Abstract
In flowering plants, the tapetum, the innermost layer of the anther, provides both nutrient and lipid components to developing microspores, pollen grains, and the pollen coat. Though the programmed cell death of the tapetum is one of the most critical and sensitive steps for fertility and is affected by various environmental stresses, its regulatory mechanisms remain mostly unknown. Here we show that autophagy is required for the metabolic regulation and nutrient supply in anthers and that autophagic degradation within tapetum cells is essential for postmeiotic anther development in rice. Autophagosome-like structures and several vacuole-enclosed lipid bodies were observed in postmeiotic tapetum cells specifically at the uninucleate stage during pollen development, which were completely abolished in a retrotransposon-insertional OsATG7 (autophagy-related 7)-knockout mutant defective in autophagy, suggesting that autophagy is induced in tapetum cells. Surprisingly, the mutant showed complete sporophytic male sterility, failed to accumulate lipidic and starch components in pollen grains at the flowering stage, showed reduced pollen germination activity, and had limited anther dehiscence. Lipidomic analyses suggested impairment of editing of phosphatidylcholines and lipid desaturation in the mutant during pollen maturation. These results indicate a critical involvement of autophagy in a reproductive developmental process of rice, and shed light on the novel autophagy-mediated regulation of lipid metabolism in eukaryotic cells.
Keywords: anther, autophagy, male sterility, pollen development, rice
Introduction
Metabolic regulation and a supply of nutrients are essential for developmental processes in both plants and animals. In flowering plants, the anther consists of 4 distinct cell layers comprising the epidermis, endothecium, middle layer, and the tapetum. The tapetum is known to act as a supplier of metabolites and nutrients to developing microspores, pollen grains, and the pollen coat.1 Lipid bodies containing triacylglycerols (TAGs) in the tapetum are also necessary for pollen maturation and pollen-tube elongation as a supplier of lipid components,2,3 and some factors involved in the regulation of lipid metabolism in the anther have been suggested by recent bioinformatic and transcriptomic analyses.4,5 Though the programmed cell death (PCD) of the tapetum6 is one of the most critical and sensitive steps for fertility and is affected by various environmental stresses, their regulatory mechanisms remain mostly unknown.
Autophagy, a major catabolic pathway in eukaryotic cells, plays a role in the recycling of proteins and metabolites, including lipids, by delivering cytoplasmic components and organelles to degradation compartments such as lysosomes and vacuoles7,8 and is involved in many physiological processes including abiotic and biotic stress responses in plants.9,10 Lipophagy is a type of selective autophagy that has been suggested to contribute to lipid metabolism through the breakdown of intracellular lipids. Defects in lipophagy have been linked to important metabolic disorders such as fatty liver, obesity, and atherosclerosis in animals.11,12 However, the direct involvement of lipophagy during development processes as well as autophagy-mediated regulation of lipid metabolism has not previously been addressed in all eukaryotes.
Autophagy is essential for developmental processes in many eukaryotes, including preimplantation in mice and dauer development in nematodes.13-15 In plants, autophagy has been suggested to be involved in senescence and fertile floret development under the nutrient-starved conditions.16-18 However, the life cycle of the autophagy-defective mutants in Arabidopsis is normal, and direct involvement of autophagy during normal reproductive development has not yet been addressed in plants.10 Though the atg6 mutant of Arabidopsis shows gamete male sterility,19 ATG6 is one of the components of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex and the mutant phenotype is similar to that of other mutants of the PtdIns3K complex,20 indicating that the sterile phenotype of atg6 is attributed to other functions of PtdIns3K than autophagy.
In the present study, we have discovered that autophagy-defective mutants of rice show complete sporophytic male sterility. Multidisciplinary analyses including molecular genetics, electron microscopy, and lipidomics revealed that autophagy in tapetum cells of anthers plays critical roles in pollen maturation and male reproductive development in rice.
Results and Discussion
Isolation of autophagy-deficient mutants in rice
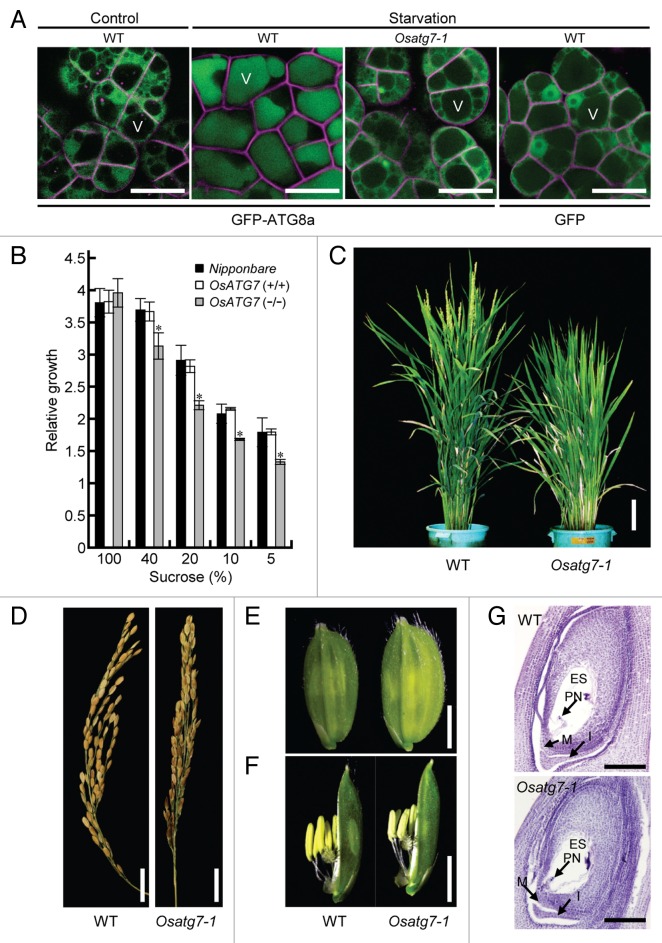
To analyze the physiological role of autophagy in developmental processes in rice, we isolated a mutant (Osatg7-1) of OsATG7 (locus ID: Os01g0614900, Fig. S1A), a sole rice homolog of ATG7, from retrotransposon Tos17-insertional mutant pools.21 We evaluated autophagy activity in the Osatg7-1 mutant using biochemical and imaging approaches. ATG7 is specific to autophagy and possesses E1-like activity in the ATG12 conjugation system that is essential for autophagosome biogenesis.22 First, the OsATG12–ATG5 conjugate was detected in the wild type but not the mutant using immunoblot analysis with an anti-OsATG5 antibody (Fig. S1C). The conjugate formation was restored by introduction of the wild-type OsATG7 gene but not the OsATG7 (C to S) mutant gene, which contains a mutation in the catalytic site and is predicted to be enzymatically inactive (Fig. S1D). Second, to monitor autophagy in vivo, we generated wild-type and Osatg7-1 cultured cells expressing the green fluorescent protein (GFP)-ATG8 fusion protein, a marker for autophagosomal membranes.23,24 Under sucrose-starved conditions, most GFP-ATG8 was delivered into the vacuolar lumen in the wild-type cells, whereas no GFP-ATG8 was detected in the vacuole in the Osatg7-1 mutant (Fig. 1A). Furthermore, cultured cells of Osatg7-1 showed more severe growth retardation under sucrose starvation than the wild type (Fig. 1B), which is consistent with the phenotypes of autophagy-deficient Arabidopsis atg mutants24-26 and indicates that autophagy is defective in the Osatg7-1 mutant.
Figure 1. The Osatg7-1 mutant exhibits a sterility phenotype. (A) In vivo imaging of autophagy in cultured rice cells. The accumulation of GFP-ATG8 in vacuoles in the presence of concanamycin A in wild-type and Osatg7-1 mutant cells under sucrose-starved conditions. Scale bar: 20 μm. V, vacuole. Data are representative of 3 experiments. FM4-64 probe (magenta color) was used to label the plasma membrane of cultured cells. (B) The effects of autophagy disruption on cell growth under sucrose starvation in cultured rice cells. The growth of cultured cells is shown for 7 d. The relative growth level at 0 d (fresh weight, 0.5 g) was standardized as 1. Data are the mean ± standard error (SE) for 3 independent experiments. *P < 0.01; significantly different from the controls. (C) The Osatg7-1 mutant shows slower heading. Plants were grown for 90 d. Scale bar: 10 cm. (D–F) Comparison of wild-type and Osatg7-1 mutant phenotypes at various reproductive stages; (D) panicles, (E) spikelet, and (F) flower organs. The Osatg7-1 mutant exhibited a sterile phenotype. (G) Ovary development appeared normal in the Osatg7 mutant. Ovaries from the wild type and Osatg7-1 mutant at the flowering stage were stained with hematoxylin and observed under a microscope. Scale bar: 100 μm. ES, embryo sac; PN, polar nucleus; M, micropyle; I, integument.
Quantitative polymerase chain reaction (qPCR) analysis showed that OsATG7 messenger RNA (mRNA) was expressed in mature leaves, shoots, roots, and suspension-cultured cells, as well as reproductive tissues (Fig. S1B). We also consulted the microarray-expression database (Rice XPro; http://ricexpro.dna.affrc.go.jp/GGEP/index.html), which showed OsATG7 expression throughout several developmental stages. Recent expression profiling using a laser microdissection-mediated microarray revealed expression of OsATG7 in the tapetum,5 suggesting that OsATG7 is expressed throughout the life cycle and that the Osatg7-1 mutant is deficient in autophagy in all developmental tissues including the tapetum.
Critical role of autophagy in rice reproductive development and its significance in crop production
Next, we investigated the effect of autophagy disruption on growth and development in rice. The rates of seed germination and vegetative growth of the Osatg7-1 mutant plants were comparable to those of wild type when grown in a greenhouse or paddy field (Fig. S2A). By contrast, the Osatg7-1 plants grown in a paddy field showed slower heading (Fig. 1C; Fig. S2B) and flowering (Fig. S2C and S2D) accompanying the delay of anther development including pollen maturation (data not shown), and had slightly larger spikelets (Fig. 1E and F). Surprisingly, unlike Arabidopsis atg mutants, the Osatg7-1 mutant exhibited a sterile phenotype (Fig. 1D; Fig. S3A; Table S1) and limited anther dehiscence under normal growth conditions (Fig. S3B). The stomium, an essential structure for anther dehiscence,27 did not show significant difference between the wild type and the mutant (data not shown), suggesting that the defects in anther dehiscence is attributed to other critical steps. These Osatg7-1 mutant phenotypes were complemented by the expression of OsATG7 (Fig. S3B–S3D). We also identified another mutant allele (Osatg7-2) of OsATG7 (3A-03442; cv Donjing) and a mutant line defective in OsATG9 (2D-00675; cv Hwayoung) from the T-DNA-tag lines of the POSTECH Biotech Center (Kyungbuk, Korea; Fig. S1A), both of which were sterile (Fig. S4A and S4B), indicating that autophagy plays critical roles in rice reproductive development. Overall, the present findings using the autophagy-defective mutants demonstrate the important role of autophagy in plant reproductive development and crop production.
Autophagy is required for male reproductive development in rice
To characterize the fertility phenotype of the Osatg7-1 mutant, we performed reciprocal crossing analysis. Mutant plants produced seeds when fertilized with the pollen of wild type (cv NB; Table S2). Moreover, the structure of the ovary at the flowering stage was similar between the wild type and the mutant (Fig. 1G), indicating that the sterility phenotype is caused by the male gametophyte.
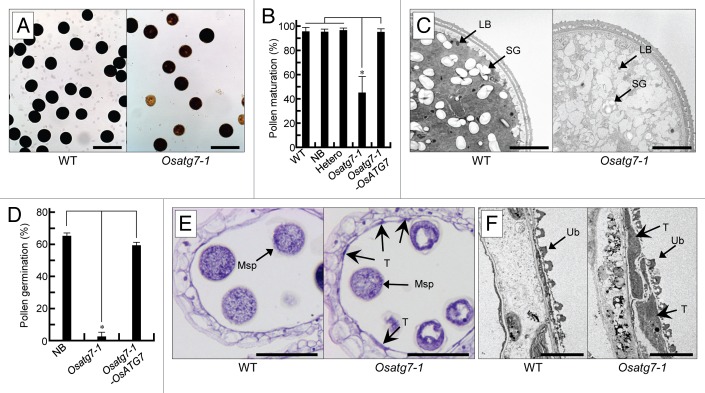
We then assessed pollen maturation. Mature pollen stained by iodine accounted for approximately 95% and 45% of the total pollen in the wild type and mutant, respectively (Fig. 2A and B). Undeveloped mutant pollen was confirmed by toluidine blue staining (Fig. S5A). At the flowering stage, the coat structure of many pollen grains in Osatg7-1 was undeveloped (Fig. S5B) and there was less accumulation of lipid bodies in Osatg7-1 mutant than wild-type pollen (Fig. 2C), indicating that the defects in Osatg7-1 pollen maturation occur at the flowering stage. Some mutant pollen was stained by the iodine reagent, and the pollen coat appeared normal. We therefore checked the germination activity of mutant pollen, and observed germination in only 0.74% of the Osatg7-1 pollen grains compared with more than 60% of wild-type grains (Fig. 2D; Fig. S5C). These results suggest that the pollens of autophagy-defective mutants are premature due to critical defects in the anther during pollen maturation. The ratio of mature pollens in the heterozygous plants was similar to that in the wild type (Fig. 2B), and the pollination of the wild-type (cv NB) stigmas with heterozygous pollens led to a normal segregation rate in the next generation (Table S3). These results suggest that the immature pollen phenotype of the Osatg7-1 mutant is attributed to defects in parental tissues or organs. The autophagosome-like structures observed in the tapetum cells were not detected in the pollen during developmental processes (Fig. 2C and data not shown).
Figure 2. Autophagy is required for male reproductive development in rice. (A) Pollen grains from the wild-type and Osatg7-1 mutant stained with I2-KI solution. Scale bar: 100 μm. (B) Viable pollen grains shown in (A) were quantified. Data are mean ± SD; n = 3 independent samples. *P < 0.01; significantly different from the controls. (C) The ultrastructure of pollen in the wild-type and Osatg7-1 mutant at the flowering stage. Scale bar: 5 μm. LB, lipid body; SG, starch granule. (D) In vitro pollen germination rate. Pollen grains from NB, Osatg7-1, and complementation line (Osatg7-1-OsATG7) anthers at the mature stage were assayed. Data are mean ± SD; n = 3 independent samples. *P < 0.01; significantly different from the controls. (E) Transverse section analysis of wild-type and Osatg7-1 anthers stained with hematoxylin at the flowering stage. Scale bar: 50 μm. (F) Ultrastructure of the tapetum in the wild-type and Osatg7-1 mutant at the flowering stage. Scale bar: 3 μm. Msp, microspore; T, tapetum.
Autophagy in postmeiotic tapetum cells in rice
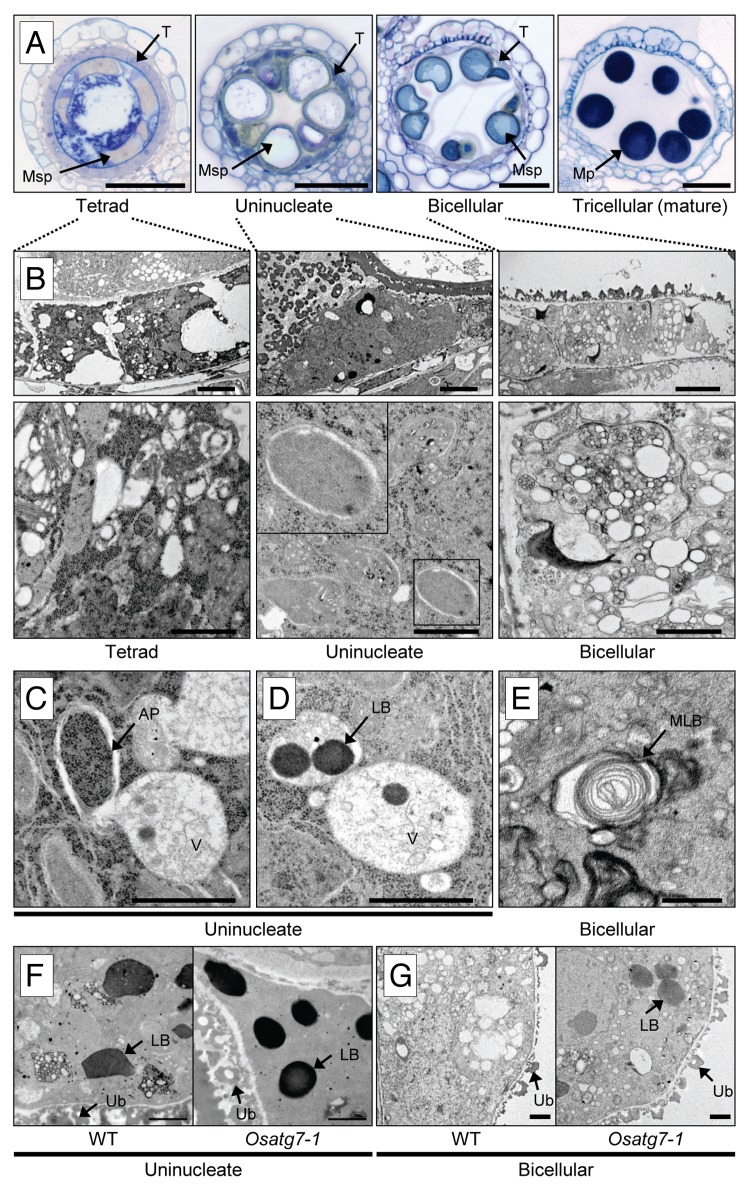
The tapetum acts as a supplier of metabolites and nutrients to developing microspores and pollen grains.1 To investigate whether autophagy takes place in the rice tapetum under normal growth conditions, we performed transmission electron microscopy (TEM) analysis (Fig. 3). As shown in Figure 3B, no obvious autophagosome-like structures were detected in the cytoplasm at the tetrad stage in rice anthers. By contrast, at the uninucleate stage, autophagosome-like structures were clearly observed in the cytoplasm (Fig. 3B and C). Numerous vacuole-enclosed dense globular bodies, lipid bodies, were also found at the uninucleate stage (Fig. 3D). Thereafter, at the bicellular stage, some multilamellar bodies, presumably intermediate structures of autophagosomes,28 were observed in the cytoplasm (Fig. 3E). Moreover, vacuoles fused with the lipid bodies that could be degraded by autophagy, appeared in the tapetum (Fig. 3B). Finally, the tapetum disappeared at the mature stage (Fig. 3A). These results suggest induction of autophagy at the uninucleate stage in postmeiotic tapetum cells in rice.
Figure 3. Autophagy occurs in tapetal cells during the male reproductive phase. (A) Transverse section analysis during rice anther development. Samples taken at the 4 stages of anther development, and stained with toluidine blue. MP, mature pollen; Msp, microspore; T, tapetum. Scale bar: 50 μm. (B–E) The ultrastructure of the tapetum during anther development by TEM analysis. (B) Autophagosome-like structures and many vacuole-enclosed lipid bodies were observed in the cytoplasm at the uninucleate stage. Thereafter, at the bicellular stage, many characteristic intermediate structures including vacuoles fused with lipid bodies appeared in the tapetum. Scale bar: (upper) 3 μm, (lower) 1 μm. (C–E) Close-up images of the cell in (B). Scale bar: 1 μm. AP, autophagosome; LB, lipid body; V, vacuole; MLB, multilamellar body. (F andG) Ultrastructure of the tapetum at reproductive stages in the wild-type and Osatg7-1 mutant. Samples taken at uninucleate (1n; F) and bicellular (2n; G) stages of anther development, respectively. Scale bar: 1 μm. LB, lipid body; Ub, Ubisch body.
We next performed TEM analysis to determine the dynamics of autophagy in the tapetum during pollen maturation. At the tetrad stage, no obvious differences were apparent in the intracellular structure between the wild type and Osatg7-1 mutant (Fig. 3; Fig. S6). In the uninucleate stage, however, few double-membrane autophagosome-like structures and the vacuole-enclosed dense globular bodies were observed in the cytoplasm of the mutant (Fig. 3F and G; Fig. S6). More lipid body-like structures remained in the cytoplasm in the mutant than in the wild type at the bicellular stage (Fig. 3F and G). Intracellular organelles such as plastids and mitochondria, which have been reported to be degraded by autophagy,18 were also clearly observed in the cytoplasm of the mutant even at the bicellular stage (Fig. 3B; Fig. S6). These results indicate that autophagy is active in the tapetum, which may be involved in the degradation of intracellular components such as plastids and lipid bodies during pollen maturation.
Involvement of autophagy in the regulation of lipid metabolism and nutrient supply in anthers
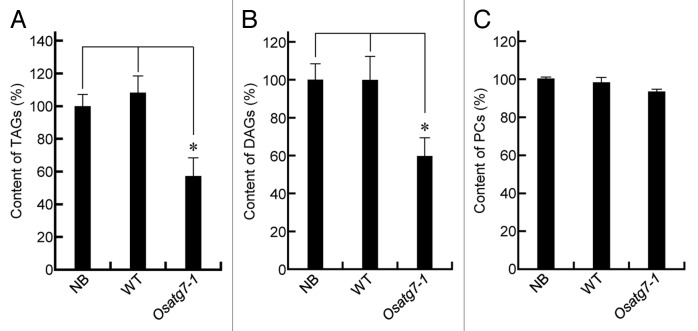
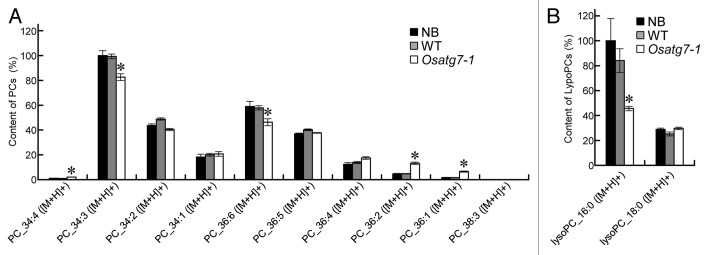
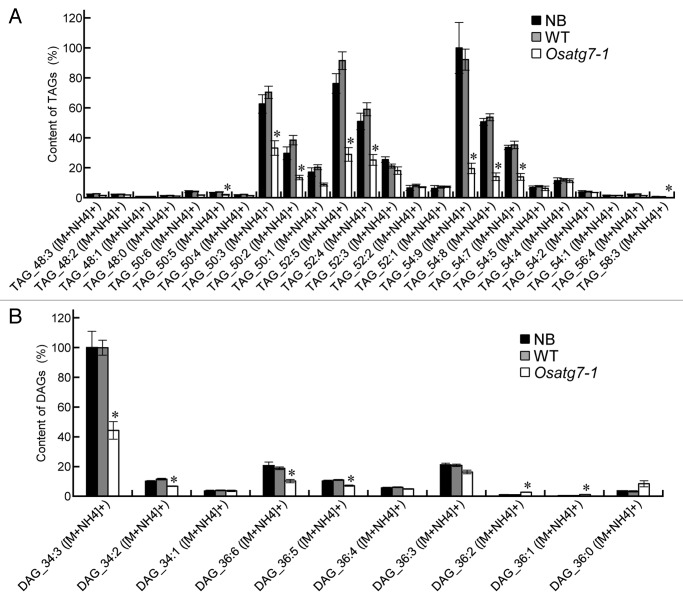
Lipid bodies inside the pollen grains are necessary for its maturation and pollen-tube elongation.2,3 In fact, fewer lipid bodies and starch grains were accumulated in the pollen grains of Osatg7-1 mutant than the wild type at the flowering stage (Fig. 2C; Fig. S5B), which is consistent with reduced pollen germination activity in the mutant (Fig. 2D). To investigate the roles of tapetal autophagy in lipid metabolism of pollen grains at the flowering stage, crude lipid fractions were extracted from the mature anthers mainly occupied by pollen grains, and subjected to lipid profiling by LC-MS.29 TAGs are the main components stored in lipid bodies within pollen grains,30 and essential for normal pollen development.31 The amounts of TAGs and diacylglycerols (DAGs), intermediary metabolites in TAG synthesis, were significantly lower in the mutant than in the wild type (Fig. 4A and B). Desaturation of acyl groups of TAGs involves editing of phosphatidylcholine (PC) through the Lands cycle.32 Interestingly, the total amount of PCs was similar between the wild type and the mutant (Fig. 4C), whereas the amount of highly desaturated PCs (34:3 and 36:6) was significantly lower in the mutant than the wild type (Fig. 5A). The level of lysoPC (16:0), a possible intermediary metabolite in the Lands cycle pathway, also decreased in the mutant (Fig. 5B), suggesting that autophagy may play roles in the PC editing through the Lands cycle pathway. The decrease in some highly desaturated TAGs and DAGs in the Osatg7-1 mutant (Fig. 6) can be accounted for by impairment in the PC editing. On the other hand, the level of de novo DAG (34:1), synthesized by the Kennedy pathway,32 was comparable to those of the wild type (Fig. 6B). These results suggest that tapetal autophagy contributes to lipid metabolism of pollen grains, especially the Lands cycle-mediated PC editing and desaturation in rice anthers, which affects the pollen maturation including coat formation.
Figure 4. Autophagy contributes to lipid metabolism during tapetum and pollen maturation. (A–C) Levels of individual lipid molecules (A) integrated value of TAGs, (B) integrated value of DAGs, and (C) integrated value of PCs in the wild-type and Osatg7-1 mutant were expressed as relative values against the sum of the peak areas of lipid molecules with the same polarity in the wild type. Samples (mature anthers) were taken from each plant. *P < 0.01; significantly different from the control.
Figure 5. Composition of phosphatidylcholines (PCs) and lysophosphatidylcholines (LysoPCs) of the anthers at the flowering stage. Levels of individual lipid molecules (A) PC, and (B) LysoPC in the wild-type and Osatg7-1 mutant were expressed as relative values against the sum of the peak areas of lipid molecules with the same polarity in the wild type. Samples (mature anthers) were taken from each plant. *P < 0.01; significantly different from the control.
Figure 6. Composition of triacylglycerols (TAGs) and diacylglycerols (DAGs) of the anthers at the flowering stage. Levels of individual lipid molecules (A) TAG, and (B) DAG in the wild-type and Osatg7-1 mutant were expressed as relative values against the sum of the peak areas of lipid molecules with the same polarity in the wild type. Samples (mature anthers) were taken from each plant. *P < 0.01; significantly different from the control.
A lipid transfer protein of rice, anther specific protein 6 (OsC6), is specifically expressed in tapetal cells and is able to transport lipid molecules such as fatty acids from tapetal cells to developing microspores. The RNA interference (RNAi)-suppression lines of OsC6 contain more lipid bodies in tapetal cells than in the control during pollen maturation, and show decreased pollen fertility.33 These phenotypes of the OsC6-suppressed plants were similar in part to those of Osatg7-1 mutant, suggesting that the immature pollen phenotype of Osatg7-1 mutant is attributed to the defects in the transport of materials including lipids from the tapetum cells to developing microspores.
Lipophagy has recently been suggested to be involved in lipid droplet degradation in animals.11 However, the direct involvement of lipophagy during developmental processes as well as autophagy-mediated regulation of lipid metabolism had remained mostly unknown in plants. The present results suggest that autophagy in the tapetum may be involved in the degradation of lipid bodies and regulation of lipid metabolism during pollen development, and shed light on the importance of autophagy-mediated regulation of lipid metabolism in development not only in plants but in all eukaryotic cells. Autophagy has also recently been suggested to contribute to the turnover of peroxisomes,34 which are known to be involved in the regulation of lipid metabolisms.35 Possible involvement of autophagy in the regulation of lipid metabolism is an emerging important future subject of research.
Possible involvement of autophagy in the degradation and programmed cell death of the tapetum
During pollen development, the tapetum is degraded to supply metabolites, nutrients, and sporopollenin precursors to developing microspores. Defects in the degradation of the tapetum cause abnormal formation of the pollen coat and pollen grains, resulting in severe male sterility.1 The Osatg7-1 mutant showed limited anther dehiscence (Fig. S3B), which could also contribute to its sterility.
The dense thin layer connected with the orbicules was only observed by both the transverse section and TEM analysis in the Osatg7-1 mutant even at the flowering stage (Fig. 2E and F, arrows), suggesting that the tapetal cell layer thoroughly disappeared in the wild-type, whereas partially remained in the mutant. It has been proposed that the tapetum is degraded by PCD during the later stages of pollen maturation in plants.1,6 Autophagy is also involved in PCD in animals.36,37 Autophagic cell death is characterized by the occurrence of double-membrane autophagosomes within the dying cells that remove the cell remnants.38 In Drosophila melanogaster, destruction of the salivary glands and digestive tract is mediated by a dramatic upregulation of autophagy prior to and during cell death during metamorphosis.39 Autophagy may therefore contribute, at least in part, to tapetum degradation in rice. Future detailed TEM/imaging analyses of various tapetum developmental stages under environmental stress conditions in the Osatg7 mutants may reveal a novel mechanism for autophagy-mediated PCD and its physiological significance in rice.
Concluding Remarks
In hybrid rice breeding, male sterility lines with cytoplasmic and/or nuclear mutations are of agricultural importance for the production of hybrids to improve yield.40 We here demonstrated that autophagy-defective mutants of rice show complete sporophytic male sterility, and suggest the involvement of tapetum autophagy in the regulation of lipid metabolism and nutrient supply in anthers. Importantly, the life cycle of the autophagy-defective mutants in Arabidopsis were found to be normal. The most significant difference of the tapetum between monocots and dicots is that monocots do not form tapetosomes for lipid transport in tapetal cells, while dicots have lipidic tapetosomes. It may suggest a critical difference in the development of lipidic components in the pollen grains between monocots and dicots. Future TEM/imaging analyses of tapetum in various developmental stages in other plant species including Arabidopsis along with genetic analyses may reveal a novel function of autophagy in lipid metabolism and its physiological significance during postmeiotic anther development.
Materials and Methods
Plant materials and growth conditions
Surface-sterilized seeds of rice, Oryza sativa L. cv NB, Donjing and Hwayoung, were germinated on MS medium41 containing 0.8% agar and grown for 10 d in a growth chamber under long-day conditions (16 h light/8 h darkness, 28 °C). Seedlings were transplanted into soil and grown in a greenhouse (16 h light/8 h darkness, 28 °C and 60% humidity) or paddy field.
To generate cultured cells of Osatg7-knockout and wild type (WT), in which the insertion of Tos17 in OsATG7 was removed by heterozygous segregation, seeds were placed onto callus-inducing medium. The calli were suspension-cultured at 25 °C in a liquid L medium42 containing 2,4-D (0.5 mg L−1) (Wako Pure Chemical, 119-0563) in the dark, and subcultured in fresh medium every 7 d. Cells were filtered through a 20-mesh screen every 2 wk to produce fine aggregates. In our experiments, NB, Donjing, Hwayoung, and WT plants as well as cultured cells were used as controls.
TEM analysis
Samples were fixed in a 0.05 M cacodylate buffer (pH 7.4) containing 2% glutaraldehyde and 4% paraformaldehyde for 3 h in an ice bath. Specimens were rinsed in the same buffer and post-fixed in 2% osmium tetroxide for 3 h on ice. Dehydrated specimens were embedded in epoxy resin (Epon 812; Nisshin EM, 341). Ultrathin sections (70 to 80 nm) were made with an LKB-2088 ULTROTOME V (LKB) using a diamond knife, and mounted on 200-mesh copper grids (Nisshin EM, 2632). Sections were stained with 2% uranyl acetate and lead citrate, and observed with a JEM-1200EX transmission electron microscope (JEOL Ltd.) operating at 80 kV.
RNA isolation and RT-PCR
Total RNA was isolated using Sepasol reagent (Nacalai Tesque, 30486-56) in accordance with the manufacturer’s protocol, and quantified using a spectrophotometer. First-strand complementary DNA (cDNA) was synthesized from 3 μg total RNA with an oligo-dT primer (Life Technologies, 18418-012) and reverse transcriptase (Promega, M3682). PCR amplification was performed with an initial denaturation at 95 °C for 3 min followed by 30 cycles of incubation at 95 °C for 60 s, 55 °C for 90 s, and 72 °C for 40 s using specific primers for OsATG7. OsActin1 (locus ID: Os03g0718100) was used as a quantitative control.43 Aliquots of individual PCR products were resolved by agarose gel electrophoresis, and visualized using ethidium bromide staining and exposure to UV light.
OsATG-null mutants
Large populations (39,744 lines) of rice (O. sativa cv NB) mutants generated by Tos17-mediated mutagenesis21 were made available by the National Institute of Agrobiological Resources (NIAS; Tsukuba, Japan). Tos17-mediated mutagenesis details have been described previously.21 Using this technique, we isolated one mutant line (NC7558) by PCR screening. Tos17 insertion in the mutant (Osatg7-1) was confirmed by DNA gel-blot analysis, and its exact position was determined by sequencing. Plants homozygous for the Osatg7-null allele were confirmed by the isolation of total RNA followed by reverse transcription (RT)-PCR using the following primer set: forward primer 5′-TCAAGCTGGA CGTCCTCGG-3′ and reverse primer 5′-TTGCTTCAGG CACATAATCA GG-3′. OsActin1 was used as a quantitative control.43
Another allele (3A-03442; cv Donjing) and a mutant line defective in OsATG9 were also isolated from the T-DNA-tag lines of the POSTECH Biotech Center.44,45 The primer (5′-GTGGTAGTAA GAATGGAACT CAC-3′) annealing to the T-DNA left border was used together with the OsATG7-specific (5′-AACAATTGTC CAGTCCCAGG-3′) and OsATG9-specific (5′-AATGCATCGT AGCCCTTTTG-3′) primers for the identification of putative mutant lines. T-DNA insertions in the mutants were confirmed by DNA gel-blot analysis, and exact insertion positions were determined by DNA sequencing.
Establishment of transgenic rice cell lines stably expressing GFP-AtATG8a
The GFP-AtATG8a fusion construct (pBI121 binary vector) was kindly provided by K Yoshimoto (Institut National de la Recherche Agronomique, Paris, France). The construct was introduced into rice calli using Agrobacterium-mediated transformation.46 Transformed calli were screened by hygromycin selection (50 μg/mL; Nacalai Tesque, 07296-24); transgenic plants were then regenerated. Transgenic cell lines derived from T2 plants were used for various analyses.
In vivo imaging of autophagy
Six-d-old cultured rice cells expressing the GFP-AtATG8a protein were incubated in a sucrose-free medium42 for 3 d, and then incubated for an additional 16 h in the presence of 1 μM concanamycin A (Sigma, C9705). The control was set at time 0.
All images were observed using an LSM 5 EXCITER confocal fluorescence microscope with a 40 × objective lens (Carl Zeiss). For all experiments, laser intensity was adjusted to the lowest level that retained a significant signal-to-noise ratio. The fluorescent styryl membrane probe FM 4-64 (Life Technologies, T-13320) was kept as a 17-mM stock solution in sterile water, and used at a final concentration of 4.25 μM. Cultured rice cells were treated with FM 4-64 for 10 min and washed twice with the wash buffer at room temperature to label the plasma membrane.
Immunoblot analyses
A rabbit polyclonal anti-OsATG5 antibody was generated to detect the OsATG12–ATG5 conjugate. The coding region of the amino (N)-terminal half domain of OsATG5 (M1-C190) was amplified using the following sequence-specific primers: M1-C190F, 5′-CACCATGGCG GCGCAGCGCG ACGA-3′; and M1-C190R, 5′ -GCAATCCTCT TCGAATGGTC-3′. A fusion protein consisting of the domain fused to the histidine-tag in the pDEST17 vector (Life Technologies, 11803-012) was transformed into E. coli BL21-AI (Life Technologies, C6070-03). Inclusion bodies with the recombinant protein were obtained after induction at 37 °C for 6 h, and were resolved using a preparative 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel. A ground polyacrylamide gel slice of the fusion protein was checked using tandem mass spectrometry (MS/MS) analysis, and used to immunize rabbits by intradermal injections.
For immunoblotting analysis, crude protein samples were separated by 10% SDS-PAGE gel, and blotted onto a PVDF membrane. The membrane was blocked in 1 × TTBS buffer (10 mM TRIS-HCl, 150 mM NaCl, and 0.05% Tween-20, pH 7.5) with 5% nonfat milk overnight at 4 °C. Blots were incubated with the affinity-purified anti-OsATG5, and then with horseradish peroxidase (HRP)-linked anti-rabbit immunoglobulin G. Bands were detected using chemiluminescent HRP substrate (Merck Millipore, WBKLS0050) and the chemiluminescent analyzer, LAS3000 (GE Healthcare).
Pollen viability
Pollen maturity was investigated with I2-KI staining (0.2% iodine and 2% potassium iodine) for starch accumulation. More than 200 pollen grains were counted in each examination under bright-field microscopy.
Pollen-germination assay
Pollen grains from anthers at the mature stage during flowering were immediately placed on Ishikawa’s liquid germination medium47 [0.01% H3BO3, 0.1% Ca(NO3)2 · 4 H2O, and 15% sucrose in sterilized water (pH 5.7)] and incubated at room temperature in a humid chamber. After 1 h, more than 200 pollen grains were counted in each examination under bright-field microscopy. Pollen grains that developed normal tubes longer than the grain diameter were considered as “germinated.”
Growth assay of rice cultured cells
The cell-growth assay was performed as described by Kurusu et al.48 Seven-d-old cultured cells were transferred to a 50-mL tube containing sucrose-free L medium and preincubated for 3 h. Cells (fresh weight, 0.5 g) were transferred to L medium42 with standard (100%) or low sucrose conditions (40, 20, 10, and 5%). After culturing for 0 and 7 d, the fresh weight of cells was measured.
Real-time RT-PCR quantification
Total RNA was extracted from various tissues of rice plants as well as cultured cells. Real-time RT-PCR assays were performed as described by Kurusu et al.49 Real-time PCR was performed using an ABI PRISM 7300 sequence detection system (Life Technologies) with SYBR Green real-time PCR Master Mix (Toyobo, QPS-201) and the following OsATG7-specific primers: OsATG7-RealF, 5′-GATCGTGACA GCCCAAAAGC A-3′; and OsATG7-RealR, 5′-GACCAAGTGG ACCCTCACTG C-3′.50 Relative mRNA levels were calculated using the standard curve method and normalized to corresponding OsEF1α (locus ID: Os03g0177500) gene levels.51 Standard samples of known template amounts were used to quantify PCR products.
Complementation analysis
The PAC clone P0410E01 encompassing the OsATG7 locus was obtained from NIAS. A DNA fragment (9.5 kb) of the OsATG7 genome region was amplified by PCR using the PAC clone as a template and the following primer sets (KpnI and SmaI sites are underlined): KpnI-OsATG7F, 5′-cggggtacc GTCACAACAC TTACATCGGG TTTCATTGC-3′; and SmaI-OsATG7R, 5′-aaacccggg GAAGAGAGGA AGAAGCAGAG GACGC-3′. The resulting product was cut with KpnI and SmaI, and inserted into the KpnI-ApaI site of the pPZP2H-lac binary vector.52 To make the C to S mutant, the following primer sets were used for PCR amplification: OsATG7F(C910S), 5′-GAACACTGGA TCAACAAAGT ACAGTAACAC GGCCT-3′; and OsATG7R(C910S), 5′-AGGCCGTGTT ACTGTACTTT GTTGATCCAG TGTTC-3′. The construct and control vector were introduced into Osatg7 calli using Agrobacterium-mediated transformation.46 Transformed calli were screened by hygromycin selection (50 μg/mL); transgenic calli were then regenerated. Transformed cultured cells and plants were used for various analyses.
Lipidome analysis
Lipidome analyses were performed as described by Okazaki et al.29 High-resolution electrospray ionization mass spectra were acquired in both positive and negative ion modes. Unless stated otherwise, all the lipid profile data was analyzed based on the data recorded in positive ion mode, except for fatty acids which are detected in negative ion mode. Peak areas of individual lipid molecules were calculated based on the m/z values of their molecular-related ions. The ions used for the calculation of peak areas are as follows: [M+H]+ for PG, [M+NH4]+ for PE, [M+H]+ for lysoPC, [M+NH4]+ for TAG, and [M–H]– for fatty acid.
Statistical analyses
Significance was determined using an unpaired Student t test at P < 0.01.
Supplementary Material
Glossary
Abbreviations:
- DAGs
diacylglycerols
- GFP
green fluorescent protein
- NB
Nipponbare
- OsC6
anther specific protein 6
- PCD
programmed cell death
- PCs
phosphatidylcholines
- PtdIns3K
phosphatidylinositol 3-kinase
- TAGs
triacylglycerols
- TEM
transmission electron microscopy
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank K Yoshimoto for providing the vector containing GFP-AtATG8a, G An for providing the T-DNA lines, H Ohta for helpful discussion and critical comments, and Y Kitagawa, K Kawamura, Y Maeno, and A Ikeda for technical assistance. This work was supported, in part, by Grants-in-Aid for Challenging Exploratory Research Nos. 23658061 and 25660049 and Grants-in-Aid for Scientific Research on Priority Area Nos. 21117516 and 23117718 from MEXT, Japan.
References
- 1.Ariizumi T, Toriyama K. . Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 2011; 62:437 - 60; http://dx.doi.org/ 10.1146/annurev-arplant-042809-112312; PMID: 21275644 [DOI] [PubMed] [Google Scholar]
- 2.Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. . Acyl-lipid metabolism. Arabidopsis Book 2010; 8:e0133; http://dx.doi.org/ 10.1199/tab.0133; PMID: 22303259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy DJ. . The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 2012; 249:541 - 85; http://dx.doi.org/ 10.1007/s00709-011-0329-7; PMID: 22002710 [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Wu H, Yang M. . Microscopy and bioinformatic analyses of lipid metabolism implicate a sporophytic signaling network supporting pollen development in Arabidopsis.. Mol Plant 2008; 1:667 - 74; http://dx.doi.org/ 10.1093/mp/ssn027; PMID: 19825571 [DOI] [PubMed] [Google Scholar]
- 5.Huang MD, Wei FJ, Wu CC, Hsing YI, Huang AH. . Analyses of advanced rice anther transcriptomes reveal global tapetum secretory functions and potential proteins for lipid exine formation. Plant Physiol 2009; 149:694 - 707; http://dx.doi.org/ 10.1104/pp.108.131128; PMID: 19091874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papini A, Mosti S, Brighigna L. . Programmed-cell-death events during tapetum development of angiosperms. Protoplasma 1999; 207:213 - 21; http://dx.doi.org/ 10.1007/BF01283002 [DOI] [Google Scholar]
- 7.Mizushima N, Yoshimori T, Ohsumi Y. . The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107 - 32; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005; PMID: 21801009 [DOI] [PubMed] [Google Scholar]
- 8.Avin-Wittenberg T, Honig A, Galili G. . Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 2012; 249:285 - 99; http://dx.doi.org/ 10.1007/s00709-011-0296-z; PMID: 21660427 [DOI] [PubMed] [Google Scholar]
- 9.Hanamata S, Kurusu T, Okada M, Suda A, Kawamura K, Tsukada E, Kuchitsu K. . In vivo imaging and quantitative monitoring of autophagic flux in tobacco BY-2 cells. Plant Signal Behav 2012; 8:e22510; http://dx.doi.org/ 10.4161/psb.22510; PMID: 23123450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshimoto K. . Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 2012; 53:1355 - 65; http://dx.doi.org/ 10.1093/pcp/pcs099; PMID: 22764279 [DOI] [PubMed] [Google Scholar]
- 11.Dong H, Czaja MJ. . Regulation of lipid droplets by autophagy. Trends Endocrinol Metab 2011; 22:234 - 40; http://dx.doi.org/ 10.1016/j.tem.2011.02.003; PMID: 21419642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu K, Czaja MJ. . Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ 2013; 20:3 - 11; http://dx.doi.org/ 10.1038/cdd.2012.63; PMID: 22595754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N, Komatsu M. . Autophagy: renovation of cells and tissues. Cell 2011; 147:728 - 41; http://dx.doi.org/ 10.1016/j.cell.2011.10.026; PMID: 22078875 [DOI] [PubMed] [Google Scholar]
- 14.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. . Autophagy is essential for preimplantation development of mouse embryos. Science 2008; 321:117 - 20; http://dx.doi.org/ 10.1126/science.1154822; PMID: 18599786 [DOI] [PubMed] [Google Scholar]
- 15.Meléndez A, Levine B. . Autophagy in C. elegans.. WormBook 2009; 1 - 26; http://dx.doi.org/ 10.1895/wormbook.1.147.1; PMID: 19705512 [DOI] [PubMed] [Google Scholar]
- 16.Chung T, Suttangkakul A, Vierstra RD. . The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol 2009; 149:220 - 34; http://dx.doi.org/ 10.1104/pp.108.126714; PMID: 18790996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghiglione HO, Gonzalez FG, Serrago R, Maldonado SB, Chilcott C, Curá JA, Miralles DJ, Zhu T, Casal JJ. . Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J 2008; 55:1010 - 24; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03570.x; PMID: 18547393 [DOI] [PubMed] [Google Scholar]
- 18.Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, Ohsumi Y, Hanson MR, Mae T. . Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol 2008; 148:142 - 55; http://dx.doi.org/ 10.1104/pp.108.122770; PMID: 18614709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiki Y, Yoshimoto K, Ohsumi Y. . An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol 2007; 143:1132 - 9; http://dx.doi.org/ 10.1104/pp.106.093864; PMID: 17259285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Kim ES, Choi Y, Hwang I, Staiger CJ, Chung YY, Lee Y. . The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol 2008; 147:1886 - 97; http://dx.doi.org/ 10.1104/pp.108.121590; PMID: 18515640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirochika H. . Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 2001; 4:118 - 22; http://dx.doi.org/ 10.1016/S1369-5266(00)00146-1; PMID: 11228433 [DOI] [PubMed] [Google Scholar]
- 22.Li F, Vierstra RD. . Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 2012; 17:526 - 37; http://dx.doi.org/ 10.1016/j.tplants.2012.05.006; PMID: 22694835 [DOI] [PubMed] [Google Scholar]
- 23.Nakatogawa H, Ichimura Y, Ohsumi Y. . Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007; 130:165 - 78; http://dx.doi.org/ 10.1016/j.cell.2007.05.021; PMID: 17632063 [DOI] [PubMed] [Google Scholar]
- 24.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. . Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004; 16:2967 - 83; http://dx.doi.org/ 10.1105/tpc.104.025395; PMID: 15494556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. . The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana.. J Biol Chem 2002; 277:33105 - 14; http://dx.doi.org/ 10.1074/jbc.M204630200; PMID: 12070171 [DOI] [PubMed] [Google Scholar]
- 26.Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. . Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 2005; 138:2097 - 110; http://dx.doi.org/ 10.1104/pp.105.060673; PMID: 16040659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsui T, Omasa K, Horie T. . Mechanism of Anther Dehiscence in Rice (Oryza sativa L.). Ann Bot 1999; 84:501 - 6; http://dx.doi.org/ 10.1006/anbo.1999.0943 [DOI] [Google Scholar]
- 28.Hariri M, Millane G, Guimond MP, Guay G, Dennis JW, Nabi IR. . Biogenesis of multilamellar bodies via autophagy. Mol Biol Cell 2000; 11:255 - 68; http://dx.doi.org/ 10.1091/mbc.11.1.255; PMID: 10637306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki Y, Otsuki H, Narisawa T, Kobayashi M, Sawai S, Kamide Y, Kusano M, Aoki T, Hirai MY, Saito K. . A new class of plant lipid is essential for protection against phosphorus depletion. Nat Commun 2013; 4:1510; http://dx.doi.org/ 10.1038/ncomms2512; PMID: 23443538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HU, Hsieh K, Ratnayake C, Huang AH. . A novel group of oleosins is present inside the pollen of Arabidopsis.. J Biol Chem 2002; 277:22677 - 84; http://dx.doi.org/ 10.1074/jbc.M109298200; PMID: 11929861 [DOI] [PubMed] [Google Scholar]
- 31.Zhang M, Fan J, Taylor DC, Ohlrogge JB. . DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009; 21:3885 - 901; http://dx.doi.org/ 10.1105/tpc.109.071795; PMID: 20040537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Shen W, Kazachkov M, Chen G, Chen Q, Carlsson AS, Stymne S, Weselake RJ, Zou J. . Metabolic interactions between the Lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell 2012; 24:4652 - 69; http://dx.doi.org/ 10.1105/tpc.112.104604; PMID: 23150634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D. . OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 2010; 154:149 - 62; http://dx.doi.org/ 10.1104/pp.110.158865; PMID: 20610705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honig A, Avin-Wittenberg T, Ufaz S, Galili G. . A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 2012; 24:288 - 303; http://dx.doi.org/ 10.1105/tpc.111.093112; PMID: 22253227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linka N, Esser C. . Transport proteins regulate the flux of metabolites and cofactors across the membrane of plant peroxisomes. Front Plant Sci 2012; 3:3; http://dx.doi.org/ 10.3389/fpls.2012.00003; PMID: 22645564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. . Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 2004; 6:1221 - 8; http://dx.doi.org/ 10.1038/ncb1192; PMID: 15558033 [DOI] [PubMed] [Google Scholar]
- 37.Tsujimoto Y, Shimizu S. . Another way to die: autophagic programmed cell death. Cell Death Differ 2005; 12:Suppl 2 1528 - 34; http://dx.doi.org/ 10.1038/sj.cdd.4401777; PMID: 16247500 [DOI] [PubMed] [Google Scholar]
- 38.Gump JM, Thorburn A. . Autophagy and apoptosis: what is the connection?. Trends Cell Biol 2011; 21:387 - 92; http://dx.doi.org/ 10.1016/j.tcb.2011.03.007; PMID: 21561772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meléndez A, Neufeld TP. . The cell biology of autophagy in metazoans: a developing story. Development 2008; 135:2347 - 60; http://dx.doi.org/ 10.1242/dev.016105; PMID: 18567846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ouyang Y, Chen J, Xie W, Wang L, Zhang Q. . Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol Biol 2009; 70:341 - 57; http://dx.doi.org/ 10.1007/s11103-009-9477-y; PMID: 19277876 [DOI] [PubMed] [Google Scholar]
- 41.Murashige T, Skoog F. . A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 1962; 15:473 - 92; http://dx.doi.org/ 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- 42.Kuchitsu K, Kikuyama M, Shibuya N. . N-acetylchitooligosaccharides, biotic elicitors for phytoalexin production, induce transient membrane depolarization in suspension-cultured rice cells. Protoplasma 1993; 174:79 - 81; http://dx.doi.org/ 10.1007/BF01404046 [DOI] [Google Scholar]
- 43.Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. . Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 2002; 43:1096 - 105; http://dx.doi.org/ 10.1093/pcp/pcf156; PMID: 12407188 [DOI] [PubMed] [Google Scholar]
- 44.Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, et al. . T-DNA insertional mutagenesis for functional genomics in rice. Plant J 2000; 22:561 - 70; http://dx.doi.org/ 10.1046/j.1365-313x.2000.00767.x; PMID: 10886776 [DOI] [PubMed] [Google Scholar]
- 45.Jeong DH, An S, Park S, Kang HG, Park GG, Kim SR, Sim J, Kim YO, Kim MK, Kim SR, et al. . Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J 2006; 45:123 - 32; http://dx.doi.org/ 10.1111/j.1365-313X.2005.02610.x; PMID: 16367959 [DOI] [PubMed] [Google Scholar]
- 46.Hiei Y, Ohta S, Komari T, Kumashiro T. . Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 1994; 6:271 - 82; http://dx.doi.org/ 10.1046/j.1365-313X.1994.6020271.x; PMID: 7920717 [DOI] [PubMed] [Google Scholar]
- 47.Lin SY, Ikehashi H, Yanagihara S, Kawashima A. . Segregation distortion via male gametes in hybrids between Indica and Japonica or wide-compatibility varieties of rice (Oryza sativa L). Theor Appl Genet 1992; 84:812 - 8; PMID: 24201479 [DOI] [PubMed] [Google Scholar]
- 48.Kurusu T, Sakurai Y, Miyao A, Hirochika H, Kuchitsu K. . Identification of a putative voltage-gated Ca2+ -permeable channel (OsTPC1) involved in Ca2+ influx and regulation of growth and development in rice. Plant Cell Physiol 2004; 45:693 - 702; http://dx.doi.org/ 10.1093/pcp/pch082; PMID: 15215504 [DOI] [PubMed] [Google Scholar]
- 49.Kurusu T, Hamada J, Nokajima H, Kitagawa Y, Kiyoduka M, Takahashi A, Hanamata S, Ohno R, Hayashi T, Okada K, et al. . Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol 2010; 153:678 - 92; http://dx.doi.org/ 10.1104/pp.109.151852; PMID: 20357140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia K, Liu T, Ouyang J, Wang R, Fan T, Zhang M. . Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res 2011; 18:363 - 77; http://dx.doi.org/ 10.1093/dnares/dsr024; PMID: 21795261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jain M, Nijhawan A, Tyagi AK, Khurana JP. . Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 2006; 345:646 - 51; http://dx.doi.org/ 10.1016/j.bbrc.2006.04.140; PMID: 16690022 [DOI] [PubMed] [Google Scholar]
- 52.Fuse T, Sasaki T, Yano M. . Ti-Plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol 2001; 18:219 - 22; http://dx.doi.org/ 10.5511/plantbiotechnology.18.219 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.