Abstract
The filamentous ascomycete Podospora anserina is a well-established aging model in which a variety of different pathways, including those involved in the control of respiration, ROS generation and scavenging, DNA maintenance, proteostasis, mitochondrial dynamics, and programmed cell death have previously been demonstrated to affect aging and life span. Here we address a potential role of autophagy. We provide data demonstrating high basal autophagy levels even in strains cultivated under noninduced conditions. By monitoring an N-terminal fusion of EGFP to the fungal LC3 homolog PaATG8 over the lifetime of the fungus on medium with and without nitrogen supplementation, respectively, we identified a significant increase of GFP puncta in older and in nitrogen-starved cultures suggesting an induction of autophagy during aging. This conclusion is supported by the demonstration of an age-related and autophagy-dependent degradation of a PaSOD1-GFP reporter protein. The deletion of Paatg1, which leads to the lack of the PaATG1 serine/threonine kinase active in early stages of autophagy induction, impairs ascospore germination and development and shortens life span. Under nitrogen-depleted conditions, life span of the wild type is increased almost 4-fold. In contrast, this effect is annihilated in the Paatg1 deletion strain, suggesting that the ability to induce autophagy is beneficial for this fungus. Collectively, our data identify autophagy as a longevity-assurance mechanism in P. anserina and as another surveillance pathway in the complex network of pathways affecting aging and development. These findings provide perspectives for the elucidation of the mechanisms involved in the regulation of individual pathways and their interactions.
Keywords: aging, autophagy, ATG1, ATG8, Podospora anserina
Introduction
Biological aging is a complex process controlled by stochastic, environmental, and genetic traits. A multitude of theories have been put forward to explain aging. Common to a number of theories1-7 is an impact of the age-dependent accretion of damage leading to cumulative functional impairments and ultimately to cellular death. The accumulation of damaged cellular components strongly depends on the rate of reactions leading to damage and the persistence of the resulting material. The first process can be modulated by controlling the abundance of reactive oxygen species (ROS) via both their generation and by scavenging processes. Persistence of damaged compounds depends on the effectiveness of different types of surveillance systems including repair, degradation, and remodeling of damaged components (for a review see ref. 8). Autophagy is one of the pathways leading to the degradation of damaged biomolecules and organelles.
Podospora anserina is an established aging model to study basic mechanisms of organismic aging. This filamentous ascomycete is characterized by a limited, short life span which is depending on the genetic constitution and on growth conditions.9 Aging of individuals can be followed macroscopically, since hyphal morphology changes during senescence of this fungus. P. anserina is accessible to experimental manipulation both by classical genetics as well as by genetic engineering.10-12 The consequences of such manipulations can be analyzed by established techniques.13 Over the years, various pathways including those linked to DNA instability and maintenance,14-16 copper homeostasis,17,18 and ROS scavenging19-21 have been demonstrated to affect aging. More recently, different quality control pathways were investigated and found to be effective at different levels. In particular, it has been found that overexpression of PaLon encoding a mitochondrial matrix protease leads to an increased healthy life span22 while the deletion of the gene shortens life span.23 PaCLPP, another mitochondrial matrix protease, and PaIAP, the iAAA protease of the inner mitochondrial membrane of P. anserina, are involved in temperature-dependent protein quality control.24,25 Finally, the manipulation of reactions of the apoptotic cell death machinery, that brings the life of P. anserina to an end, has been found to strongly affect life span.26-28
In a recent computational study, published data have been used to integrate different pathways involved in the control of the quality of mitochondria. In this mathematical model, the generation of molecular damage, spread of damage, biogenesis of cellular components, mitochondrial fission and fusion, and mitophagy, as a type of selective autophagy, are modeled and simulated. The results indicate that the analyzed pathways effectively cooperate in a regulated way to keep mitochondria functional over time.29,30 While the role of mitochondrial dynamics on aging was first reported in P. anserina31 and the impact of the generation of damage has been previously demonstrated,32 the impact of autophagy on aging has not been investigated yet although autophagy has been studied in the context of vegetative incompatibility between strains of different genotypes and found to protect cells against death.33-35
Here, we address the impact of autophagy on aging and life span of P. anserina. We report the adaptation and use of tools developed for autophagy studies in yeast and higher eukaryotes and show that autophagy acts as a longevity-assurance mechanism.
Results
The abundance of autophagosomes increases under nitrogen starvation and during aging
In a first series of experiments, we set out to investigate whether autophagy is induced by nitrogen starvation and during aging of P. anserina. For these analyses, a Gfp-PaAtg8 strain was generated, which expresses an N-terminal fusion of the GFP protein to PaATG8, the fungal ATG8 ortholog (in mammals MAP1LC3, hereafter referred to as LC3). Labeling of ATG8/LC3 allows the visualization of the translocation of the fusion protein from the cytosol to the autophagosome to the vacuole, where the final degradation of the autophagosome takes place in fungi.
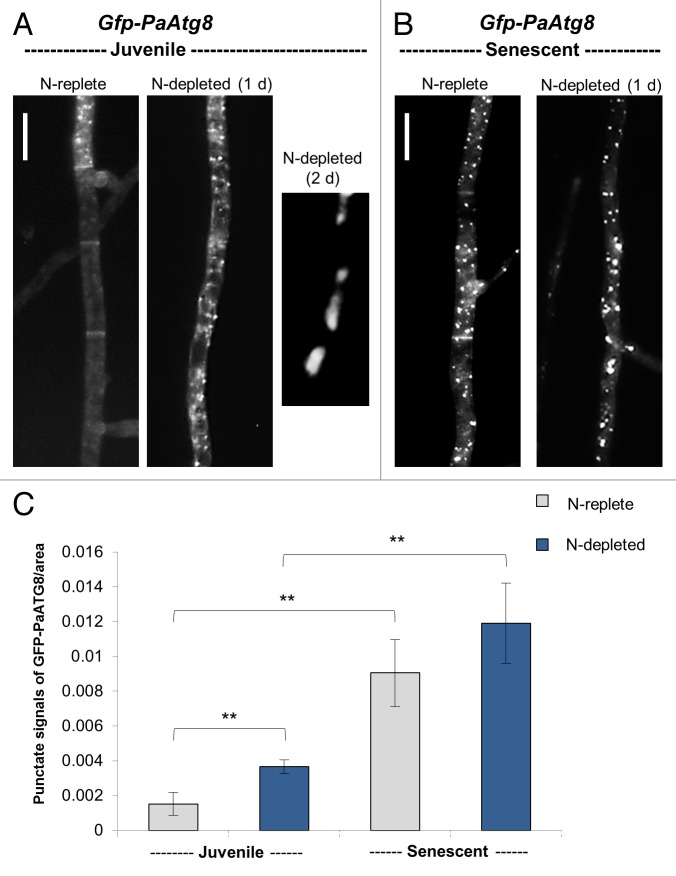
Fluorescence microscopy revealed that in juvenile strains (4 d old, grown on nitrogen-replete medium) GFP-PaATG8 was mostly diffusely distributed within the cytoplasm. During nitrogen starvation on medium lacking urea as a nitrogen source, a well-known regime to induce autophagy in other systems,36-40 the fusion protein is localized in bright punctate structures in nearly all hyphae (Fig. 1A), indicating that autophagy is induced under these conditions also in P. anserina. Although autophagosomes are visible, the GFP signal is not seen in the vacuole after 1 d on nitrogen-replete medium, an observation consistent with earlier findings of Pinar et al.41 in Aspergillus nidulans. Surprisingly, and in contrast to A. nidulans, in P. anserina also under nitrogen depletion the GFP-stained vacuoles are not detectable after this short period of time. However, prolonged incubation on this medium leads to the accumulation of the GFP signal in the vacuole (Fig. 1A).
Figure 1. Comparative fluorescence microscopy analysis of juvenile vs. senescent P. anserina Gfp-PaAtg8 strains. (A) GFP-PaATG8 distributes diffusely to the cytoplasm in the juvenile (4 d old) hyphae on M2 medium while the number of punctate autophagosome-like structures is strongly increased during 1 d incubation on nitrogen-depleted medium (M2-N). Longer incubation (2 d) leads to the delivery of GFP-PaATG8 to vacuoles and further degradation. (B) In senescent cultures (20 d), GFP-PaATG8 localizes to punctate autophagosome-like structures grown on M2-N and also on M2 medium indicating that autophagy increases with age. Scale bar: 10 μm. (C) Quantification of punctate autophagosome-like structures per area for hyphae of juvenile and senescent P. anserina cultures, grown on M2 and M2-N. Error bars correspond to the standard error (n = 10). P values were determined between juvenile and senescent wild type grown on M2 medium, respectively M2-N medium and also between juvenile wild-type strains grown on M2 and M2-N medium by 2-tailed Wilcoxon rank-sum test (P < 0.01). (A–C): Nitrogen-replete: M2 medium supplemented with nitrogen (0.5 g/L urea); nitrogen-depleted: M2-medium without nitrogen.
Next, we analyzed whether or not autophagy is upregulated during aging of P. anserina. We compared the localization of the fusion protein in young (4 d old) and senescent (20 d old) Gfp-PaAtg8 strains grown on nitrogen-replete (M2), as well as on nitrogen-depleted medium (M2-N), and quantified autophagosomes in the different cultures. In old cultures, a strong translocation of the fusion protein to punctate autophagosome-like structures was observed suggesting a strong upregulation of autophagy during aging (Fig. 1B and C). In senescent cultures, N-starvation was found to lead only to a slight increase of autophagosomes in comparison to senescent cultures grown on nitrogen-replete M2 medium (Fig. 1B and C). For visualization of the vacuoles, differential interference contrast microscopy (DIC) was used in addition to fluorescence microscopy (Fig. S1A and S1B).
The autophagy-dependent degradation of a GFP fusion protein increases during aging
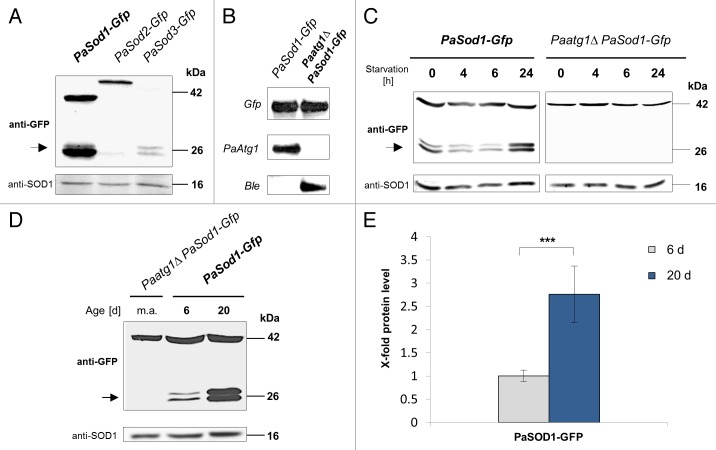
The determination of punctate GFP-ATG8 signals as a measure for the intensity of autophagy has to be viewed critically. An increase in number of autophagosomes may reflect an upregulation of autophagy but as well it may result from an accumulation of autophagosomes due to an impairment of their translocation to the vacuole and the subsequent degradation. Consequently, in order to discriminate between the 2 possibilities, we used a modified experimental approach that originally was developed to monitor mitophagy in yeast.42,43 In this approach, in principle, the fate of a GFP fusion protein is followed. Upon degradation via autophagy, the processed GFP remains stable and can be detected by western blot analysis. To test whether this assay also works in P. anserina, 3 fusion proteins with different cellular localizations were analyzed: PaSOD1-GFP, PaSOD2-GFP, and PaSOD3-GFP. The corresponding strains are described in Zintel et al.20 PaSOD1-GFP is localized in the cytosol, PaSOD2-GFP in the perinuclear ER and PaSOD3-GFP in mitochondria. Total protein extracts were subjected to western blot analysis with a GFP antibody. As expected, the most prominent signal of processed GFP is found in PaSod1-Gfp, since this protein is highly abundant in total protein extracts. As a mitochondrial protein PaSOD3 is under-represented in total protein extracts of the PaSod3-Gfp strain. Thus, only the processed GFP is visible. Interestingly, while the PaSOD2-GFP is abundantly found in the extracts of the PaSod2-Gfp strain, no processed GFP is detectable (Fig. 2A). This suggests that the cytosolic as well as the mitochondrial PaSOD isoforms are degraded via autophagy but not the ER-located and presumably secreted PaSOD2.
Figure 2. Autophagy-induced protein degradation during aging. (A) Western blot analysis of total protein extracts from different PaSod-Gfp strains. GFP processing was monitored by immunoblotting with anti-GFP and anti-SOD1 (loading control) antibody. (B) Southern blot analysis of HindIII digested genomic DNA from PaSod1-Gfp and the Paatg1Δ PaSod1-Gfp double mutant. The Gfp-specific hybridization probe in the upper part detects a ~9.5 kb fragment containing Gfp in both strains. The PaAtg1-specific hybridization probe in the middle part detects the 6.5 kb fragment of the PaAtg1 gene only in the genomic DNA of the PaSod1-Gfp strain. The gene encoding the phleomycin resistance gene (Ble) is only present as a 6.5 kb fragment in the genomic DNA of the double mutant (lower part). (C) Monitoring autophagy by western blot analysis using the cytosolic protein PaSOD1-GFP during starvation. Wild-type (WT) and Paatg1Δ strains expressing PaSOD1-GFP were cultured in CM medium then shifted to CM-N medium for 0, 4, 6, and 24 h. GFP processing was monitored by immunoblotting with anti-GFP and anti-SOD1 (loading control) antibody. The positions of molecular mass markers are indicated on the right. (D) Monitoring autophagy by western blot using the cytosolic protein PaSOD1-GFP of juvenile and senescent cultures. Six and 20 d old wild-type (WT), and middle-aged (m.a.) Paatg1Δ strains expressing PaSod1-Gfp were cultured in CM medium for 2 d. GFP processing was monitored by immunoblotting with anti-GFP and anti-SOD1 antibody (loading control). The positions of molecular mass markers are indicated on the right. (E) The GFP protein levels of the PaSod1-Gfp strains (n = 7) were normalized to the level of SOD1, and the protein amount present in the 6 d old strain was set to 1. The 20 d old strain possesses a nearly 5-fold higher protein amount than the 6 d old PaSod1-Gfp strain. Error bars correspond to the standard error. P values were determined between juvenile (6 d old) and senescent (20 d old) PaSod1-Gfp (P < 0.001) by 2-tailed Wilcoxon rank-sum test. The arrow marks the processed GFP. (A), (C–E): CM medium = supplemented with nitrogen (NH4Cl: 3.7 g/L); CM-N medium = CM medium without nitrogen.
To verify that the occurrence of processed GFP is indeed depending on a functional autophagy machinery, a strain lacking PaATG1, the serine/threonine kinase at the start of the autophagy-induction cascade was constructed (Fig. S2A). Previously, Pinan-Lucarré et al.34 have reported, that in a P. anserina strain lacking PaATG1, autophagy does not occur. To follow the fate of PaSOD1-GFP in the genetic background of the Paatg1 deletion, a double mutant was constructed (Fig. 2B). Total protein extracts of this strain (Paatg1Δ PaSod1-Gfp) were isolated and analyzed by western blot analysis using GFP antibodies. As expected, in the genetic background of a Paatg1 deletion strain, no processed GFP is visible even under nitrogen-depleted conditions (Fig. 2C). Thus, the occurrence of processed GFP depends on autophagy and the amount of GFP is a measure for autophagy in P. anserina. Obviously, P. anserina is characterized by a high level of basal autophagy because even under conditions with sufficient nitrogen a pronounced band of the processed GFP is visible in the PaSod1-Gfp strain.
Next, we tested the effect of nitrogen starvation on the amount of processed GFP. Short-time nitrogen starvation (4 to 6 h) results in a decline in the amount of processed GFP, while prolonged nitrogen starvation (24 h) leads to a substantial increase (Fig. 2C). It is possible that this effect results from the activity of other factors, like an increased proteasome activity or simply because this short period of time is not sufficient to induce a nitrogen starvation response. During aging, a pronounced and significant increase in the amount of processed GFP can be detected in strains of old age (i.e., 20 d), strongly supporting the microscopic observations with the Gfp-PaAtg8 strain that aging in P. anserina is accompanied by enhanced autophagy (Fig. 2D and E).
Autophagy is involved in development and functions as a longevity-assurance mechanism
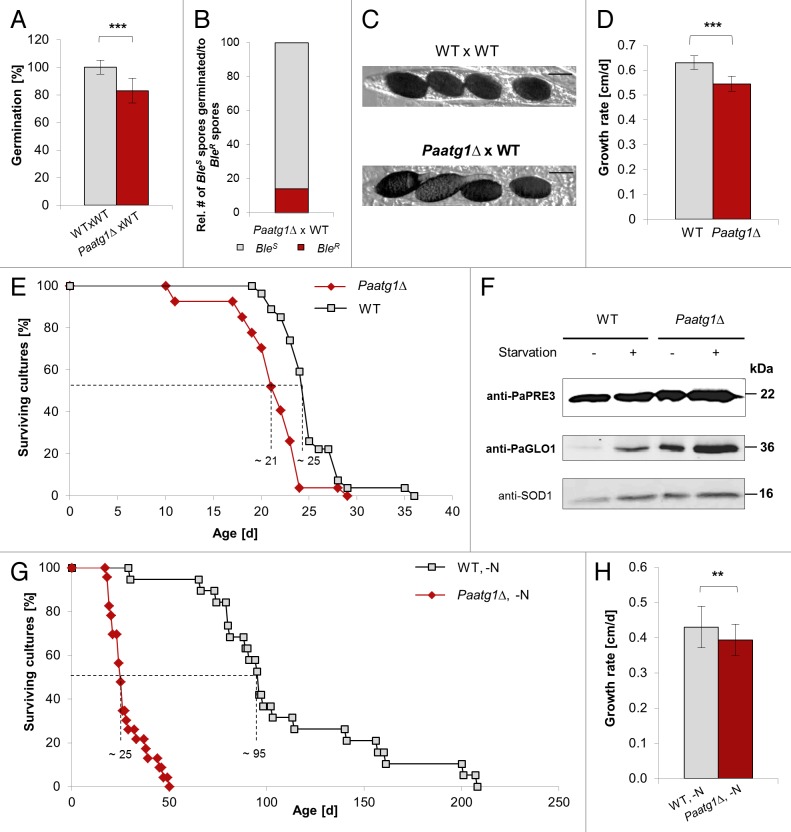
In an earlier publication34 the Paatg1Δ mutant was studied in detail with respect to its role in the vegetative incompatibility reaction between fungal strains of unlike genotype. In this study several developmental defects of the mutant were observed compared with the wild type including a slightly reduced growth rate, a lower density of aerial hyphae on rich media, a decreased pigmentation of the mycelium and the inability to form protoperithecia (the female reproductive organ) leading to female sterility. These different phenotypic characteristics were fully restored by complementation with a wild-type copy of the Paatg1 gene.34 In our study, we constructed an independent Paatg1 deletion strain based on a protocol described in Hamann et al.12 (Fig. S2A) and observed similar phenotypic characteristics of the mutant as described earlier (Fig. S2B and S2C). In addition, we tested the germination capability of monokaryotic spores isolated from a cross between 2 wild-type strains, and a cross between the wild type and the deletion mutant (Fig. 3A). The spores of the latter cross showed a significant decrease in germination rate. To test whether both, the wild-type spores as well as the spores carrying the deletion are affected, the resistance of the germinated spores was determined. From 236 germinated ascospores of a cross of the wild-type strain and Paatg1Δ, theoretically 50% (= 118) should possess the allele with the deletion, detectable as phleomycin resistance. However, only 33 (= 14% of all spores) formed phleomycin-resistant mycelia and are therefore carrying the Paatg1 deletion (Fig. 3B). This is a smaller number than the expected 50% phleomycin-resistant offspring, indicating impairment in germination of the Paatg1Δ spores. As described before, the deletion strain is virtually female sterile, however, male fertility is not impaired, since spermatization of wild-type mycelium with Paatg1Δ spermatia (the male gametes) does not significantly reduce the amount of fruiting bodies (Fig. S2D).
Figure 3. Characterization of the Paatg1∆ strain. (A) The germination rate of ascospores from perithecia of fertilized Paatg1Δ (n = 10) and WT cultures (n = 10). P values (P < 0.001) were determined in comparison with the wild-type sample by 2-tailed Wilcoxon rank-sum test. (B) Determination of the genotype (BleR resistance) of ascospores of a Paatg1Δ × WT cross (n = 10). Colonies grown from germinated ascospores were transferred to phleomycin containing BMM medium. Grey/bright = spores germinated (BleS); red/dark = BleR spores germinated with Paatg1 deletion background. (C) Ascospore phenotype of spores from WT × WT and WT × Paatg1Δ crosses. Scale bar: 20 µm. (D) Growth rates of the WT (n = 27) and the Paatg1 deletion strain (n = 27; P < 0.001). P values were determined in comparison with the wild-type sample by 2-tailed Wilcoxon rank-sum test. (E) Life span of monokaryotic wild-type (n = 27; median life span = ~25 d) and Paatg1Δ (n = 27; median life span = ~21 d; P < 0.001) isolates on M2 medium at 27 °C. P values were determined in comparison with the wild-type sample by 2-tailed Wilcoxon rank-sum test. (F) Western blot analysis of total protein extracts from the Paatg1 deletion mutant and WT strains grown on CM or CM-N media, respectively. Incubation with anti-PaPRE3 (corresponding to the β1 subunit of the 20S proteasome) and anti-PaGLO1 antibody (glyoxalase 1) showed different protein amounts in the WT and Paatg1Δ strain and in response to nitrogen starvation compared with anti-SOD1 (loading control). (G) Life span of monokaryotic wild-type (n = 11; median life span = ~80 d) and Paatg1Δ (n = 20; median life span = ~25 d; P < 0.001) isolates at 27 °C on M2-N medium. P values were determined in comparison with the wild-type sample by 2-tailed Wilcoxon rank-sum test. (H) Growth rates of the WT (n = 11) and the Paatg1 deletion strain (n = 20; P < 0.01) on M2-N medium at 27 °C. P values were determined in comparison with the wild-type sample by 2-tailed Wilcoxon rank-sum test. M2 medium: supplemented with nitrogen (0.5 g/L urea); N: M2-medium without nitrogen.
The ascospores themselves display clear morphological aberrancies. Since spores are only formed in crosses between the wild type and Paatg1Δ, in each ascus the wild-type as well as the mutant allele is equally present. If no crossing-over occurs during meiosis between the centromer and the Paatg1 locus (prereduction), 2 of the 4 dikaryotic ascospores contain exclusively the wild-type allele, while the other 2 contain the Paatg1Δ allele. These latter 2 spores are bigger, darker and possess a clearly visible appendix (Fig. 3C). Normally, the appendix becomes hyaline during ripening of the spores and this process is delayed in the mutant, which implies that autophagy is involved in this process.
In accordance with the findings of Pinan-Lucarré et al.,34 we also observed a slight but significant reduction of the growth rate of the Paatg1Δ strain (Fig. 3D). Next, we analyzed the growth rate of the Paatg1Δ strain compared with wild type during heat stress (37 °C) and exogenous oxidative stress conditions applied by paraquat treatment, a widely used redox cycler to stimulate superoxide production. Heat stress applied to either 3 or 17 d old Paatg1Δ isolates results in the same growth retardation as with wild-type isolates of the same age (Fig. S3A and S3B). In contrast, after paraquat treatment, 3 d as well as 17 d old Paatg1Δ isolates displayed a significantly increased sensitivity against higher paraquat concentrations (from 200 µM on in 3 d old cultures and from 250 µM on in 17 d old cultures) compared with the corresponding wild-type isolates (Fig. S3C and S3D). This increased sensitivity against oxidative stress is not unexpected, since autophagy is important for the clearance of damaged proteins.
The increase of autophagosomes in old P. anserina cultures suggests a role of autophagy in aging. In order to investigate this possibility further, we analyzed the impact of a Paatg1 deletion on life span. We found that the genetic disruption of the autophagy pathway leads to a significant decrease in life span even under noninduced conditions, which is an indication for the important role of selective autophagy in P. anserina (Fig. 3E). Moreover, the absence of autophagy and nitrogen limitation results in a slight upregulation of PaPRE3 (proteasome core particle subunit β1) (Fig. 3F), suggesting that the proteasome-dependent protein degradation acts as an alternative, compensatory mechanism in the mutant.
Recently, a link between autophagy and the glyoxalase system was reported.44 This pathway is involved in the detoxification of methylglyoxal, a byproduct of glycolysis which modifies proteins, lipids, and DNA. Therefore, in order to test whether failure to undergo autophagy leads to impairments in the degradation of damaged proteins, the glyoxalase I (PaGLO1) abundance was determined. Indeed, at the protein level, we found a slight upregulation in the abundance of PaGLO1 in the deletion mutant (Fig. 3F).
Since nitrogen starvation is known to induce autophagy, we next tested the impact of nitrogen starvation on the life span of the P. anserina wild-type and the Paatg1∆ strains. We found that both strains are characterized by an increased life span under nitrogen-depleted conditions. Remarkedly, this increase is much more pronounced in the wild-type background. The median life span in the Paatg1∆ strain was 21 d on M2 (Fig. 3E) and 25 d on M2-N (Fig. 3G) compared with a median life span of WT 25 d (Fig. 3E) on M2, and 95 d on M2-N (Fig. 3G). The growth rate of the deletion mutant under nitrogen depletion is slightly decreased compared with the wild type (Fig. 3H). The life span of WT is nearly 4-fold increased under nitrogen starvation, suggesting that the induction of autophagy indeed is highly beneficial for P. anserina and providing additional data supporting a longevity assurance function of autophagy.
Finally we investigated the effect of rapamycin on life span of P. anserina. This immunosuppressant drug is known to selectively inhibit the serine/threonine kinase target of rapamycin (TOR) and thereby stimulating autophagy in a variety of organisms comparable to starvation.45-48 First, we tested the effect of the drug on the growth rate of the fungus (Fig. S4A). Addition of rapamycin to the growth medium resulted in a dose-dependent reduction in growth rate indicating that the drug is taken up by the hyphae from the medium. Next, we determined the median life span on media containing different amounts of rapamycin. A clear life span increasing effect was observed in medium containing 10 and 20 ng/ml rapamycin (Fig. S4B). Surprisingly, lack of PaATG1 does not prevent life-span extension by rapamycin demonstrating that this effect is not autophagy-dependent (Fig. S4C).
Discussion
Autophagy is a cellular degradation and recycling process that is highly conserved in eukaryotes. It is strictly regulated and affects various processes and pathways including the adaptation to starvation, turnover of damaged organelles, immunity and modulation of host defense, cell growth, aging, and development.49-58 Depending on the cellular conditions, excessive autophagy may lead to a special kind of programmed cell death, referred to as the “autophagic cell death.”59,60 However, this term describes cell death accompanied by autophagy, not necessarily the execution of cell death by autophagy.61 Overall experimental data rather support the prosurvival role of autophagy, since it has only very rarely been shown that downregulation of autophagy genes reduces cell death, while a huge number of studies describe an acceleration of cell death upon genetic suppression of autophagy.61
In filamentous fungi it has been shown that the deletion of basic autophagy related genes like Atg1, Atg4, or Atg8 affect morphogenesis and development leading to a decrease in the formation of aerial hyphae, impaired conidiation, delayed spore germination, and defects in the formation of sexual organs and fruiting bodies.33,34,62-65 Moreover, SmATG7 involved in the ATG8 and ATG12 conjugation pathway as well as the autophagic kinases SmVPS34 and SmVPS15 are required for viability in Sordaria macrospora.66,67 The results of our current study with P. anserina, which identified impairments in female fertility, spore differentiation, and germination in an atg1 deletion strain are consistent with these earlier findings and provide additional evidence for an essential role of autophagy in fungal development.68
With its activity to degrade damaged molecules and impaired organelles, autophagy is a component of a network of pathways involved in quality control.8,69 In the past, we have demonstrated a number of different pathways to be part of such a network to affect aging and life span control in P. anserina (reviewed in refs. 8, 69, and 70). Until now, we had no experimental evidence for such an impact of autophagy. The microscopic identification of increased autophagosome numbers in senescent cultures and the biochemical demonstration of an increased degradation of GFP-labeled SOD1 during aging, reported in the current study, provides the first evidence. Here, we report for the first time, an impact of impaired autophagy on aging and life span of the fungal aging model P. anserina. This role is supported by the observation that the life span of the Paatg1 deletion strain is not increased under nitrogen starvation to the same extent as in the wild type. Similarly, it is reported that the inactivation of different autophagy relevant genes like unc-51, bec-1, or Igg-1 (Atg1, Beclin1, Lc3 in mammals) lead to a reduced life span in C. elegans.48,71-75
The effects on aging, observed after perturbation of the autophagy machinery, can partially be explained by impairments in the degradation of damaged cellular components. However, a known “crosstalk” with the ubiquitin-proteasome system complicates the situation. While macroautophagy and the ubiquitin-proteasome system were formerly regarded as 2 separate cellular pathways with distinct functions, it is now clear that a number of proteins can be degraded by both autophagy and the proteasome.76-79 Recent work suggests that the 2 systems “communicate” with each other under certain circumstances.80-83 Different studies with yeast, Drosophila, and mammalian cells report that proteasome inhibition leads to the upregulation of autophagy.83-90 Moreover, the observed increase in glyoxalase 1 (lactoylglutathione lyase, PaGLO1) is indicative for increased molecular stress due to a lack of autophagy, identifying a link of this cellular protection pathway to autophagy. In P. anserina, the glyoxalase system is relevant for aging.91 In mammals, Glo1 is transcriptionally controlled by NRF2 (also known as NFE2L2) and represents a stress-responsive system that protects proteins and DNA from damage.92
Another relevant link is the interaction of autophagy to the activity of TOR, a central regulator of multiple cellular responses (reviewed in ref. 93). TOR inhibits autophagy via phosphorylation of ATG13 and is itself inactivated by low amino acid levels. In yeast it has been previously described that autophagy deficiency leads to the reduction of the total free amino acid pool and subsequent impaired protein synthesis under nutrient starvation conditions.94 Thus, autophagy deficiency can lead to TOR inactivation and thereby results in numerous pleiotropic effects. In previous studies, a life-span-prolonging effect originating from TOR inactivation via rapamycin is reported in different organisms.95-98 However, only in a few cases, this increase is unequivocally correlated with Atg genes.99,100 Surprisingly, in P. anserina the life-span-prolonging effect of rapamycin treatment of the wild type turns out to be autophagy-independent and the result of other rapamycin or TOR-influenced pathways or cellular functions, such as cell growth and proliferation, ribosome biogenesis, transcription, mRNA translation, cytoskeletal reorganization, and others.101
In our current study, we observed an induction of autophagy to occur during aging of P. anserina that is supported by a recent longitudinal genome-wide transcriptome analysis in which transcripts of autophagy-related genes have been found to increase in abundance during aging.102 This was a rather unexpected finding. In other systems like rats, mice, and human fibroblasts, a decrease in autophagy is reported.52,103-108 In contrast, in aged primary fibroblasts and in aged mouse brain, macroautophagy is increased.109 The reasons for these seemingly contradictory findings are not clearly understood but may reflect species-, tissue- and cell type-specific differences in the age-related regulation of autophagy. However, it appears to be the consensus that upregulation or maintenance of autophagy until advanced age is beneficial for cells and whole organisms by improving quality control and mitigating accumulation of altered cellular components within the aging cell.110-112 Keeping a high activity of autophagy and of other quality control pathways at very old age may, however, be impaired after passing a critical threshold of cellular damage. This scenario is supported by the observation that—unlike in mammalian systems—nitrogen limitation is not able to increase formation of autophagosomes at old age (Fig. 1B), suggesting that the capacity of the autophagy machinery is at its limit in old P. anserina cultures. Consequently, the system finally dies.
In conclusion, with the development of appropriate tools and the demonstration of a clear impact of autophagy as a longevity-assurance mechanism, as well as the earlier demonstration of other quality control pathways to be effective in aging and life-span control in P. anserina, we are now in the position to specifically address important issues with the aim to integrate autophagy into the network of quality control pathways in this organism. One question we are currently addressing is the potential contribution of mitophagy as part of the longevity-assurance mechanisms active in P. anserina. An impact of this type of selective autophagy is likely since this aging model is characterized by a strong mitochondrial etiology of aging.113,114 Another point of special interest is to elucidate the regulation of autophagy as a potential compensatory mechanism in situations when other pathways fail. Such mechanisms may explain unexpected and counterintuitive experimental data as they are found in the literature and may unravel yet unknown regulatory circuits in which autophagy and other pathways play a key role.
Materials and Methods
Cloning procedures and generation of Podospora anserina mutants
The generation of the Gfp-PaAtg8 strain was performed by a 4-fragment ligation using a Gfp-containing plasmid (pSM4). The plasmid pSM4 is based on pSM2115 which contains an eGfp gene without promoter region. Restriction of the pSM4-plasmid with NcoI (Thermo Scientific, ER0571) and NotI (Thermo Scientific, ER0591) leads to the removal of the eGfp. The PaAtg8 own promoter region (1052 bps) was amplified by PCR from a cosmid carrying the PaAtg8 genomic region116 using oligonucleotides Atg8-1 (5′-GGAAGCTTTT CGGTACTTGG GATCAG-3′, Biomers, Ulm, Germany) with restriction site for HindIII (Thermo Scientific, ER0501) and Atg8-2 (5′-TTGCCGGCGG GTTGGTTGTT GCT-3′, Biomers) with restriction site for EheI (Thermo Scientific, ER0441). Restriction sites were underlined. Together with the Gfp gene, the PaAtg8 promoter region was then ligated with the pKO6 plasmid. This plasmid contains the phleomycin resistance cassette of pKO412,22 restricted with EcoRI and Eco72I (Thermo Scientific, ER0361) ligated into EcoRI/EcoRV (Thermo Scientific, ER0271/ER0301) digested pBSSK (Agilent Technologies, 212205). Important for the creation of the Gfp-PaAtg8 plasmid is the unusual N-terminal fusion of the Gfp gene to the PaAtg8 gene, preventing proteolytical removal of Gfp by processing through the protease ATG4 during autophagy induction. Therefore Gfp had to be ligated in front of the PaAtg8 gene but behind the PaAtg8 own promoter. The open reading frame of PaAtg8 with the PaAtg8 terminator region was amplified by PCR from the cosmid using oligonucleotides Atg8-4 (5′-GGCTCGAGAA GCAATGAGGA ACAAGAGG-3′, Biomers) with restriction site for XhoI (Thermo Scientific, ER0691) (underlined) and phosphorylated Atg8-5 (5′-GGGATGAGAT CCAAGTTTA-3′, Biomers). These 2 fragments were cloned in the vector backbone of plasmid pKO6 (SalI [Thermo Scientific, ER0641], HindIII), which carries an ampicillin-resistance gene for selection in Escherichia coli and a phleomycin resistance gene for selection in P. anserina. Ligation of these fragments resulted in a plasmid called pAtg8eGfp2 which was finally used to transform P. anserina wild-type spheroplasts according to Osiewacz et al.11 and Stumpferl et al.117 Transformants were selected on phleomycin resistance (Ble) and used for fluorescence microscopy to analyze the localization of the GFP-PaATG8 fusion protein in hyphae.
The PaSod1-Gfp, PaSod2-Gfp, and PaSod3-Gfp strains used in this study for different western blot analysis are previously described by Zintel et al.20
The generation of a Paatg1 deletion strain was performed as described previously12 based on a method originally described by Chaveroche et al.118 Small flanking regions of the PaAtg1 gene were amplified using the 5′-flank oligonucleotides Paatg1-KO1-1 (5′-CCGGTACCCC ACTTTCCTACA CCACCC-3′, Biomers) and Paatg1-KO1-2 (5′-CCCTGCAGGG CTACCTGCTG ATGTTGG-3′, Biomers), introducing KpnI (Thermo Scientific, ER0521) and PstI (Thermo Scientific, ER0611) restriction sites (underlined), and the 3′-flank oligonucleotides Paatg1-KO1-3 (5′-GGACTAGTCA AAGCTGACGA TTAACG-3′, Biomers) and Paatg1-KO1-4 (5′-GGGCGGCCGCA AAAAGAAAAA CGCGC-3′, Biomers), introducing NotI and BcuI (Thermo Scientific, ER1251) restriction sites (underlined). The 5′ fragment was digested with KpnI and PstI and the 3′ fragment was digested with BcuI and NotI. Meanwhile, deletion plasmid pKO412,22 was digested with all 4 restriction enzymes (KpnI, PstI, BcuI, and NotI) and a 4-fragment ligation was performed, resulting in the plasmid patg1KO1, containing both fragments flanking a phleomycin and blasticidin resistance cassette for fungal and bacterial selection. The resistance cassette with the flanking regions was excised by restriction with NotI and KpnI and transformed into the E. coli KS272 strain, containing the plasmid pKOBEG,118 and a cosmid bearing the PaAtg1 gene.116 The homologous recombination between the flanks of the resistance cassette and the cosmid causes the generation of a Paatg1Δ cosmid, containing the phleomycin-blasticidin cassette with large flanking genomic regions. Subsequently, the Paatg1Δ cosmid was transformed into the P. anserina wild-type spheroplasts. Selection of received transformants, were realized by growth on phleomycin (Genaxxon, M3429) containing medium. Successful deletion of PaAtg1 was indicated by phleomycin resistance accompanied by hygromycin (Calbiochem, 400051) sensitivity. The correct replacement of the PaAtg1 gene was verified by Southern blot analysis using a PaAtg1-specific probe and a phleomycin-specific probe (Fig. S2A). Positive strains, lacking the PaAtg1 gene and possessing the phleomycin gene instead, were finally termed Paatg1Δ.
Transformation of P. anserina spheroplasts
Production, regeneration, and the integrative transformation of P. anserina spheroplasts were performed as previously described.11,117
P. anserina strains and cultivation
In the present study the P. anserina wild-type strain “s”,9 the newly generated Paatg1-deletion (Paatg1Δ) and the Gfp-PaAtg8 strain were used as well as the Gfp strains PaSod1-Gfp, PaSod2-Gfp, and PaSod3-Gfp20 and the newly generated PaSod1-Gfp Paatg1Δ double mutant. All strains were constructed in the genetic background of the wild-type strain “s”. Strains were grown on standard cornmeal agar (BMM) at 27 °C under constant light conditions.10 For spore germination, the BMM medium was added with 60 mM ammonium acetate and the spores incubated at 27 °C in the dark for 2 d. All used strains originated from monokaryotic ascospores isolated from irregular asci.
Fertility and germination analysis
Female fertility was assessed as previously described.25 In principle, freshly isolated monokaryotic wild-type or Paatg1Δ isolates of both mating types were allowed to overgrow the surface of M2 agar (M2 medium: 0.25 g/L KH2PO4, 0.3 g/L K2HPO4, 0.25 g/L MgSO4 × 7 H2O, 0.5 g/L urea and 10 g/L yellow dextrin. Addition of 2.5 mg/L biotin, 50 mg/L thiamine, 5 mg/L citric acid × 1 H2O, 5 mg/L ZnSO4 × 7 H2O, 1 mg/L Fe(NH4)2 (SO4)2 × 6 H2O, 2.5 mg/L CuSO4 × 5 H2O, 25 mg/L MnSO4 × 1 H2O, 50 mg/L Na2MoO4 × 2 H2O and 50 mg/L H3BO3 after sterilization of the basal medium) plates at 27 °C under constant light. Spermatia (corresponding to male gametes) of these strains with the opposite mating type were harvested by flooding the plates with 5 ml of sterile water. The suspension was diluted 1:1 with sterile H2O and drops of 100 μl were pipetted onto mycelium of each wild type and Paatg1Δ. After 5 min, the drops were removed carefully and the strains were incubated for 4 d at 27 °C under constant light. The number of perithecia was counted and the resulting values were divided by the area of the drop. The number of perithecia developing on plates overgrown with the wild-type “s” were set to 100% fertility.
To determine the germination rate of spores from wild-type (s− × s+) crosses and wild-type × Paatg1Δ crosses (female sterility of the Paatg1Δ strain allowed only fertilization of Paatg1Δ− × s+, respectively Paatg1Δ+ × s−) monokaryotic spores were isolated from irregular asci. After 2 d incubation on germination medium (BMM medium added with 60 mM ammonium acetate) at 27 °C in the dark, the number of germinated spores was counted.
Southern blot analysis
Isolation of total DNA of P. anserina was realized by the protocol developed by Lecellier and Silar for rapid extraction of nucleic acids from filamentous fungi.119 DNA digestion, gel electrophoresis, and Southern blotting were performed according to standard protocols. For Southern blot hybridization and detection, digoxigenin-labeled hybridization probes (DIG DNA Labeling and Detection Kit, Roche Applied Science, 11175033910) were used according to the manufacturer’s protocol. The PaAtg1-specific hybridization probe was amplified by PCR using the oligonucleotides Atg1-3 (5′-TCAGCCATCT ACTTCAGC-3′, Biomers) and Atg1-4 (5′-TGCTTGTACT TGACCTCG-3′, Biomers). The PaAtg8-specific hybridization probe was amplified by PCR using the oligonucleotides Atg8-for (5′-CCCGCCAAAA TGAGATCC-3′, Biomers) and Atg8-rev (5′-CCAAAGGTGT TCTCGCCC-3′, Biomers) and the respective cosmids as template. As a hybridization probe specific for the phleomycin resistance gene (Ble), the 1293 bp BamHI-fragment (ER0051, Thermo Scientific) of the plasmid pKO4 was used. The 737 bp BcuI-/NcoI-fragment of the plasmid pSM4 was used as a hybridization probe specific for the Gfp gene.
Growth rate and life-span determination
Life span and growth rate of monokaryotic isolates on M2 medium were determined as described.120 The life span of P. anserina is defined as the time period (d) of linear hyphal growth whereas the growth rate is defined as the measured growth (cm) per time period (d). To determine the life span and growth rate under nitrogen starvation, the monokaryotic isolates were placed on race tubes containing M2 medium lacking the nitrogen source urea (M2-N). The determination of growth rates under stress conditions was performed on M2 medium added with different concentrations (0, 10, 50, 100, 200 and 250 µM) of paraquat (Sigma-Aldrich, 856177) at 27 °C, by incubation of the strains under heat stress conditions (37 °C) at constant light or on M2 medium to which different concentrations (0, 2.5, 5, 10, and 20 ng/mL) of rapamycin (Sigma-Aldrich, R8781) is added.
Light sheet-based fluorescence microscopy (LSFM)
In order to image P. anserina with the LSFM, pieces of P. anserina cultures (Gfp-PaAtg8: 4 respectively 20 d old) were grown on a coverslip, immersed in M2 or M2-N medium, and incubated in a “wet chamber” for 1 d at 27 °C and under constant light. The “wet chamber” consists of a petri dish filled with sterile wet tissue paper. Imaging was performed with a light sheet based fluorescence microscope, in its implementation monolithic digitally scanned light sheet-based fluorescence microscope (mDSLM) developed in Ernst Stelzer’s laboratory (Frankfurt, Germany). It is an improved and compact version of the DSLM described in Keller et al.121 The specimen was prepared as following. First, a 5 mm × 5 mm glass coverslip was glued with nail polish onto the custom mDSLM specimen holder (Fig. S5). Shortly before imaging, a 5 mm × 5 mm square portion of mycelium was cut with a scalpel out of the agarose slab. The square portion was then deposited onto the glass coverslip. In order to ensure a stable adhesion between glass and specimen, a small quantity of liquid low-melting agarose was deposited on the glass coverslip. Finally, the mounted specimen was vertically inserted in the mDSLM chamber containing the M2 or M2-N medium. The fluorescence in the specimen was excited by a light sheet generated by scanning a thin (FWHM—full width at half maximum—of the beam waist: 2 μm) laser light beam. The light sheet illuminates the specimen from the side. In order to resolve individual autophagosomes in the hyphae, we employed a pair of CZ Plan-Neofluar 5×/0.16 objective lens (Carl Zeiss Microscopy, Jena, Germany) for the illumination (thickness of light sheet ~2 μm) and a CZ Plan-Apochromat 63× /1.0 W objective lens (Carl Zeiss Microscopy) for the detection. Green fluorescent protein (GFP) was excited at 488 nm by a diode laser and detected between 500 and 550 nm. Images were acquired by a CCD camera (pixel pitch 6.45 μm, Neo, Andor, Ireland). The specimen mounted on the mDSLM holder was placed vertically in the medium-filled chamber close to the common focal point of the 2 excitation and detection objective lenses. Image stacks consisting on average of 100 16-bit-TIFF (tagged image file format) individual images (slices) were recorded. Each slice in a stack represents a plane of the 3-dimensional specimen volume. The spacing between the slices was 0.4 µm. The image stacks were first deconvolved by employing the parallel iterative deconvolution plugin of the freeware image software Fiji (http://fiji.sc/Fiji), by choosing the WPL algorithm and performing 5 iterations. The absence of deconvolution artifacts was verified by visual inspection of the de-blurred stacks. The point-spread function (PSF) was calculated with the Fiji plugin diffraction PSF 3D by inserting the experimental imaging parameters. The deconvolved stacks were segmented by employing the manual threshold function of Fiji. The resulting segmented stacks were processed with the open 3D morphological plugin. Next, a z-projection of the stacks was generated by choosing the sum slices option. Finally, the total number of autophagosomes in the stack was determined from the z-projected stack with the analyze particles function. The area of each individual hypha was determined manually with the segmented line selection tool. Finally, the density of autophagosome was calculated for each image stack.
Wide-field fluorescence and differential interference contrast (DIC) imaging
A Carl Zeiss Axio Imager.Z2 upright microscope equipped with a CZ AxioCamMR3 (image size 1388 × 1040, pixel pitch 6.45 µm, 12 bits) (Carl Zeiss Microscopy) and the ZEN 2012 (blue edition) acquisition software was employed for imaging. A CZ oil immersion Plan-Apochromat 63× 1.4 N.A. DIC objective lens objective lens (Carl Zeiss Microscopy) was used for both wide-field fluorescence and DIC imaging. After properly setting the Köhler illumination, the matching DIC-Prisma II HC 63× (Carl Zeiss Microscopy, 426924-9030-000) as well as the DIC-analyzer were inserted. Both the DIC shift and azimuth were adjusted in order to achieve maximum contrast. The GFP fluorescence was excited with a Colibri.2 LED light source, equipped with a 474/28 band-pass excitation filter (Carl Zeiss Microscopy). The filter set HE BFP/GFP/HcRed (Carl Zeiss Microscopy, 489062-9901-000) was employed.
Western blot analysis
For extraction of total protein extracts, mycelia from different P. anserina strains was allowed to overgrow a cellophane foil covered M2 surface for 2 d at 27 °C and constant light. Afterwards, the grown mycelia was transferred into CM-liquid medium (CM medium: 1 g/L KH2PO4, 0.5 g/L KCL, 0.5 g/L MgSO4 × 7 H2O, 10 g/L glucose-monohydrate, 3.7 g/L NH4Cl, 2 g/L trypton, 2 g/L yeast-extract, 0.001 g/L ZnSO4, 0.001 g/L FeCl2, 0.001 g/L MnCl) (nitrogen re- or depleted) for 2 further d at 27 °C under constant light and shaking conditions. Harvested mycelia were pulverized in liquid nitrogen and the protein was isolated from the powder as described.91 100–300 μg total protein extracts of P. anserina strains, were fractionated by 2-phase SDS-PAGE (12% separating gels) according to the standard protocol.28 After electrophoresis, proteins were transferred to PVDF membranes (Millipore, IPFL00010). Blocking, antibody incubation, and washing steps were performed according to the Odyssey western blot analysis handbook (LIC-OR Biosciences, Bad Homburg, Germany). The following primary antibodies were used: Anti-GFP (mouse, 1:10000 dilution, Sigma-Aldrich, G6795), anti-PaGLO1 (1: 2000 dilution, NEP, Frankfurt, Germany) raised against a specific peptide ([AC]-CVQNERFADKANF-[OH]) of PaGLO1 (glyoxalase1) previously described in Scheckhuber et al.,91 anti-SOD1 (1:2000 dilution) from Biomol Stressgen (Hamburg, Germany) (SOD100) and Anti-PaPRE3 (1:2500 dilution) raised against a specific synthetic peptide ([H]-LYLPDTDYKVRHEN-[OH]; Sigma) of PaPRE3 (corresponding to the β1 subunit of the 26S proteasome). In all analyses, secondary antibodies conjugated with IRDye 680 (1:15000 dilution, goat anti-mouse 680RD: 926-68070; goat anti-rabbit 680LT: LIC-OR Biosciences, 926-68021) or IRDye CW 800 (1:15000 dilution, goat anti-mouse 800: 926-32210; goat anti-rabbit 800: LIC-OR Biosciences, 926-3221) were used. The Odyssey infrared scanner (LIC-OR Biosciences) was used for detection and quantification using the manufacturer’s software.
Statistical analysis
For statistical analyses of life span, growth rate, and female fertility as well as germination rate 2-tailed Mann-Whitney-Wilcoxon U test was used. The respective samples were compared with the appropriate wild-type sample. For all analyses the minimum level of statistical significance was set at P < 0.05 (not significant different means P > 0.05; significant different (*) means P < 0.05; high significant different (**) means P < 0.01; very high significant different (***) means P < 0.001).
Supplementary Material
Glossary
Abbreviations:
- Atg
autophagy-related gene
- Ble
phleomycin resistance gene
- BMM
Biomalt Maize Medium
- bp
base pairs
- CM
complete medium
- DIC
differential interference contrast
- GFP
green fluorescent protein
- PaGLO1
P. anserina glyoxalase 1
- Hyg
hygromycin-resistance gene
- kDa
kilodalton
- LC3
microtubule-associated protein 1A/1B-light chain 3
- mDSLM
monolithic digitally scanned light sheet-based fluorescence microscope
- PCR
polymerase chain reaction
- PE
phosphatidylethanolamine
- PaPRE3
P. anserina proteasome core particle subunit β1
- ROS
reactive oxygen species
- PaSOD
P. anserina superoxide dismutase
- TOR
target of rapamycin kinase
- wt
wild type
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank the Goethe University for financial support via the Frankfurt Autophagy Network (FAN) program. In addition, the work was funded by the German Federal Ministry for Education and Research (BMBF) through the GerontoMitoSys project (FKZ 0315584A). This paper is dedicated to Karl Esser (Bochum) on the occasion of his 90th birthday.
References
- 1.Harman D. . Aging: a theory based on free radical and radiation chemistry. J Gerontol 1956; 11:298 - 300; http://dx.doi.org/ 10.1093/geronj/11.3.298; PMID: 13332224 [DOI] [PubMed] [Google Scholar]
- 2.Shringarpure R, Davies KJ. . Protein turnover by the proteasome in aging and disease. Free Radic Biol Med 2002; 32:1084 - 9; http://dx.doi.org/ 10.1016/S0891-5849(02)00824-9; PMID: 12031893 [DOI] [PubMed] [Google Scholar]
- 3.Krtolica A, Campisi J. . Cancer and aging: a model for the cancer promoting effects of the aging stroma. Int J Biochem Cell Biol 2002; 34:1401 - 14; http://dx.doi.org/ 10.1016/S1357-2725(02)00053-5; PMID: 12200035 [DOI] [PubMed] [Google Scholar]
- 4.Finkel T, Holbrook NJ. . Oxidants, oxidative stress and the biology of ageing. Nature 2000; 408:239 - 47; http://dx.doi.org/ 10.1038/35041687; PMID: 11089981 [DOI] [PubMed] [Google Scholar]
- 5.Mandavilli BS, Santos JH, Van Houten B. . Mitochondrial DNA repair and aging. Mutat Res 2002; 509:127 - 51; http://dx.doi.org/ 10.1016/S0027-5107(02)00220-8; PMID: 12427535 [DOI] [PubMed] [Google Scholar]
- 6.Beckman KB, Ames BN. . The free radical theory of aging matures. Physiol Rev 1998; 78:547 - 81; PMID: 9562038 [DOI] [PubMed] [Google Scholar]
- 7.Weinert BT, Timiras PS. . Invited review: Theories of aging. J Appl Physiol (1985) 2003; 95:1706 - 16; PMID: 12970376 [DOI] [PubMed] [Google Scholar]
- 8.Fischer F, Hamann A, Osiewacz HD. . Mitochondrial quality control: an integrated network of pathways. Trends Biochem Sci 2012; 37:284 - 92; http://dx.doi.org/ 10.1016/j.tibs.2012.02.004; PMID: 22410198 [DOI] [PubMed] [Google Scholar]
- 9.Rizet G. . is Impossibility of obtaining uninterrupted and unlimited multiplication of the ascomycete Podospora anserina. (in English).. C R Hebd Seances Acad Sci 1953; 237:838 - 40; PMID: 13107134 [PubMed] [Google Scholar]
- 10.Esser K. Podospora anserina. In: King RC, ed. Handbook of Genetics. New York: Plenum Press, 1974; 531-551. [Google Scholar]
- 11.Osiewacz HD, Skaletz A, Esser K. . Integrative transformation of the ascomycete Podospora anserina: identification of the mating-type locus on chromosome VII of electrophoretically separated chromosomes. Appl Microbiol Biotechnol 1991; 35:38 - 45; http://dx.doi.org/ 10.1007/BF00180633; PMID: 1367277 [DOI] [PubMed] [Google Scholar]
- 12.Hamann A, Krause K, Werner A, Osiewacz HD. . A two-step protocol for efficient deletion of genes in the filamentous ascomycete Podospora anserina.. Curr Genet 2005; 48:270 - 5; http://dx.doi.org/ 10.1007/s00294-005-0018-1; PMID: 16160832 [DOI] [PubMed] [Google Scholar]
- 13.Osiewacz HD, Hamann A, Zintel S. . Assessing organismal aging in the filamentous fungus Podospora anserina.. Methods Mol Biol 2013; 965:439 - 62; http://dx.doi.org/ 10.1007/978-1-62703-239-1_29; PMID: 23296676 [DOI] [PubMed] [Google Scholar]
- 14.Borghouts C, Kimpel E, Osiewacz HD. . Mitochondrial DNA rearrangements of Podospora anserina are under the control of the nuclear gene grisea. Proc Natl Acad Sci U S A 1997; 94:10768 - 73; http://dx.doi.org/ 10.1073/pnas.94.20.10768; PMID: 9380708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soerensen M, Gredilla R, Müller-Ohldach M, Werner A, Bohr VA, Osiewacz HD, Stevnsner T. . A potential impact of DNA repair on ageing and lifespan in the ageing model organism Podospora anserina: decrease in mitochondrial DNA repair activity during ageing. Mech Ageing Dev 2009; 130:487 - 96; http://dx.doi.org/ 10.1016/j.mad.2009.05.003; PMID: 19486911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kück U, Kappelhoff B, Esser K. . Despite mtDNA polymorphism the mobile intron (plDNA) of the COI gene is present in ten different races of Podospora anserina.. Curr Genet 1985; 10:59 - 67; http://dx.doi.org/ 10.1007/BF00418494 [DOI] [Google Scholar]
- 17.Borghouts C, Scheckhuber CQ, Stephan O, Osiewacz HD. . Copper homeostasis and aging in the fungal model system Podospora anserina: differential expression of PaCtr3 encoding a copper transporter. Int J Biochem Cell Biol 2002; 34:1355 - 71; http://dx.doi.org/ 10.1016/S1357-2725(02)00078-X; PMID: 12200031 [DOI] [PubMed] [Google Scholar]
- 18.Osiewacz HD, Nuber U. . GRISEA, a putative copper-activated transcription factor from Podospora anserina involved in differentiation and senescence. Mol Gen Genet 1996; 252:115 - 24; http://dx.doi.org/ 10.1007/BF02173211; PMID: 8804410 [DOI] [PubMed] [Google Scholar]
- 19.Borghouts C, Scheckhuber CQ, Werner A, Osiewacz HD. . Respiration, copper availability and SOD activity in P. anserina strains with different lifespan. Biogerontology 2002; 3:143 - 53; http://dx.doi.org/ 10.1023/A:1015696404723; PMID: 12075133 [DOI] [PubMed] [Google Scholar]
- 20.Zintel S, Schwitalla D, Luce K, Hamann A, Osiewacz HD. . Increasing mitochondrial superoxide dismutase abundance leads to impairments in protein quality control and ROS scavenging systems and to lifespan shortening. Exp Gerontol 2010; 45:525 - 32; http://dx.doi.org/ 10.1016/j.exger.2010.01.006; PMID: 20080171 [DOI] [PubMed] [Google Scholar]
- 21.Zintel S, Bernhardt D, Rogowska-Wrzesinska A, Osiewacz HD. . PaCATB, a secreted catalase protecting Podospora anserina against exogenous oxidative stress. Aging (Albany NY) 2011; 3:768 - 81; PMID: 21865610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luce K, Osiewacz HD. . Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat Cell Biol 2009; 11:852 - 8; http://dx.doi.org/ 10.1038/ncb1893; PMID: 19543272 [DOI] [PubMed] [Google Scholar]
- 23.Adam C, Picard M, Déquard-Chablat M, Sellem CH, Hermann-Le Denmat S, Contamine V. . Biological roles of the Podospora anserina mitochondrial Lon protease and the importance of its N-domain. PLoS One 2012; 7:e38138; http://dx.doi.org/ 10.1371/journal.pone.0038138; PMID: 22693589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer F, Weil A, Hamann A, Osiewacz HD. . Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain. Nat Commun 2013; 4:1397; http://dx.doi.org/ 10.1038/ncomms2397; PMID: 23360988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weil A, Luce K, Dröse S, Wittig I, Brandt U, Osiewacz HD. . Unmasking a temperature-dependent effect of the P. anserina i-AAA protease on aging and development. Cell Cycle 2011; 10:4280 - 90; http://dx.doi.org/ 10.4161/cc.10.24.18560; PMID: 22134244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamann A, Brust D, Osiewacz HD. . Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina.. Mol Microbiol 2007; 65:948 - 58; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05839.x; PMID: 17627766 [DOI] [PubMed] [Google Scholar]
- 27.Brust D, Daum B, Breunig C, Hamann A, Kühlbrandt W, Osiewacz HD. . Cyclophilin D links programmed cell death and organismal aging in Podospora anserina.. Aging Cell 2010; 9:761 - 75; http://dx.doi.org/ 10.1111/j.1474-9726.2010.00609.x; PMID: 20626725 [DOI] [PubMed] [Google Scholar]
- 28.Brust D, Hamann A, Osiewacz HD. . Deletion of PaAif2 and PaAmid2, two genes encoding mitochondrial AIF-like oxidoreductases of Podospora anserina, leads to increased stress tolerance and lifespan extension. Curr Genet 2010; 56:225 - 35; http://dx.doi.org/ 10.1007/s00294-010-0295-1; PMID: 20306265 [DOI] [PubMed] [Google Scholar]
- 29.Figge MT, Reichert AS, Meyer-Hermann M, Osiewacz HD. . Deceleration of fusion-fission cycles improves mitochondrial quality control during aging. PLoS Comput Biol 2012; 8:e1002576; http://dx.doi.org/ 10.1371/journal.pcbi.1002576; PMID: 22761564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Figge MT, Osiewacz HD, Reichert AS. . Quality control of mitochondria during aging: is there a good and a bad side of mitochondrial dynamics?. Bioessays 2013; 35:314 - 22; http://dx.doi.org/ 10.1002/bies.201200125; PMID: 23359437 [DOI] [PubMed] [Google Scholar]
- 31.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nyström T, Osiewacz HD. . Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol 2007; 9:99 - 105; http://dx.doi.org/ 10.1038/ncb1524; PMID: 17173038 [DOI] [PubMed] [Google Scholar]
- 32.Gredilla R, Grief J, Osiewacz HD. . Mitochondrial free radical generation and lifespan control in the fungal aging model Podospora anserina.. Exp Gerontol 2006; 41:439 - 47; http://dx.doi.org/ 10.1016/j.exger.2006.01.010; PMID: 16530367 [DOI] [PubMed] [Google Scholar]
- 33.Pinan-Lucarré B, Paoletti M, Dementhon K, Coulary-Salin B, Clavé C. . Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina.. Mol Microbiol 2003; 47:321 - 33; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03208.x; PMID: 12519185 [DOI] [PubMed] [Google Scholar]
- 34.Pinan-Lucarré B, Balguerie A, Clavé C. . Accelerated cell death in Podospora autophagy mutants. Eukaryot Cell 2005; 4:1765 - 74; http://dx.doi.org/ 10.1128/EC.4.11.1765-1774.2005; PMID: 16278443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinan-Lucarré B, Paoletti M, Clavé C. . Cell death by incompatibility in the fungus Podospora.. Semin Cancer Biol 2007; 17:101 - 11; http://dx.doi.org/ 10.1016/j.semcancer.2006.11.009; PMID: 17204431 [DOI] [PubMed] [Google Scholar]
- 36.Tsukada M, Ohsumi Y. . Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae.. FEBS Lett 1993; 333:169 - 74; http://dx.doi.org/ 10.1016/0014-5793(93)80398-E; PMID: 8224160 [DOI] [PubMed] [Google Scholar]
- 37.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. . Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci U S A 1996; 93:12304 - 8; http://dx.doi.org/ 10.1073/pnas.93.22.12304; PMID: 8901576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klionsky DJ, Emr SD. . Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717 - 21; http://dx.doi.org/ 10.1126/science.290.5497.1717; PMID: 11099404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Huang WP, Stromhaug PE, Klionsky DJ. . Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 2002; 277:763 - 73; http://dx.doi.org/ 10.1074/jbc.M109134200; PMID: 11675395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meijer AJ. . Amino acid regulation of autophagosome formation. Methods Mol Biol 2008; 445:89 - 109; http://dx.doi.org/ 10.1007/978-1-59745-157-4_5; PMID: 18425444 [DOI] [PubMed] [Google Scholar]
- 41.Pinar M, Pantazopoulou A, Peñalva MA. . Live-cell imaging of Aspergillus nidulans autophagy: RAB1 dependence, Golgi independence and ER involvement. Autophagy 2013; 9:1024 - 43; http://dx.doi.org/ 10.4161/auto.24483; PMID: 23722157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanki T, Klionsky DJ. . Mitophagy in yeast occurs through a selective mechanism. J Biol Chem 2008; 283:32386 - 93; http://dx.doi.org/ 10.1074/jbc.M802403200; PMID: 18818209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanki T, Kang D, Klionsky DJ. . Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy 2009; 5:1186 - 9; http://dx.doi.org/ 10.4161/auto.5.8.9854; PMID: 19806021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H, Yu S, Zhang H, Xu J. . Angiogenesis impairment in diabetes: role of methylglyoxal-induced receptor for advanced glycation endproducts, autophagy and vascular endothelial growth factor receptor 2. PLoS One 2012; 7:e46720; http://dx.doi.org/ 10.1371/journal.pone.0046720; PMID: 23056421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. . Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 1992; 119:301 - 11; http://dx.doi.org/ 10.1083/jcb.119.2.301; PMID: 1400575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda T, Ohsumi Y. . Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998; 273:3963 - 6; http://dx.doi.org/ 10.1074/jbc.273.7.3963; PMID: 9461583 [DOI] [PubMed] [Google Scholar]
- 47.Scott RC, Schuldiner O, Neufeld TP. . Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 2004; 7:167 - 78; http://dx.doi.org/ 10.1016/j.devcel.2004.07.009; PMID: 15296714 [DOI] [PubMed] [Google Scholar]
- 48.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. . A role for autophagy in the extension of lifespan by dietary restriction in C. elegans.. PLoS Genet 2008; 4:e24; http://dx.doi.org/ 10.1371/journal.pgen.0040024; PMID: 18282106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogov VV, Suzuki H, Fiskin E, Wild P, Kniss A, Rozenknop A, Kato R, Kawasaki M, McEwan DG, Löhr F, et al. . Structural basis for phosphorylation-triggered autophagic clearance of Salmonella.. Biochem J 2013; 454:459 - 66; http://dx.doi.org/ 10.1042/BJ20121907; PMID: 23805866 [DOI] [PubMed] [Google Scholar]
- 50.Kraft C, Deplazes A, Sohrmann M, Peter M. . Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 2008; 10:602 - 10; http://dx.doi.org/ 10.1038/ncb1723; PMID: 18391941 [DOI] [PubMed] [Google Scholar]
- 51.van der Klei IJ, Yurimoto H, Sakai Y, Veenhuis M. . The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim Biophys Acta 2006; 1763:1453 - 62; http://dx.doi.org/ 10.1016/j.bbamcr.2006.07.016; PMID: 17023065 [DOI] [PubMed] [Google Scholar]
- 52.Cuervo AM, Bergamini E, Brunk UT, Dröge W, Ffrench M, Terman A. . Autophagy and aging: the importance of maintaining “clean” cells. Autophagy 2005; 1:131 - 40; http://dx.doi.org/ 10.4161/auto.1.3.2017; PMID: 16874025 [DOI] [PubMed] [Google Scholar]
- 53.Vellai T. . Autophagy genes and ageing. Cell Death Differ 2009; 16:94 - 102; http://dx.doi.org/ 10.1038/cdd.2008.126; PMID: 19079287 [DOI] [PubMed] [Google Scholar]
- 54.Rubinsztein DC, Mariño G, Kroemer G. . Autophagy and aging. Cell 2011; 146:682 - 95; http://dx.doi.org/ 10.1016/j.cell.2011.07.030; PMID: 21884931 [DOI] [PubMed] [Google Scholar]
- 55.Levine B. . Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005; 120:159 - 62; PMID: 15680321 [DOI] [PubMed] [Google Scholar]
- 56.Schmid D, Münz C. . Innate and adaptive immunity through autophagy. Immunity 2007; 27:11 - 21; http://dx.doi.org/ 10.1016/j.immuni.2007.07.004; PMID: 17663981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levine B, Kroemer G. . Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/ 10.1016/j.cell.2007.12.018; PMID: 18191218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mizushima N, Levine B. . Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823 - 30; http://dx.doi.org/ 10.1038/ncb0910-823; PMID: 20811354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamann A, Brust D, Osiewacz HD. . Apoptosis pathways in fungal growth, development and ageing. Trends Microbiol 2008; 16:276 - 83; http://dx.doi.org/ 10.1016/j.tim.2008.03.003; PMID: 18440231 [DOI] [PubMed] [Google Scholar]
- 60.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. . Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8:741 - 52; http://dx.doi.org/ 10.1038/nrm2239; PMID: 17717517 [DOI] [PubMed] [Google Scholar]
- 61.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, et al. , Nomenclature Committee on Cell Death 2009. . Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 2009; 16:3 - 11; http://dx.doi.org/ 10.1038/cdd.2008.150; PMID: 18846107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu XH, Lu JP, Zhang L, Dong B, Min H, Lin FC. . Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell 2007; 6:997 - 1005; http://dx.doi.org/ 10.1128/EC.00011-07; PMID: 17416896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kikuma T, Ohneda M, Arioka M, Kitamoto K. . Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae.. Eukaryot Cell 2006; 5:1328 - 36; http://dx.doi.org/ 10.1128/EC.00024-06; PMID: 16896216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M, Rhodes JC, Askew DS. . Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus.. Eukaryot Cell 2007; 6:2437 - 47; http://dx.doi.org/ 10.1128/EC.00224-07; PMID: 17921348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voigt O, Pöggeler S. . Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora.. Autophagy 2013; 9:33 - 49; http://dx.doi.org/ 10.4161/auto.22398; PMID: 23064313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nolting N, Bernhards Y, Pöggeler S. . SmATG7 is required for viability in the homothallic ascomycete Sordaria macrospora.. Fungal Genet Biol 2009; 46:531 - 42; http://dx.doi.org/ 10.1016/j.fgb.2009.03.008; PMID: 19351563 [DOI] [PubMed] [Google Scholar]
- 67.Voigt O, Herzog B, Jakobshagen A, Pöggeler S. . Autophagic kinases SmVPS34 and SmVPS15 are required for viability in the filamentous ascomycete Sordaria macrospora. Microbiol Res 2014; 169:128 - 38; http://dx.doi.org/ 10.1016/j.micres.2013.07.012; PMID: 23953726 [DOI] [PubMed] [Google Scholar]
- 68.Pollack JK, Harris SD, Marten MR. . Autophagy in filamentous fungi. Fungal Genet Biol 2009; 46:1 - 8; http://dx.doi.org/ 10.1016/j.fgb.2008.10.010; PMID: 19010432 [DOI] [PubMed] [Google Scholar]
- 69.Luce K, Weil AC, Osiewacz HD. Mitochondrial protein quality control systems in aging and disease. In: Tavernarakis N, ed. Protein metabolism and homeostasis in aging. New York: Springer Science+Buisiness Media, LLC Landes Bioscience, 2010; 108-125 [DOI] [PubMed] [Google Scholar]
- 70.Osiewacz HD, Bernhardt D. . Mitochondrial quality control: impact on aging and life span - a mini-review. Gerontology 2013; 59:413 - 20; http://dx.doi.org/ 10.1159/000348662; PMID: 23615432 [DOI] [PubMed] [Google Scholar]
- 71.Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. . Autophagy genes are essential for dauer development and life-span extension in C. elegans.. Science 2003; 301:1387 - 91; http://dx.doi.org/ 10.1126/science.1087782; PMID: 12958363 [DOI] [PubMed] [Google Scholar]
- 72.Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. . Autophagy regulates ageing in C. elegans.. Autophagy 2007; 3:93 - 5; PMID: 17204841 [DOI] [PubMed] [Google Scholar]
- 73.Jia K, Levine B. . Autophagy is required for dietary restriction-mediated life span extension in C. elegans.. Autophagy 2007; 3:597 - 9; PMID: 17912023 [DOI] [PubMed] [Google Scholar]
- 74.Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. . The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy 2008; 4:870 - 3; PMID: 18728385 [DOI] [PubMed] [Google Scholar]
- 75.Tóth ML, Sigmond T, Borsos E, Barna J, Erdélyi P, Takács-Vellai K, Orosz L, Kovács AL, Csikós G, Sass M, et al. . Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans.. Autophagy 2008; 4:330 - 8; PMID: 18219227 [DOI] [PubMed] [Google Scholar]
- 76.Cuervo AM, Hu W, Lim B, Dice JF. . IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell 1998; 9:1995 - 2010; http://dx.doi.org/ 10.1091/mbc.9.8.1995; PMID: 9693362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De Domenico I, Vaughn MB, Li L, Bagley D, Musci G, Ward DM, Kaplan J. . Ferroportin-mediated mobilization of ferritin iron precedes ferritin degradation by the proteasome. EMBO J 2006; 25:5396 - 404; http://dx.doi.org/ 10.1038/sj.emboj.7601409; PMID: 17082767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kidane TZ, Sauble E, Linder MC. . Release of iron from ferritin requires lysosomal activity. Am J Physiol Cell Physiol 2006; 291:C445 - 55; http://dx.doi.org/ 10.1152/ajpcell.00505.2005; PMID: 16611735 [DOI] [PubMed] [Google Scholar]
- 79.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. . Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 2003; 278:25009 - 13; http://dx.doi.org/ 10.1074/jbc.M300227200; PMID: 12719433 [DOI] [PubMed] [Google Scholar]
- 80.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. . Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383 - 435; http://dx.doi.org/ 10.1152/physrev.00030.2009; PMID: 20959619 [DOI] [PubMed] [Google Scholar]
- 81.Park C, Cuervo AM. . Selective autophagy: talking with the UPS. Cell Biochem Biophys 2013; 67:3 - 13; http://dx.doi.org/ 10.1007/s12013-013-9623-7; PMID: 23709310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Korolchuk VI, Menzies FM, Rubinsztein DC. . A novel link between autophagy and the ubiquitin-proteasome system. Autophagy 2009; 5:862 - 3; PMID: 19458478 [DOI] [PubMed] [Google Scholar]
- 83.Korolchuk VI, Menzies FM, Rubinsztein DC. . Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 2010; 584:1393 - 8; http://dx.doi.org/ 10.1016/j.febslet.2009.12.047; PMID: 20040365 [DOI] [PubMed] [Google Scholar]
- 84.Chen Q, Ding Q, Thorpe J, Dohmen RJ, Keller JN. . RNA interference toward UMP1 induces proteasome inhibition in Saccharomyces cerevisiae: evidence for protein oxidation and autophagic cell death. Free Radic Biol Med 2005; 38:226 - 34; http://dx.doi.org/ 10.1016/j.freeradbiomed.2004.10.019; PMID: 15607905 [DOI] [PubMed] [Google Scholar]
- 85.Ding Q, Dimayuga E, Martin S, Bruce-Keller AJ, Nukala V, Cuervo AM, Keller JN. . Characterization of chronic low-level proteasome inhibition on neural homeostasis. J Neurochem 2003; 86:489 - 97; http://dx.doi.org/ 10.1046/j.1471-4159.2003.01885.x; PMID: 12871590 [DOI] [PubMed] [Google Scholar]
- 86.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. . Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 2007; 171:513 - 24; http://dx.doi.org/ 10.2353/ajpath.2007.070188; PMID: 17620365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iwata A, Riley BE, Johnston JA, Kopito RR. . HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 2005; 280:40282 - 92; http://dx.doi.org/ 10.1074/jbc.M508786200; PMID: 16192271 [DOI] [PubMed] [Google Scholar]
- 88.Rideout HJ, Lang-Rollin I, Stefanis L. . Involvement of macroautophagy in the dissolution of neuronal inclusions. Int J Biochem Cell Biol 2004; 36:2551 - 62; http://dx.doi.org/ 10.1016/j.biocel.2004.05.008; PMID: 15325592 [DOI] [PubMed] [Google Scholar]
- 89.Pan T, Kondo S, Zhu W, Xie W, Jankovic J, Le W. . Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol Dis 2008; 32:16 - 25; http://dx.doi.org/ 10.1016/j.nbd.2008.06.003; PMID: 18640276 [DOI] [PubMed] [Google Scholar]
- 90.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. . HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007; 447:859 - 63; http://dx.doi.org/ 10.1038/nature05853; PMID: 17568747 [DOI] [PubMed] [Google Scholar]
- 91.Scheckhuber CQ, Mack SJ, Strobel I, Ricciardi F, Gispert S, Osiewacz HD. . Modulation of the glyoxalase system in the aging model Podospora anserina: effects on growth and lifespan. Aging (Albany NY) 2010; 2:969 - 80; PMID: 21212464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xue M, Rabbani N, Momiji H, Imbasi P, Anwar MM, Kitteringham N, Park BK, Souma T, Moriguchi T, Yamamoto M, et al. . Transcriptional control of glyoxalase 1 by Nrf2 provides a stress-responsive defence against dicarbonyl glycation. Biochem J 2012; 443:213 - 22; http://dx.doi.org/ 10.1042/BJ20111648; PMID: 22188542 [DOI] [PubMed] [Google Scholar]
- 93.Neufeld TP. . TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol 2010; 22:157 - 68; http://dx.doi.org/ 10.1016/j.ceb.2009.11.005; PMID: 20006481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Onodera J, Ohsumi Y. . Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 2005; 280:31582 - 6; http://dx.doi.org/ 10.1074/jbc.M506736200; PMID: 16027116 [DOI] [PubMed] [Google Scholar]
- 95.Aris JP, Alvers AL, Ferraiuolo RA, Fishwick LK, Hanvivatpong A, Hu D, Kirlew C, Leonard MT, Losin KJ, Marraffini M, et al. . Autophagy and leucine promote chronological longevity and respiration proficiency during calorie restriction in yeast. Exp Gerontol 2013; 48:1107 - 19; http://dx.doi.org/ 10.1016/j.exger.2013.01.006; PMID: 23337777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, et al. . Rapamycin slows aging in mice. Aging Cell 2012; 8:1371 - 82; PMID: 22587563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. . Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009; 460:392 - 5; PMID: 19587680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harrison B, Tran TT, Taylor D, Lee SD, Min KJ. . Effect of rapamycin on lifespan in Drosophila. Geriatr Gerontol Int 2010; 10:110 - 2; http://dx.doi.org/ 10.1111/j.1447-0594.2009.00569.x; PMID: 20102391 [DOI] [PubMed] [Google Scholar]
- 99.Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA Jr., Aris JP. . Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy 2009; 5:847 - 9; PMID: 19458476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. . Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster.. Cell Metab 2010; 11:35 - 46; http://dx.doi.org/ 10.1016/j.cmet.2009.11.010; PMID: 20074526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarbassov DD, Ali SM, Sabatini DM. . Growing roles for the mTOR pathway. Curr Opin Cell Biol 2005; 17:596 - 603; http://dx.doi.org/ 10.1016/j.ceb.2005.09.009; PMID: 16226444 [DOI] [PubMed] [Google Scholar]
- 102.Philipp O, Hamann A, Servos J, Werner A, Koch I, Osiewacz HD. . A genome-wide longitudinal transcriptome analysis of the aging model Podospora anserina.. PLoS One 2013; 8:e83109; http://dx.doi.org/ 10.1371/journal.pone.0083109; PMID: 24376646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dice JF. . Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem 1982; 257:14624 - 7; PMID: 7174658 [PubMed] [Google Scholar]
- 104.Cuervo AM, Dice JF. . Age-related decline in chaperone-mediated autophagy. J Biol Chem 2000; 275:31505 - 13; http://dx.doi.org/ 10.1074/jbc.M002102200; PMID: 10806201 [DOI] [PubMed] [Google Scholar]
- 105.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. . Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem 2006; 281:14474 - 85; http://dx.doi.org/ 10.1074/jbc.M600364200; PMID: 16522639 [DOI] [PubMed] [Google Scholar]
- 106.Kaushik S, Arias E, Kwon H, Lopez NM, Athonvarangkul D, Sahu S, Schwartz GJ, Pessin JE, Singh R. . Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep 2012; 13:258 - 65; http://dx.doi.org/ 10.1038/embor.2011.260; PMID: 22249165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, et al. . Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A 2010; 107:14164 - 9; http://dx.doi.org/ 10.1073/pnas.1009485107; PMID: 20660724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vittorini S, Paradiso C, Donati A, Cavallini G, Masini M, Gori Z, Pollera M, Bergamini E. . The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J Gerontol A Biol Sci Med Sci 1999; 54:B318 - 23; http://dx.doi.org/ 10.1093/gerona/54.8.B318; PMID: 10496537 [DOI] [PubMed] [Google Scholar]
- 109.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. . Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 2009; 28:889 - 901; http://dx.doi.org/ 10.1038/emboj.2009.29; PMID: 19229298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. . Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila.. Autophagy 2008; 4:176 - 84; PMID: 18059160 [DOI] [PubMed] [Google Scholar]
- 111.Yang W, Hekimi S. . Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans.. Aging Cell 2010; 9:433 - 47; http://dx.doi.org/ 10.1111/j.1474-9726.2010.00571.x; PMID: 20346072 [DOI] [PubMed] [Google Scholar]
- 112.Schiavi A, Torgovnick A, Kell A, Megalou E, Castelein N, Guccini I, Marzocchella L, Gelino S, Hansen M, Malisan F, et al. . Autophagy induction extends lifespan and reduces lipid content in response to frataxin silencing in C. elegans.. Exp Gerontol 2013; 48:191 - 201; http://dx.doi.org/ 10.1016/j.exger.2012.12.002; PMID: 23247094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Osiewacz HD. . Mitochondrial functions and aging. Gene 2002; 286:65 - 71; http://dx.doi.org/ 10.1016/S0378-1119(01)00804-6; PMID: 11943461 [DOI] [PubMed] [Google Scholar]
- 114.Osiewacz HD. . Mitochondrial quality control in aging and lifespan control of the fungal aging model Podospora anserina.. Biochem Soc Trans 2011; 39:1488 - 92; http://dx.doi.org/ 10.1042/BST0391488; PMID: 21936839 [DOI] [PubMed] [Google Scholar]
- 115.Pöggeler S, Masloff S, Hoff B, Mayrhofer S, Kück U. . Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr Genet 2003; 43:54 - 61; PMID: 12684845 [DOI] [PubMed] [Google Scholar]
- 116.Osiewacz HD. . A versatile shuttle cosmid vector for the efficient construction of genomic libraries and for the cloning of fungal genes. Curr Genet 1994; 26:87 - 90; http://dx.doi.org/ 10.1007/BF00326309; PMID: 7954902 [DOI] [PubMed] [Google Scholar]
- 117.Stumpferl SW, Stephan O, Osiewacz HD. . Impact of a disruption of a pathway delivering copper to mitochondria on Podospora anserina metabolism and life span. Eukaryot Cell 2004; 3:200 - 11; http://dx.doi.org/ 10.1128/EC.3.1.200-211.2004; PMID: 14871950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chaveroche MK, Ghigo JM, d’Enfert C. . A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans.. Nucleic Acids Res 2000; 28:E97; http://dx.doi.org/ 10.1093/nar/28.22.e97; PMID: 11071951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lecellier G, Silar P. . Rapid methods for nucleic acids extraction from Petri dish-grown mycelia. Curr Genet 1994; 25:122 - 3; http://dx.doi.org/ 10.1007/BF00309536; PMID: 8087879 [DOI] [PubMed] [Google Scholar]
- 120.Kunstmann B, Osiewacz HD. . Over-expression of an S-adenosylmethionine-dependent methyltransferase leads to an extended lifespan of Podospora anserina without impairments in vital functions. Aging Cell 2008; 7:651 - 62; http://dx.doi.org/ 10.1111/j.1474-9726.2008.00412.x; PMID: 18616635 [DOI] [PubMed] [Google Scholar]
- 121.Keller PJ, Schmidt AD, Santella A, Khairy K, Bao Z, Wittbrodt J, Stelzer EH. . Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat Methods 2010; 7:637 - 42; http://dx.doi.org/ 10.1038/nmeth.1476; PMID: 20601950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.