Abstract
Macroautophagy (hereafter autophagy) is a regulated intracellular process during which cytoplasmic cargo engulfed by double-membrane autophagosomes is delivered to the vacuole or lysosome for degradation and recycling. Atg8 that is conjugated to phosphatidylethanolamine (PE) during autophagy plays an important role not only in autophagosome biogenesis but also in cargo recruitment. Conjugation of PE to Atg8 requires processing of the C-terminal conserved glycine residue in Atg8 by the Atg4 cysteine protease. The Arabidopsis plant genome contains 9 Atg8 (AtATG8a to AtATG8i) and 2 Atg4 (AtATG4a and AtATG4b) family members. To understand AtATG4’s specificity toward different AtATG8 substrates, we generated a unique synthetic substrate C-AtATG8-ShR (citrine-AtATG8-Renilla luciferase SuperhRLUC). In vitro analyses indicated that AtATG4a is catalytically more active and has broad AtATG8 substrate specificity compared with AtATG4b. Arabidopsis transgenic plants expressing the synthetic substrate C-AtAtg8a-ShR is efficiently processed by endogenous AtATG4s and targeted to the vacuole during nitrogen starvation. These results indicate that the synthetic substrate mimics endogenous AtATG8, and its processing can be monitored in vivo by a bioluminescence resonance energy transfer (BRET) assay. The synthetic Atg8 substrates provide an easy and versatile method to study plant autophagy during different biological processes.
Keywords: Arabidopsis, AtATG8, AtATG4 cysteine protease, citrine yellow fluorescence protein, Renilla luciferase
Autophagy is an evolutionarily conserved process to remove or recycle cellular components during growth, development, and various stress responses. Autophagosomes, which are double-membrane compartments formed during autophagy, carry cargo material to the vacuole or lysosomes. Biogenesis of the autophagosome requires multiple components including the Atg4 cysteine protease and the substrate, the ubiquitin-like protein Atg8. Unlike yeast, the Arabidopsis genome encodes 2 Atg4 (AtATG4a and AtATG4b) and 9 Atg8 (AtATG8a to AtATG8i) isoforms. Expansion of AtATG4 and AtATG8 members in plants suggests that different members may participate in activating autophagy in different tissues during various biological processes. Recently, we developed a unique approach to understand differential enzyme activity of AtATG4s. For this, we generated a new and versatile AtATG8 synthetic substrate, C-AtATG8-ShR. This substrate contains a pH stable variant of yellow fluorescent protein citrine (C) at the N terminus of AtATG8 and a plant optimized Renilla luciferase SuperhRLUC (ShR) at the C terminus (Fig. 1). This synthetic substrate when processed by AtATG4 cysteine proteases at the conserved C-terminal glycine residue in AtATG8 will result in the generation of C-AtATG8 and ShR byproducts. This system provides a fast and easy assay to differentiate the processed ShR byproduct from the full-length uncleaved substrate on a native polyacrylamide gel by applying the luciferase substrate coelenterazine (CLZ) directly onto the gel. We refer to this assay as native gel assay of SuperhRLUC catalytic activity (NASCA).
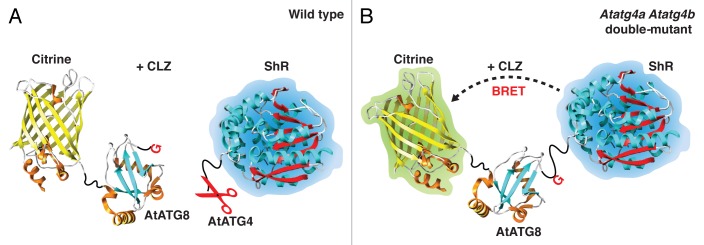
Figure 1. Illustration of the BRET-based AtATG8 synthetic substrate C-AtATG8-ShR. A synthetic substrate, C-AtATG8-ShR, was engineered to contain citrine (C) and a plant-optimized Renilla luciferase (SuperhRLUC, ShR) fused to the N and the C terminus of AtATG8, respectively. Intramolecular bioluminescence resonance energy transfer (BRET) from ShR to citrine occurs in the presence of the luciferase substrate, coelenterazine (CLZ). This synthetic substrate can be processed by the cysteine protease AtATG4 in the wild-type cells (A). Cleavage of the synthetic substrate by AtATG4 results in separation of the energy donor, ShR, from the energy acceptor, citrine, resulting in a decreased BRET ratio. However, in Atatg4a Atatg4b double-mutant cells, the proximity of citrine and ShR allows transfer of resonance energy generated by ShR to citrine, resulting in excitation of the latter (B).
The NASCA assay demonstrated preferential substrate selectivity of AtATG4a and AtATG4b to the 9 AtATG8 isoforms. The results indicated preferential AtATG4a-mediated cleavage of AtATG8a, AtATG8c, AtATG8d, and AtATG8i. The other AtATG8 isoforms were processed with similar efficiency by the 2 AtATG4 proteins. Therefore, AtATG4a has predominant activity in processing and production of certain mature AtATG8 isoforms. This hints at a potential selective function for the different AtATG8 isoforms in planta during different biological processes. Interestingly, the differential processing activity of AtATG4s might not be due to different AtATG8 substrate affinity, but rather due to higher catalytic activity of AtATG4a. However, human ATG4s show preferential enzyme activity by differential LC3/Atg8 substrate affinity. ATG4 proteases from different organisms show high sequence similarity at the amino acid level. In fact, similar to the human ATG4 studies, hydrogen peroxide reversibly inhibits AtATG4 enzyme activity in vitro. However, AtATG4 fails to process a human Atg8 homolog, LC3A, in the NASCA assay, suggesting that biochemical properties of Atg4 proteases might be distinct among homologs from different organisms.
Our synthetic substrate C-AtATG8-ShR is also useful to monitor autophagy in vivo. Since the absorption spectrum (λmax = 516 nm) of citrine in C-AtATG8-ShR overlaps with the emission spectrum of ShR (λmax = 475 nm), in the presence of CLZ, bioluminescence resonance energy transfer occurs within a single molecule (Fig. 1). In nonautophagy-inducing conditions, proximity between citrine and ShR will allow significant BRET to occur. In autophagy-inducing conditions, the synthetic substrate C-AtATG8-ShR will be cleaved by AtATG4s and therefore the BRET ratio (yellow fluorescence/blue luminescence) will be significantly lower. We were able to use Arabidopsis transgenic plants expressing the synthetic substrate C-AtATG8a-ShR to monitor the rate of autophagy during nitrogen starvation. In addition, this substrate also allowed us to visualize autophagosomes in the vacuole. Our analyses of transgenic wild-type and Atatg4a Atatg4b double-mutant plants expressing C-AtATG8a-ShR indicated that processing and delivery of the synthetic substrate to the vacuole requires endogenous AtATG4 function. Therefore, transgenic Arabidopsis expressing our new synthetic substrate C-AtATG8-ShR provides a great tool for studying autophagy at a whole plant level at temporal and spatial resolution.
Currently, the precise role of each AtATG8 family member in autophagosome biogenesis and cargo recruitment is unknown. Modification of the synthetic substrate described here with different fluorescent proteins should facilitate simultaneous monitoring of different AtATG8 family members during growth, development, and different biotic and abiotic stress responses in plants. In addition, the system is amenable for high throughput screening for regulators of autophagy in Arabidopsis.
Glossary
Abbreviations:
- NASCA
native gel assay of SuperhRLUC catalytic activity
- BRET
bioluminescence resonance energy transfer
- CLZ
coelenterazine
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is supported by the University of California, Davis, and the National Science Foundation Molecular and Cellular Biosciences grant NSF-MCB-1355459 (to SPD-K). National Science Foundation Grant NSF-IOS-1258135 funded to Dr Drakakaki supported EP.