Abstract
Microfluidic human organ models, microphysiology systems (MPS), are currently being developed as predictive models of drug safety and efficacy in humans. To design and validate MPS as predictive of human safety liabilities requires safety data for a reference set of compounds, combined with in vitro data from the human organ models. To address this need, we have developed an internet database, the MPS database (MPS-Db), as a powerful platform for experimental design, data management, and analysis, and to combine experimental data with reference data, to enable computational modeling. The present study demonstrates the capability of the MPS-Db in early safety testing using a human liver MPS to relate the effects of tolcapone and entacapone in the in vitro model to human in vivo effects. These two compounds were chosen to be evaluated as a representative pair of marketed drugs because they are structurally similar, have the same target, and were found safe or had an acceptable risk in preclinical and clinical trials, yet tolcapone induced unacceptable levels of hepatotoxicity while entacapone was found to be safe. Results demonstrate the utility of the MPS-Db as an essential resource for relating in vitro organ model data to the multiple biochemical, preclinical, and clinical data sources on in vivo drug effects.
Key words: : computational toxicology, in vitro methods, high throughput, liver, multi-organ models
Introduction
Microfluidic human organ models, microphysiology systems (MPS),1–6 have the potential to dramatically improve the prediction of drug safety and efficacy in humans early in the discovery and development process. In addition, combining MPS into multiorgan systems,2 including the ultimate goal of a human on-a-chip,2,7 will further enable the prediction of safety liabilities that result from organ interactions, such as liver metabolism that produces metabolites that are toxic to other organs. However, all models are simplified with respect to reality, and the human on-a-chip is a conceptual design for modeling the functions of major human organs. To develop and validate MPS as predictive of human safety liabilities will require data on human organ safety for a reference set of compounds. In the end, the degree to which human organ models are able to predict human safety liabilities will be as much determined by the content of the reference data as it is by the sophistication of the MPS and their readouts. Furthermore, disease models are now being incorporated into the MPS using a variety of cells including cell lines, primary cells, and induced pluripotent stem cells (iPSCs), and a robust database is required to manage the experimental designs and analyses. We have developed an internet database, the MicroPhysiology Systems Database (MPS-Db), as a powerful platform to design experiments as well as to manage, analyze, and combine experimental data with external data and ultimately to computationally model the data. The present study demonstrates the capability of the MPS-Db in early safety testing using a human liver MPS.
The discordance between preclinical and clinical liver toxicity can be explained by the differences in testing protocols between animal and humans and by species-dependent toxicokinetics of drug metabolism and drug clearance. Animals are tested at drug doses several orders of magnitude higher than human exposure, which confounds the extrapolation from high dose testing to low-dose exposure since many drugs display nonlinear dose response effects.8 Well-documented differences in the kinetics of phase 1 and phase 2 metabolism exist between animal species, particularly for clinically relevant drug metabolizing phase 1 cytochrome P-450 isoforms CYP1A, -2C, -2D, and -3A.9 These points illustrate the difficulty predicting human liver toxicity from animal in vivo studies. It is generally accepted that animal models can have limited ability to predict in vivo human risk10 and efficacy of drugs for many diseases,11, 12 but what evidence is there to suggest in vitro human models would be better?
There is a bewildering number of factors that confound predicting human liver toxicity from in vitro assays: incorrect or insufficient biochemical assays; use of cells adapted to growth in an artificial environment; lack of, or inappropriate, 3D cellular architecture; lack of metabolically competent liver cells; exclusion of systemic pharmacokinetics; lack of the in vivo cell–cell interactions within an organ; lack of organ–organ interactions; and lack of systemic endocrine modulation to name a few. However, there is evidence that in vitro assays coupled with computational models can improve identification of compound risks. For instance, a human liver cell line (HepG2) profile consisting of 12 multiplexed cell features and 3 time points correctly classified 72% of compounds with known clinical toxicities into moderate or high-risk categories out of a 136-test compound set.13,14 Pfizer, Inc. (Groton, CT) used two in vitro assays for BSEP and mitochondrial inhibition to correctly categorize 79% of DILI compounds in a set of 72 compounds that were clinically classified into most, less, or no concern for drug induced liver injury (DILI). Compounds having dual BSEP and mitochondrial inhibitions were classified into the “most or less” likely DILI concern groups, with none of them incorrectly assigned into the “No DILI” concern category.15 These three examples suggest that robust human in vitro to in vivo predictions can be achieved when the right biological models, the right biological assays or endpoints, and the right computational approaches are used. In both cases, a key element in establishing the predictive capability was the availability of clinical data for the reference compounds.
An area of active research is the development of human organ models, multicellular 3D perfused organ systems for the purpose of studying physiology, drug safety, and liver diseases.2,16–18 Recently, a four-cell human liver MPS (the SQL-SAL) was shown to correctly identify clinically relevant drug hepatotoxic responses that are driven through the interaction between parenchymal hepatocytes and competent immune- or fibrotic-responsive cells while under microfluidic flow.19,20 Even simpler cocultures of hepatocytes with Kupffer, endothelial, and stellate cells,21 human bone marrow mesenchymal stem cells22 and other cells in static or perfusion cultures23 provide clear evidence that more in vivo–like tissue models under more physiological conditions improves the health and functions of hepatocytes, leading to expectations that the predictive power of such in vitro systems should also improve. To realize those expectations will require human exposure data for compounds that impact organ functions to use as a reference for evaluating and validating in vitro models, and developing computational models that translate the MPS data to the in vivo human.
The MPS-Db described in this article was developed with open source tools and is hosted on the internet for broad access to researchers.24 The MPS-Db is intended to be a critical tool when integrated into the experimental organ model workflow using MPS models or even microplate-based systems (Fig. 1). The MPS-Db is designed to manage any data generated by MPS (e.g., Fig. 1A) including functional imaging and biochemical and mass spectrometry (MS) data (Fig. 1B and C), and to integrate that data with multiple sources of human exposure data accessed from a variety of external databases through the MPS-Db (Fig. 1D). To illustrate the use of the MPS-Db we present an example study comparing the MPS data resulting from the exposure of the liver SQL-SAL to tolcapone and entacapone, with reference biochemical, clinical, and post-marketing data integrated into the MPD-Db.
FIG. 1.
Overview of the platforms and data types for studying human physiology, disease models, and drug safety testing. MPS and microplate-based devices containing many organ models: (A) Data is collected from the organ model via (B) high content imaging readouts of transmitted light contrast and fluorescent biosensors; and (C) biochemical and mass spectrometry readouts. The multiplexed data is uploaded into (D) the Microphysiology Systems Database (MPS-Db) to design studies, manage data, and associate with data from the external data sources shown.
Tolcapone and entacapone are selective catechol-O-methyl transferase (COMT) inhibitors used to treat Parkinson's disease. Tolcapone was introduced in the European Union in 1997 and in the United States in 1998. Soon after introduction to the market, incidents of fatal hepatotoxicity were reported.21 Initially, tolcapone was withdrawn in European and Canadian markets and a black box label attached in the United States. Eventually new data emerging from patients receiving tolcapone in the United States following restrictive United States Food and Drug Administration (FDA) guidelines for liver toxicity produced enough evidence and allowed tolcapone to be reintroduced into the European market. Entacapone, approved by the European Medicines Agency in 1998 and by the FDA in 1999, has remained on the market without incidents of hepatotoxicity. These two compounds were chosen to be evaluated in the SQL-SAL as a representative pair of marketed drugs because they are: (a) structurally similar compounds intended for the same therapeutic application; (b) were found safe or had an acceptable risk in preclinical and clinical trials; and (c) tolcapone induced unacceptable levels of hepatotoxicity while entacapone was found to be safe to the liver during post market use.25 The hypothesis was that the readouts from the SQL-SAL liver model would detect liver toxicity with tolcapone but not entacapone. A secondary hypothesis was that the SQL-SAL readouts that were most indicative of tolcapone liver toxicity should relate to its clinical toxicity indications.
Materials and Methods
MPS-Db data model
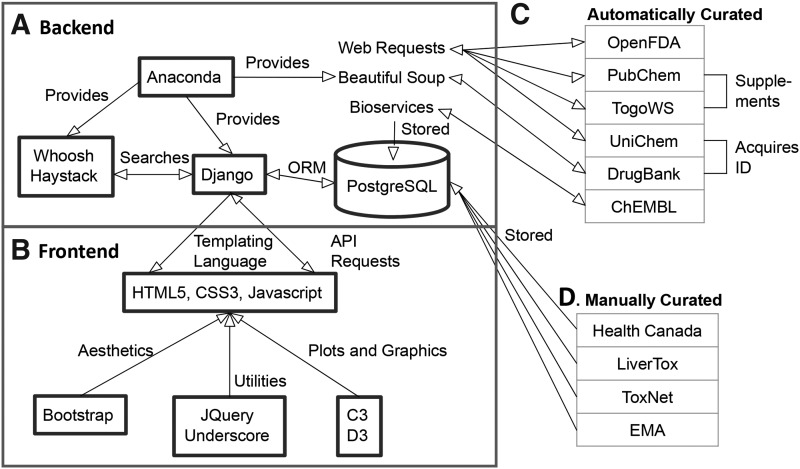
Figure 2 illustrates the architecture of the MPS-Db website, including the components and their interactions. At the heart of the MPS-Db is the Django26 framework and a PostgreSQL27 database. The Django framework consists of a series of “apps” that describe objects that consist of a number of fields, referred to as “models.” Django models and their information are stored as tables in the PostgreSQL database. At its highest level, the Django apps for the MPS-Db consist of: Assays, Bioactivities, Cell samples, Compounds, Drug Trials, and Microdevices, which define the associated MPS database tables.
FIG. 2.
Block diagram of the MPS-Db. (A) The MPS-Db “Backend” is constructed using the Django framework, Anaconda Python, and a PostgreSQL database. (B) The “frontend” (the website) is constructed using HTML, CSS3, and Javascript. (C) Automatically curated data is acquired from the listed internet accessible databases and cached locally in the PostgreSQL database. (D) Manually curated data is acquired from the listed sources, entered manually, and reviewed to supplement the automatically acquired data.
The Assays app is designed to manage the MPS study data, which is entered by users. This app stores assay metadata, raw data, and results from studies. Assay data is modularized in a hierarchical model of the format: Study →Setup → Readout → Results. For example, a Study, the highest tier, acts as a container for the rest of the data and describes an experiment or set of experiments with some defined purpose. A Setup captures data on how a single device was prepared. This includes information on the cell samples, the type of device, and the compound treatments. A Readout contains the list of assays performed in a given Setup and the raw data from those assays. A Result summarizes the data from a Readout with textual conclusions and/or pertinent calculations such as half-life and IC50. The design of the models supports microfluidic devices and microplates, treatment with compounds, and multiple time points.
The Bioactivities app includes bioassay data for MPS-Db compounds extracted from PubChem28,29 and ChEMBL.30,31 The data includes Assays, Targets, ChEMBL Bioactivities, and PubChem Bioactivities. The Cell samples app contains models that describe Cell samples, Cell types, and Cell subtypes. A Cell sample is a particular lot of cells of a defined Cell type, with its organ of origin (e.g., liver stellate cells), and Cell subtype, which describes the cell supply (e.g., primary fresh, cryopreserved, cell line, iPSC-derived). The Compounds app contains the reference compounds and their chemical and pharmacological properties acquired from ChEMBL and DrugBank.32,33 DrugBank data includes a list of targets that the compound may interact with. Users can also manually add summaries and additional properties. The Drug Trial app comprises manually curated and automatically acquired preclinical, clinical, and post-marketing data for compounds. Finally, the Microdevices app describes the physical characteristics of microfluidic devices, microplates, and the components of the organ models for which they are used. In addition to a description of the organ model, protocols used in their preparation can be uploaded.
Data sources, extraction, and curation
ChEMBL, a chemical database maintained by the European Bioinformatics Institute (EBI), was chosen as a data source because it primarily contains data on compounds exhibiting biological activity. We access ChEMBL's chemical and assay data using the ChEMBL web service wrapper provided by the Python34 package Bioservices. Most of the compounds in the MPS Database are linked to ChEMBL entries. All compounds that do have a corresponding ChEMBL entry rely on ChEMBL for the majority of their chemical properties (molecular weight, pKa, etc.). When entering new compounds, a curator supplies a ChEMBL ID that is used to automatically acquire the compound data. Compound bioactivities are automatically refreshed every quarter. Because of the curation methods, the bioactivity data in ChEMBL is not stored with standardized units. To be useful in developing predictive models, standard data units are needed. We have mitigated this problem by creating a lookup table to standardize the units.
Additional bioassay data is pulled from PubChem. PubChem, like ChEMBL, catalogues compounds and their respective chemical properties and bioactivities. The National Center for Biotechnology Information (NCBI) maintains PubChem. To acquire PubChem's bioactivity data, we use their RESTful service called the PubChem Power User Gateway (PUG). PUG is queried and its returned data parsed with Python's native web request libraries. In order to get a PubChem compound ID (CID) for each compound, PUG is queried for the compound entry that best matches the compound's name. If that fails, PUG is queried for the entry with a matching InChIKey. Given a CID, PUG produces data for the assays and bioactivities linked to that compound. However, PUG does not generate enough information about assay targets for our purposes. To acquire this desired target data, we use the cross-database API provided by the Database Center for Life Science (DBCLS) called TogoWS.35 Some of the PubChem data collides with ChEMBL, so duplicate targets and assays are consolidated. Like the ChEMBL activity data, the PubChem data is updated automatically every quarter. PubChem's bioactivities are standardized to be in micromolar units (μM), so it was not necessary to employ a lookup table.
DrugBank is referenced to acquire data for pharmacologically active compounds. DrugBank is hosted by the University of Alberta and The Metabolomics Innovation Centre. DrugBank IDs for compounds are acquired using UniChem, a RESTful web service for cross-referencing database entries.36 As DrugBank does not have an API for programmatically accessing data, it was decided that we should leverage web scraping to obtain data. Web scraping entails pulling all of the content from a web page and parsing it. We used the Python library Beautiful Soup to perform web scrapes. DrugBank is scraped for all compounds with a ChEMBL ID.
Adverse event data is acquired from OpenFDA, a public database (currently in beta test) of adverse event reports released by the FDA.37 Python's native web request tools are used to query OpenFDA's API with each compound name. The most frequent adverse events in general and the most frequent adverse events related to specific organs are then acquired. OpenFDA adheres to naming standards outlined by the Medical Dictionary for Regulatory Activities (MedDRA).38 MedDRA terms are organized hierarchically. OpenFDA uses the second most specific level of terms, called Preferred Terms, to name adverse events. Adverse events are associated with the most pertinent organs, using MedDRA Preferred Terms. Currently, adverse events are linked to the liver, kidney, and intestine, with additional organ linking in progress. The stored OpenFDA data is updated automatically every week. For front-end visualization of the adverse events, we use JQuery39 to pull from OpenFDA when the page loads. The data from OpenFDA includes the monthly count for each event (e.g., nausea, ALT increase, liver failure, etc.) from 2003 to present. The adverse event counts from OpenFDA were normalized to the frequency of drug usage using data from the National Ambulatory Medical Care Survey (NAMCS) and the National Hospital Medical Ambulatory Care Survey (NHAMCS), which are conducted by the Centers for Disease Control and Prevention's National Center for Health Statistics.40,41 The survey contains data on reports from patients on medication therapy. For example, in 2004 there were medication use reports from more than 585 million office visits comprising more than 1 billion drug mentions.41
As there is not a standard online source of clinical data, most of the preclinical and clinical data is manually curated. Sources for this information include the following databases: Health Canada Drug Product Database, European Medicines Agency, the LiverTox Clinical and Research Information on Drug Induced Liver Injury database, and ToxNet (Toxicology Data Network) HSDB database. The amount of information available varies considerably from compound to compound, often dependent on the date of compound approval, the individual pharmaceutical or Biotech companies, and the requirements of the regulatory agency. Older compounds approved before the widespread use of electronic data submissions can be difficult to locate, so the data is spread into various research articles. Health Canada and the European medicines agency publish preclinical toxicity and pharmacokinetics but the FDA does not, with exceptions to animal reproductive and developmental studies. On the converse, Health Canada database does not maintain product literature from compounds removed from the market. The inconsistency between submission types and the lack of equivalent data for all compounds is a liability that limits exclusive dependency on manual curation for predictive toxicology.
Implementation of the MPS-Db website
The back-end code for the website is written in Python using the Anaconda42 distribution (Continuum Analytics) to more easily manage our dependencies. The central component of the back-end is the Django web application framework. The Django framework provides methods to organize, manipulate, and query data in the database, manage URL dispatch, transmit data to the front-end using Python, and manage user permissions. With its object-relational mapper, Django communicates with PostgreSQL, a free and open source object-relational database management system that adheres to SQL standards.27 The PostgreSQL database is used to manage all the data in the MPS-Db.
The front-end code for the website, at its core, relies on HTML5, CSS3, and Javascript. Through its templating language, Django dynamically generates HTML to be served to the user. In instances where the page needs to be updated without a refresh, an AJAX request is sent to Django so that it can return the desired data in the JSON format. The JQuery and Underscore.js libraries are used to add utility functions and user interface improvements.43 Twitter Bootstrap44 is used to modify the appearance of the website and to enable a responsive design, which involves changing a page's content so that it conforms to the user's screen size. This permits users to view the site on devices like cell phones and tablets without the need to resize the content to make it legible. D3.js45 and C3.js,46 data visualization libraries, are used to generate dendrograms, heatmaps, line plots, and bar graphs. Finally, we use the JQuery library DataTables47 to create instantly searchable and sortable tables for displaying various sets of data.
Results
Study design
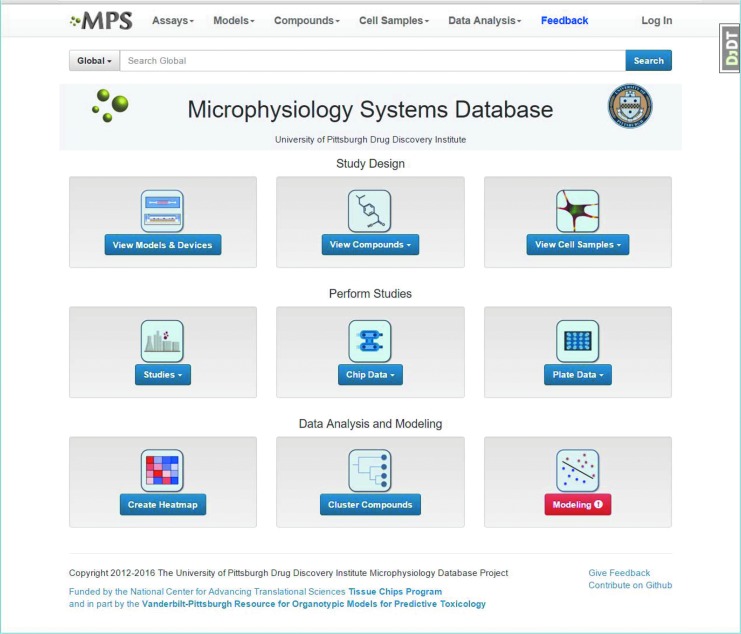
Tolcapone and Entacapone are structurally similar drugs that are used to treat Parkinson's disease. Entacapone has proven to be relatively safe, while tolcapone has caused fatal hepatotoxicity and therefore carries a black box warning.21,25 Although these findings are well known, we wanted to determine whether we could identify a more quantitative measure of the risk resulting from the use of tolcapone compared with entacapone, relate that risk to measurements made in the SQL-SAL MPS, and in the process demonstrate the use of the MPS-Db. The MPS-Db is an open, internet accessible website with a simple icon-driven interface (Fig. 3). The icons on the home page are functionally organized according the standard workflow: Study Design → Study Performance → Data Analysis and Modeling. In this study, the comparison of entacapone and tolcapone, the Study Design, started with the evaluation of the data for the compounds of interest. In other studies, compounds might be selected from the database based on drug, molecular, or biological properties.
FIG. 3.
The Microphysiology Database home page. The MPS-Db website is organized functionally into Study Design, Study Performance, and Data Analysis and Modeling. There is also a global MPS-Db search function to find information on a compound or component of interest.
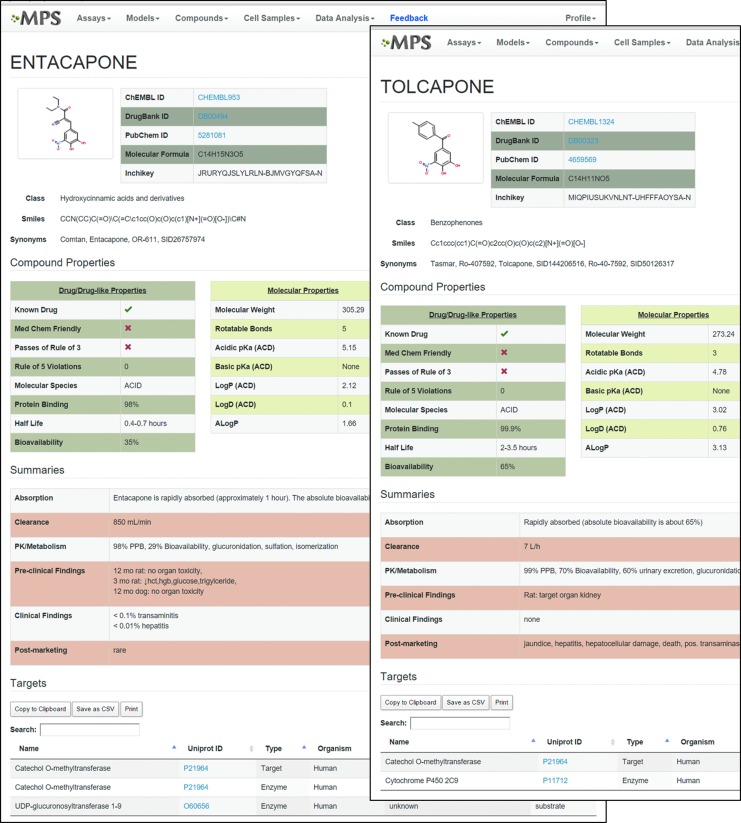
Figure 4 illustrates the compound pages from the MPS-Db, which include molecular, chemical, and druglike properties, along with summaries of the absorption, clearance, PK, and preclinical and clinical properties, as well as some known targets (see full pages in Supplementary Fig. S1A and B; Supplementary Data are available online at www.liebertpub.com/aivt). Structurally the compounds are similar, though tolcapone is somewhat more hydrophobic than entacapone (LogP = 3.02 vs. 2.12). Although both compounds pass the Lipinksi rule of 5, neither compound is MedChem friendly and both fail the rule of 3,48 but only tolcapone carries a black box warning. Tolcapone also has about a five-fold longer half-life and a two-fold higher bioavailability. No liver toxicity was identified in preclinical studies of the two compounds. Post market clinical use identified tolcapone liver liability, while entacapone has relatively few liabilities. The summary data for these two compounds shows that the physical properties, computational “drug-likeness” measure, preclinical studies, and even the clinical trial data can fail to identify the toxic compound between structurally similar chemical species. However, the organization of this information in the MPS-Db as shown in Figure 4 provides a single view of the data and facilitates the identification of gaps that would be needed to complete the picture, and in this case separate the hepatotoxic tolcapone from the liver-safe entacapone.
FIG. 4.
Examples from the MPS-Db compound summary pages for entacapone and tolcapone. The compound data sources are listed along with the structure and chemical descriptors from ChEMBL. Drug/Druglike properties are combined from ChEMBL and DrugBank; the Molecular Properties come from ChEMBL; and the Summaries of PK, preclinical, clinical, and post-marketing data are manually abstracted from data in the MPS-Db. At the bottom are targets and enzymes listed in DrugBank. Full pages are available in Supplementary Figures S1A and B.
To explore the liabilities associated with entacapone and tolcapone in sufficient detail and address the gaps in the information found by reviewing the compound characteristics, the MPS-Db provides adverse event (AE) data from the OpenFDA database37 and normalizes the AE frequency by the frequency of drug use reported in surveys by the CDC.49 Figure 5 compares the AE frequencies for entacapone and tolcapone. There are a much higher number of AE reports for entacapone (lighter bars); however, because tolcapone carries a black box warning and is used much less frequently, it was necessary to consider the frequency of AEs relative to the frequency of use. After normalizing to the use frequency (darker bars) it is clear that tolcapone has more frequent liabilities in the liver (ALT, AST, and Hepatic Failure), while the rate of AEs in the kidney, intestine, and brain are similar to entacapone (Fig. 5). To explore the liver AEs in more detail we reviewed all the liver AEs extracted from the OpenFDA database (Fig. 6). Of the 70 liver AEs in the OpenFDA database, 18 have been reported with a frequency ≥1/10,000 mentions. The frequency of reporting relative to usage, for 17 of the 18, is much higher for tolcapone than entacapone (only reports of hepatic function abnormal is about 20% lower for tolcapone compared with entacapone). There is a broad range of reports of AE in the human liver for tolcapone, but it may be that many are secondary effects. Although the frequency of an AE does not necessarily reflect the magnitude or seriousness of the event, the much higher frequency of ALT reports over AST reports likely indicates a greater effect on ALT over AST, implying an increase in hepatocellular damage. Clinicians consider ALT the specific marker for hepatocyte damage compared to AST, which is elevated from damage to liver, kidney, heart, skeletal muscle, and red blood cells.50 The increase in normalized ALT frequency is consistent with the increased reports of deaths, hepatotoxicity, and hepatic necrosis with tolcapone treatments.
FIG. 5.
Comparison of selected adverse event data for entacapone and tolcapone. Plot of most frequent AEs in human liver, kidney, intestine, and brain. Solid bars, associated with the left axis, indicate the frequency of AEs relative to the frequency of use, while the light bars, associated with the right axis, indicate the number of AE reports. In the liver (the first three pairs of bars) the relative frequency of AE is much higher for tolcapone compared with entacapone, while for the other organs the frequencies are similar for tolcapone and entacapone. AE, adverse event.
FIG. 6.
Relative liver adverse event frequencies for entacapone and tolcapone. Out of 70 liver AEs in the MPS-Db, there were 18 for tolcapone with ≥1/10,000 mentions. The AEs are divided into two groups, with the first 7 indicating changes in enzymes and secreted substances and the other 11 AEs indicating other abnormal liver functions. For all 18 AEs except hepatic function abnormal (for which tolcapone ∼1, entacapone ∼1.2), the relative frequency for entacapone was significantly lower than for tolcapone.
Although the focus of the MPS-Db is on human organ models, the database also includes preclinical data from animal studies, principally in rats, that can be accessed from external databases such as ChEMBL for direct comparison to human data. There are many rat studies of the inhibition of COMT in the MPS-Db (currently 20 for each compound), the primary target for tolcapone and entacapone.51 Tolcapone is a more potent inhibitor than entacapone (IC50 = 2.2 nM vs. 12.8 nM) and a much more effective inhibitor, inhibiting 92 ± 11% compared to 57 ± 31% for entacapone in rats (data not shown but available in the MPS-Db). In humans, tolcapone also has a two- to five-fold lower clearance rates relative to entacapone (Fig. 7 and Supplementary Fig. S2).
FIG. 7.
Comparison of human bioactivities for tolcapone and entacapone in the MPS-Db. The human bioactivities were retrieved from ChEMBL. The clearance (CL) and enzyme inhibition or modulation data with <30 uM is shown on a portion of the page. The full page is available in Supplementary Figure S2.
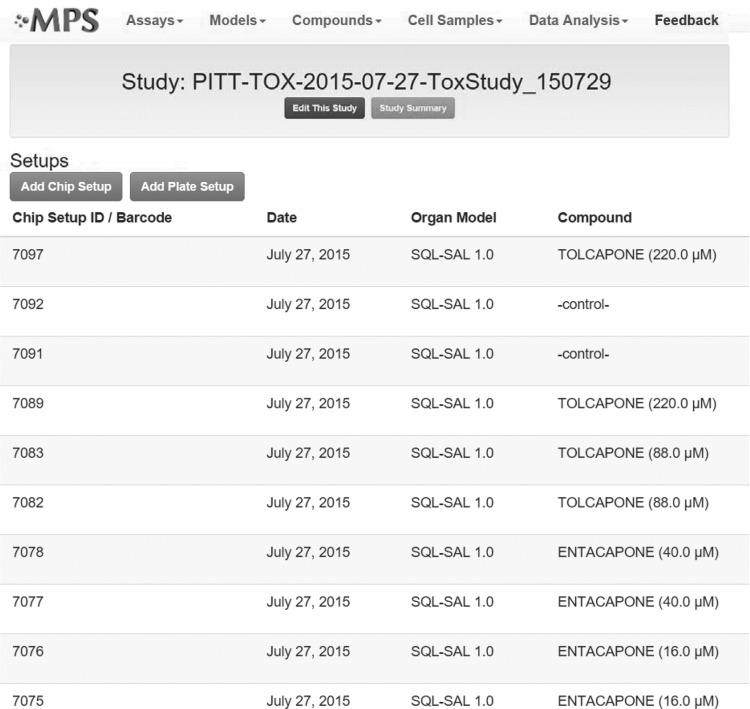
These reference data suggest that tolcapone leads to increased cellular damage in the human liver relative to entacapone, resulting in more AEs, which should be reflected in a human liver model. To address these hypotheses, a study was configured in the MPS-Db as shown in Figures 8 and 9. Because the adverse events are the most severe in the liver, we selected the SQL-SAL 1.0 liver model for the comparison of tolcapone and entacapone human liver effects. The primary features of the SQL-SAL model are illustrated in Figure 8 and published by Vernetti et al.19 This study consisted of 10 chips, comprising the two compounds at two concentrations and a control, all in duplicate, as depicted in Figure 9. Each chip was configured with the SQL-SAL 1.0 liver model and maintained under flow for the duration of the study, 21 days.
FIG. 8.
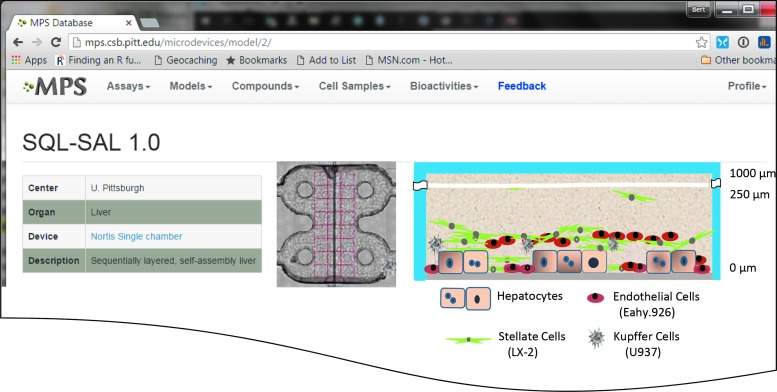
The SQL-SAL 1.0 liver model. For each organ model the MPS-Db stores the device, configuration of the model, a link to the device, and an illustration of the model. The design of the SQL-SAL model includes a layer of hepatocytes on a collagen-fibronectin substrate with NPCs in the matrix above.
FIG. 9.
Defining the study for the analysis of entacapone and tolcapone in the SQL-SAL. Each compound is run at two concentrations and in duplicate chips, along with duplicate negative control chips. Configuring the study in the MPS-Db sets up the import of the resulting data and the subsequent analysis.
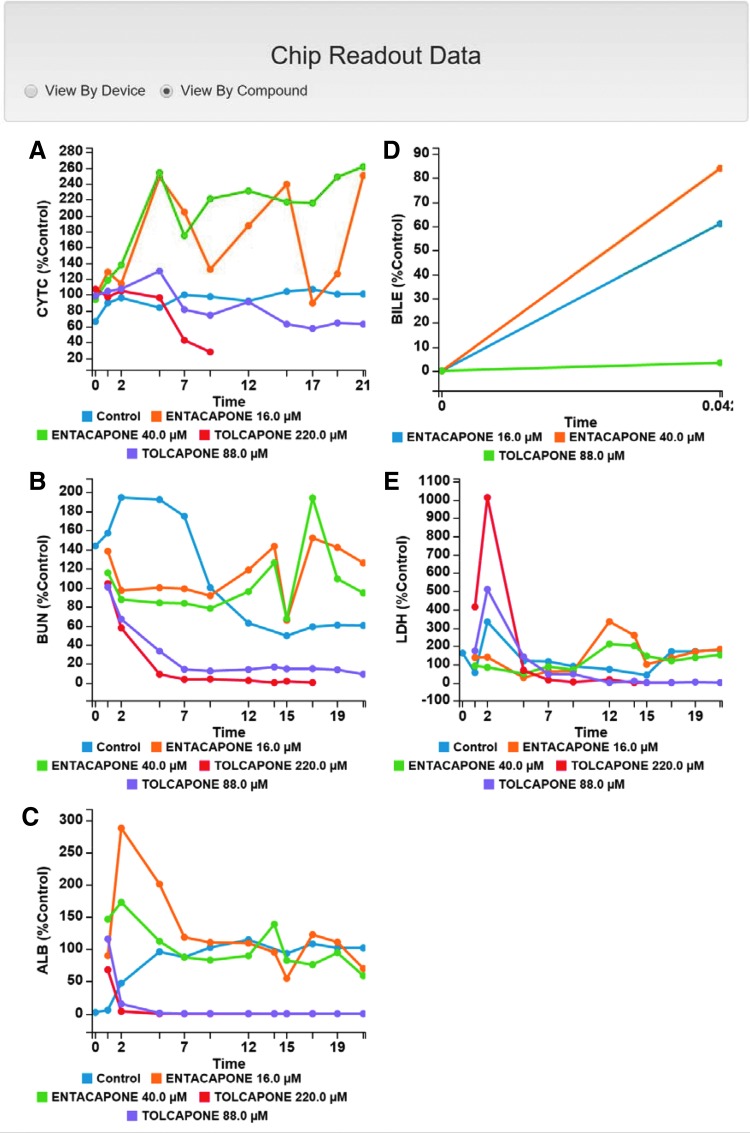
Data was collected for five readouts: timed media concentrations of albumin, urea, and LDH and cytochrome-C biosensor intensity in the mitochondria. A single measurement for bile canalicular efflux was collected.19 The data sets were uploaded to the MPS-Db for analysis and comparison with the reference data. Figure 10 shows the data plots that are presented on the Study Summary page of the MPS-Db. Dose and time-dependent effects on the apoptosis biosensor, urea, albumin, and LDH release were found for tolcapone, but not for entacapone. Tolcapone at 220 μM (10 × Cmax) induced a 75% loss of CytC intensity in 9 days, a 90% decrease in urea secretion in 5 days, a 90% decrease in albumin production in 2 days, a 10-fold peak increase in LDH concentration at 2 days (Fig. 10A–C and E). Due to the rapid loss of cell viability, bile efflux was not measured in these chips. Tolcapone at 88 μM (4 × Cmax) had a reduced effect, inducing a 25% reduction in CytC intensity over 9 days, an 80% reduction in urea secretion over 7 days, a 90% reduction in albumin secretion in 2 days, a five-fold increase in LDH concentration at 2 days, and a complete inhibition of bile efflux. Entacapone at 16 μM (4 × Cmax) or 40 μM (10 × Cmax) had little or no effect on CytC intensity, urea secretion, or albumin secretion. Both concentrations induced a two- to three-fold peak increase in LDH concentration at 12 days. For both concentrations the bile efflux was 60–90% of control. The decrease in albumin and peak LDH release were the earliest indicators of impaired hepatocyte health, responding in the first 2 days even at just 4 × Cmax. The decrease in CytC intensity and urea secretion were much slower, taking about 5–7 days. The slowest measured response was the LDH release induced by entacapone, taking 12 days to the peak.
FIG. 10.
Plots of the temporal readouts as displayed on the MPS-Db Study Summary page. After importing data into the MPS-Db, a graphical view of the results is provided for review and further analysis. In this study, there are five readouts (time in days). (A) Tolcapone at 220 μM induced cytochrome C release from mitochondria, a precursor to induction of apoptosis (T½ = 6.5 days), while at 88 μM tolcapone the signal was similar to the control. Entacapone at 40 μM and 16 μM showed only an accumulation or condensation of cytochrome C in the mitochondria. (B) Tolcapone (220 μM and 88 μM) induced a rapid reduction in urea production (BUN) over 2–7 days while entacapone did not significantly affect the urea. (C) Tolcapone (220 μM and 88 μM) induced a rapid loss of albumin secretion while entacapone had no effect. (D) Tolcapone (88 μM) completely blocked bile efflux. (E) Tolcapone (220 μM and 88 μM) induced rapid (2 day) release of LDH while entacapone induced LDH release only after about 12 days.
A complete summary of the reference data and the measured data is shown in the compound Summary Report from the MPS-Db (Fig. 11 and Supplementary Fig. S3). Manually curated preclinical data from rat studies indicates no organ toxicity for entacapone and only kidney toxicity for tolcapone. Clinical trial data indicates a very low frequency of liver adverse reactions for entacapone, and post-market data found serious liver adverse events for tolcapone. The time course for the five measured liver functions in this study are shown in the report as “sparklines.”52 A sparkline is a small line chart, typically drawn without axes or coordinates, that presents the general shape of the variation in the measured parameter, in this case over time. Sparklines are a compact method to display graphical representations of the trends in the data in Figure 10.
FIG. 11.
MPS-Db Compound Summary page displays the properties and results for entacapone and tolcapone. The MPS-Db provides a tabular summary of the reference data and measured data for comparison and analysis. The preclinical and clinical findings, as well as the PK/metabolism, are manually curated summaries from MPS-Db sources or other sources. The Organ Model Results are sparkline plots of the time course of the measured value over the interval in the heading. The full page without overlay is available in Supplementary Figure S3.
Discussion
One of the major challenges associated with the development of human organ models as surrogates for testing the safety and efficacy of new drugs is establishing quantitative relationships between the effects observed in the model and human health. The goal of MPS is to mimic the human response of the patient in vitro. MPS provides an opportunity to read out more detailed molecular characteristics of the biological system than is possible in vivo and thereby enhance our understanding of organ function and the mechanisms underlying adverse events. However, correlating MPS readouts with clinical readouts can be difficult due to differences in measurement methods.17 To enable interpretation, readouts from MPS must be mapped to clinical diagnostics, requiring computational models that translate data from MPS to their clinical significance. The MPS-Db is designed to capture and aggregate data from organ models and associate that data with reference data from chemical, biochemical, preclinical, clinical, and post-marketing sources in order to support the design, development, validation, and interpretation of organ models. The goal of the MPS-Db is to facilitate the acquisition of sufficient experimental and reference data for compounds with known human exposure results to enable the development of computational models that predict the effects of human exposure to unknown or new compounds. We are currently building the necessary reference dataset in order to initiate computational modeling as we previous published for a fixed multiparameter, endpoint platform.14
The MPS-Db was developed using open source tools to ensure its accessibility in the research community, and is organized into 3 functional components, Study Design, Study Performance, and Data Analysis and Modeling (Fig. 3). Study design includes selecting or designing organ models, selecting compounds and exploring compound properties, and selecting cells with which to construct the model. In this study, the SQL-SAL liver model was used. The MPS-Db includes the design of the model (Fig. 8), along with the protocol for assembling and using it. The MPS-Db also contains the cell types and batch numbers to enable comparison of compound activity or cell performance across individual chips.
Reference data for tolcapone and entacapone
For this study, tolcapone and entacapone were chosen to be evaluated in the SQL-SAL as a pair of marketed drugs with differential liver toxicity to test the hypothesis that the readouts from the SQL-SAL liver model would detect liver toxicity with tolcapone but not entacapone. A secondary hypothesis was that the SQL-SAL readouts that were most indicative of tolcapone activity should relate to its clinical toxicity indications. To characterize the known differences between tolcapone and entacapone we evaluated the reference data in the MPS-Db which consisted of biochemical/pharmacological data from ChEMBL and PubChem (Fig. 4), manually curated data from clinical trials (Figs. 4 and 11), and normalized adverse event data from OpenFDA (Figs. 5 and 6). Both compounds target and inhibit COMT. Both compounds are extensively metabolized in the human liver.53 Tolcapone is also listed as an inhibitor of CyP450 2c9 in DrugBank, while entacapone is not. The MPS-Db provides links to the source data it contains, making it easy to find the original DrugBank source for the inhibition, the SuperCYP database.54 In SuperCYP both tolcapone and entacapone are listed as inhibitors of CyP450 2c9, even though DrugBank only lists the inhibition of CyP450 2c9 for tolcapone. This is not an uncommon problem with databases, and one of the main reasons that the MPS-Db maintains links to the source data for verification as well as for further investigation.
The main biochemical/pharmacological differences were that tolcapone has a five-fold longer half-life, almost two-fold greater bioavailability, and about seven-fold lower clearance rate. The fact that tolcapone persists longer in vivo due to fewer clearance mechanisms may contribute to its overall toxicity but does not give insight into the mechanism of toxicity (MOT). In preclinical rat studies, neither compound exhibited significant liver toxicity, providing only the insight that rat is not a good model for the human MOT. Clinical findings from manually curated data indicated significant liver adverse events including hepatocellular damage, positive transaminases, hepatitis, and death with tolcapone but not entacapone (Fig. 4). Although manually curated data provides insights that may be useful for identifying potential MOTs, the anecdotal nature and low availability of clinical data limits its use as a basis for establishing a correlation between MPS readouts and clinical results. To establish a more quantitative measure of effects in humans we used AE report statistics from the OpenFDA database (Figs. 5 and 6). The OpenFDA database provides data on the frequency of AEs, but not the frequency of use for the drugs. Drugs with severe AEs are generally used less frequently than “clean” drugs, so the total number of AEs can be misleading. Also, AEs are associated with all drugs that were mentioned in the report, so some common drugs like aspirin and acetaminophen are associated with many AEs. For example, the number of “ALT Increased” AEs in OpenFDA for aspirin, entacapone, and tolcapone are 76, 476, 431, and 38, respectively. Therefore, it was necessary to normalize the AE count to the frequency of drug use, with the NAMCS data from the CDC.41 The NAMCS data is based on drug mentions during interviews of patients coming into clinics, and then projected to overall usage. While it may not be entirely accurate, it is a large dataset with over 1 billion drug mentions in 2004. So the relative frequency of individual AEs associated with tolcapone and entacapone are only estimates, but the frequencies appear to be consistent with the manually curated data and are certainly based on much larger sample sizes than the articles, clinical trial reports, and case reports, and provide a more quantitative basis of comparison. The most frequently reported AE for tolcapone was “ALT increased,” followed by “AST increased,” in a roughly 2:1 ratio. More frequent reports of ALT increases are accepted by clinicians as evidence of liver toxicity.
Comparison of tolcapone and entacapone in an MPS
When tolcapone and entacapone were tested in the SQL-SAL devices, dose and time dependent effects on the apoptosis biosensor, albumin, urea, and LDH release were found for tolcapone at 88 μM (4 × Cmax) and 220 μM (40 × Cmax) but not for 16 μM (4 × Cmax) or 40 μM (38 × Cmax) entacapone (Fig. 10). Decreases in albumin, urea, and LDH were the earliest indicators for impaired hepatocyte health followed by increased numbers of apoptotic cells in tolcapone treated SQL-SAL. Bile efflux was 97% inhibited by 88 μM tolcapone, a finding consistent with earlier reports.55 The higher concentration of tolcapone produced excessive toxicity by day 5; the time bile efflux is reestablished in the in vitro liver and therefore was not tested. The hepatotoxicity finding is consistent with clinical use of tolcapone but not with the structurally related compound entacapone. Although both compounds are extensively metabolized in the liver, tolcapone metabolism includes significant nitro-reduction whereas entacapone does not; tolcapone is cleared by glucuronidation whereas entacapone is cleared by glucuronidation and sulfation, suggesting that metabolic pathway differences may be related to tolcapone toxicity.56
Treatment at 10 × and 40 × Cmax levels found dose-dependent increases in LDH leakage, decreases in albumin and urea synthesis, increased apoptosis, and inhibition of bile efflux in SQL-SAL devices treated with tolcapone compared to treatment with vehicle or 10 × and 40 × entacapone (Fig. 10). One possible reason the SQL-SAL discriminated between entacapone and tolcapone was simply due to treating the human organoid at higher exposure levels for extended periods of time. Although rising dose studies were used to determine the maximum tolerated dose (MTD) in phase 1 clinical trials, extending the dose beyond first evidence of toxicity is not possible. However, one of the benefits of the human organ models like SQL-SAL is that it is possible to treat above the human MTD provided by traditional clinical biochemical and metabolic data.
The pattern of enzyme release during clinical manifestation of liver damage from tolcapone treatment was hepatocellular in nature with no accompanying inflammatory responses. In the in vitro model used here, dose-dependent increase in LDH release was found soon after the initiation of exposure, but cell death as determined by the apoptosis biomarker required prolonged treatment. A modest spike in LDH activity was found after 10 days of treatment to entacapone but with no accompanying changes in albumin, urea, bile efflux inhibition, or activation of hepatocyte apoptosis. We can speculate this may be an adaptive rather than an adverse response of the in vitro liver, similar to the self-eliminating elevation in transaminases sometimes found in the clinic for entacapone and other drugs such as erythromycin. This will be investigated in the future.
Why the MPS-Db is essential to the successful application of MPS
There are several reasons why an MPS-Db is needed. (1) Storing, managing, and comparing data between organ models is complicated by the complexity of the models. Comparing and interpreting data between models, or even from run-to-run in the same model, requires capturing information about the model (e.g., device characteristics, cell types, media, flow rate, extracellular matrix material, etc.) along with the measurement data. (2) The reference “truth” data for validation and computational modeling is difficult to access. However, there are multiple sources of data on the biochemical, preclinical, clinical, and post-marketing effects of drugs and other chemicals, including journals and a wide array of databases. However, each data source only has a piece of the required data. ChEMBL contains bioactivity data, the Health Canada Drug Product Database, and ClinicalTrials.gov, and other sources contain clinical information, but it is mostly embedded in reports and requires manual extraction. The FDA has established the OpenFDA database, which is a tremendous source of adverse event data, but there is not a standard user interface, and the data is not normalized to the frequency of usage, so it cannot be directly used to compare drugs. Once the data has been extracted and normalized, it becomes a more useful resource. (3) Data contained in databases sometimes contains transcription errors (e.g., the wrong units), comes from different experimental sources (e.g., different species or tissue types), or is in different units and therefore needs to be reviewed, corrected, and validated. In comparing the two compounds in this study, several inconsistencies in the ChEMBL data and the PubChem data were discovered and had to be corrected in the MPS-Db. Corrections are annotated for future reference. (4) Comparing the performance of MPS, whether they are different designs or different versions of the same design, requires a reference standard. The MPS-Db will facilitate establishing standard reference data sets for comparing organ models. (5) The MPS-Db provides a means to compare data across organ systems. MPS are currently being integrated into multiorgan systems, but even in the absence of directly coupled organs, combinations of data from multiple organs can be used to evaluate the clinical translation of the data.
Future goal and prospects for the MPS-Db
One of the major goals of the MPS-Db is to provide a platform for developing computational models that predict human clinical outcomes from MPS data, as well as to perform pharmacokinetic analyses. On the home page (Fig. 3) there is a section for Data Analysis and Modeling that includes Heatmaps, Clustering, and Modeling. While there is sufficient reference data for generating useful heatmaps and applying clustering in order to identify compounds with similar bioactivities—and/or preclinical, clinical, or post-marketing effects—the necessary MPS data is only now being generated and uploaded to the database. However, as demonstrated here, the MPS-Db is a tool that can be used today for selecting and testing compounds, as well as MPS, and for translating microphysiological systems data from the in vitro model to the in vivo human. We project that MPS models and the MPS-Db will become an integral part of the application of quantitative systems pharmacology (QSP) to drug discovery and development.57
Software development processes evolve rapidly and trends can change. To take advantage of improving functionality we have used components with active maintenance and ongoing developments. Furthermore, online data sources may be updated or become inaccessible. The MPS-Db has been designed to cache data locally to protect against the loss of a data source, and to automatically update data on a regular basis to take advantage of improvements to the content of the sources. The principal data sources are widely used, well established, and supported databases that should remain accessible for a long time.
We believe the MPS-Db provides a valuable resource for users involved in developing or using human organ models for prediction of safety and efficacy. The database brings together resources that are each accessible individually, but it is the combination that makes them more useful. For example, the OpenFDA database provided by the FDA does not have a graphical user interface, is not adjusted for frequency of drug use, and does not link adverse events to a particular organ. To address these limitations, we developed a user interface, used the NAMCS data to normalize the adverse event frequencies, and used the MEDRA terms in the OpenFDA to identify the relevant organ. Similarly, the integration of experimental design and data management provides value to the laboratory scientist for managing studies.
The MPS-Db is available online now for browsing the reference data and selected MPS datasets. The source code for the MPS-Db is managed on GitHub.com as a repository named MPS-Database-Server. Comments and issue reports are monitored regularly and will be prioritized for development. Individuals interested in participating in the development can contact the development team via GitHub. The MPS-Db code is freely available to nonprofit researchers who can download the current version from GitHub, and will be made available to corporations for a nominal fee to help support maintenance and extension of the MPS-Db. The MPS-Db content will be provided as a PostgreSQL database to nonprofit organizations, and for a license fee to for-profit corporations. Interested parties should contact the authors.
Supplementary Material
Acknowledgments
The authors are grateful to Ahmet Bakan, PhD, formerly of the Computational and Systems Biology Department at the University of Pittsburgh, for his initial design and development of a prototype of the Microphysiology Systems database. Many of his designs and concepts persist in the current version of the database. We thank members of the UPDDI for experimental support and discussions including Nina Senutovich, Richard DeBiasio, David Zaidins, Felipe Lee-Montiel, Subin George, and Celeste Reese. We gratefully acknowledge support for this project from the National Institutes of Health (5UH3TR00503, 1S10-OD01226, and 3UH3TR00503-04S1) and the U.S. Environmental Protection Agency (EPA STAR 83573601).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 2015:14;248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med (Maywood) 2014:239;1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature 2014:507;181–189 [DOI] [PubMed] [Google Scholar]
- 4.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014:32;760–772 [DOI] [PubMed] [Google Scholar]
- 5.Vunjak-Novakovic G. Advanced methods for tissue engineering and regenerative medicine. Methods 2015:84:1–2 [DOI] [PubMed] [Google Scholar]
- 6.Griffith LG, Wells A, Stolz DB. Engineering liver. Hepatology. 2014:60;1426–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esch MB, King TL, Shuler ML. The role of body-on-a-chip devices in drug and toxicity studies. Annu Rev Biomed Eng. 2011:13;55–72 [DOI] [PubMed] [Google Scholar]
- 8.Suvorov A, Takser L. Facing the challenge of data transfer from animal models to humans: the case of persistent organohalogens. Environ Health 2008:7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol 2006:2;875–894 [DOI] [PubMed] [Google Scholar]
- 10.Olson H, Betton G, Robinson D, et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol 2000:32;56–67 [DOI] [PubMed] [Google Scholar]
- 11.Mak IWY, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. American Journal of Translational Research 2014:6;114–118 [PMC free article] [PubMed] [Google Scholar]
- 12.Greek R, Menache A. Systematic reviews of animal models: methodology versus epistemology. International Journal of Medical Sciences 2013:10;206–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giuliano KA, Gough AH, Taylor DL, et al. Early safety assessment using cellular systems biology yields insights into mechanisms of action. J Biomol Screen 2010:15;783–797 [DOI] [PubMed] [Google Scholar]
- 14.Vernetti L, Irwin W, Giuliano KA, et al. Cellular systems biology applied to preclinical safety testing: a case study of cellCiphr(TM) profiling. In: Drug Efficacy, Safety, and Biologics Discovery: Emerging Technologies and Tools. Ekins S, Xu JJ, eds; 53–73. New Jersey: Wiley & Sons; 2009 [Google Scholar]
- 15.Aleo MD, Luo Y, Swiss R, et al. Human drug-induced liver injury severity is highly associated with dual inhibition of liver mitochondrial function and bile salt export pump. Hepatology. 2014:60;1015–1022 [DOI] [PubMed] [Google Scholar]
- 16.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 2015:14;248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capulli AK, Tian K, Mehandru N, et al. Approaching the in vitro clinical trial: engineering organs on chips. Lab Chip 2014:14;3181–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutherland ML, Fabre KM, Tagle DA. The National Institutes of Health Microphysiological Systems Program focuses on a critical challenge in the drug discovery pipeline. Stem Cell Res Ther 2013:4 Suppl 1:I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernetti LA, Senutovitch N, Boltz R, et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med (Maywood) 2016:241;101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senutovitch N, Vernetti L, Boltz R, et al. Fluorescent protein biosensors applied to microphysiological systems. Exp Biol Med (Maywood) 2015:240;795–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeiffer E, Kegel V, Zeilinger K, et al. Featured Article: Isolation, characterization, and cultivation of human hepatocytes and non-parenchymal liver cells. Exp Biol Med (Maywood) 2015:240;645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebelo SP, Costa R, Silva MM, et al. Three-dimensional co-culture of human hepatocytes and mesenchymal stem cells: improved functionality in long-term bioreactor cultures. J Tissue Eng Regen Med. 2015. [Epub ahead of print]; doi: 10.1002/term.2099 [DOI] [PubMed] [Google Scholar]
- 23.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol 2008:26;120–126 [DOI] [PubMed] [Google Scholar]
- 24.Microphysiology Systems Database. http://mps.csb.pitt.edu (Accessed March1, 2016)
- 25.McBurney RN, Hines WM, Von Tungeln LS, et al. The liver toxicity biomarker study: phase I design and preliminary results. Toxicol Pathol 2009:37;52–64 [DOI] [PubMed] [Google Scholar]
- 26.Django [computer program]. Version 1.8.8. www.djangoproject.com/: Django Software Foundation; 2016 [Google Scholar]
- 27.PostgreSQL [computer program]. Version 9.3.10. www.postgresql.org/: The PostgreSQL Global Development Group; 2016 [Google Scholar]
- 28.Kim S, Thiessen PA, Bolton EE, et al. PubChem Substance and Compound databases. Nucleic Acids Res 2016:44:D1202–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Suzek T, Zhang J, et al. PubChem BioAssay: 2014 update. Nucleic Acids Res 2014:42;D1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bento AP, Gaulton A, Hersey A, et al. The ChEMBL bioactivity database: an update. Nucleic Acids Res 2014:42;D1083–D1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 2012:40;D1100–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knox C, Law V, Jewison T, et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res 2011:39;D1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wishart DS, Knox C, Guo AC, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 2006:34;D668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Python [computer program]. Version 2.7.11. www.python.org/: Python Software Foundation; 2016 [Google Scholar]
- 35.Katayama T, Nakao M, Takagi T. TogoWS: integrated SOAP and REST APIs for interoperable bioinformatics web services. Nucleic Acids Res 2010:38;W706–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers J, Davies M, Gaulton A, et al. UniChem: a unified chemical structure cross-referencing and identifier tracking system. J Cheminform. 2013:5;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.OpenFDA [computer program]. Version. https://open.fda.gov/: U.S. Food and Drug Administration; 2016 [Google Scholar]
- 38.Brown EG, Wood L, Wood S. The medical dictionary for regulatory activities (MedDRA). Drug Saf 1999:20;109–117 [DOI] [PubMed] [Google Scholar]
- 39.JQuery [computer program]. Version 1.11.2. https://jquery.com/: The JQuery Foundation; 2016 [Google Scholar]
- 40.Centers for Disease Control and Prevention. Ambulatory Health Care Data. www.cdc.gov/nchs/ahcd.htm (Accessed March1, 2016)
- 41.Woodwell DA, Cherry DK. National Ambulatory Medical Care Survey: 2002 summary. Adv Data 2004:1–44 [PubMed] [Google Scholar]
- 42.Anaconda [computer program]. Version 2-2.4.0. Anaconda Software Distribution (https://continuum.io): Continuum Analytic; 2015 [Google Scholar]
- 43.Underscore [computer program]. Version 1.8.3. http://underscorejs.org/
- 44.Bootstrap [computer program]. Version 3.3.1. http://getbootstrap.com/; 2015
- 45.D3 [computer program]. Version 3.5.12. https://d3js.org/: Mike Bostock; 2015 [Google Scholar]
- 46.C3 [computer program]. Version 0.4.10. http://c3js.org/: Masayuki Tanaka; 2014 [Google Scholar]
- 47.DataTables [computer program]. Version 1.10.4. www.datatables.net/: SpryMedia Ltd; 2016 [Google Scholar]
- 48.Congreve M, Carr R, Murray C, et al. A 'rule of three' for fragment-based lead discovery? Drug Discov Today 2003:8;876–877 [DOI] [PubMed] [Google Scholar]
- 49.Hsiao CJ, Cherry DK, Beatty PC, et al. National Ambulatory Medical Care Survey: 2007 summary. Natl Health Stat Report 2010:1–32 [PubMed] [Google Scholar]
- 50.Giannini EG, Testa R, Savarino V. Liver enzyme alteration: a guide for clinicians. CMAJ 2005:172:367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tai CH, Wu RM. Catechol-O-methyltransferase and Parkinson's disease. Acta Med Okayama 2002:56;1–6 [DOI] [PubMed] [Google Scholar]
- 52.Tufte ER. Beautiful evidence. Cheshire, Conn.: Graphics Press; 2006 [Google Scholar]
- 53.Benet LZ, Broccatelli F, Oprea TI. BDDCS applied to over 900 drugs. AAPS J 2011:13;519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Preissner S, Kroll K, Dunkel M, et al. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res 2010:38:D237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgan RE, van Staden CJ, Chen Y, et al. A multifactorial approach to hepatobiliary transporter assessment enables improved therapeutic compound development. Toxicol Sci 2013:136;216–241 [DOI] [PubMed] [Google Scholar]
- 56.Kalgutkar AS. Role of bioactivation in idiosyncratic drug toxicity: Structure–toxicity relationships. Advances in bioactivation research. New York: Springer; 2008:1–29 [Google Scholar]
- 57.Stern A, Schurdak M, Bahar I, et al. A Perspective on Implementing a Quantitative Systems Pharmacology Platform for Drug Discovery and the Advancement of Personalized Medicine. J Biomol Screen 2016: [Epub ahead of print]; pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.