (2 × 106 HeLa cells per mouse) all mice were randomly divided into 4 groups (n = 8 per group): 1) Vehicle (normally fed, receiving daily i.p saline), 2) OC (normally receiving daily i.p 30 mg/kg of OC), 3) caloric restriction (CR) (fed with 70% of their normal food intake, receiving daily i.p. saline), and 4) OC+caloric restriction (calorie-restricted mice receiving daily i.p. 30 mg/kg of OC). At the end of the experiment (5 wk after tumor implantation), the mice were sacrificed and the tumor weight of each animal was analyzed.
Immunohistochemistry
Tumors were resected immediately after euthanasia and fixed in 10% neutral buffered paraformaldehyde at 4 °C for 48 h. Selected samples were embedded in paraffin, sectioned and stained with hematoxylin and eosin (Sinopharm Chemical Reagent Co., Ltd.), cleaved CASP3 (Biosynthesis Biotech, bs-0087R), LC3B (Sigma, L7543), SQSTM1/p62 (Medical and Biological Laboratories, PM045), CTSB (Abcam, 2487-1) and CTSD (Abcam, 3547-1). The primary antibodies were used at 1:100 for cleaved CASP3, 1:200 for LC3, 1:1000 for SQSTM1, 1:500 for CTSB and 1:100 for CTSD. The sections were finally mounted with D.P.X mountant (Sigma, 317616) for histology analysis.
Statistical analysis
All results were expressed as means ± SD of 3 independent experiments. Statistical analyses were performed using the Student 2-tailed t test. Values of *P < 0.05 were considered to be significant.
Supplementary Material
Glossary
Abbreviations:
- AO
acridine orange
- ATG5
autophagy-related 5
- ATG7
autophagy-related 7
- BAFA1
bafilomycin A1
- CASP3
caspase 3, apoptosis-related cysteine peptidase
- CTSB
cathepsin B
- CTSD
cathepsin D
- GFP
green fluorescent protein
- GK
guttiferone K
- HCQ
hydroxychloroquine
- MAP1LC3/LC3
microtubule-associated protein 1 light chain 3
- MEF
mouse embryonic fibroblast
- OC
oblongifolin C
- PARP1
poly (ADP-ribose) polymerase
- PPAPs
polycyclic polyprenylated acylphloroglucinols
- siRNA
short interfering RNA
- SQSTM1
sequestosome 1
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
We would like to thank Professor Kevin Ryan for providing the MEF atg7−/− cell line. This work was supported by the National Natural Science Foundation of China (No. 21272135 and No. 81173485), the Overseas High-caliber Personnel Foundation of Shenzhen (No. KQC201109050084A) and the Innovation Program of Shanghai Municipal Education Commission (No. 13ZZ093).
References
- 1.Xie Z, Klionsky DJ. . Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102 - 9; http://dx.doi.org/ 10.1038/ncb1007-1102; PMID: 17909521 [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein DC, Mariño G, Kroemer G. . Autophagy and aging. Cell 2011; 146:682 - 95; http://dx.doi.org/ 10.1016/j.cell.2011.07.030; PMID: 21884931 [DOI] [PubMed] [Google Scholar]
- 3.Carew JS, Nawrocki ST, Cleveland JL. . Modulating autophagy for therapeutic benefit. Autophagy 2007; 3:464 - 7; PMID: 17495516 [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. . Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069 - 75; http://dx.doi.org/ 10.1038/nature06639; PMID: 18305538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenfeldt MT, Ryan KM. . The role of autophagy in tumour development and cancer therapy. Expert Rev Mol Med 2009; 11:e36; http://dx.doi.org/ 10.1017/S1462399409001306; PMID: 19951459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathew R, Karantza-Wadsworth V, White E. . Role of autophagy in cancer. Nat Rev Cancer 2007; 7:961 - 7; http://dx.doi.org/ 10.1038/nrc2254; PMID: 17972889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, Kroemer G. . Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ 2009; 16:87 - 93; http://dx.doi.org/ 10.1038/cdd.2008.131; PMID: 18806760 [DOI] [PubMed] [Google Scholar]
- 8.Levine B. . Cell biology: autophagy and cancer. Nature 2007; 446:745 - 7; http://dx.doi.org/ 10.1038/446745a; PMID: 17429391 [DOI] [PubMed] [Google Scholar]
- 9.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. . Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003; 100:15077 - 82; http://dx.doi.org/ 10.1073/pnas.2436255100; PMID: 14657337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, et al. . Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007; 9:1142 - 51; http://dx.doi.org/ 10.1038/ncb1634; PMID: 17891140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. . Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006; 8:688 - 99; http://dx.doi.org/ 10.1038/ncb1426; PMID: 16799551 [DOI] [PubMed] [Google Scholar]
- 12.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. . Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672 - 6; http://dx.doi.org/ 10.1038/45257; PMID: 10604474 [DOI] [PubMed] [Google Scholar]
- 13.Yang ZJ, Chee CE, Huang S, Sinicrope FA. . The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011; 10:1533 - 41; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0047; PMID: 21878654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carew JS, Kelly KR, Nawrocki ST. . Autophagy as a target for cancer therapy: new developments. Cancer Manag Res 2012; 4:357 - 65; PMID: 23091399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Klionsky DJ. . Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr 2007; 27:19 - 40; http://dx.doi.org/ 10.1146/annurev.nutr.27.061406.093749; PMID: 17311494 [DOI] [PubMed] [Google Scholar]
- 16.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. . In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 2004; 15:1101 - 11; http://dx.doi.org/ 10.1091/mbc.E03-09-0704; PMID: 14699058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, et al. . Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A 2007; 104:19023 - 8; http://dx.doi.org/ 10.1073/pnas.0709695104; PMID: 18024584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N, Yoshimori T, Levine B. . Methods in mammalian autophagy research. Cell 2010; 140:313 - 26; http://dx.doi.org/ 10.1016/j.cell.2010.01.028; PMID: 20144757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445 - 544; http://dx.doi.org/ 10.4161/auto.19496; PMID: 22966490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. . p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603 - 14; http://dx.doi.org/ 10.1083/jcb.200507002; PMID: 16286508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Man S, Gao W, Wei C, Liu C. . Anticancer drugs from traditional toxic Chinese medicines. Phytother Res 2012; 26:1449 - 65; PMID: 22389143 [DOI] [PubMed] [Google Scholar]
- 22.Han QB, Xu HX. . Caged Garcinia xanthones: development since 1937. Curr Med Chem 2009; 16:3775 - 96; http://dx.doi.org/ 10.2174/092986709789104993; PMID: 19747141 [DOI] [PubMed] [Google Scholar]
- 23.Anantachoke N, Tuchinda P, Kuhakarn C, Pohmakotr M, Reutrakul V. . Prenylated caged xanthones: chemistry and biology. Pharm Biol 2012; 50:78 - 91; http://dx.doi.org/ 10.3109/13880209.2011.636176; PMID: 22196584 [DOI] [PubMed] [Google Scholar]
- 24.Yi T, Yi Z, Cho SG, Luo J, Pandey MK, Aggarwal BB, Liu M. . Gambogic acid inhibits angiogenesis and prostate tumor growth by suppressing vascular endothelial growth factor receptor 2 signaling. Cancer Res 2008; 68:1843 - 50; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-5944; PMID: 18339865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Chen W. . Gambogic acid is a novel anti-cancer agent that inhibits cell proliferation, angiogenesis and metastasis. Anticancer Agents Med Chem 2012; 12:994 - 1000; http://dx.doi.org/ 10.2174/187152012802650066; PMID: 22339063 [DOI] [PubMed] [Google Scholar]
- 26.Richard JA, Pouwer RH, Chen DY. . The chemistry of the polycyclic polyprenylated acylphloroglucinols. Angew Chem Int Ed Engl 2012; 51:4536 - 61; http://dx.doi.org/ 10.1002/anie.201103873; PMID: 22461155 [DOI] [PubMed] [Google Scholar]
- 27.Simpkins NS. . Adventures in bridgehead substitution chemistry: synthesis of polycyclic polyprenylated acylphloroglucinols (PPAPs). Chem Commun (Camb) 2013; 49:1042 - 51; http://dx.doi.org/ 10.1039/c2cc37914g; PMID: 23229029 [DOI] [PubMed] [Google Scholar]
- 28.Feng C, Zhou LY, Yu T, Xu G, Tian HL, Xu JJ, Xu HX, Luo KQ. . A new anticancer compound, oblongifolin C, inhibits tumor growth and promotes apoptosis in HeLa cells through Bax activation. Int J Cancer 2012; 131:1445 - 54; http://dx.doi.org/ 10.1002/ijc.27365; PMID: 22116711 [DOI] [PubMed] [Google Scholar]
- 29.Kan WL, Yin C, Xu HX, Xu G, To KK, Cho CH, Rudd JA, Lin G. . Antitumor effects of novel compound, guttiferone K, on colon cancer by p21Waf1/Cip1-mediated G(0) /G(1) cell cycle arrest and apoptosis. Int J Cancer 2013; 132:707 - 16; http://dx.doi.org/ 10.1002/ijc.27694; PMID: 22733377 [DOI] [PubMed] [Google Scholar]
- 30.Han QB, Tian HL, Yang NY, Qiao CF, Song JZ, Chang DC, Luo KQ, Xu HX. . Polyprenylated xanthones from Garcinia lancilimba showing apoptotic effects against HeLa-C3 cells. Chem Biodivers 2008; 5:2710 - 7; http://dx.doi.org/ 10.1002/cbdv.200890225; PMID: 19089829 [DOI] [PubMed] [Google Scholar]
- 31.Huang SX, Feng C, Zhou Y, Xu G, Han QB, Qiao CF, Chang DC, Luo KQ, Xu HX. . Bioassay-guided isolation of xanthones and polycyclic prenylated acylphloroglucinols from Garcinia oblongifolia. J Nat Prod 2009; 72:130 - 5; http://dx.doi.org/ 10.1021/np800496c; PMID: 19113969 [DOI] [PubMed] [Google Scholar]
- 32.Xia ZX, Zhang DD, Liang S, Lao YZ, Zhang H, Tan HS, Chen SL, Wang XH, Xu HX. . Bioassay-guided isolation of prenylated xanthones and polycyclic acylphloroglucinols from the leaves of Garcinia nujiangensis. J Nat Prod 2012; 75:1459 - 64; http://dx.doi.org/ 10.1021/np3003639; PMID: 22871217 [DOI] [PubMed] [Google Scholar]
- 33.Bartlett BJ, Isakson P, Lewerenz J, Sanchez H, Kotzebue RW, Cumming RC, Harris GL, Nezis IP, Schubert DR, Simonsen A, et al. . p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 2011; 7:572 - 83; http://dx.doi.org/ 10.4161/auto.7.6.14943; PMID: 21325881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carew JS, Medina EC, Esquivel JA 2nd, Mahalingam D, Swords R, Kelly K, Zhang H, Huang P, Mita AC, Mita MM, et al. . Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med 2010; 14:2448 - 59; http://dx.doi.org/ 10.1111/j.1582-4934.2009.00832.x; PMID: 19583815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon YH, Cho KS, Hwang JJ, Lee SJ, Choi JA, Koh JY. . Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Invest Ophthalmol Vis Sci 2010; 51:6030 - 7; http://dx.doi.org/ 10.1167/iovs.10-5278; PMID: 20574031 [DOI] [PubMed] [Google Scholar]
- 36.Vázquez CL, Colombo MI. . Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol 2009; 452:85 - 95; http://dx.doi.org/ 10.1016/S0076-6879(08)03606-9; PMID: 19200877 [DOI] [PubMed] [Google Scholar]
- 37.Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S. . Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy 2007; 3:154 - 7; PMID: 17204842 [DOI] [PubMed] [Google Scholar]
- 38.Kaminskyy V, Zhivotovsky B. . Proteases in autophagy. Biochim Biophys Acta 2012; 1824:44 - 50; http://dx.doi.org/ 10.1016/j.bbapap.2011.05.013; PMID: 21640203 [DOI] [PubMed] [Google Scholar]
- 39.Guha S, Padh H. . Cathepsins: fundamental effectors of endolysosomal proteolysis. Indian J Biochem Biophys 2008; 45:75 - 90; PMID: 21086720 [PubMed] [Google Scholar]
- 40.Levine B, Klionsky DJ. . Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463 - 77; http://dx.doi.org/ 10.1016/S1534-5807(04)00099-1; PMID: 15068787 [DOI] [PubMed] [Google Scholar]
- 41.Bergamini E, Cavallini G, Donati A, Gori Z. . The anti-ageing effects of caloric restriction may involve stimulation of macroautophagy and lysosomal degradation, and can be intensified pharmacologically. Biomed Pharmacother 2003; 57:203 - 8; http://dx.doi.org/ 10.1016/S0753-3322(03)00048-9; PMID: 12888255 [DOI] [PubMed] [Google Scholar]
- 42.Harhaji-Trajkovic L, Arsikin K, Kravic-Stevovic T, Petricevic S, Tovilovic G, Pantovic A, Zogovic N, Ristic B, Janjetovic K, Bumbasirevic V, et al. . Chloroquine-mediated lysosomal dysfunction enhances the anticancer effect of nutrient deprivation. Pharm Res 2012; 29:2249 - 63; http://dx.doi.org/ 10.1007/s11095-012-0753-1; PMID: 22538436 [DOI] [PubMed] [Google Scholar]
- 43.Grifantini K. . Understanding pathways of calorie restriction: a way to prevent cancer?. J Natl Cancer Inst 2008; 100:619 - 21; http://dx.doi.org/ 10.1093/jnci/djn142; PMID: 18445812 [DOI] [PubMed] [Google Scholar]
- 44.White E. . Role of the metabolic stress responses of apoptosis and autophagy in tumor suppression. Ernst Schering Found Symp Proc 2007; 23 - 34; PMID: 18811051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin S, White E. . Role of autophagy in cancer: management of metabolic stress. Autophagy 2007; 3:28 - 31; PMID: 16969128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. . Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21:1621 - 35; http://dx.doi.org/ 10.1101/gad.1565707; PMID: 17606641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carew JS, Espitia CM, Esquivel JA 2nd, Mahalingam D, Kelly KR, Reddy G, Giles FJ, Nawrocki ST. . Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J Biol Chem 2011; 286:6602 - 13; http://dx.doi.org/ 10.1074/jbc.M110.151324; PMID: 21148553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, Lynch JP, Uehara T, Sepulveda AR, Davis LE, et al. . Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A 2012; 109:8253 - 8; http://dx.doi.org/ 10.1073/pnas.1118193109; PMID: 22566612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yue W, Hamaï A, Tonelli G, Bauvy C, Nicolas V, Tharinger H, Codogno P, Mehrpour M. . Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy 2013; 9:714 - 29; http://dx.doi.org/ 10.4161/auto.23997; PMID: 23519090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Hu SE, Tan SH, Cao R, Chen Y, Xia D, Zhu X, Yang XF, Ong CN, Shen HM. . Andrographolide sensitizes cisplatin-induced apoptosis via suppression of autophagosome-lysosome fusion in human cancer cells. Autophagy 2012; 8:338 - 49; http://dx.doi.org/ 10.4161/auto.18721; PMID: 22302005 [DOI] [PubMed] [Google Scholar]
- 51.Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. . Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 2011; 13:453 - 60; http://dx.doi.org/ 10.1038/ncb2204; PMID: 21394080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. . TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429 - 33; http://dx.doi.org/ 10.1126/science.1204592; PMID: 21617040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganley IG, Wong PM, Gammoh N, Jiang X. . Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 2011; 42:731 - 43; http://dx.doi.org/ 10.1016/j.molcel.2011.04.024; PMID: 21700220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turk B, Stoka V, Rozman-Pungercar J, Cirman T, Droga-Mazovec G, Oresić K, Turk V. . Apoptotic pathways: involvement of lysosomal proteases. Biol Chem 2002; 383:1035 - 44; http://dx.doi.org/ 10.1515/BC.2002.112; PMID: 12437086 [DOI] [PubMed] [Google Scholar]
- 55.Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. . Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 2012; 8:200 - 12; http://dx.doi.org/ 10.4161/auto.8.2.18554; PMID: 22252008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gondi CS, Rao JS. . Cathepsin B as a cancer target. Expert Opin Ther Targets 2013; 17:281 - 91; http://dx.doi.org/ 10.1517/14728222.2013.740461; PMID: 23293836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benes P, Vetvicka V, Fusek M. . Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol 2008; 68:12 - 28; http://dx.doi.org/ 10.1016/j.critrevonc.2008.02.008; PMID: 18396408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frlan R, Gobec S. . Inhibitors of cathepsin B. Curr Med Chem 2006; 13:2309 - 27; http://dx.doi.org/ 10.2174/092986706777935122; PMID: 16918357 [DOI] [PubMed] [Google Scholar]
- 59.Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, Bidwell BN, Edgington L, Wang L, Moin K, et al. . Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res 2012; 72:1199 - 209; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-2759; PMID: 22266111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.