Abstract
Background and purpose
A rapid learning approach has been proposed to extract and apply knowledge from routine care data rather than solely relying on clinical trial evidence. To validate this in practice we deployed a previously developed decision support system (DSS) in a typical, busy clinic for non-small cell lung cancer (NSCLC) patients.
Material and methods
Gender, age, performance status, lung function, lymph node status, tumor volume and survival were extracted without review from clinical data sources for lung cancer patients. With these data the DSS was tested to predict overall survival.
Results
3919 lung cancer patients were identified with 159 eligible for inclusion, due to ineligible histology or stage, non-radical dose, missing tumor volume or survival. The DSS successfully identified a good prognosis group and a medium/poor prognosis group (2 year OS 69% vs. 27/30%, p < 0.001). Stage was less discriminatory (2 year OS 47% for stage I–II vs. 36% for stage IIIA–IIIB, p = 0.12) with most good prognosis patients having higher stage disease. The DSS predicted a large absolute overall survival benefit (~40%) for a radical dose compared to a non-radical dose in patients with a good prognosis, while no survival benefit of radical radiotherapy was predicted for patients with a poor prognosis.
Conclusions
A rapid learning environment is possible with the quality of clinical data sufficient to validate a DSS. It uses patient and tumor features to identify prognostic groups in whom therapy can be individualized based on predicted outcomes. Especially the survival benefit of a radical versus non-radical dose predicted by the DSS for various prognostic groups has clinical relevance, but needs to be prospectively validated.
Keywords: Lung cancer, Rapid learning, Decision support system, Radiotherapy
Lung cancer has a global incidence of 1.6 million with 1.4 million lung cancer deaths per year [1]. In the developed world, 85% of cases are non-small cell lung cancer (NSCLC) patients and 44% present with non-metastatic disease, for whom primary radical surgery or radiotherapy, in combination with chemotherapy, is indicated [2]. The outcome of inoperable NSCLC undergoing radical (chemo)radiotherapy is bleak with two-year overall survival (2 year-OS) rates reported as approximately 50% in early stage (stage I–II) patients [3] and 25% in locally advanced cancers (stage IIIA–IIIB) [4]. To improve survival, more individualized radiotherapy schedules have been proposed with encouraging results [5].
To inform decision making and enable individualized therapy a validated prediction of expected outcome is needed for an individual patient. This can be provided via clinical decision support systems (DSS) [6]. A rapid learning health-care system, where we can learn from each patient to guide practice, has been proposed to extract knowledge, such as a DSS, from routine clinical care data rather than solely relying on evidence from clinical trials [7,8]. Previous rapid learning work has resulted in a DSS (www.predictcancer.org) which can predict overall survival in non-small cell lung cancer patients treated with radical (chemo) radiotherapy [9]. This DSS might be used in a clinic as an objective guide to individualize treatment, and discussions with patients, according to prognosis.
To validate a DSS, input features and outcome data from the local care population (called the clinical cohort in this report) are needed. However capturing and extracting data requires significant resources, which limits the acceptance of DSSs and rapid learning in general. If successfully extracted, such new data cannot only be used for validation but also to identify improvements and to assess clinical relevance by investigating if and how the DSS might change local practice.
In this study, we hypothesize that it is possible to deploy a rapid learning environment which can be used to validate the above DSS. Our aim is to apply rapid learning in practice, to see if this novel approach is feasible in a typical, busy cancer center, to test the limits and areas of improvement and to determine the clinical relevance of the DSS.
Materials and methods
Patients
All lung cancer patients in whom electronic routine care data were available at Liverpool and Macarthur Cancer Therapy Centres, Sydney, Australia were selected for consideration for the clinical cohort. This is an integrated cancer center which multi-disciplinary treats approximately 2000 patients per year with radiotherapy and records data in an oncology information system (MOSAIQ, Elekta, Stockholm, Sweden). A previously published [9] DSS was learned from a cohort consisting of 322 consecutive lung cancer patients from MAASTRO Clinic, The Netherlands using 2-norm support vector machines, using two year overall survival, calculated from the start of the RT, as the outcome measure. This cohort is called the training cohort in this report. The study was approved by the internal ethics review boards of both institutes.
To match the clinical cohort to the training cohort the following inclusion and exclusion criteria were applied. Inclusion: non-small cell lung cancer; stage I–IIIB; prescribed radiation dose 4500 cGy or higher with at least one fraction of radiotherapy administered. Exclusion: small cell lung cancer, stage IV, prescribed radiation less than 4500 cGy. Two additional criteria were applied to the clinical cohort: no missing data for the outcome feature (2 year-OS) and the tumor volume (the sum of local and regional gross tumor volume in cubic centimeters). The first is needed to validate the DSS, the second is the most sensitive feature of the DSS for which imputation was deemed too unreliable. Missing data were allowed for the other DSS input features being gender, ECOG performance status, forced expiratory volume in 1 s (FEV1), number of positive lymph node stations on a PET scan (PLNS); these were imputed before validation (see below).
Data sources and extraction of the clinical cohort
The oncology information system and treatment planning system (XiO, Elekta, Stockholm, Sweden) were used as the data sources for the clinical cohort. From the treatment planning system a custom bulk export script was used to extract all CT-delineated structures and their volume as DICOM objects which were stored in an open source java-based PACS (DCM4CHEE) [10]. An open source java-based data integration platform (Kettle, Pentaho, Orlando, FL, USA) was used to extract and store the relevant features as statements on patients in the form of triples (e.g., Patient1-hasGenderMale) to allow maximal flexibility in the data model [11].
Data quality, missing data and imputation
Data found in the clinical data sources were assumed to be correct and complete. No review or update of clinical sources was performed. The underlying hypothesis is that the amount of data will compensate sufficiently for any data quality issues that may or may not be present. The quality of the data extraction process was checked via spot crosschecks with the source systems and with a spreadsheet from previous work that contained data manually collected on a subset of patients. No errors were found.
Clinical data sources often have missing data elements. In this study these were imputed using a Bayesian network. Such a network can leverage existing data to infer missing data (e.g., a N0 stage patient is likely to have zero positive lymph nodes on a PET scan) [12]. This network was learned from the training cohort (Hugin Researcher, Hugin Expert, Aalborg, Denmark) and used to impute ECOG performance status, FEV1 and PLNS.
Estimating DSS performance and statistical considerations
The DSS was implemented and validated in Matlab (Mathworks, Natick, MA, USA). After imputation of missing data elements, the predicted 2 year-OS in the clinical cohort was compared with actual outcome. The predictive performance of the DSS was assessed in terms of discrimination and calibration [13]. Discrimination between those reaching and those not reaching two year survival was tested using the area-under-the-curve (AUC) of the receiver operating characteristic curve and the Matthews correlation coefficient at 50% predicted probability of survival. From previous work, cut-offs in terms of survival probability were defined to divide the training cohort into three prognostic groups (poor, medium and good prognosis consisting of 25%, 50% and 25% of patients respectively). The same cut-offs were applied to the clinical cohort and the difference in survival of the three prognostic groups was tested using Kaplan–Meier survival curves and a logrank test. Calibration refers to the agreement between observed outcomes and predictions and was assessed using a calibration plot with the same three prognostic groups.[14] A significance level of 0.05 was used with a Bonferroni–Holm correction for multiple comparisons between the two cohorts and between multiple survival curves.
Role of the funding source
All funding sources were public and had no role in any of the following areas: study design; collection, analysis, and interpretation of data; writing of the report; decision to submit the paper for publication.
Results
Data extraction & imputation
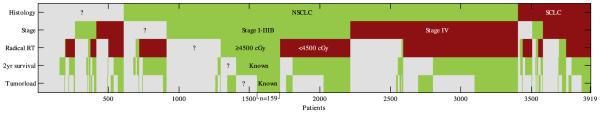
In total 3919 patients (1995–2013) were identified as lung cancer patients of which 1301 had known stage I–IIIB NSCLC with 419 known to have received radical radiotherapy (>4500 cGy). Ultimately 159 patients were eligible for inclusion in the clinical cohort. In Fig. 1 a diagram is given which shows the reasons for exclusion of patients. Since tumor volume was only recorded after the introduction of 3D treatment planning and 2 year-OS had to be known, the clinical cohort only included patients treated between September 2002 and August 2011. The characteristics of the clinical and training cohort are given in Table 1. The clinical cohort had statistically significant larger tumor volumes. Staging and lung function where somewhat different compared to the training cohort, but these did not reach statistical significance. The clinical cohort also had significant missing data for PLNS (100% missing) and FEV1 (60%). Interestingly the Bayesian network imputed a worse general condition (poorer ECOG and lower FEV1) for patients with lower stage disease than for patients with higher stage disease (not shown).
Fig. 1.
Diagram showing the reduction in the number of eligible patients when applying the inclusion and exclusion criteria (histology, stage and radical RT) and when excluding patients with missing data elements (2 year survival and tumor volume). A total of 3919 had a diagnosis of lung cancer in the source system, of these 159 were eligible for inclusion in the clinical cohort. Green indicates patients meeting the inclusion criteria or having a known data element, red patients were excluded because of an exclusion criteria. Gray indicates patients in which a criteria or data element was unknown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Patient characteristics.
| Training cohort | Clinical cohort | |||
|---|---|---|---|---|
| Site | Netherlands | Australia | ||
| # Patients | 322 | 159 | ||
|
Age (years)
Gender |
Mean ± SD | 69 ± 10 | 68 ± 11 | p = NS |
| Male | 249 (77%) | 113 (71%) | p = NS | |
| Female | 73 (23%) | 46 (29%) | p = NS | |
| Stage | ||||
| I | 73 (23%) | 26 (16%) | p = NS | |
| II | 29 (9%) | 26 (16%) | p = NS | |
| IIIA | 81 (25%) | 62 (39%) | p = NS | |
| IIIB | 134 (42%) | 45 (28%) | p = NS | |
| Missing | 5 (2%) | 0a (0%) | ||
| ECOG | ||||
| 0 | 94 (29%) | 49 (31%) | p = NS | |
| 1 | 169 (53%) | 72 (45%) | p = NS | |
| ≥2 | 52 (16%) | 13 (8%) | p = NS | |
| Missing | 7 (2%) | 25 (16%) | ||
| FEV1 (%) | ||||
| Mean ± SD | 70 ± 24 | 77 ± 20 | p = NS | |
| Missing | 48 (15%) | 95 (60%) | ||
| Tumor volume (cc) | ||||
| Mean ± SD | 106 ±113 | 161 ± 147 | p < 0.001 | |
| Median | 77 | 106 | ||
| Missing | 36 (11%) | 0a (0%) | ||
| PLNS | ||||
| 0 | 143 (44%) | N/A | ||
| 1 | 59 (18%) | N/A | ||
| 2 | 44 (14%) | N/A | ||
| 3 | 31 (10%) | N/A | ||
| ≥4 | 30 (9%) | N/A | ||
| Missing | 15 (5%) | 159b (100%) | ||
| 2 year OS | ||||
| Yes | 103 (32%) | 58 (36%) | p = NS | |
| No | 219 (68%) | 101 (64%) | p = NS | |
| Missing | 0 (0%) | 0a (0%) |
ECOG: Performance status as defined by Eastern Cooperative Oncology Group; FEV1: forced expiratory volume in 1 s; tumor volume = gross tumor volume of the primary tumor and involved nodes; PLNS = number of positive lymph node stations on a PET scan; 2 year OS = two year overall survival after the start of radiotherapy. p-Values corrected for multiple comparisons using Bonferroni–Holm.
Patients with missing data were excluded from the clinical cohort.
Is not recorded in routine clinical practice in the Australian center.
DSS performance
The AUC of the DSS in the clinical cohort was 0.69, which is lower than the AUC found in the training cohort (AUC 0.75). The receiver operating curve showed that the DSS performed better in the clinical cohort at higher operating points (medium-good prognosis,) than at lower operating points (poor prognosis). The Matthews correlation coefficient at 50% predicted survival was 0.35 with false negatives the main limit of the model at this operating point.
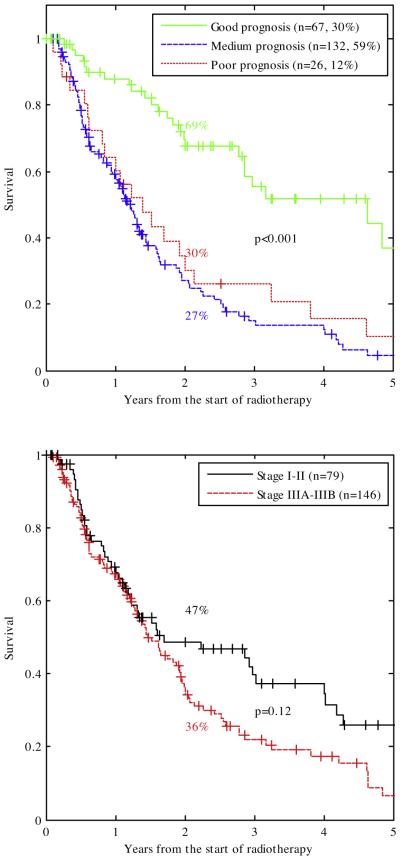
The Kaplan–Meier curves for the good, medium and poor prognosis group are given in Fig. 2(top). This shows that a good prognosis group was successfully identified (2 year-OS 69%, p < 0.001), but that the survival difference between the medium and poor prognosis group (2 year-OS 30% and 27%, p = NS) could not be reproduced in the clinical cohort. More than half of the good prognosis group consisted of stage IIIA–IIIB patients. Compared to the training cohort, the clinical cohort had fewer patients in the poor prognosis group (25% vs. 12%).
Fig. 2.
Top: Kaplan–Meier curves of the clinical cohort (including censored survival data so numbers are higher than Table 1) stratified by good, medium and poor prognostic score from the training cohort [9]. The two year overall survival for these three groups were 69%, 27% and 30% respectively. The separation between the good and medium/bad prognosis group was highly significant (p < 0.001). More than half of the good prognosis group comprised advanced stage patients (20 stage I, 10 stage II, 27 stage IIIA and 10 stage IIIB patients). Unlike what was found in previous work, the medium and poor prognosis group of the clinical cohort did not show a significant difference in survival. Bottom: Kaplan–Meier curves of the clinical cohort including censored survival data stratified by TNM stage. The difference between stage I–II and stage IIIA–IIIB did not reach significance (2 year-OS 47% vs. 36%, p = 0.12). p-Values corrected for multiple comparisons using Bonferroni–Holm.
Fig. 2(bottom) shows that TNM stage I–II and IIIA–IIIB could also discriminate between a good (2 year OS 47%) and poor prognosis group (2 year OS 36%), but that discrimination was not as good as the DSS and did not reach statistical significance (p = 0.12).
The calibration plot is given in Fig. 3. The calibration was not different from ideal in the medium prognosis group, but survival was higher than predicted in the poor and good prognosis groups.
Fig. 3.
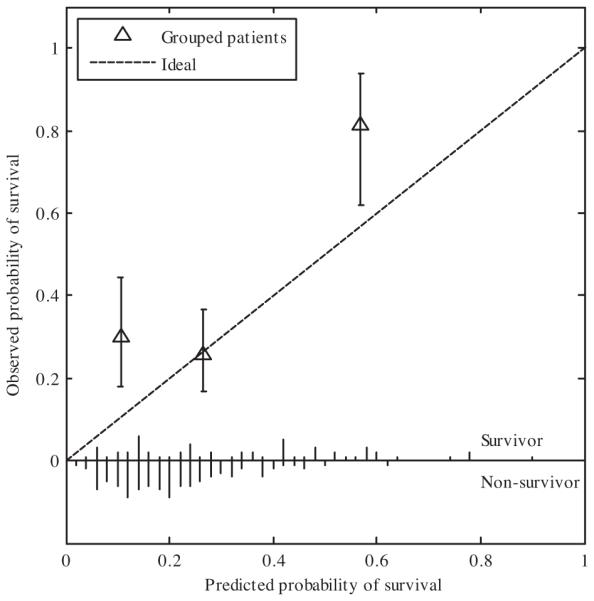
Calibration plot of the DSS for the clinical cohort using three prognosis groups. Each point represents the predicted and observed probability for the group. The error bar is the 95% confidence interval. On the axis the number of survivors and non-survivors are shown per 2% interval predicted probability. The calibration curve of the medium prognosis group is not different from the ideal curve, in the good and poor prognosis group a higher than predicted survival is observed.
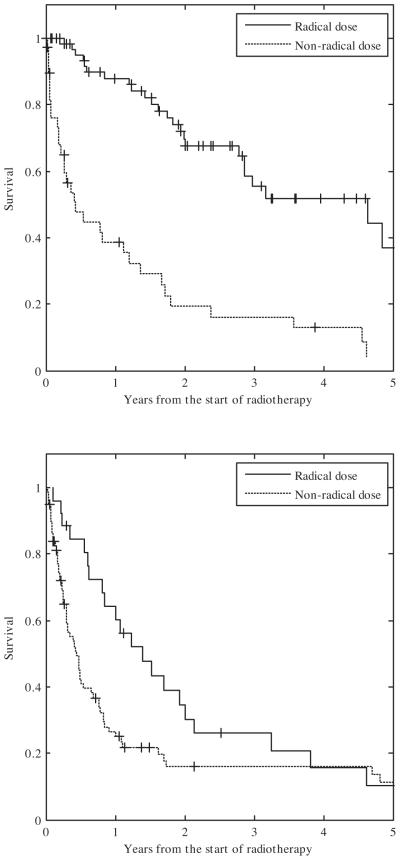
In Fig. 4 the Kaplan–Meier survival curves are given of patients treated to a radical dose vs. a non-radical dose for the good and poor prognosis groups. In both groups the patients receiving a radical radiation dose are seen to have a better survival than those not receiving a radical dose. This effect is more pronounced in the good prognosis group than in the poor prognosis group.
Fig. 4.
Kaplan–Meier curves of the patients treated with a radical vs. those treated with a non-radical radiation dose. Top: good prognosis patients. Bottom: poor prognosis patients.
Discussion
Rapid learning has been proposed as a framework whereby “routinely collected real-time clinical data drive the process of scientific discovery, which becomes a natural outgrowth of patient care.” [8,15]. In this study, we have demonstrated a practical application of these ideas. We deployed a rapid leaning environment in which clinical data were extracted and used to validate DSS, itself learned on clinical data from another center in another country. We believe that achieving interoperability and leveraging data from many centers will be necessary to achieve sufficient data volume and variety for valuable knowledge extraction.
The Achilles heel of rapid learning is data quality. Unlike data from prospective trials, clinical data have errors, bias and are often missing. This might lead to a “garbage in garbage out” qualification of the knowledge gained in rapid learning and might prevent one from answering even the most basic questions. This study demonstrates that the situation is not that bleak. Clinical data from “front-line” physicians are at least able to successfully validate a DSS that can identify patients who are more likely to survive than others (Fig. 2).
We used the data ‘as is’ without any effort to improve the quality of the data. This was deliberate, as it is not feasible to do this for the large number of patients in a busy clinic. This can be viewed as both a weakness and a strength of this study. The weakness is that the underlying assumption of data gaps and errors being random may not be valid. The strength is that this study shows that even from routine clinical data with no quality checks there is something to learn.
Starting from close to 4000 patients, 419 patients were identified who met the basic inclusion criteria and only 159 were included in the clinical cohort, which is sobering given the mild criteria applied in this study, although this “rapid learning” inclusion is still much higher than the number of patients in clinical trials which is estimated to be 2–3% in Australia [16]. Fig. 1 shows that besides the histology and staging distribution, both of which were in line with expected levels [2], only a minority of patients with non-metastatic disease were treated with a radical dose. Such insights and the ability to detect variations in local practice are an important feature of a rapid learning environment.
Missing data led to a large reduction in the number of eligible patients as well as required imputation in the eligible patients (Table 1). A relatively easy way to avoid missing data is the use of naming conventions [17]. Other data elements such as FEV1, ECOG status and PLNS are often implicitly known based on the available free text information, but significant extra time is required to document these data in a structured manner. For these novel data capture or data transfer methods may be considered. Some of these missing data are unavoidable due to patients treated before the era of CT simulation or too recently for the outcome. These reasons are clearly time dependent which raises the concern that, due to the inevitable correlation of time with practice, this exclusion is biased. There is some evidence that this is true. The clinical cohort contained mostly patients from 2006 to 2011 as earlier patients had often tumor volume missing and later patients did not have enough follow-up. In a validation analysis (not shown) these later patients (2011–2013) were more often stage I patients and less often stage IIIB patients perhaps indicating early diagnoses and/or other changes in practice over time. Another possibility of bias is the staging method. As the training cohort came from 2002 to 2006 and the clinical cohort primarily from 2006 to 2011 and they came from different institutes it cannot be excluded that the more recent clinical cohort was differently and more correctly staged due to more modern staging methods and/or that the institutes used different staging methods with one of the two perhaps superior. As an example, Table 1 shows that the training cohort had 42% stage IIIB patients versus 28% in the clinical cohort. It cannot be excluded that in the training cohort inferior staging methods where used and that part of these stage IIIB patients are in fact stage IV patients that were incorrectly staged. There are no straightforward answers to the problem of biased and biased-missing data. But having more patients from a wide range of eras and institutions in combination with domain expertise will help in detecting bias. Bayesian methods are probably the best way of modeling bias [18] and, for instance, can be used to model the probability that the staging is correct depending on the staging method used.
A method for imputation is needed as missing data are unavoidable. In previous work we showed that Bayesian networks are excellent to handle missing data [12]. The observation that the Bayesian network imputed a better general condition (low ECOG and high FEV1) for higher stage patients may be explained by the selection of lower stage patients in good general condition for surgery. This may also explain that stage is not a very good predictor of survival in inoperable patients as was observed previously [19] and can also be seen in Fig. 2(bottom). This point is further emphasized by the observation that most patients in the good prognosis group were stage IIIA–IIIB patients. Although it seems better than mean imputation, the imputation of the Bayesian Network is still far from perfect. In a validation analysis (results not shown) the imputed ECOG status and FEV1 were compared to patients with observed ECOG and FEV1. As it is based on the training cohort, the Bayesian Network has a strong preference for the most common ECOG status (ECOG 1) of the training cohort and predicts a low FEV1 in stage I patients which is not observed in the clinical cohort. In future work learning better and more extensive networks based on more patients may make this imputation more generalizable and better.
We applied a DSS that was learned from a completely independent training cohort and can predict survival in non-small lung cancer patients. The DSS had a reasonable discriminatory performance in the clinical cohort and was able to identify patients with a good prognosis but could not discriminate between a medium and poor prognosis. The observed AUC (0.69) is better than physician predicted outcomes (AUC 0.56) as was recently shown in a prospective study [20].
The cut-off for radical radiation in this study was taken as 45 Gy based on the idea that such a dose was intended to be radical at the time it was given. However one could question if a different cut-off such as 60 Gy, which currently has the highest level of evidence, would not have been better. To check this, a validation analysis (not shown) was done after including only patients with 60 Gy or more in the clinical cohort. This did not change any of the conclusions which might be a result of the rather flat dose–response curve if one assumes that a biological equivalent dose of 70 Gy is needed to control 50% of the tumor [21] and that a dose of 60 Gy in once-daily fractions has a biological equivalence of less than 50 Gy.
In the calibration analysis, we showed that patients in both the good and poor prognosis survived longer than predicted (Fig. 3). Possible explanations for the limited performance of the DSS are that (a) clinical practice at the Australian centers is different from the Netherlands in that patients with comorbidities and suboptimal general condition were more likely to be treated with palliative radiotherapy (Fig. 1) and (b) there are confounding features not included in the model at the moment due to the limited training set (e.g., concurrent chemotherapy, details of palliative care) which have a positive influence on survival. Including the Australian cohort in the training set would not only allow the model to learn from these patients (e.g., to model the effect of radical vs. non-radical dose) but also allow more features to be included without the risk of overfitting due to the increased size of the training set. But the addition of the Australian cohort to the training cohort to learn an updated DSS would also require another external cohort for its validation. Such a rapid learning network that allows addition of diverse datasets from centers worldwide to learn new, improve existing and validate DSS is the subject of ongoing research.
The clinical relevance of this work lies in the individualization of therapy. From a guideline perspective the inoperable, non-metastatic NSCLC population is homogenous with concurrent chemoradiation as its preferred treatment. However many patients are not meeting the highly selective criteria for clinical trials from which guidelines are derived. The patient may not be considered fit enough for concurrent chemoradiation or a radical radiotherapy dose may be considered too toxic due to the tumor size or location. A DSS learned and validated using rapid learning approaches such as presented here, support these clinical decisions objectively and can be used to direct resources to the patients that might benefit most from them. The good-prognosis patients in whom currently a non-radical dose is given, can be expected to especially benefit from advanced technologies such as intensity modulated and image guided radiotherapy in order to feasibly treat these patients with a radical dose. The comparison given in Figs. 3 and 4 (top) also gives some indication of the expected and testable survival benefit: The model predicted two year overall survival is about 60% for good prognosis patients if they are given a radical dose (Fig. 3) while the clinical cohort suggests that a non-radical dose leads to a survival of only about 20% in good prognosis patients (Fig. 4 (top)).
Vice versa, one can question if poor prognosis patients should receive long course radiotherapy to a radical dose as the DSS does not predict a survival benefit compared to palliative radiotherapy (Figs. 3 and 4 (bottom)). Although rapid learning thus give guidance in these often difficult decisions, the use of a DSS and resulting practice changes should be rigorously and prospectively tested to validate that these practice changes indeed result in better outcomes. This prospective validation is the subject of future work where in a randomized seamless Phase II–III design patients will be treated with or without the help of a DSS predicting survival (and toxicity based on previous work) with the primary Phase II endpoint being a change in practice between the two arms and Phase III the primary endpoint overall survival which, if successful, would lead to level I evidence for the use of a DSS in these patients.
The previous insights on the clinical relevance and the limitations of the DSS have been made possible by the inclusion of data from a site that treats patients differently than the cohort from which the DSS was learned. This is a general characteristic of rapid learning. Only by combining diverse datasets from a wide variety of patient populations and health care practices will the size and diversity of the data be sufficient to perform clinically relevant rapid learning. In our rapid learning vision, new data from additional sources and/or new patients are used to continuously improve and validate knowledge and change practice.
In future work the rapid learning environment described in this report, will be deployed at more hospitals to improve the DSS in the following manner: first this work clearly shows the DSS needs a better accuracy and generalizability especially in patients with a poor prognosis. Second, to increase its clinical relevance, the DSS should predict both survival and quality-of-life affecting toxicities of the various treatments. Third, the DSS should be able to handle missing data rather than relying on imputation during pre-processing, as was done in the present study. Given these requirements, Bayesian networks seem to be the ideal machine learning approach for this type of work [12,22] and learning and validating these is the subject of future work.
Finally the efforts described in this report need to be linked to other rapid learning initiatives, such as CancerLinQ of the American Society of Clinical Oncology, to achieve a true multi-disciplinary rapid learning environment [23].
Acknowledgements
This research was financially supported by the Australian Department of Health and Ageing (DoHA), Better Access to Radiation Oncology (BARO) initiative, euroCAT (IVA Interreg, www.eurocat.info), CTMM (AIRFORCE project, grant 030-103, TraIT project, grant 05T-401), EU 6th and 7th framework program (METOXIA, EURECA, ARTFORCE), Kankeronderzoekfonds Limburg from the Health Foundation Limburg and the Dutch Cancer Society (KWF UM 2011-5020, KWF UM 2009-4454). None of these funding sources had any involvement in study design, in the collection, analysis and interpretation of data, in the writing of the manuscript or in the decision to submit the manuscript for publication. We would like to thank Ms. Nasreen Kaadan and Dr. Ariyanto Pramana for providing data for cross checking.
Footnotes
Conflicts of interest statement
Andre Dekker: No Conflicts of interest.
Shalini Vinod: No Conflicts of interest.
Lois Holloway: No Conflicts of interest.
Cary Oberije: No Conflicts of interest.
Armia George: No Conflicts of interest.
Gary Goozee: No Conflicts of interest.
Geoff P. Delaney: No Conflicts of interest.
Philippe Lambin: No Conflicts of interest.
David Thwaites: No Conflicts of interest.
References
- [1].Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- [2].Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- [3].Smith SL, Palma D, Parhar T, Alexander CS, Wai ES. Inoperable early stage nonsmall cell lung cancer: comorbidity, patterns of care and survival. Lung Cancer. 2011;72:39–44. doi: 10.1016/j.lungcan.2010.07.015. [DOI] [PubMed] [Google Scholar]
- [4].O’Rourke N, Macbeth F. Is concurrent chemoradiation the standard of care for locally advanced non-small cell lung cancer? A review of guidelines and evidence. Clin Oncol. 2010;22:347–55. doi: 10.1016/j.clon.2010.03.007. [DOI] [PubMed] [Google Scholar]
- [5].De Ruysscher D, van Baardwijk A, Steevens J, Botterweck A, Bosmans G, Reymen B, et al. Individualised isotoxic accelerated radiotherapy and chemotherapy are associated with improved long-term survival of patients with stage III NSCLC: a prospective population-based study. Radiother Oncol. 2012;102:228–33. doi: 10.1016/j.radonc.2011.10.010. [DOI] [PubMed] [Google Scholar]
- [6].Lambin P, van Stiphout RGPM, Starmans MHW, Rios-Velazquez E, Nalbantov G, Aerts HJWL, et al. Predicting outcomes in radiation oncology–multifactorial decision support systems. Nat Rev Clin Oncol. 2013;10:27–40. doi: 10.1038/nrclinonc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sullivan R, Peppercorn J, Sikora K, Zalcberg J, Meropol NJ, Amir E, et al. Delivering affordable cancer care in high-income countries. Lancet Oncol. 2011;12:933–80. doi: 10.1016/S1470-2045(11)70141-3. [DOI] [PubMed] [Google Scholar]
- [8].Abernethy AP, Etheredge LM, Ganz PA, Wallace P, German RR, Neti C, et al. Rapid-learning system for cancer care. J Clin Oncol. 2010;28:4268–74. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dehing-Oberije C, Yu S, De Ruysscher D, Meersschout S, Van Beek K, Lievens Y, et al. Development and external validation of prognostic model for 2-year survival of non-small-cell lung cancer patients treated with chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2009;74:355–62. doi: 10.1016/j.ijrobp.2008.08.052. [DOI] [PubMed] [Google Scholar]
- [10].Warnock MJ, Toland C, Evans D, Wallace B, Nagy P. Benefits of using the DCM4CHE DICOM archive. J Digit Imaging. 2007;20:125–9. doi: 10.1007/s10278-007-9064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pathak J, Kiefer RC, Chute CG. Using linked data for mining drug–drug interactions in electronic health records. Stud Health Technol Inform. 2013;192:682–6. [PMC free article] [PubMed] [Google Scholar]
- [12].Jayasurya K, Fung G, Yu S, Dehing-Oberije C, De Ruysscher D, Hope A, et al. Comparison of Bayesian network and support vector machine models for two-year survival prediction in lung cancer patients treated with radiotherapy. Med Phys. 2010;37:1401–7. doi: 10.1118/1.3352709. [DOI] [PubMed] [Google Scholar]
- [13].Bouwmeester W, Zuithoff NPA, Mallett S, Geerlings MI, Vergouwe Y, Steyerberg EW, et al. Reporting and methods in clinical prediction research: a systematic review. PLoS Med. 2012;9:e1001221. doi: 10.1371/journal.pmed.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21:128–38. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lambin P, Petit SF, Aerts HJWL, van Elmpt WJC, Oberije CJG, Starmans MHW, et al. From population to voxel-based radiotherapy: exploiting intra-tumour and intra-organ heterogeneity for advanced treatment of non-small cell lung cancer. Radiother Oncol. 2009;2010:145–52. doi: 10.1016/j.radonc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- [16].Grand MM, O’Brien PC. Obstacles to participation in randomised cancer clinical trials: a systematic review of the literature. J Med Imaging Radiat Oncol. 2012;56:31–9. doi: 10.1111/j.1754-9485.2011.02337.x. [DOI] [PubMed] [Google Scholar]
- [17].Santanam L, Hurkmans C, Mutic S, van Vliet-Vroegindeweij C, Brame S, Straube W, et al. Standardizing naming conventions in radiation oncology. Int J Radiat Oncol Biol Phys. 2012;83:1344–9. doi: 10.1016/j.ijrobp.2011.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Luta G, Ford MB, Bondy M, Shields PG, Stamey JD. Bayesian sensitivity analysis methods to evaluate bias due to misclassification and missing data using informative priors and external validation data. Cancer Epidemiol. 2013;37:121–6. doi: 10.1016/j.canep.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dehing-Oberije C, De Ruysscher D, van der Weide H, Hochstenbag M, Bootsma G, Geraedts W, et al. Tumor volume combined with number of positive lymph node stations is a more important prognostic factor than TNM stage for survival of non-small-cell lung cancer patients treated with (chemo) radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70:1039–44. doi: 10.1016/j.ijrobp.2007.07.2323. [DOI] [PubMed] [Google Scholar]
- [20].Oberije C, Nalbantov G, Dekker A, Boersma L, Borger J, Reymen B, et al. A prospective study comparing the predictions of doctors versus models for treatment outcome of lung cancer patients: a step toward individualized care and shared decision making. Radiother Oncol J Eur Soc Ther Radiol Oncol. 2014 doi: 10.1016/j.radonc.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Van Baardwijk A, Bosmans G, Bentzen SM, Boersma L, Dekker A, Wanders R, et al. Radiation dose prescription for non-small-cell lung cancer according to normal tissue dose constraints: an in silico clinical trial. Int J Radiat Oncol Biol Phys. 2008;71:1103–10. doi: 10.1016/j.ijrobp.2007.11.028. [DOI] [PubMed] [Google Scholar]
- [22].Oh JH, Craft J, Al Lozi R, Vaidya M, Meng Y, Deasy JO, et al. A Bayesian network approach for modeling local failure in lung cancer. Phys Med Biol. 2011;56:1635–51. doi: 10.1088/0031-9155/56/6/008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sledge G, Miller R, Hauser R. CancerLinQ and the future of cancer care. Am Soc Clin Oncol Educ Book. 2012;2013:430–4. doi: 10.14694/EdBook_AM.2013.33.430. [DOI] [PubMed] [Google Scholar]