An important adaptation of cells to proteasome inhibition is the induction of new proteasomes via the transcription factor Nrf1 [1, 2], which is produced as a precursor bound to the endoplasmic reticulum (ER) through its N-terminus. Nrf1 was reported to require proteolytic processing to enter the nucleus [3]. Increased proteasome production is induced by low concentrations of proteasome inhibitors that reduce proteolysis by <50%. Surprisingly, we found that proteasome induction and Nrf1 processing to its shorter form (which we estimated to be 75kDa [2]) were suppressed by high concentrations of inhibitors that markedly reduce proteasome activity [4]. This unusual bimodal concentration-dependence implied that some proteasome function was necessary for Nrf1 processing. Because we found that Nrf1 processing also required ubiquitin conjugation [2], we proposed that the Nrf1 processing is catalyzed by partially-inhibited proteasomes [2]. However, Vangala et al present compelling evidence that conversion of the ER-bound Nrf1 to the shorter form, which they described as 110kDa, is independent of proteasomes and was not blocked by high concentrations of proteasome inhibitors [5]. Therefore, we investigated the basis for these different results. The studies presented below show that: 1) we and Vangala et al studied the same processed form of Nrf1, whose actual molecular weight appears to be 90–95kDa; 2) as we reported, high concentrations of inhibitors suppressed proteasome induction and accumulation of processed Nrf1 in soluble lysates, but not by preventing its processing, and instead by causing the processed Nrf1 to aggregate. 3) Others recently showed that Nrf1 is actually cleaved by an unusual endoprotease, Ddi1/Ddi2 [6], which requires ubiquitination and associates with proteasomes.
To determine whether the processed forms reported by the two labs [2, 5] are the same, we employed the anti-Nrf1 antibody and proteasome inhibitor used by Vangala et al, and confirmed our earlier observations that a single processed Nrf1 band (our p75) accumulated in lysates of HEK293A cells after 16h treatment with 0.05–0.5μM carfilzomib (CFZ), but was suppressed up to 90% by ≥1μM (Fig 1A, S1A). We also used the approach of Vangala et al. [5] to cause Nrf1 to accumulate as the glycosylated precursor using a p97 inhibitor. Upon removal of the inhibitor, we also observed Nrf1 processing to a single band (their p110) (Fig 1A, S1B). Despite their different reported sizes, the processed forms from these distinct experiments migrated identically on SDS-PAGE (Fig 1A), and only a single band was detected by either antibody [7]. This band must have resulted from cleavage of Nrf1’s N-terminus because it also was generated from Nrf1 bearing an N-terminal HA-tag (i.e. by removal of this N-terminal tag (Fig S1D)).
Figure 1. When proteasomes are completely inhibited, Nrf1 can still be processed, but becomes insoluble and leaves solution.
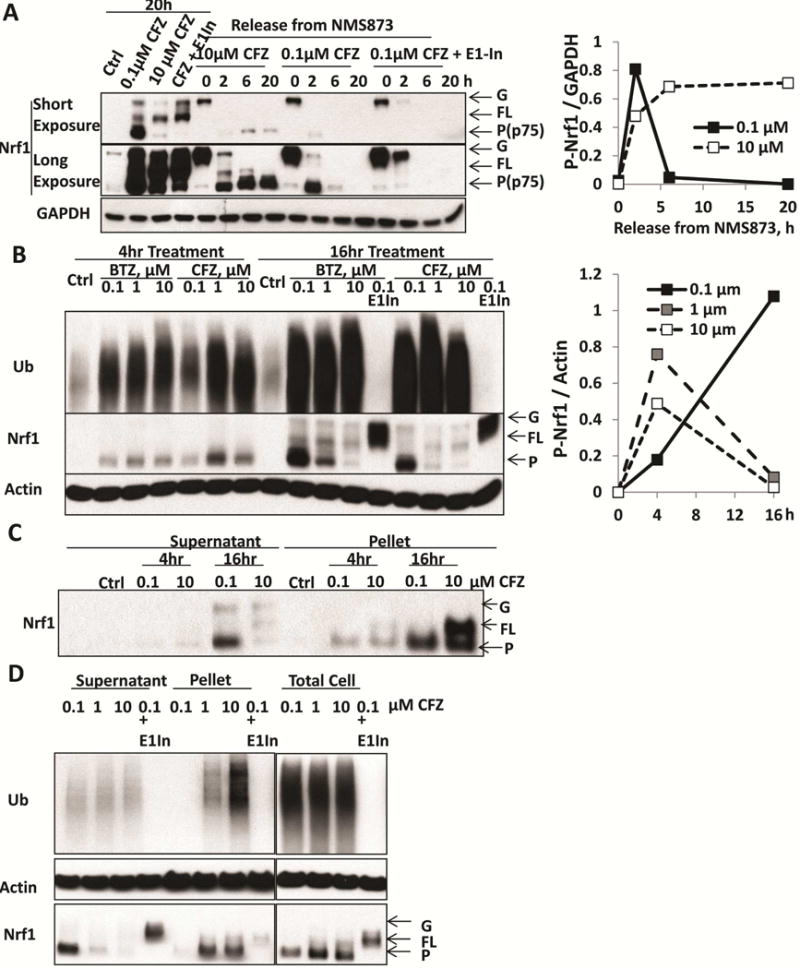
A. HEK293A cells were treated for 20h with 0.1 or 10 μM carfilzomib (CFZ), or with a combination of 0.1 μM CFZ and 0.5 μM E1 inhibitor (ML00603997, E1-In). In addition, to monitor the processing of glycosylated precursor (G-Nrf1), the cells were first treated with the p97 inhibitor NMS873 (10 μM) for 2h to cause endogenous Nrf1 to accumulate as the glycosylated precursor. NMS873 was then removed and the cells cultured for 2, 6, and 20h in the presence of cycloheximide to inhibit new protein synthesis (Supplemental Fig S1B). Following NMS873 removal, cells were treated with 0.1 or 10 μM CFZ, or with a combination of 0.1 μM CFZ and 0.5 μM E1-inhibitor in order to determine whether processing of glycosylated Nrf1 required proteasome activity (completely blocked by 10 μM CFZ) or ubiquitination (by E1-In). The levels of glycosylated (G), deglycosylated full-length (FL), and processed (P) forms (also termed p75 [2]) of Nrf1 were assayed by western blotting (Left). Densitometry was performed to quantify the levels of processed (P-Nrf1, p75) in cells undergoing NMS873-release with co-treatment of 0.1 or 10 μM CFZ and normalized with GAPDH in right panel.
B. HEK293F cells were treated with 0.1, 1, or 10 μM BTZ or CFZ for 4 or 16h. These cells were also treated with 0.1 μM BTZ or CFZ together with 0.5 μM E1-In for 16h. The levels of glycosylated, full-length, and processed forms of Nrf1 in the cell lysate were measured by western blotting (Left). Quantifications of the levels of processed Nrf1 (P-Nrf1, normalized to actin) in CFZ-treated cells are shown in the right panel.
C. HEK293F cells were treated with 0.1 or 10 μM CFZ for 4 or 16h. The 10,000 × g pellet fraction of the cells after lysis with buffer containing 1% TX-100 was collected by centrifugation for 10min and solubilized in 2% SDS. The levels of G, FL, P forms of Nrf1 in this pellet fraction were measured by western blotting.
D. HEK293F cells were treated for 16h with 0.1, 1, or 10 μM CFZ, or a combination of 0.1 μM CFZ and 0.5 μM E1 inhibitor. The levels of G, FL, P forms of Nrf1 in the lysate and pellet fractions, or in the total cell protein (by directly lysis of cells in 2% SDS) were measured by Western blotting.
The molecular weights determined by SDS-PAGE are only approximations affected by the standards and conditions used. These factors presumably caused different MW estimates for processed Nrf1 (We used SeeBlue Plus2, ThermoFisher; the standards and conditions used by Vangala et al. were not indicated). To more accurately estimate its size, we used multiple standards and compared the mobility of processed Nrf1 with other proteins. It migrates slightly faster than p97, but slower than PI3 Kinase p85 subunit, indicating an actual size of 90–95kDa (Fig S1E). Thus, the two laboratories studied the same 90–95 kDa fragment of Nrf1, which can be generated when proteasomes are completely inhibited (Fig S1C).
This ability of cells to generate processed Nrf1 after complete proteasome inhibition seems to contradict our reproducible findings that 16h treatment with high concentrations of proteasome inhibitors consistently suppressed the accumulation of processed Nrf1 in our lysates and proteasome induction (Fig S1A). We therefore tested whether the duration of inhibition affects Nrf1 processing. At 4h, the level of processed Nrf1 in cell lysates was highest with 1μM BTZ or CFZ and only slightly less with 10μM (Fig 1B), as shown in [5]. However, after 16h treatment, the level of processed Nrf1 was greatly suppressed by 1 or 10 μM BTZ or CFZ (Fig 1B) below the levels at 4h. Since processed Nrf1 is not degraded under these conditions (Fig 1A), its disappearance from the lysate with 16h treatment is probably due to its precipitation. Accordingly, between 4 and 16h, a large amount of the processed form built up in the 10,000×g pellet in cells lysed in buffer containing 1% TX-100 (Fig 1C), and much more accumulated in the insoluble fraction with 1 or 10μM CFZ than with 0.1μM (Fig 1D). However, when cells were treated with high concentrations of CFZ and lysed in 2% SDS to solubilize such aggregates, no clear decrease in cellular content of processed Nrf1 was evident (Fig 1D). Our previous immunostaining of cells treated for 16h with high inhibitor concentrations had also indicated that Nrf1 aggregates in the cytosol.
Although we confirmed our prior finding that high concentrations of proteasome inhibitors block the accumulation of processed Nrf1, this effect is by blocking Nrf1 processing. Instead with high inhibitor concentrations, Nrf1 was processed, but accumulated with time as aggregates. While processed Nrf1 remained largely soluble with 0.1 uM CFZ, it was rapidly degraded (t½=50min) (Fig 1A). Thus, the undegraded Nrf1 is prone to aggregate. Once insoluble (as occurred following prolonged strong proteasome inhibition), Nrf1 seems inactive in transcription, since the induction of proteasome subunits is suppressed under these conditions [2]. In related studies, we recently observed that combining different proteasome inhibitors to block multiple peptidase activities and reduce proteolysis further also prevented compensatory induction of new proteasomes, presumably by increasing Nrf1 aggregation (Weyburn et al. submitted).
These findings clearly indicate that Nrf1 processing is catalyzed by a non-proteasomal endoprotease. This enzyme must be unusual since we confirmed our observation [2] that Nrf1 processing requires ubiquitin conjugation (Fig 1A, 1D). An unusual protease that can bind ubiquitin was recently described by Lehrbach and Ruvkun, who found that processing of the Nrf1 homolog in C. elegans requires Ddi1 [6], and by Murata and colleagues, who showed that its human homolog, Ddi2, is required for Nrf1 processing (personal communication). Ddi1/2 contain an aspartyl protease domain resembling that in retroviruses [8], but they also are UbL/UbA proteins that bind to both ubiquitinated proteins and proteasomes [9, 10]. We recently confirmed that HAP1 cells lacking Ddi2 cannot process Nrf1 (Fig S2). Therefore, Ddi2 is clearly the protease cleaving Nrf1, and most likely Nrf1 ubiquitination is required for its association with Ddi2.
Supplementary Material
Document S1. Two figures and experimental procedures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes two figures and experimental procedures and can be found with this article online at *bxs.
Author Contributions
Conceptualization, Z.S. and A.L.G.; Methodology, Z.S. and A.L.G.; Investigation, Z.S.; Writing–Original Draft, Z.S. and A.L.G.; Writing–Review & Editing, Z.S. and A.L.G.; Funding Acquisition, A.L.G.; Resources, A.L.G.; Supervision, A.L.G.
References
- 1.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Molecular cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Current biology: CB. 2014;24:1573–1583. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steffen J, Seeger M, Koch A, Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Molecular cell. 2010;40:147–158. doi: 10.1016/j.molcel.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. The Journal of biological chemistry. 2006;281:8582–8590. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 5.Vangala JR, Sotny F, Kruger E, Deshaies RJ, Radhakrishnan SK. Nrf1 can be processed and activated in a proteasome-independent manner. Current biology: CB. 2016 doi: 10.1016/j.cub.2016.08.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehrbach NJ, Ruvkun G. Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. eLIFE. 2016 doi: 10.7554/eLife.17721. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radhakrishnan SK, den Besten W, Deshaies RJ. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLIFE. 2014;3:e01856. doi: 10.7554/eLife.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sirkis R, Gerst JE, Fass D. Ddi1, a eukaryotic protein with the retroviral protease fold. Journal of molecular biology. 2006;364:376–387. doi: 10.1016/j.jmb.2006.08.086. [DOI] [PubMed] [Google Scholar]
- 9.Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. Journal of molecular biology. 2006;356:1027–1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Nowicka U, Zhang D, Walker O, Krutauz D, Castaneda CA, Chaturvedi A, Chen TY, Reis N, Glickman MH, Fushman D. DNA-damage-inducible 1 protein (Ddi1) contains an uncharacteristic ubiquitin-like domain that binds ubiquitin. Structure. 2015;23:542–557. doi: 10.1016/j.str.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Two figures and experimental procedures.


