Abstract
Extracellular vesicles have created great interest as possible source of biomarkers for different biological processes and diseases. Although the biological function of these vesicles is not fully understood, it is clear that they participate in the removal of unnecessary cellular material and act as carriers of various macromolecules and signals between the cells. In this report, we analyzed the proteome of extracellular vesicles secreted by primary hepatocytes. We used one- and two-dimensional liquid chromatography combined with data-independent mass spectrometry. Employing label-free quantitative proteomics, we detected significant changes in vesicle protein expression levels in this in vitro model after exposure to well-known liver toxins (galactosamine and Escherichia coli-derived lipopolysaccharide). The results allowed us to identify candidate markers for liver injury. We validated a number of these markers in vivo, providing the basis for the development of novel methods to evaluate drug toxicity. This report strongly supports the application of proteomics in the study of extracellular vesicles released by well-controlled in vitro cellular systems. Analysis of such systems should help to identify specific markers for various biological processes and pathological conditions.
Keywords: LC–MS, Label-free quantitation, Microvesicles, Exosomes, Hepatocytes, Liver
1. Introduction
The qualitative and quantitative analysis of (sub)proteomes is an important step toward better understanding of diverse biological functions, and is one of the greatest challenges in the field of proteomics. Mass spectrometry quantitation is already widely applied in comparative studies of protein expression but the majority of the relative quantitative methods use isotopic labeling. Such analytical schemas involve multiple sample preparation steps to incorporate the label either metabolically or chemically [1–5]. One important limitation of labeling approaches is that the number of available tags might not be sufficient for the simultaneous discrimination of multiple analytes [6,7]. Recently, label-free LC–MS quantitation methods have been increasingly employed to compare the levels of various proteins under different conditions. Some quantitative, label-free LC–MS-based strategies for the profiling of complex protein mixtures have been reported. These strategies rely either on spectral counting methods [8,9] or on the direct measurement of signal intensity [10–14]. Label-free LC methods for the quantitative analysis of proteins have been recently reviewed [15,16]. In contrast to label-based techniques, label-free methods are not restricted by the number of samples; however, more care has to be taken to minimize experimental variation, mainly involving the sample preparation stage.
The successful application of quantitative proteomics in biomedicine is difficult because of the complexity and dynamic range of protein samples derived from various tissues and body fluids. Recently, extracellular cell-secreted vesicles (EVs) [17] were recognized as a novel biological material with reduced protein complexity and created interest as a potential source of disease biomarkers. These vesicles fall mainly into two groups, depending on their size, origin, and the mechanism of their release: the endosome-derived vesicles named “exosomes” and the vesicles shed from plasma membranes, referred to as ectosomes or microparticles (MPs). Exosomes are intraluminal vesicles (40–150 nm) produced by inward budding of the limiting membrane of multivesicular bodies (MVB), which are the central organelles of the endocytic and secretory pathways [18]. There is a growing body of evidence that there are at least 2 different kinds of MVBs. One class that ends up in the lysosomes and another class that fuses with the plasma membrane. The latter type is responsible for releasing the exosomes into the extracellular space [19]. As a consequence of their endosomal origin, exosomes contain proteins involved in membrane transport, fusion, and MVB biogenesis, including CD9, CD63, CD81, Rab GTPases, annexins, flotillin, Alix and Tsg101. MPs are a population of vesicles that vary in size (0.1–1.0 µm), and are formed by outward budding of the cell plasma membranes in response to different stimuli. These vesicles are shed by different cell types and express a subset of cell surface proteins that depend on the cells of origin [20,21]. Although the cell biology of these two types of EVs is different, both types circulate in the adjacent extracellular space and appear in biological fluids after their release from the cells. They have been identified in human, rodent, and fetal calf sera [22–27]. They are released both by the cells of haematopoietic and non-haematopoietic origin [28,29], quiescent and activated [30], and non-transformed and tumor cells [31]. Because of their involvement in the intercellular signaling, the examination of their protein components in the healthy and diseased individuals may provide valuable markers for determining the site, type, and an extent of injury in various pathological conditions.
Our group reported the secretion of EVs by the primary hepatocytes in culture [32]. In the current report, we identified novel putative markers for liver injury in an in vitro model. We used two well-known hepatotoxins, galactosamine (galN), which causes liver injury resembling acute viral hepatitis [33], and Escherichia coli-derived lipopolysaccharide (LPS) promoting liver inflammation and damage [34–38]. Finally, by using an animal model for acute liver injury, we showed that similar protein alterations can be detected in the EVs isolated from sera. Our results provide the basis for the creation of novel, non-invasive tools to assess liver toxicity supporting the use of EVs as a biological source of disease biomarkers.
2. Experimental procedures
2.1. Reagents
All media and reagents for tissue culture were purchased from Invitrogen (Carlsbad, CA). All other reagents were of analytical grade and mainly acquired from Sigma-Aldrich (St. Louis, MO). Monoclonal antibodies were purchased from the following vendors: anti-Clusterin (clone 0.T.19) and anti-CPS1 (clone OCH1E5) from Abcam (Cambridge, UK), anti-Hsp70 (clone BRM-22) from Sigma Chemical Co. (St. Louis, MO), anti-Hsp90 (clone 68) and anti-AIP1/Alix (clone 49) from BD Biosciences (Mountain View, CA). Rabbit polyclonal antibodies were purchased from the following vendors: anti-FRIL1 (clone D-9) was purchased from Santa Cruz Biotech. Inc. (Santa Cruz, CA), anti-SLC27A2, anti-SULT1, and anti-Tsg101 from Abcam (Cambridge, UK). Goat anti-CES3 (clone M-14) and anti-COMT were from Santa Cruz Biotech., Inc. and Abcam, respectively. Horseradish peroxidase (HRP)-conjugated secondary antibody was from GE Healthcare (Buckinghamshire, UK).
2.2. Animal experimentation
All the animal experimentation was conducted in accordance with the Spanish Guide for the Care and Use of Laboratory Animals (RD 1201/2005 — BOE 21/10/05). Eight male 14-week-old Sprague–Dawley rats (body weight 300–400 g) were maintained in an environmentally controlled room at 22 °C on a 12 h light/ dark cycle and provided with standard diet (Rodent Maintenance Diet, Harlan Teklad Global Diet 2014) and water ad libitum. The rats were randomly allocated to two groups. The test group (n = 4) received an intraperitoneal injection of 1000 mg/kg/5 ml of d(+)-galactosamine (2-amino-2-deoxy-d-galactose) hydrochloride (Sigma-Aldrich) and the control group animals (n = 4) were injected with the same volume of saline solution (5 ml/kg of sterile 0.9% NaCl). Individual urine samples were obtained 6 h after injection; the animals were housed in metabolic cages for a 12 h period. The animals were sacrificed 18 h after the injection. We obtained blood and liver samples from each animal. An aliquot of 250 µL of serum from each animal was used to determine alanine transaminase (ALT) activity using Infinity™ ALT(GPT) Liquid Stable Reagent (Thermo Electron, Waltham, MA). Carboxylesterase (CES) activity was examined as previously described by Polsky-Fisher et al. [39]. The remaining serum samples were pooled and used to obtain EVs from sera of each experimental group as described below. Livers were frozen in liquid nitrogen and used to obtain protein extracts for Western blot analysis.
2.3. Primary cell culture preparation and extracellular vesicle production
Suspensions of primary rat hepatocytes were prepared as described by Seglen [40] from the livers of 14-week-old male rats, and seeded onto collagen-coated 150-mm dishes, at 20 × 106 cells per dish. Each suspension was split into three equally represented sets to be used under different conditions. One set was used as a control and incubated in the culture media (exosome-depleted DMEM, 25 mM HEPES, 10% fetal bovine serum, penicillin–streptomycin) (condition I). Two other sets were treated with culture media containing 10 mM galN (condition II) or 10 µg/ml LPS (condition III). Cells were incubated at 37 °C and 5% CO2 for 36 h and EVs secreted to the extracellular media were isolated as described below. MTT assay showed a significant impact on the hepatocytes viability ejected by these conditions (Supplementary Fig. S1) in agreement with previous reports.
2.4. Extracellular vesicle purification
EVs were isolated from the conditioned media and the rat sera pooled using the methodology previously described by Conde-Vancells et al. [32]. Briefly, samples were centrifuged at 500 ×g for 10 min and the supernatants were filtered through 0.22 µm pore filters to enrich the preparation in exosome-like vesicles. The samples were then centrifuged at 10,000 ×g and 100,000 ×g for 30 min and 60 min, respectively. While the pellet obtained after 10,000 ×g was discarded, the pellet obtained in the 100,000 ×g centrifugation was resuspended and washed with PBS, and centrifuged again at 100,000 ×g for 60 min. The final pellet of EVs was resuspended in PBS to 1/2000 of the original volume and the aliquoted solutions were stored at −80 °C. Western-blotting of the obtained EV preparations showed no significant presence of the proteins Grp78 (endoplasmic reticulum marker) or prohibitin (mitochondria marker) as reported in Conde-Vancells et al. [32] supporting that purified vesicles were not produced by cell lysis (data not showed).
2.5. Western blot analysis
Cell lysates were prepared by lysing 106 trypsinized cells for 15 min on ice in 100 µL of lysis buffer (300 mM NaCl, 50 mM Tris pH 7.4, 0.5% Triton X-100, and protease inhibitors). After centrifugation at 20,000 ×g, the supernatants were transferred to fresh Eppendorf tubes. To prepare the liver extracts, 50 mg of frozen liver tissue was homogenized in 1 mL of cold lysis buffer (20 mM Tris pH 7.5; 150 mM NaCl; 10 mM NaH2PO4; 2 mM NaF; 1 mM EDTA; 0.1 mM EDTA pH 8; 1% Triton X-100; 1 µM Na3VO4, and a cocktail of protease inhibitors) in a Precellys 24 homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France), using CK14 beads, for 23 s at 6500 rpm.
The protein concentration of the cell lysates, liver extracts, and purified EVs were determined using Bradford Protein Assay (Bio-Rad, Hercules, CA) with BSA as the standard. SDS sample buffer was added and samples were incubated for 5 min at 37 °C, 65 °C and 95 °C and separated on 4–12% pre-casted acrylamide gels (Invitrogen, Carlsbad, CA). After transfer to PVDF membranes (Millipore, Bedford, MA) and blocking overnight in 5% milk and 0.05% Tween-20 in PBS, the primary antibody was added for 1 h, followed by PBS wash and the application of the secondary HRP-conjugated antibody. Chemiluminescence detection was performed with ECL Plus Reagents (GE Healthcare).
2.6. Tryptic digestion
The proteins were extracted from the isolated vesicles by incubating the samples with 0.1% SDS in 0.5 M triethyl-ammonium bicarbonate on ice for 30 min; protein solubilization was aided by gentle pipetting and brief sonication. The insoluble materials were spun down and the protein concentration was determined using a Bradford Protein Assay kit (Bio-Rad). Each sample (50 µg of protein) was lyophilized and suspended in 50 mM ammonium bicarbonate (pH 8.5) with 0.05% RapiGest (Waters Corporation, Milford, MA), to redissolve the lyophilized peptides. The samples were incubated at 60 °C for 15 min. Each sample was reduced in the presence of 10 mM dithiothreitol at 60 °C for 30 min. The protein mixture was alkylated in the dark, with 50 mM iodoacetamide, at room temperature for 30 min. Proteolytic digestion was initiated by adding modified trypsin at a ratio of 1:10 (w:w) and incubated overnight at 37 °C. To hydrolyze the RapiGest surfactant, 2 µl of HCl was added to the sample. The sample was then incubated at 37 °C for 30 min, centrifuged for 30 min at 13,000 rpm, and the supernatant recovered.
2.7. One- and two-dimensional LC–DIA-MS proteomic analysis
The principle of a DIA acquisition is briefly explained since it is primarily used for the quantitative interpretation of LC–MS data. Post-acquisition, the peak detection program interrogates both low and elevated functions, giving a chromatographic precursor and product ion apices. This forms the basic premise of the applied DIA (LC–MSE) acquisition method; namely, the top peaks of the product ions within the elevated collision energy function have exactly the same retention time as their corresponding precursors within the low collision energy function. Moreover, the combination of high-peak capacity chromatographic separation and high-sampling rate orthogonal acceleration ToF MS maximizes the detection of all eluting species across the complete chromatographic space. An example precursor and product ion mass from extracted DIA chromatograms and the annotated elevated energy MS spectrum for the identification of a peptide from glutathione-S-transferase are shown in Supplementary Fig. S2. As mentioned-previously, the duty cycle of the mass spectrometer is maximized in a DIA strategy which increases the dynamic range of this approach in comparison with DDA-based methods [41].
Proteins were identified and quantified by direct analysis of the tryptic digest samples described above. All analyses were performed using a nanoAcquity system (Waters Corporation) and Q-ToF Premier mass spectrometer (Waters Corporation, Manchester, UK). Dried samples were resuspended in 0.1% formic acid, spiked with 50 fmol of yeast enolase digest, and analyzed by reverse-phase LC. For the one-dimensional LC–MS experiments, tryptic peptides were focused and desalted on a 5 µm Symmetry C18, 180 µm × 2 cm trapping cartridge (Waters Corporation) and further separated on a 1.7 µm BEH C18 (75 µm ID × 15 cm) analytical column (Waters Corporation). Peptides were eluted at a flow rate of 300 nL/min from the analytical column directly coupled to an electrospray ionization emitter tip (New Objective, Woburn, MA) using a 90 min gradient from 3 to 50% solvent B [99.9% acetonitrile (ACN), 0.1% FA].
Two-dimensional chromatography was performed using a 5 µm Xbridge BEH130 C18 (300 µm ID × 50 mm) first dimension column. Peptides were loaded in 20 mM ammonium formate (pH ~ 10) in water containing 0.1% FA and eluted by injecting solvent plugs of ACN (11.1, 14.5, 17.4, 20.8, 45 and 65%) [42]. After injection of a buffer plug, the peptide fraction was eluted from the SCX column and subsequently retained on a 5 µm Symmetry C18 (180 µm × 2 cm) reversed-phase trap column by dilution with water at pH 2. The second dimension separations were performed in a manner identical to 1D-LC reversed-phase separation described in the previous paragraph.
Peptides were analyzed in the positive ion mode using a Q-ToF Premier mass spectrometer, which was operated in v-mode with the resolving power of 10,000 FWHM. Prior to analyses, the ToF analyzer was calibrated using the fragment ions of [Glu1]-Fibrinopeptide B. After calibration, the data were lock mass-corrected using the doubly charged precursor ion of [Glu1]-Fibrinopeptide B (785.8426 m/z), which was acquired with sampling frequency of 30 s. Accurate LC–DIA-MS mass data were collected in a data-independent acquisition mode by alternating the energy applied to the collision cell between low and elevated collision energy state. The time of each elevated/low acquisition was 1 s with 0.1 s inter-scan delay. Low energy data were acquired at constant collision energy of 4 eV; elevated collision energy acquisitions were obtained using a 15–40 eV ramp. The RF applied to the quadrupole mass analyzer was adjusted so that the ions from m/z 320 to about 2400 Da were efficiently transmitted, ensuring that any ion with a mass below m/z 300 only arose from the dissociation in the collision cell [43]. For each assayed condition seven technical 1D-LC–DIA-MS replicates and two 2D-LC–DIA-MS technical replicates were obtained.
2.8. Data processing, database searching, and quantitation analysis
ProteinLynx Global Server version 2.4 (Waters Corporation) was used to process all acquired data. The lock mass-corrected spectra were centroided, deisotoped, and charge-state-reduced to produce a single, accurately measured monoisotopic mass for each detected precursor and product ion. The correlation of a precursor and a potential fragment ion is initially achieved by means of time alignment. Further correlation is obtained during the database searches based on the physicochemical characteristics of peptides when they undergo collision induced fragmentation [44]. Protein identifications were obtained by searching reviewed Rattus norvegicus UniProt database entries (2012_03, 7769 entries). The sequence of Saccharomyces cerevisiae enolase internal standard was appended to the database. Protein identifications from the low/high collision spectra for each sample were filtered after the search. The matching process required more than three fragment ions per peptide, seven fragment ions per protein and more than two peptides per protein. Single-peptide protein identifications were not considered. Peptide and fragment ion tolerances were determined automatically by the program (approximately 10 and 25 ppm, respectively). The allowed number of missed cleavages was 1; the fixed modification chosen was carbamidomethyl cysteine, and variable modifications, oxidation of methionine and N-terminal acetylation. The initially set maximum false discovery rate (FDR) at the protein level of the identification algorithm was 4%. However, the final FDR is minimized by applying the replication filter since false positive identifications tend to be random and as such are not reproduced in all injections. Final reporting and statistical analysis of the qualitative results was conducted with Scaffold v4.0.3 (Proteome Software, Inc., Portland, OR).
For the quantitative analysis, intensity measurements were obtained by integrating the total ion volume of each extracted, charge-state-reduced, deisotoped and mass-corrected ion across the mass-spectrometric and chromatographic space. If a particular component exists as more than a single charge-state, the corresponding area for any given monoisotopic ion is reported as the summed area from all contributing charge-states and isotopes. For relative protein quantitation, the intensity of each peptide is normalized against the intensity value of peptides of the internal standard added to the sample (50 fmol of enolase). Quantitative analyses were only performed with the 1D-LC– DIA-MS data, comparing peak area/intensity of each peptide from the control to the values for the treated samples using ProteinLynx Global Server v2.4 [44]. The quantitative results were additionally filtered. We considered only the protein identifications with good technical replication rate (at least 3 out of 7 technical replicates). The relative abundances of these proteins in controls and treated samples had to be significantly different (expressed as an upregulation probability value) [44]. Moreover, the variance of regulation, expressed as a 95% credible interval, had to be smaller than 0.05. To filter out any non-significant, non-regulated proteins, a single standard (z) score was used as a final reporting filter. This filter was derived from the mean and eighty-fifth percentile of the regulation distributions of the control vs. GalN and control vs. LPS experiments; its value was approximately ±1.3.
Condition-unique protein identifications were only considered when the estimated amount was more than 10 times the amount of the 10% percentile of the complete experimental data set comprising twenty-one LC–DIA-MS runs [45,46].
2.9. Gene Ontology analysis and networks, functional and pathway mapping
The functional annotations of the identified proteins were initially assigned using Protein Center software (http://proteincenter.proxeon.com, Proxeon Bioinformatics, Odense, Denmark). Three main types of annotations were obtained from the Gene Ontology Consortium website: cellular components, molecular functions, and biological distribution. GO Slim mapping defined specifically for Protein Center helped to reduce the multiple GO annotations to a manageable set of approximately 20 high-level terms; these were used for filtering the information into percentage estimates. The Ingenuity Pathway Analysis program (IPA) (Ingenuity Systems, Redwood City, CA) was used to find the pathways associated with the proteins identified in the mass spectrometry analysis. The program uses computational algorithms to identify local networks that are particularly enriched in the data sets. Such local networks contain the most highly connected focus proteins that specifically interact with other proteins in the network. The filters and general settings for the core analysis were set to consider all the molecules as well as direct and indirect relationships. All data sources, tissues and cell lines were taken into account and a stringent filter for molecules and relationships was chosen. Networks of focus genes were then algorithmically generated on the basis of their connectivity and ordered by the score. This score reflects the relevance of the network based on a p-value calculation, i.e., the probability that the network-eligible molecules have been found in a network by chance alone. Networks were also associated with biological functions (and/or diseases) that were most significant for the proteins in the network. Fisher’s exact test was used to calculate a p-value determining the probability that the assignment of each biological function and/or disease to that network was due to chance alone.
3. Results
3.1. Qualitative analysis of EVs released from hepatocytes
Previously, we reported an overview of proteins from rat hepatocyte EVs [32]. In that study, we used SDS-polyacrylamide matrix to increase the resolving power of the overall analysis. Each gel slice was subjected to LC–MS/MS analysis; eventually, 234 proteins were successfully identified. In the current work, we used gel-free proteomics and only 8% of the proteins detected in our previous study were not detected in the current. In our 1D-LC–DIA-MS and 2D-LC–DIA-MS analyses, we identified 412 and 557 proteins listed in Supplementary Tables S1 and S2, respectively. To maximize the power of the method we used three biological replicates and seven technical replicates were used to assess technical LC–MS variation [47].
The qualitative 1D-LC–DIA-MS and 2D-LC–DIA-MS analyses (summarized in Supplementary Tables S1 and S2) typically exceeded an identification probability of 95%, while maintaining a protein and peptide FDR of 4% and 1%, respectively. The coverage of the characterized proteins ranged from 2 to 89%, with 2 to 147 peptides per protein (Supplementary Tables S1 and S2).
In order to gain an insight into the functional roles of hepatocyte-derived EVs, clustering on molecular function GO-based categories was performed for the 557 identified proteins detected in these vesicles (Fig. S3). A significant enrichment in proteins involved in energy production and lipid, amino acid, carbohydrate, and drug metabolism was revealed. These results suggested the participation of hepatocyte-derived EVs in those processes, in agreement with the fact that hepatocytes play a central role in the metabolism of essential and harmful substances. We also observed enrichment in molecules belonging to protein synthesis, folding, modification, trafficking, and degradation categories, suggesting that EVs have a role in extracellular protein homeostasis.
3.2. Quantitative analysis of EVs released by hepatocytes
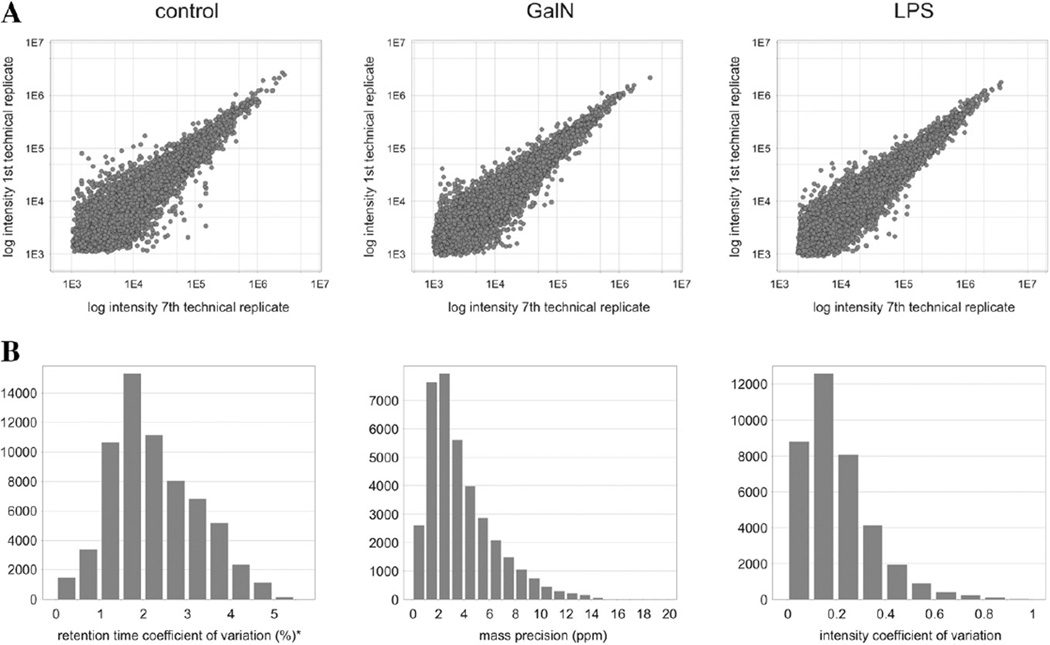
We also performed a quantitative proteomic study of EVs released by primary hepatocytes challenged with model hepatotoxins galN and LPS to detect candidate markers of liver injury. The quantitative analysis was performed using three biological replicates as described in the Experimental procedures section, using exclusively the 1D-LC–DIA-MS data and three independent preparations of primary rat hepatocytes. The quality of the DIA data was checked in terms of reproducibility as a first step in our quantitative analysis. The biological replicates allowed assessing the similarity between the animals and the reproducibility of the EVs isolation procedure, while the seven technical LC–MS replicates helped to evaluate the experimental variation during the 1D-LC–DIA-MS procedures. First, the intensity variations for all matched peptide components were evaluated by comparing the corresponding values from replicate injections of one of the biological replicas for each sample. Under ideal conditions, a binary comparison between technical replicates should provide a 45-degree diagonal intersecting the origin and displaying minimal signal variation. The resulting distribution between the first and seventh injection from each sample showed minor intensity variation (Fig. 1A). The control and galN- and LPS-treatment samples all show a high degree of technical reproducibility maintained during the 42 h of instrument operating time. The measurement statistics of the accurate mass–retention pairs (AMRT), i.e., a deconvoluted mass eluting at a given retention time, were also evaluated. The reproducibility of the retention time assigned to AMRT components across replicate injections was high, with an average centering on 2% (left panel in Fig. 1B). In addition, the median mass precision within any given AMRT bin was overall less than 2 to 3 ppm (middle panel in Fig. 1B). The median intensity variation within an AMRT bin was approximately 15% (right panel in Fig. 1B).
Fig. 1.
(A) Comparison of intensity measurements of the matched accurate mass–retention time components for the first and last technical replica of one of the biological replicas of each sample type: control, galN, and LPS. (B) Left: The average retention time coefficient of variation (CV) centered at 0.5%; Middle: The mass precision measurements (ppm) from all detected accurate mass-retention time clusters were within ±5 ppm, with the median mass measurement precision of approximately 2 ppm; Right: The relative standard deviation of the measured signal intensity of the clusters.
Although they were only used indirectly to justify thresholds for condition-unique proteins, the quantitative (molar) values of the control sample were plotted against the number of proteins to assess the technical LC–MS variation; the estimated amounts also reflect the consistency of intensity measurement. The three most abundant ions identified in a protein were used for quantitation purposes and normalized against a known concentration of a standard (50 fmol of enolase). Using this method [43], it was possible to quantify protein amounts over almost three orders of magnitude (from 0.05 ng to 50 ng), even though the majority were within two specific orders (from 0.4 ng to 40 ng) (Supplementary Fig. S4A). Furthermore, a comparison of these quantitative values and the relative values of the binary comparisons showed a linear regression trend in both instances (Supplementary Figs. S4B and C).
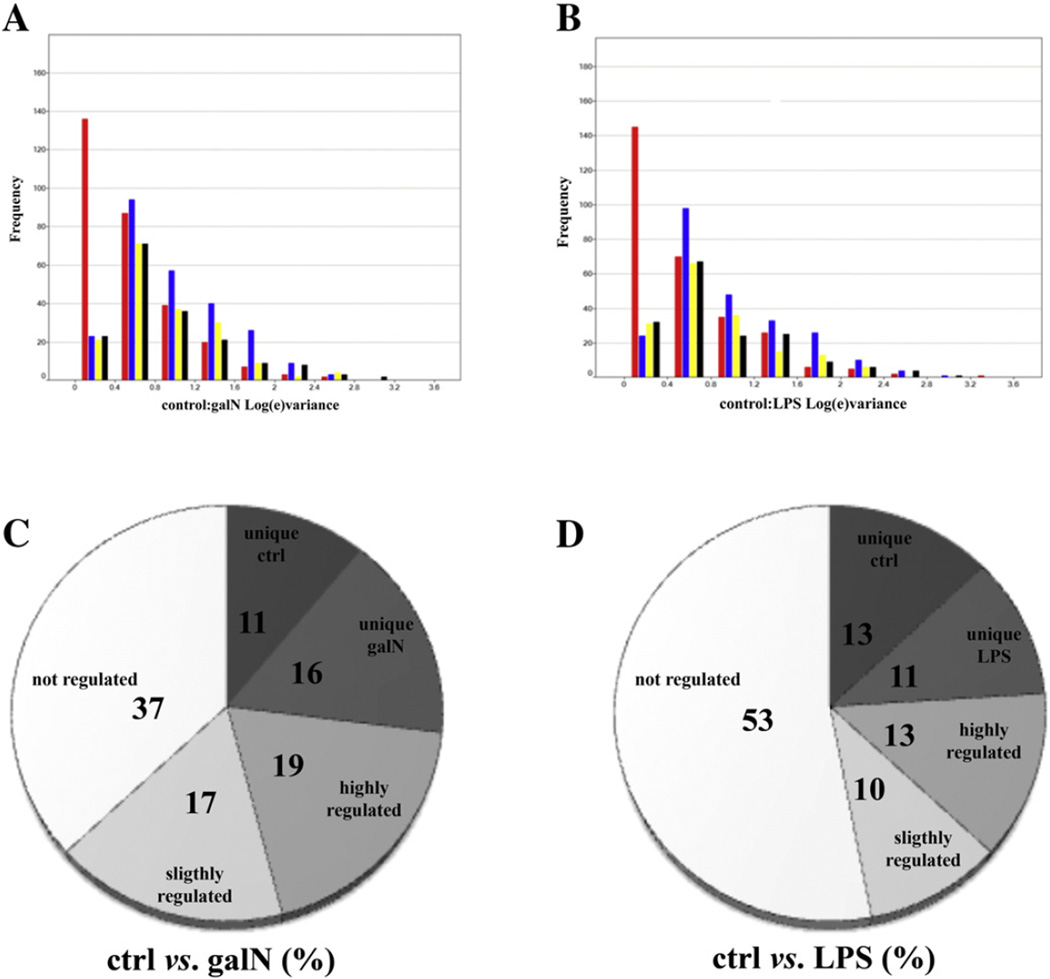
Binary comparisons of the confidence levels for the intensity measurements in all the experiments and technical replicas were made to calculate the total experimental variance. Fig. 2A and B show the confidence levels of control versus galN and control versus LPS treatment, respectively. The red bars represent all experiments, including technical and biological variability; the blue, yellow, and black bars stand for the technical variability for the first, second and third test animal, respectively. The true biological variation plus the variation due to protein extraction, sample processing, and LC–MS analysis was approximately 16% (average), with the majority of the data below 12% (median). Consequently, for the label-free quantitative data, the significance of regulation threshold was assumed to be at least 30%, to increase the likelihood that the variation was due to differences among samples and not to technical variations. Following these criteria of selection for the 412 proteins identified by 1D-LC– DIA-MS, 63% and 47% of the EVs proteins show significant regulation after galN or LPS treatment, respectively (Fig. 2C and D). This observation agrees with the degree of the toxicity of the treatment; galN treatment is more severe than LPS, leading to more profound expression level changes in the EV proteome. The relative number of substantially regulated proteins (fold-change greater than 1.5) was also greater after galN treatment (Fig. 2C). The higher number of proteins regulated in EVs in comparison with the cellular or tissue proteome itself [48–52] might be explained by the fact that EV proteome, with its lower complexity, is more sensitive to environmental changes. This phenomenon supports the hypothesis that EVs are a suitable source of biomarkers.
Fig. 2.
(A & B) Confidence levels for galN vs. control (A) or LPS vs. control (B). The red bars represent all replicas including biological and technical variation, the blue bars, the biological variance for subject A (first test animal), the yellow bars, the biological variance for subject B (second test animal), and the black bars, biological variance for subject C (third test animal). Frequency corresponds to the number of counts in the indicated interval of variance. (C & D) Percentage of the up- or down-regulated proteins after treatment with galN (C) and LPS (D). Annotation diagram (D): “not significantly regulated” stands for the proteins whose ratio is between 1 and 1.3, “slightly regulated” for the proteins with a ratio between 1.3 and 1.5, and “substantially regulated” for those with a ratio higher than 1.5. Proteins called “unique” are the proteins that were identified only under one of the conditions (control, galN, or LPS).
An overview of the regulated proteins identified and quantified in the control, galN-treated, and LPS-treated samples are shown in Fig. 3 and detailed in Tables S3 and S4 (supplementary data), with their relative fold-changes and a probability of regulation values. When a protein was only identified in one of the samples (“sample- or condition-unique identifications”) no fold-change could be calculated. For these particular protein identifications to be included in the final report, the estimated amount had to be at least 10 times greater than the 10% amount percentile of the complete dataset, otherwise, the protein would have been identified in the other conditions/samples.
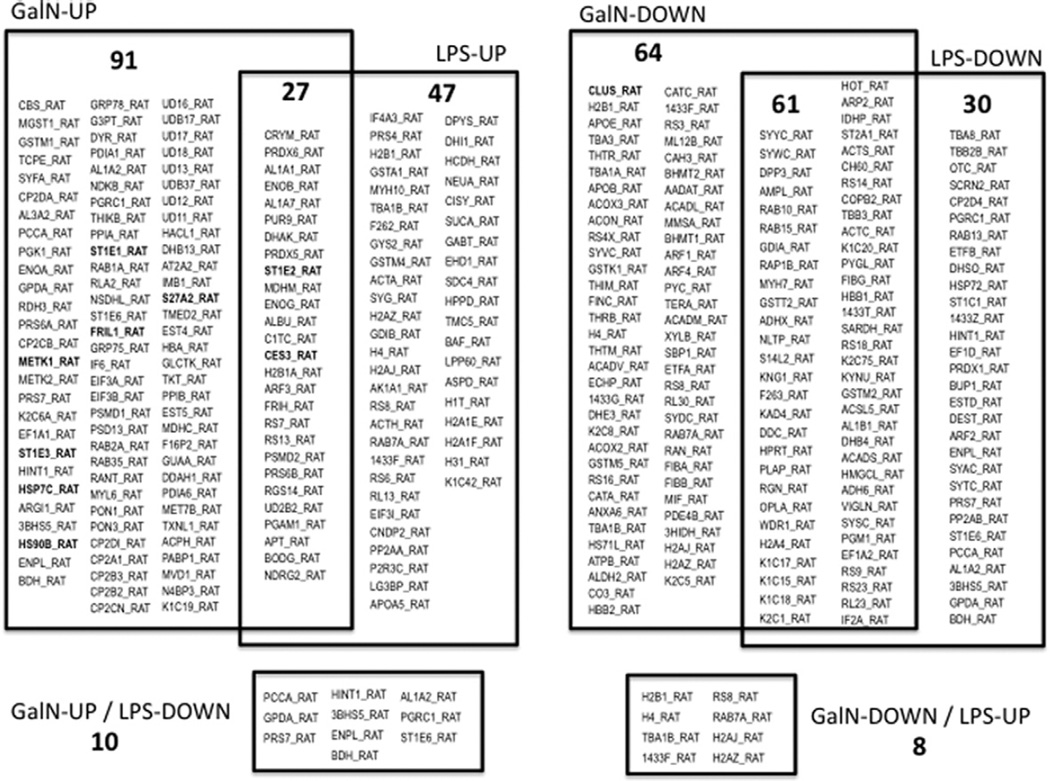
Fig. 3.
Regulation by galN or LPS treatments of hepatic EV-associated proteins. Numbers of common and treatment-specific up- or down-regulated proteins are indicated along with their IDs. A more detailed information including fold change, significance and description of the proteins are indicated in Tables S3 and S4.
Overall, 88 proteins displayed a common quantitative trend in EVs released by galN- or LPS-treated hepatocytes (Fig. 3). This set of proteins possibly reflects a general response of hepatocytes to an external injury, independently of the etiology of the insult. A significant number of these proteins are structural and regulatory cytoskeletal proteins such us keratins, actins, tubulins, actin-related protein 2 (ARP2), and small GTPases (RAP1B, RAB10 and 15). This group also includes coatomer subunit beta′ (COPB2) and ADP-ribosylation factor 3 (ARF3), involved in the regulation of vesicular trafficking. Ribosomal proteins, tRNA synthases and peptidases involved in protein synthesis and degradation, enzymes involved in carbohydrate (ENOB, ENOG, PGM1, PGAM1, C1TC, PYGL, AL1B1, ADH6, APT), lipid (ACADS, ACSL5, VIGLN, NLTP) and xenobiotic (CES3, SULT1) metabolism, and proteins involved in iron (ferritin) and redox (GSTs, PRDX5, PRDX6) homeostasis were similarly affected by both hepatotoxins. These results suggest that reorganization of the cellular morphology, vesicular trafficking, protein synthesis and degradation as well as adjustments in carbon, lipid, iron, and redox metabolisms take place in response to hepatotoxins. A number of 18 proteins were found to be regulated as an effect of both treatments, although in opposite directions and include histones, ribosomal, metabolic enzymes (GPDA, PCCA) and regulatory (14-3-3 eta) proteins (Fig. 3). There were some proteins regulated only in response to galN or LPS treatment (155 and 77, respectively). These events might represent the tuning of the cellular response to the more significant lesions generated by the two toxins; i.e., cell death (galN) and inflammation (LPS) (Fig. 3, Table S3 and S4). A GO-based enrichment IPA analysis carried out for these treatment-specific responses revealed that galN-treatment induced enzymes associated with xenobiotic metabolism. This treatment also affected a significant number of proteins involved in glutathione metabolism (Fig. S5); this result supports the mechanism described for galN-mediated cellular toxicity involving severe oxidative stress [53]. Proteasomal proteins and those involved in cell movement were more affected by LPS treatment (Fig. S6). Together, these results reflect the expected reorganization taking place in the cells in response to an external insult. The data support the suggestion that the analysis of the released EVs is a suitable non-invasive way to infer the events taking place within the living cells.
3.3. Biochemical validation of up- & down-regulated proteins in EVs from rat hepatocytes
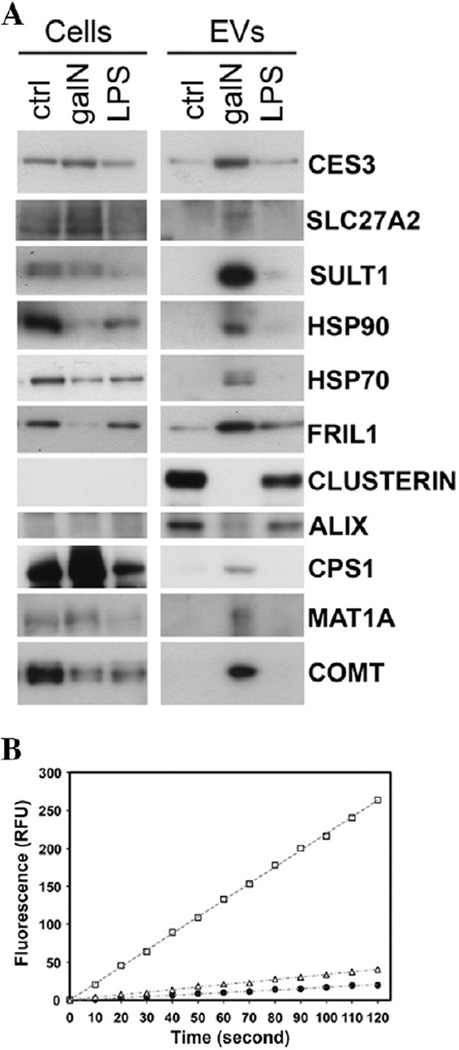
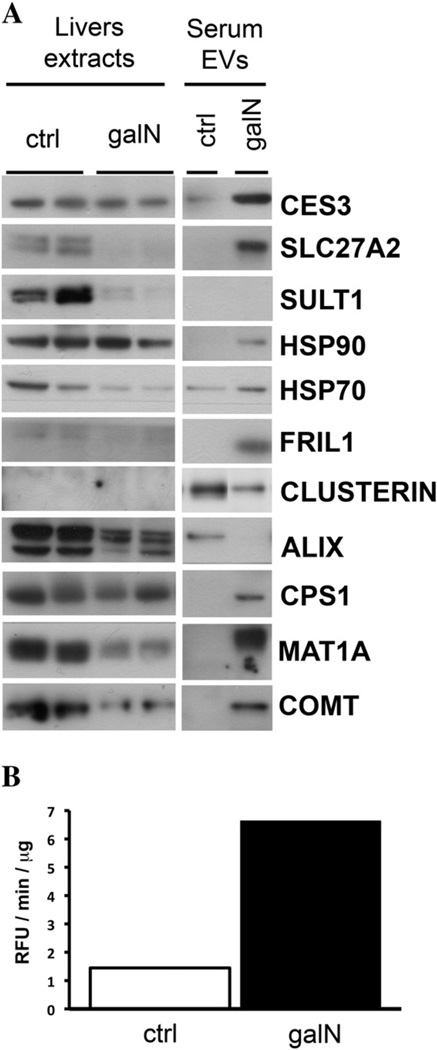
An independent suspension of primary rat hepatocytes was subjected to galN and LPS treatments, and EVs released to the medium were purified and analyzed by Western blotting, in parallel with cellular extracts (Fig. 4A). Using immunoblotting, we established that, in agreement with the proteomics data, the levels of heat shock protein Hsp90 and its partner Hsp70, ferritin (FRIL1), carboxylesterase 3 (CES3), SLC27A2, SULT1, and MAT increased in EVs after galN treatment. Also in agreement with the proteomics data were the levels of clusterin, drastically reduced by the treatment with galN. For EVs released by hepatocytes exposed to LPS an agreement between proteomics data and immunoblotting was also obtained; we observed a reduction in the levels of clusterin and increased levels of FRIL1. Western blot analysis detected significant changes in the levels of other proteins in EVs released by galN-exposed hepatocytes: ALIX, CPS1 and COMT; these changes were not detected by our quantitative proteomics approach.
Fig. 4.
Biochemical analysis of EVs released by primary cultures of rat hepatocytes exposed to galN or LPS. (A) Western blot analysis of indicated proteins was performed for cellular extracts and EVs that were secreted by these cells. Normalization was based on the amount of protein determined by Bradford assay. (B) Carboxylesterase activity of EVs secreted by primary culture of rat hepatocytes untreated (filled circles), treated with LPS (empty triangles), or galN (empty squares).
One question arising when an increase in the levels of a protein is observed is whether there is a concomitant increase in the protein activity. To establish this, we asked whether the augmented protein level of the carboxylesterase CES3 observed in EVs released by hepatocytes exposed to galN translates into an increased activity of this enzyme (Fig. 4B). Indeed, the carboxylesterase activity in these EVs was significantly higher than in the EVs released by untreated or LPS-treated hepatocytes (although a slight increment was observed in EVs from LPS-treated hepatocytes). This result suggests that EVs released by hepatocytes in response to an insult carry active proteins that might have a function in the extracellular space.
3.4. “in vitro” regulation of EV proteins are also observed using an “in vivo” model
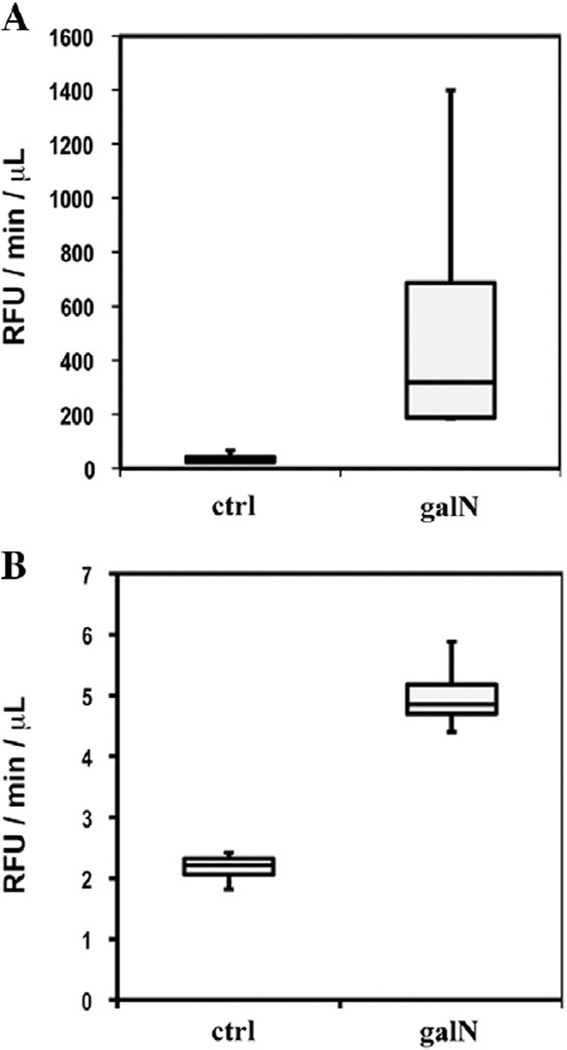
In order to evaluate whether the in vitro model used in this study reflects the in vivo situation, we analyzed EVs purified from the sera in an induced acute liver injury rat model. In this model, we tested, using Western blotting, some of the candidate markers for hepatotoxicity obtained in our proteomics approach in vitro. A group of four rats was treated with saline (control) while another group of four rats was treated with galN, as described in Experimental procedures. After 18 h, ALT activity in the sera of both groups was determined. Saline- and galN-treated rats showed ALT activity of 23.3 ± 5.3 and 6936.8 ± 1636.0 U/L, respectively, reflecting the acute liver injury induced by the galN treatment. Next, sera from same treatment groups were pooled to purify a sufficient amount of serum EVs to perform the Western blot and enzymatic analyses. Equal protein amounts from EVs of saline- or galN-treated animals were analyzed by Western blotting in parallel with representative liver extracts (Fig. 5A) prepared from these animals. CES3, SLC27A2, HSP90, HSP70 and FRIL1 were more expressed in the pooled EVs isolated from galN-treated rats, in clear agreement with the data obtained in the in vitro system. We also observed increased levels of the liver-specific proteins CPS1, MAT and COMT. The reduced amount of clusterin in EVs isolated from sera of galN-treated rats was also in agreement with in vitro results. These data indicate that the levels of these EVs-associated proteins could be used as serological indicators of liver toxicity. It is also clear that quantitative proteomics results obtained for in vitro primary hepatocyte-based models reflect the in vivo state. Remarkably, while some of the protein levels were augmented in EVs isolated from the sera, they showed no significant alterations in liver extracts (proteins CES3, HSP90, and CPS). The levels of other proteins such as SLC27A2, ALIX, MAT1A, and COMT were reduced in liver extracts (Fig. 5A), suggesting that different regulation processes apply to different proteins in response to galN-induced liver damage. Confirming our in vitro observations, the increased level of CES3 in the serum EVs from injured animals was accompanied by a significant increase in the activity of this enzyme (Fig. 5B). Finally, we evaluated the possibility of detecting an increase in the CES3 activity in the sera without purifying EVs. As shown in the box plots of Fig. 6A, the sera of the four galN-treated rats showed significantly increased activity compared to the saline-treated rats. Interestingly, we consistently detected this injury-associated augmented activity of carboxylesterase in the samples of urine collected from the animals (Fig. 6B). These results suggest that the analysis of this enzymatic activity could be used as a non-invasive indicator of drug toxicity.
Fig. 5.
Biochemical analysis of in vivo model for drug-induced toxicity. (A) Western blot analysis of indicated proteins was performed using liver extracts and pooled sera-EVs obtained from saline (ctrl)- or galN-treated rats. (B) Carboxylesterase activity of pooled EVs isolated from the sera of saline (ctrl)- and galN-treated rats.
Fig. 6.
Carboxylesterase activity in serum (A) and urine (B) samples from saline- and galN-treated rats.
4. Discussion
Response to drug toxicity and differentiation between various types of hepatic injury has been based primarily on the determination of hepatic enzymes in the blood, most notably alanine aminotransferase (ALT)/aspartate aminotransferase (AST) and alkaline phosphatase (ALP)/gamma-glutamyl transpeptidase (GGT), as indicators of liver injury (reviewed in [54]). In the present study, we performed a qualitative and quantitative label-free LC–DIA-MS proteomic analysis of EVs released by primary hepatocytes exposed to well-characterized hepatotoxins as an approach to detect novel and complementary candidate markers for liver injury. We generated an extensive protein catalog of more than 550 proteins associated with hepatocyte-released EVs. This work complements our previous research on these extracellular vesicles [32]. The enrichment in proteins involved in the pathways related to energy production, lipid and xenobiotic metabolism, and protein homeostasis points to these vesicles as important enzymatic machines providing energy, and metabolizing proteins, lipids, and drugs outside the cell. It is clear that these vesicles play an important role in the extracellular homeostasis.
In this work, one of our aims was the identification of candidate low invasive markers for liver damage. Some existing in vitro and in vivo studies using toxins such as LPS and/or galN, in combination with proteomics provided the identification of proteins and molecular mechanisms involved in the development of liver damage [48–52]. In this study, we investigated for the first time the proteome of EVs released by primary hepatocytes challenged with these two toxins. E. coli-derived lipopolysaccharide (LPS)is an activator of Nalp3 inflammasome complex [55], taking part in the key event in the development of liver inflammation [56,57]. It has been shown that LPS is mostly captured and removed from the circulation by hepatocytes in a process that activates p38 MAPK [58] and Toll-like receptor [59], both involved in inflammation-related pathways. In our study, LPS treatment affected the abundance of 165 proteins detected in EVs, including many structural and regulatory cytoskeleton-related proteins and vesicle-mediated trafficking proteins, such as actin, tubulin, small GTPases (RAB7A, RAB10, RAB13, RAB15, RAP1B, ARF2 and ARF3), coatomer subunits of COPII complex, and syndecan-4 protein, reflecting cellular changes in morphology and intracellular protein trafficking in response to this bacterial product. Interestingly, a deficiency in syndecan-4 causes high mortality in mice injected with LPS [60], confirming an important role of this protein in the response to endotoxins. Our proteomics results indicate that the levels of this protein in EVs are elevated, pointing to a possible role of these EVs in the syndecan-4-dependent response mechanism. Other proteins whose levels were significantly altered in EVs released by hepatocytes exposed to LPS were regulatory subunits of the proteasome machinery, PSMD2, PRS6B, and PRS7. It was reported that, in macrophages and glial cells, LPS induces the activity of immunoproteosome [61,62] an inducible form of the proteasome complex with a reported role in the regulation of proinflammatory cytokine production [63]. Our results indicate that a similar regulation of the proteasome complex could also apply to hepatocytes, although, to confirm this, further investigation would be necessary.
The other toxin used in this work was the amino sugar galN, which induces acute liver injury resembling human viral hepatitis [33] by a mechanism involving depletion of UTP and glutathione cellular reservoirs [64]. After the treatment of the hepatocytes with galN, significant abundance changes were detected in more than 240 proteins in the released EVs; the changes were mostly associated with redox and xenobiotic metabolism, affecting glutathione transferases, sulfotransferases, and cytochromes. Using Western blotting (Fig. 4), we validated some of these changes and observed a concordance between the quantitative label-free LC–DIA-MS data and the amount of protein established by immunodetection. Using quantitative LC–MS proteomics, we found that the levels of the enzyme CES3 were induced 3- and 1.5-fold after galN and LPS treatments, respectively. Similar trends were observed using immunoblotting (Fig. 4). The highest levels of the protein were found in the galN lane, followed by the LPS and control lanes. Similar result was obtained after analyzing ferritin light chain (FRL1) protein, whose levels, based on the proteomics results, were induced 5- and 1.5-fold after galN and LPS treatments, respectively. This was confirmed by immunoblotting results. In case of clusterin, a drastic level reduction (15-fold) in EVs released by galN-treated hepatocytes was also validated by the results obtained by immunodetection. A clear band for clusterin was observed for the control and LPS treatments; this protein was undetectable in galN-treated EVs. In summary, a good correlation was found between the quantitative label-free proteomics data and biochemical analysis results, supporting the application of this high-content technology in identifying candidate markers for the response to a specific stimulus.
Remarkably, we have observed increased levels of some liver-specific proteins such as the enzymes carbamoyl phosphate synthetase 1 (CPS1), S-adenosyl methionine synthetase 1 (MAT), and catechol-O-methyltransferase (COMT) in hepatocyte-derived EVs and serum-isolated EVs from liver-injured animals. In agreement with these results, up-regulation of MAT enzyme has been previously shown to be required for the proper liver regeneration in hepatectomized mice [65]. Although the biological significance of these proteins in hepatocyte-derived EVs remains unclear, our results suggest that the study of these proteins in plasma-circulating EVs could help to determine liver-specific damage or monitor the recovery, in a non-invasive manner. Brodsky and collaborators have isolated plasma-circulating EVs enriched in liver-origin proteins using an antibody specific to CPS1 [66,67]. They showed that in HCC, the levels of these plasma-circulating hepatic-derived EVs correlate positively with the size of liver tumors. They proposed that these hepatic EVs might be used in clinical practice as markers of the functional status of transplanted livers [66].
We also identified another protein whose levels were altered in rat hepatocyte-derived EVs: the enzyme carboxylesterase 3, with 89% homology to the human liver carboxylesterase 1. These enzymes belong to a class of esterases that are ubiquitously expressed from bacteria to man, and, as their name implies, they cleave carboxyl esters into the corresponding alcohols and carboxylic acid. However, these enzymes can also hydrolyze thioesters and carbamates [68,69]. Due to this diverse substrate specificity, these proteins are frequently referred to as ‘promiscuous’ enzymes [70]. The exact function of these enzymes is unknown since no endogenous substrate has been definitively identified. In general, it is thought that CES might play a protective role, detoxifying xenobiotics by cleaving these compounds to less toxic hydrolysis products [71]. Numerous compounds can be hydrolyzed by CES activity. This includes the illegal recreational drugs heroin and cocaine [72,73], the anticancer agents capecitabine and CPT-11 [74,75], the pyrethroid class of pesticides [76] as well as a whole host of widely used therapeutic molecules [77,78]. Pharmaceutical companies frequently add methyl or ethyl groups to candidate drug molecules via an ester linkage to improve water solubility and/or bioavailability. As a consequence, frequently prescribed medicines such as Plavix, Tamiflu, Demerol, and Ritalin all contain ester moieties and, correspondingly, are metabolized by CES activity in vivo [77–80]. Our study shows that CES3 protein is localized in extracellular vesicles and in liver injury its abundance is significantly increased in this extracellular compartment; this is accompanied by an increase in its enzymatic activity. This activity is also increased in serum and liver samples from mice treated with high doses of carbon tetrachloride but remains unchanged after acetaminophen treatment [81]. These results indicate that different modes of hepatotoxicity may be associated with different hepatotoxins, and finding specific markers could help to classify liver damage. Testing for the increase in CES activity observed in EVs isolated from serum samples, and even directly in serum and urine samples, could help to evaluate and classify chemically induced toxicity, in a low invasive manner.
In conclusion, this report strongly supports the application of proteomics in the study of extracellular vesicles released by well-controlled in vitro cellular systems to obtain novel non-invasive markers for different stimuli and diseases. In a pharmaceutical context this report also highlights the importance of the studies of the hepatocyte-released circulating EVs for the development of new drugs and to understand and minimize idiosyncratic drug toxicity.
Supplementary Material
Biological significance.
Identification of low invasive candidate marker for hepatotoxicity. Support to apply proteomics in the study of extracellular vesicles released by well-controlled in vitro cellular systems to identify low invasive markers for diseases.
Acknowledgments
We thank FAES FARMA for the support in rat experimentation procedures and sample collection. This work was supported by grants from the Fondo de Investigaciones Sanitarias (Institute of Health Carlos III, 06/0621, PS09/00526 & PI12/01064 to J.M.F.P.); Program “Ramon y Cajal” of Spanish Ministry (to J.M.F.PRYC-2007-00228); National Institute of Health Grant R01 AT004896 (to S.C.L. and J.M. M.); and Centro de Investigación Biomédica en Red en el Área temática de Enfermedades Hepáticas y Digestivas (CIBERehd) funded by the Institute of Health Carlos III.
Abbreviations
- EV
extracellular vesicle
- LC–MS
liquid chromatography coupled to mass spectrometry
- MP
microparticle
- ALT
alanine transaminase
- CES
carboxylesterase
- MVB
multivesicular body
- LPS
lipopolysaccharide
- galN
d-galactosamine
- AMRT
accurate mass measurement retention time pair
- GO
Gene Ontology
- DDA
data-dependent analysis
- DIA
data-independent analysis
- ACN
acetonitrile
- RFU
relative fluorescence unit
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2014.04.008.
Transparency document
The Transparency document associated with this article can be found, in the online version.
REFERENCES
- 1.Pierce A, Unwin RD, Evans CA, Griffiths S, Carney L, Zhang L, et al. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol Cell Proteomics. 2008;7:853–863. doi: 10.1074/mcp.M700251-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;267:12. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 3.Bonzon-Kulichenko E, Pérez-Hernández D, Núñez E, Martínez-Acedo P, Navarro P, Trevisan-Herraz M, et al. A robust method for quantitative high-throughput analysis of proteomes by 18O labeling. Mol Cell Proteomics. 2011:10. doi: 10.1074/mcp.M110.003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prokhorova TA, Rigbolt KTG, Johansen PT, Henningsen J, Kratchmarova I, Kassem M, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol Cell Proteomics. 2009;8:959–970. doi: 10.1074/mcp.M800287-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Luo X, Hu H, Wang R, Sun Y, Zeng R, et al. Integrative proteomics and tissue microarray profiling indicate the association between overexpressed serum proteins and non-small cell lung cancer. PLoS One. 2012;7:e51748. doi: 10.1371/journal.pone.0051748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahn H, Wittke S, Zürbig P, Raedler TJ, Arlt S, Kellmann M, et al. Peptide fingerprinting of Alzheimer’s disease in cerebrospinal fluid: identification and prospective evaluation of new synaptic biomarkers. PLoS One. 2011;6:e26540. doi: 10.1371/journal.pone.0026540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 9.Li X-j, Yi EC, Kemp CJ, Zhang H, Aebersold R. A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Mol Cell Proteomics. 2005;4:1328–1340. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 11.Silva JC, Denny R, Dorschel C, Gorenstein MV, Li G-Z, Richardson K, et al. Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome. Mol Cell Proteomics. 2006;5:589–607. doi: 10.1074/mcp.M500321-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li G-Z, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 13.Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Radulovic D, Jelveh S, Ryu S, Hamilton TG, Foss E, Mao Y, et al. Informatics platform for global proteomic profiling and biomarker discovery using liquid chromatography-tandem mass spectrometry. Mol Cell Proteomics. 2004;3:984–997. doi: 10.1074/mcp.M400061-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez-Suárez E, Whetton AD. The application of quantification techniques in proteomics for biomedical research. Mass Spectrom Rev. 2013;32:1–26. doi: 10.1002/mas.21347. [DOI] [PubMed] [Google Scholar]
- 16.Bantscheff M, Lemeer S, Savitski M, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 17.Simons M, Raposo G. Exosomes—vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Simons M, Raposo G. Exosomes — vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Gasser O, Hess C, Miot S, Deon C, Sanchez JC, Schifferli JA. Characterisation and properties of ectosomes released by human polymorphonuclear neutrophils. Exp Cell Res. 2003;285:243–257. doi: 10.1016/s0014-4827(03)00055-7. [DOI] [PubMed] [Google Scholar]
- 21.Jimenez JJ, Jy W, Mauro LM, Soderland C, Horstman LL, Ahn YS. Endothelial cells release phenotypically and quantitatively distinct microparticles in activation and apoptosis. Thromb Res. 2003;109:175–180. doi: 10.1016/s0049-3848(03)00064-1. [DOI] [PubMed] [Google Scholar]
- 22.Almqvist N, Lonnqvist A, Hultkrantz S, Rask C, Telemo E. Serum-derived exosomes from antigen-fed mice prevent allergic sensitization in a model of allergic asthma. Immunology. 2008;125:21–27. doi: 10.1111/j.1365-2567.2008.02812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 24.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 25.Piccin A, Murphy WG, Smith OP. Circulating microparticles: pathophysiology and clinical implications. Blood Rev. 2007;21:157–171. doi: 10.1016/j.blre.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Shet AS. Characterizing blood microparticles: technical aspects and challenges. Vasc Health Risk Manag. 2008;4:769–774. doi: 10.2147/vhrm.s955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 28.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 30.Pap E, Pallinger E, Pasztoi M, Falus A. Highlights of a new type of intercellular communication: microvesicle-based information transfer. Inflamm Res. 2009;58:1–8. doi: 10.1007/s00011-008-8210-7. [DOI] [PubMed] [Google Scholar]
- 31.Wieckowski E, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36:247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- 32.Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M, et al. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res. 2008;7:5157–5166. doi: 10.1021/pr8004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keppler D, Lesch R, Reutter W, Decker K. Experimental hepatitis induced by d-galactosamine. Exp Mol Pathol. 1968;9:279–290. doi: 10.1016/0014-4800(68)90042-7. [DOI] [PubMed] [Google Scholar]
- 34.Han DW. Intestinal endotoxemia as a pathogenetic mechanism in liver failure. World J Gastroenterol. 2002;8:961–965. doi: 10.3748/wjg.v8.i6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaido T, Yamaoka S, Seto S, Funaki N, Kasamatsu T, Tanaka J, et al. Continuous hepatocyte growth factor supply prevents lipopolysaccharide-induced liver injury in rats. FEBS Lett. 1997;411:378–382. doi: 10.1016/s0014-5793(97)00744-8. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Wang Y, Zhang H, Jia B, Wang D, Li H, et al. Yohimbine enhances protection of berberine against LPS-induced mouse lethality through multiple mechanisms. PLoS One. 2012;7:e52863. doi: 10.1371/journal.pone.0052863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrison DC, Ryan JL. Endotoxins and disease mechanisms. Annu Rev Med. 1987;38:417–432. doi: 10.1146/annurev.me.38.020187.002221. [DOI] [PubMed] [Google Scholar]
- 38.Ponzetti K, King M, Gates A, Anwer MS, Webster CR. Cyclic AMP-guanine exchange factor activation inhibits JNK-dependent lipopolysaccharide-induced apoptosis in rat hepatocytes. Hepat Med. 2010;2010:1–11. doi: 10.2147/HMER.S7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polsky-Fisher SL, Cao H, Lu P, Gibson CR. Effect of cytochromes P450 chemical inhibitors and monoclonal antibodies on human liver microsomal esterase activity. Drug Metab Dispos. 2006;34:1361–1366. doi: 10.1124/dmd.106.009704. [DOI] [PubMed] [Google Scholar]
- 40.Seglen PO. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 41.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 42.Gilar M, Olivova P, Daly AE, Gebler JC. Two-dimensional separation of peptides using RP-RP-HPLC system with different pH in first and second separation dimensions. J Sep Sci. 2005;28:1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
- 43.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Li G-Z, Vissers JPC, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 45.Yu Y, Shen H, Yu H, Zhong F, Zhang Y, Zhang C, et al. Systematic proteomic analysis of human hepotacellular carcinoma cells reveals molecular pathways and networks involved in metastasis. Mol Biosyst. 2011;7:1908–1916. doi: 10.1039/c0mb00265h. [DOI] [PubMed] [Google Scholar]
- 46.Pitarch A, Nombela C, Gil C. Prediction of the clinical outcome in invasive Candidiasis patients based on molecular fingerprints of five anti-Candida antibodies in serum. Mol Cell Proteomics. 2011:10. doi: 10.1074/mcp.M110.004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levin Y. The role of statistical power analysis in quantitative proteomics. Proteomics. 2011;11:2565–2567. doi: 10.1002/pmic.201100033. [DOI] [PubMed] [Google Scholar]
- 48.Banerjee A, Russell WK, Jayaraman A, Ramaiah SK. Identification of proteins to predict the molecular basis for the observed gender susceptibility in a rat model of alcoholic steatohepatitis by 2-D gel proteomics. Proteomics. 2008;8:4327–4337. doi: 10.1002/pmic.200700368. [DOI] [PubMed] [Google Scholar]
- 49.Lorenz O, Parzefall W, Kainzbauer E, Wimmer H, Grasl-Kraupp B, Gerner C, et al. Proteomics reveals acute pro-inflammatory and protective responses in rat Kupffer cells and hepatocytes after chemical initiation of liver cancer and after LPS and IL-6. Proteomics Clin Appl. 2009;3:947–967. doi: 10.1002/prca.200800173. [DOI] [PubMed] [Google Scholar]
- 50.Lu Y, Bao X, Sun T, Xu J, Zheng W, Shen P. Triptolide attenuate the oxidative stress induced by LPS/D-GalN in mice. J Cell Biochem. 2012;113:1022–1033. doi: 10.1002/jcb.23434. [DOI] [PubMed] [Google Scholar]
- 51.Lv S, Wang JH, Liu F, Gao Y, Fei R, Du SC, et al. Senescence marker protein 30 in acute liver failure: validation of a mass spectrometry proteomics assay. BMC Gastroenterol. 2008;8:17. doi: 10.1186/1471-230X-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Ariza A, Lopez-Sanchez LM, Gonzalez R, Corrales FJ, Lopez P, Bernardos A, et al. Altered protein expression and protein nitration pattern during d-galactosamine-induced cell death in human hepatocytes: a proteomic analysis. Liver Int. 2005;25:1259–1269. doi: 10.1111/j.1478-3231.2005.01172.x. [DOI] [PubMed] [Google Scholar]
- 53.Sun F, Hamagawa E, Tsutsui C, Sakaguchi N, Kakuta Y, Tokumaru S, et al. Evaluation of oxidative stress during apoptosis and necrosis caused by d-galactosamine in rat liver. Biochem Pharmacol. 2003;65:101–107. doi: 10.1016/s0006-2952(02)01420-x. [DOI] [PubMed] [Google Scholar]
- 54.Ramaiah SK. Preclinical safety assessment: current gaps, challenges, and approaches in identifying translatable biomarkers of drug-induced liver injury. Clin Lab Med. 2011;31:161–172. doi: 10.1016/j.cll.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Ganz M, Csak T, Nath B, Szabo G. Lipopolysaccharide induces and activates the Nalp3 inflammasome in the liver. World J Gastroenterol. 2011;17:4772–4778. doi: 10.3748/wjg.v17.i43.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burdette D, Haskett A, Presser L, McRae S, Iqbal J, Waris G. Hepatitis C virus activates interleukin-1beta via caspase-1-inflammasome complex. J Gen Virol. 2011;93:235–246. doi: 10.1099/vir.0.034033-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Csak T, Ganz M, Pespisa J, Kodys K, Dolganiuc A, Szabo G. Fatty acid and endotoxin activate inflammasomes in mouse hepatocytes that release danger signals to stimulate immune cells. Hepatology. 2011;54:133–144. doi: 10.1002/hep.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scott MJ, Billiar TR. Beta2-integrin-induced p38 MAPK activation is a key mediator in the CD14/TLR4/MD2-dependent uptake of lipopolysaccharide by hepatocytes. J Biol Chem. 2008;283:29433–29446. doi: 10.1074/jbc.M803905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S, Gallo DJ, Green AM, Williams DL, Gong X, Shapiro RA, et al. Role of toll-like receptors in changes in gene expression and NF-kappa B activation in mouse hepatocytes stimulated with lipopolysaccharide. Infect Immun. 2002;70:3433–3442. doi: 10.1128/IAI.70.7.3433-3442.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishiguro K, Kadomatsu K, Kojima T, Muramatsu H, Iwase M, Yoshikai Y, et al. Syndecan-4 deficiency leads to high mortality of lipopolysaccharide-injected mice. J Biol Chem. 2001;276:47483–47488. doi: 10.1074/jbc.M106268200. [DOI] [PubMed] [Google Scholar]
- 61.Reis J, Guan XQ, Kisselev AF, Papasian CJ, Qureshi AA, Morrison DC, et al. LPS-induced formation of immunoproteasomes: TNF-alpha and nitric oxide production are regulated by altered composition of proteasome-active sites. Cell Biochem Biophys. 2011;60:77–88. doi: 10.1007/s12013-011-9182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stohwasser R, Giesebrecht J, Kraft R, Muller EC, Hausler KG, Kettenmann H, et al. Biochemical analysis of proteasomes from mouse microglia: induction of immunoproteasomes by interferon-gamma and lipopolysaccharide. Glia. 2000;29:355–365. [PubMed] [Google Scholar]
- 63.Angeles A, Fung G, Luo H. Immune and non-immune functions of the immunoproteasome. Front Biosci. 2012;17:1904–1916. doi: 10.2741/4027. [DOI] [PubMed] [Google Scholar]
- 64.Coen M, Hong YS, Clayton TA, Rohde CM, Pearce JT, Reily MD, et al. The mechanism of galactosamine toxicity revisited; a metabonomic study. J Proteome Res. 2007;6:2711–2719. doi: 10.1021/pr070164f. [DOI] [PubMed] [Google Scholar]
- 65.Chen L, Zeng Y, Yang H, Lee TD, French SW, Corrales FJ, et al. Impaired liver regeneration in mice lacking methionine adenosyltransferase 1A. FASEB J. 2004;18:914–916. doi: 10.1096/fj.03-1204fje. [DOI] [PubMed] [Google Scholar]
- 66.Brodsky SV, Facciuto ME, Heydt D, Chen J, Islam HK, Kajstura M, et al. Dynamics of circulating microparticles in liver transplant patients. J Gastrointestin Liver Dis. 2008;17:261–268. [PubMed] [Google Scholar]
- 67.Butler SL, Dong H, Cardona D, Jia M, Zheng R, Zhu H, et al. The antigen for Hep Par 1 antibody is the urea cycle enzyme carbamoyl phosphate synthetase 1. Lab Invest. 2008;88:78–88. doi: 10.1038/labinvest.3700699. [DOI] [PubMed] [Google Scholar]
- 68.Potter PM, Wadkins RM. Carboxylesterases — detoxifying enzymes and targets for drug therapy. Curr Med Chem. 2006;13:1045–1054. doi: 10.2174/092986706776360969. [DOI] [PubMed] [Google Scholar]
- 69.Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu Rev Pharmacol Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 70.Redinbo MR, Potter PM. Mammalian carboxylesterases: from drug targets to protein therapeutics. Drug Discov Today. 2005;10:313–325. doi: 10.1016/S1359-6446(05)03383-0. [DOI] [PubMed] [Google Scholar]
- 71.Hatfield MJ, Potter PM. Carboxylesterase inhibitors. Expert Opin Ther Pat. 2011;21:1159–1171. doi: 10.1517/13543776.2011.586339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hatfield MJ, Tsurkan L, Hyatt JL, Yu X, Edwards CC, Hicks LD, et al. Biochemical and molecular analysis of carboxylesterase-mediated hydrolysis of cocaine and heroin. Br J Pharmacol. 2010;160:1916–1928. doi: 10.1111/j.1476-5381.2010.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pindel EV, Kedishvili NY, Abraham TL, Brzezinski MR, Zhang J, Dean RA, et al. Purification and cloning of a broad substrate specificity human liver carboxylesterase that catalyzes the hydrolysis of cocaine and heroin. J Biol Chem. 1997;272:14769–14775. doi: 10.1074/jbc.272.23.14769. [DOI] [PubMed] [Google Scholar]
- 74.Humerickhouse R, Lohrbach K, Li L, Bosron WF, Dolan ME. Characterization of CPT-11 hydrolysis by human liver carboxylesterase isoforms hCE-1 and hCE-2. Cancer Res. 2000;60:1189–1192. [PubMed] [Google Scholar]
- 75.Quinney SK, Sanghani SP, Davis WI, Hurley TD, Sun Z, Murry DJ, et al. Hydrolysis of capecitabine to 5′-deoxy-5-fluorocytidine by human carboxylesterases and inhibition by loperamide. J Pharmacol Exp Ther. 2005;313:1011–1016. doi: 10.1124/jpet.104.081265. [DOI] [PubMed] [Google Scholar]
- 76.Crow JA, Borazjani A, Potter PM, Ross MK. Hydrolysis of pyrethroids by human and rat tissues: examination of intestinal, liver and serum carboxylesterases. Toxicol Appl Pharmacol. 2007;221:1–12. doi: 10.1016/j.taap.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi D, Yang J, Yang D, LeCluyse EL, Black C, You L, et al. Anti-influenza prodrug oseltamivir is activated by carboxylesterase human carboxylesterase 1, and the activation is inhibited by antiplatelet agent clopidogrel. J Pharmacol Exp Ther. 2006;319:1477–1484. doi: 10.1124/jpet.106.111807. [DOI] [PubMed] [Google Scholar]
- 78.Sun Z, Murry DJ, Sanghani SP, Davis WI, Kedishvili NY, Zou Q, et al. Methylphenidate is stereoselectively hydrolyzed by human carboxylesterase CES1A1. J Pharmacol Exp Ther. 2004;310:469–476. doi: 10.1124/jpet.104.067116. [DOI] [PubMed] [Google Scholar]
- 79.Tang M, Mukundan M, Yang J, Charpentier N, LeCluyse EL, Black C, et al. Antiplatelet agents aspirin and clopidogrel are hydrolyzed by distinct carboxylesterases, and clopidogrel is transesterificated in the presence of ethyl alcohol. J Pharmacol Exp Ther. 2006;319:1467–1476. doi: 10.1124/jpet.106.110577. [DOI] [PubMed] [Google Scholar]
- 80.Zhang J, Burnell JC, Dumaual N, Bosron WF. Binding and hydrolysis of meperidine by human liver carboxylesterase hCE-1. J Pharmacol Exp Ther. 1999;290:314–318. [PubMed] [Google Scholar]
- 81.Huang TL, Villalobos SA, Hammock BD. Effect of hepatotoxic doses of paracetamol and carbon tetrachloride on the serum and hepatic carboxylesterase activity in mice. J Pharm Pharmacol. 1993;45:458–465. doi: 10.1111/j.2042-7158.1993.tb05576.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.