Abstract
The human microbiome substantially affects many aspects of human physiology, including metabolism, drug interactions and numerous diseases. This realization, coupled with ever-improving nucleotide sequencing technology, has precipitated the collection of diverse data sets that profile the microbiome. In the past 2 years, studies have begun to include sufficient numbers of subjects to provide the power to associate these microbiome features with clinical states using advanced algorithms, increasing the use of microbiome studies both individually and collectively. Here we discuss tools and strategies for microbiome studies, from primer selection to bioinformatics analysis.
Human-associated microbial communities are implicated in a variety of diseases. Altered microbiota (the microorganisms that are present in a community) have been linked to vaginosis1, obesity2 and inflammatory bowel disease (IBD)3,4, among other maladies5. Although the importance of the human microbiota and the human microbiome has been hypothesized for some time, recent advances in the technology used to identify and to analyse components of the microbiome have substantially improved our knowledge of the microbial communities that are associated with various habitats, including humans.
The amount of variability in the microbiota and in the microbiomes both within a human subject and between different subjects is immense, and projects such as the Human Microbiome Project6,7 and MetaHIT8 have already done much to define this variability. Few microbial species are shared by most people at appreciable abundance levels9, at least among the well-sampled human skin, gut, oral and genital communities. Immense diversity is even observed between serial samples that have been taken from the same site in the same person9,10. Although human body sites and individuals do have distinct signatures, the differences in community composition across body sites are large, and within a body site, differences across individuals are, in general, much larger than differences between a single individual’s community across time11.
The importance of the human microbiome is thus immense, and this emerging field is rife with opportunities for discovery. Approaches to microbiome research are increasingly diverse, so here we present an outline of the many investigations that identify microorganisms (community surveys) and genes (shotgun metagenomics, referred to hereafter simply as ‘metagenomics’) that are present in the human-associated microbial communities and relate these communities to the host phenotype. Investigations into the RNAs, proteins and metabolites that are present — processes that are referred to as metatranscriptomics, metaproteomics and metabolomics, respectively — can also provide valuable insights, especially when they are combined with community surveys and metagenomic data12,13,14. In this Review, we primarily focus on DNA-based approaches, as they have been the primary means of interrogating the microbiome in recent investigations.
DNA-based microbiome studies
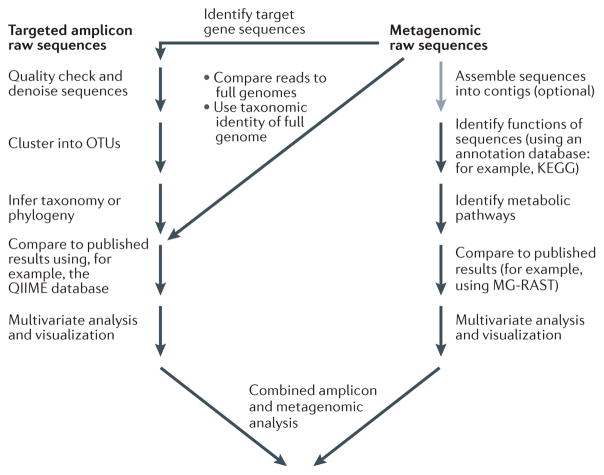
DNA-based microbiome studies frequently fall into one of two categories. Targeted amplicon studies focus on one or a few marker genes and use these markers to reveal the composition and diversity of the microbiota. Other studies use an entire metagenomic approach. This is sometimes referred to as shotgun metagenomics owing to the randomness with which genomic sequences are obtained. FIGURE 1 provides an overview of both study types and how they may be combined. Metagenomics approaches have the advantage of providing much richer data on the functional potential present in microbial communities. However, compared to targeted amplicon studies, they sacrifice resolution into the composition (that is, the identity of the microorganisms present) of those communities. Both approaches are useful, and we consider them both in this Review.
Figure 1. Bioinformatics analysis of microbiome sequence data.
Although variations exist, we show typical analysis paths for both targeted amplicon analysis (for example, analysis of 16S rDNA) and metagenomic analysis (for example, shotgun amplification). In targeted amplicon studies (left branch), raw sequences are usually passed through quality filtering and denoising algorithms to minimize the effects of sequencing artefacts. The resulting filtered sequence reads are clustered into operational taxonomic units (OTUs), representing similar organisms, and the phylogeny and taxonomic identity (when the organisms closely resemble named taxonomic groups) are inferred. At this stage, it is possible to incorporate sequence data from other relevant studies, or the data can be treated individually. The abundance of the various OTUs is then subjected to a variety of multivariate analyses and visualization procedures to elucidate the structure and patterns of the microbial communities. In metagenomic studies (right branch), the raw sequence fragments are sometimes assembled into contiguous sequences (contigs). The functional potential of those sequences is then typically assessed using a functional annotation database. The results are used to identify important metabolic pathways and are compared to the results of other metagenomic studies. The processed data are then subjected to multivariate analyses and visualizations, and they are often combined with the results of microbial profiling. Note that there are several opportunities for obtaining targeted gene data (similar to those produced in 16S rDNA gene surveys) in metagenomic studies, as indicated by the step labelled ‘identify target gene sequences’. KEGG, Kyoto Encyclopedia of Genes and Genomes; MG-RAST, Metagenomics Rapid Annotation using Subsystems Technology.
We begin by addressing how researchers can investigate various constituents of a microbial community, such as eukaryotes, viruses and various groups of bacteria. We then cover the processing of biological samples and DNA extraction, followed by DNA sequencing and the methodological options that are available. We continue by providing some discussion about the bioinformatics software used to analyse the sequencing data that emanate from both targeted amplicon and metagenomic sequencing studies, and we conclude with our outlook on the near future of this rapidly evolving field. The aim of this article is to provide a guide to the experimental designs and analytical tools used in microbiome studies and to discuss the important decisions that experimenters face when conducting investigations into the microbiome.
Selecting microbial genetic targets
Most microbial community studies include targeted amplicon sequencing of phylogenetically informative markers, such as the ribosomal small subunit (16S ribosomal DNA (rDNA)) gene. This allows researchers to compare the identities of the microorganisms that are present in the communities of interest. One advantage of rRNA genes is that ribosomes, and thus 16S rDNA, are present in all living organisms, whereas other commonly used markers have a limited taxonomic distribution. Furthermore, ribosomal genes contain both slowly evolving regions that can be used to design broad-spectrum PCR primers and fast-evolving regions that can be used to classify organisms at finer taxonomic levels (for example, at the family or genus levels), although species-level resolution might be unfeasible using this information alone. Another advantage that the 16S rDNA gene provides over other potential marker genes is the availability of several large databases of reference sequences and taxonomies, such as greengenes15, SILVA16 and the Ribosomal Database Project17. Although 16S rDNA is the most predominately used marker, the internal transcribed spacer region of the rRNA gene can be useful for some taxonomic groups, such as fungi, especially when resolution below the genus level is needed18.
Choice of universal PCR primer
There is a large choice of PCR primers (even for the widely used 16S rDNA), each of which has advantages and disadvantages. PCR primers should therefore be carefully chosen to take into account the taxonomic coverage desired, the extent of phylogenetic information generated by the fragment, the compatibility of the fragment length with the sequencing platform and the degree of specificity for amplifying microbial sequences compared to host sequences. For instance, the widely used 16S rDNA primer pair F27–R338 is highly specific for bacteria (as opposed to archaea and eukaryotes) but lacks sensitivity for taxa such as Bifidobacterium19, which is an important member of the gut microbiota. Furthermore, a complete bacterial phylum, Verrucomicrobia, is so poorly amplified by the F27–R338 primer set that this group was perceived to be a rare soil microorganism when it is, in fact, a dominant taxon that constitutes more than 20% of soil community members20.
In addition to minimizing amplification bias, an optimal primer set should amplify a region that is informative both taxonomically and phylogenetically, depending on the desired analyses. One way of assessing the taxonomic usefulness of the various hypervariable regions of the 16S rDNA gene is to compare the taxonomic assignments of short fragments (for example, 100–250 bp) in these regions against those of the full 16S rDNA gene. This type of analysis demonstrated that the 16S rDNA hypervariable region V6 poorly replicates full-length taxonomic assignments compared with the V4 or V2 region21. An illustrative cartoon on the effects of primer choice is shown in FIG. 2.
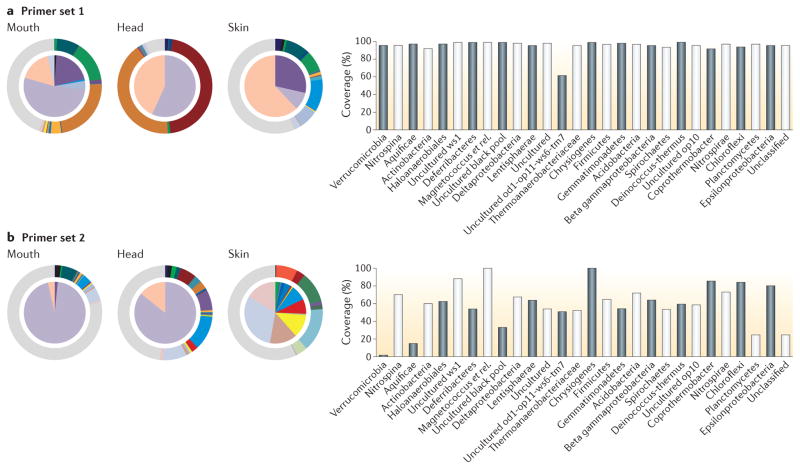
Figure 2. Effects of primer choice in targeted amplicon sequencing.
In choosing primers, there is often a trade-off between being broadly inclusive and avoiding biases for or against specific groups, which might occur owing to variation in sequence conservation between lineages. Therefore, changes that increase the representation of one group may cause another group to drop out. This figure shows predicted taxonomic coverage for two 16S rDNA primer pairs: carefully selected universal primers (a) and commonly used primers with high bias (b). This results in variations in the observed taxonomic composition of communities owing to primer sensitivity, as shown in the pie charts for the communities in the mouth, the head and the skin and in the histograms on the right, representing the primer pairs’ sensitivity to various microbial phyla (the effect continues to finer-level taxa, such as genera (not shown)). As no primer set is without some degree of bias, it is important that a priori knowledge of the target microbiota is used along with knowledge of a primer’s performance when choosing a primer set. However, primer set 1 (which amplifies the F515–R806 fragment) is an especially good choice in terms of avoiding bias and allowing for good representation of known bacterial and archaeal groups. It has been adopted by the Earth Microbiome Project, among other projects. Data taken from REF. 9.
In addition to the taxonomic coverage of a primer set and the usefulness of its amplicon, primer selection should also take into consideration what is known about the constituents of a target community. For instance, the universal primer set F515–R806 amplifies a broad range of bacterial and archaeal phyla and would be a prudent choice for a diverse community, such as a soil sample for the purpose of mapping the entire bacterial and archaeal community. However, this primer set poorly amplifies Propionibacterium, a common skin microorganism, and another primer set such as F27–R338 could be a better choice for this particular sample type22.
Amplifying eukaryotic and viral communities
High-throughput analyses of eukaryotic communities and parasites in host-associated environments are currently limited23,24 and have generally been targeted to specific taxonomic groups, such as fungi25,26. Eukaryotes generally make up a small proportion of the biomass and so fully incorporating this class of organism into microbiome studies will require addressing the challenges that are presented by the low abundance of eukaryotes relative to bacteria, as well as addressing those challenges that are presented by the difficulty of avoiding amplification of host DNA. These are probably the reasons why many current studies have focused on specific taxa. One possible solution to avoid amplifying host DNA is to use universal primers together with blocking probes that are specific to the host sequence27. As more community-wide data are generated, it will be interesting to see whether the presence or absence of specific eukaryotic taxa is more or less informative than studying the overall community composition.
Viruses also have a major role in shaping the human microbiome. A typical healthy human carries an abundance of viral particles (~1012 by some estimates)28, consisting mainly of bacteriophages but also of a substantial number of eukaryotic viruses29. Although an individual’s gut virome varies substantially over the first few weeks of life30, the virome of adults seems stable over time; this is remarkable in light of the strong variation between the viromes of different individuals31. This pattern may be similar to the rapid changes that are observed in the bacterial microbiota early in life32,33 followed by relative stability in adulthood9,11. The effects of these patterns in the human virome are mostly not understood, although certain bacteriophages in other animals are beneficial to the host34.
The lack of a gene that is universally present in all viruses means that amplicon-based studies cannot be used to characterize the virome in its totality. Instead, clade-specific viral genes can be used to identify and resolve viral subtypes from DNA sequences28,35. In order to characterize the virome comprehensively, researchers can isolate virus-like particles before metagenomic sequencing31. Elucidating the important effects of the human virome is confounded by the fact that most human virome sequences remain ‘unknown’ when compared to public databases, such as the nr database from the US National Center for Biotechnology Information (NCBI)36. The challenges of annotating the functional roles of unknown organisms are therefore particularly acute in virome studies.
Processing of biological samples and DNA
Sample collection
The rate-limiting step in obtaining samples of human-associated microorganisms is the approval of a human study protocol and the recruitment of subjects before the actual collection of the samples; the method of collection depends on the goals of the study and the willingness of the subjects to be sampled. Methods for collection are generally less controversial than other aspects of microbiome studies, but a few suggestions are presented in BOX 1 to make collecting samples (and, by extension, study design) more straightforward.
Box 1. Recommendations for sample collection.
The level of invasiveness of the sampling procedure should always be minimized when designing a study of human-associated microbiota. For instance, skin-associated communities can be sampled by swabbing106,107, scraping and performing a punch biopsy108, each of which represents a different level of discomfort for the subject. Fortunately, each method produces a similar picture of the microbial community108; as sufficient amounts of bacterial DNA can be recovered for 16S rDNA analysis by the less invasive swab method, punch biopsies should only be considered in circumstances in which deeper layers of the dermis must be studied. This logic extends to sampling other body sites. Mouth- and gut-associated microorganisms can easily be collected on swabs, which pose little risk of physical discomfort to the subject. However, when the study is focused on the lining of the gastrointestinal tract, there is little choice but to collect samples by endoscopy, as faecal samples are not sufficiently similar to the intestinal mucosa109. When designing a study, we suggest erring on the side of collecting as many samples as possible, as the increased statistical power of larger samples sizes cannot be overlooked when the difficulties of gaining approval and recruiting subjects represent a substantial barrier. Longitudinal studies, in particular, can provide much needed insights into the temporal variability of the microbial communities that are associated with specific body sites by using more samples from few individuals9,11,110.
DNA extraction
There has been some debate about which method results in the best picture of the microbial community37. However, most extraction methods use the same basic steps (cellular lysis, removal of non-DNA macromolecules and collection of the DNA) to separate the DNA from the cellular components and the environmental matrix. In general, the lysis step receives the most scrutiny, as the intensity of the lysis can result in bias towards a particular taxonomic group38. To avoid this, many studies use a combination of chemical, physical and mechanical means to lyse cells efficiently, and we suggest that all three means should be used to lyse cells in complex microbial assemblages. Incubating samples with detergents and/or enzymes followed by bead beating is a commonly used lysis method1,39,40, but there are also modified protocols for commercially available extraction kits that use a strong base, a high temperature and bead beating9,39,41. Non-DNA macromolecules are then physically separated from the DNA, and the DNA is then concentrated into a volume that is suitable for downstream applications. However, we should add that no extraction protocol works equally well on all sample types nor produces completely unbiased results: for example, researchers studying soil microorganisms have debated the ‘best’ method of DNA extraction for decades.
Minimizing contamination
Regardless of the extraction protocol used, steps must be taken to minimize contamination from a ‘foreign’ source. This will involve using sensible precautions (such as operating in as sterile an environment as possible and using clean equipment) and using commercially available kits or laboratory reagents that have been thoroughly tested to be free of contaminating DNA. Likewise, care should be taken to minimize contamination during sample collection. People collecting samples should wear gloves to prevent their own microbiota from contaminating samples, and they should adhere to an overall strategy of collecting samples in order of increasing biomass (for example, skin, then mouth and then faecal) to prevent the contamination of the lower biomass samples with those that are known to contain many microorganisms. After they have been collected, samples can be stored at various temperatures for short periods without there being a noticeable impact on the bacterial community42,43. However, the long-term effects of various storage conditions on human-associated microorganisms are not well described, so it is best to extract samples as soon as possible after collection or to use consistent storage conditions within and across studies. As the field of human microbiome research is still young, we have the chance to be early adopters of emerging standards in sample preparation and to use largely homogenous methods to allow more convenient analysis of results across studies.
DNA sequencing methodologies
Following successful extraction of DNA from the communities of interest, there remains the important step of obtaining sequences from that DNA. Microbiome studies that use 16S rDNA-based taxonomic profiling, as well as shotgun metagenomic studies, have used several sequencing technologies, including capillary (Sanger) sequencing (such as the Applied Biosystems 3730xl DNA analyser), pyrosequencing (such as the Roche 454 Genome Sequencer GS, FLX and FLX Titanium) and Illumina’s clonal arrays (such as the Illumina GAIIx and HiSeq2000). To our knowledge, no microbial community study has been published using other sequencing technologies. As shown in TABLE 1, each technology has distinct characteristics, and getting the most out of each technology requires striking the best balance between insert size, read length, depth, sequence accuracy, usability and cost.
Table 1.
Comparison of sequencing technologies
| Read length | Maximum insert size | Run time (hours (h) or days (d)) | Reads per run | Relative cost factor (per Mb) | Scale of reads per sample | Scale of samples per run | Raw error rate (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Insertions | Deletions | Mismatches | ||||||||
| ABI 3730 | 800 b | >1 Kb | 2 h | 96 | 100 | 102 | 101 | 0.001 | ≪0.1 | ≪0.1 | ≪0.1 |
| 454 FLX Titanium | 300–400 b | 800 b | 9 h | 106 | 1 | 103 | 102 | 1 | < 1 | ≪ 0.1 | ≪ 1 |
| 454 FLX+ | 500–600 b | 1200 b | 23 h | 106 | 0.7 | 103 | 102 | ||||
| Illumina GAIIx | 76–101 b | 500 b | 6–9 d | 4 × 108 | 0.1 | 105–106 | 103–104 | <1 | ≪1 | ≪1 | <1 |
| Illumina HiSeq 2000 | 101–151 b | 500 b | 9–15 d | 3 × 109 | 0.002 | 105–106 | 103–104 | ||||
| Illumina MiSeq | 36–151 b | 500 b | 4h–27 h | 107 | 0.06 | 104 | 102 | ||||
| PacBio | 1100 b | >1 Kb | 1.5 h | 3.5 × 107 | 1.5 | 103 | 101 | 15 | 13 | 1 | 1 |
| IonTorrent | 200 b | 400 b | 2–3 h | 1.5 × 106–3 × 106 | 0.4 | 103 | 102 | 2 | 1 | 1 | <1 |
The table shows a comparison of characteristics and average error rates and types of various sequencing technologies, both those that are commonly used in microbiome studies and promising but less-established technologies (MiSeq, PacBio, IonTorrent). The estimated relative cost factor presented here (relative to 454 Titanium sequencing) includes only sequencing costs and not the many other time and money costs that are involved in microbiome studies. Note that values are based on capabilities at the time of submission and that the next-generation sequencing technologies are likely to change quickly.
Taxonomic profiling studies
The first taxonomic profiling studies were based on the Sanger sequencing method, starting with the short 5S rRNA gene and then using increasingly long fragments of the 16S rDNA, ultimately leading up to near-full-length 16S rDNA sequences44. The study of increasing fragment lengths has been made easier by the recent use of high-throughput sequencing techniques (FIG. 3). These techniques allow more samples to be sequenced at a higher depth and lower cost, albeit at shorter read length (TABLE 1).
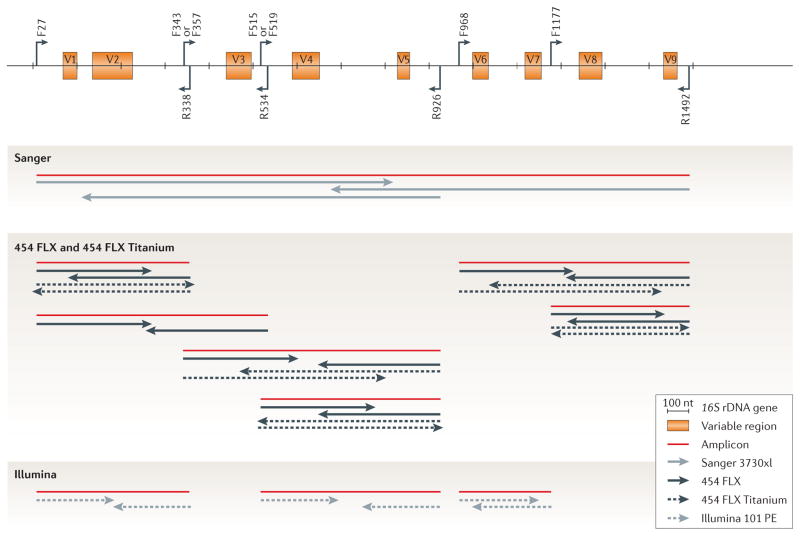
Figure 3. How to get the most taxonomic information out of each sequencing technology.
Taxonomic profiling consists of generating an amplicon (in red) of the (partial) 16S ribosomal RNA (rRNA) gene (top) with selected PCR primers, followed by sequencing that amplicon with a preferred technology (grey arrows): Sanger ABI 3730xl, 454 (FLX and FLX Titanium) and Illumina 101 paired-end (PE) sequencing technologies are compared in the figure. Arrows emanating from the schematic 16S gene represent common forward (F) and reverse (R) primers, and the orange boxes denote the hypervariable regions (V1–9), which are known to be far less conserved than the surrounding sequence. Technologies differ in the maximum allowable amplicon size and read length (TABLE 1) and therefore result in a different view of the community. To increase overall length and/or quality, Sanger- and Illumina-based strategies involve sequencing amplicons in both directions; in Sanger sequencing, there is also the option of using a third read. By contrast, a preferred 454-based strategy sequences the amplicons in a single direction owing to the lack of standard paired-end sequencing and loss of pairing information. Getting the most taxonomic information requires careful selection of primers, 16S rDNA windows and technologies in order to obtain the most data27,47. The long length of Sanger reads diminishes the need for careful selection of amplicon primer pairs, which have been shown to have a large role in taxonomic assignment and community comparison results with 454 data. A variety of options exist, and studies such as the one described in REF. 47 provide suggestions on which sets to choose. Short read effects are exacerbated further in Illumina sequencing, and choosing amplicon size such that overlapping paired-end reads occur is an important consideration.
In principle, longer sequences should yield higher resolution for applications such as classification45, defining novel taxa44 or identifying specific biomarker organisms based on a culture-independent approach46. However, changes in community composition can typically be assessed on the basis of a gene fragment that is as small as 100 bp47. The drop in the length of sequenced fragments resulted in the need to select a shorter region of the 16S rDNA as a proxy, although no consensus has been reached regarding the single best region of 16S rDNA to assay (as noted above, several regions of the 16S rDNA are almost equally effective).
Within 5 years, the read lengths obtained by 454 pyrosequencing kits evolved from 100 bases (using the GS platform) to >250 bases (using the FLX platform) and then to 500 bases (using the FLX Titanium platform). Now these kits allow sequencing of almost the entire length of amplicons, which, because of the current chemistry of this technology, were thought to be limited to <600 bases (TABLE 1). This approach can span multiple hypervariable and conserved regions, increasing the performance at different phylogenetic depths.
More recently, the promise of high depth at low cost introduced by the Illumina platform has resulted in the development of different strategies to compensate for the limitation posed by the shorter read lengths. Depending on the preferred region of 16S rDNA and amplification primers, fragments between ~100 and ~300 bases can be subjected to paired-end sequencing with read lengths of 76, 101 or 125 bases48–53. Some of these approaches result in overlapping reads, increasing the total fragment length and sequence quality, whereas others result in a gap in the middle of the sequence. However, all approaches potentially allow much larger-scale studies than are possible with the 454 pyrosequencing technology. The switch from Sanger sequencing to parallel pyrosequencing has resulted in a widespread increase to thousands of reads per sample and to hundreds of samples per run. An Illumina-based approach gains two to three orders of magnitude, achieving millions of reads per sample and thousands of samples per run (TABLE 1).
This continued evolution in the number of reads per run leads to an increasing need for higher levels of automation in sample preparation and for improved software tools to handle the resulting massive data sets. The decreased cost per sample and per nucleotide sequence has allowed immensely deep coverage of hundreds of samples simultaneously52. It has also introduced a plethora of new hurdles to overcome, from decreased taxonomic classification sensitivity owing to short read lengths and sequencing errors54 to increased time and financial costs of sample preparation, which can be mitigated through new approaches51.
Functional profiling studies
Sequencing needs are somewhat different for metagenomic functional profiling approaches compared to the typical 16S-rDNA-based taxonomic-profiling approaches described in the previous section. Initial studies involved sequencing clones of large-insert libraries (for example, bacterial artificial chromosomes) that were derived from genomic DNA extracted from a microbial community55. Despite the great use of contextual data generated by these long fragments, such directed sequencing has largely been replaced by cheaper, high-throughput shotgun sequencing, both for metagenomics and for single-organism genomics. Ideally, large contiguous pieces of DNA would be generated that contain operons as well as phylogenetic markers or even that contain complete microbial genomes of previously uncultivated species.
The first example of using a large-scale, random shotgun sequencing approach is the reconstruction of a simple environmental community making up an acid mine biofilm. This study involved 78 million bases of long Sanger reads and led to several near-complete genomes of the key organisms that are present56. However, metagenomics studies have also undergone substantial changes owing to the introduction of new sequencing technologies. Increasingly more complex communities have now been tackled, making it far more difficult to sample the full breadth of a population at a sufficient depth to result in large contiguous sequences, something that is possible in low-diversity environments, such as in acid mine drainage. Using next-generation sequencing to increase the amount of data up to billions of bases per sample (as was recently done in a large-scale microbiome study of the human gut8) still shows limitations in generating high-quality de novo assemblies: fewer than half of the short (75-base) reads could be assembled into contigs that were larger than 500 bases, and most of them were smaller than 2 kb. This depth issue is even more pronounced in those sample types that are not dominated by the microbial community and that contain large fractions of host DNA (for example, human mucosal sites can contain over 80% human DNA).
Although terabase metagenomic data sets will be available in the near future, the challenge of metagenomic assembly is unlikely to be solely addressed by increasing the depth of coverage, and improvements will depend on both the nature and the quality of the sequence data used as input and/or on the introduction of novel sequencing technologies. Approaches that are used and/or considered for metagenomic studies currently include: increasing read lengths by creating composite reads from short, overlapping paired-end sequences57; applying a hybrid approach that combines abundant short reads patched with longer reads (for a first-generation application, see REF. 58; future applications could leverage the longer reads from the PacBio RS platform); or obtaining long-range connectivity using jumping libraries59. All of these approaches are heavily dependent on advancements in computational algorithms.
Error rates
An important distinction between genome sequencing — which drives the development of many sequencing methods — and microbiome sequencing is that in genome sequencing, the error rates are less important. In genome sequencing, each region of the genome is sequenced many times; by contrast, in microbiome sequencing, each fragment (derived from either amplicon or metagenomic sequencing) may be sequenced only once, and this intrinsic variation, which is conflated with the sequencing error on each read, is itself of interest. The effect of raw sequencing error rates on the observed microbial diversity is potentially great, as every sequencing error could be portrayed as arriving from a novel organism. This effect is now typically evaluated on the basis of a synthetic, or ‘mock’, community that is created by pooling genomic DNA or cloned 16S rDNA fragments of multiple isolates60,61. Ideally, near-full-length 16S rDNA gene sequences are available at reference quality for the organisms in the mock community; this allows a description of the effect of raw sequencing error rates on the number of operational taxonomic units (OTUs) and a calculation of the actual error rates and error types62.
Research on reducing all types of errors — including both sequencing errors and PCR-based chimeric sequence formation63 — is an active area of study, especially with the regular introduction of novel technologies that require faster algorithms for downstream data analysis. In general, sequences that are suspected of including a high level of sequencing errors are discarded in an initial filtering step. Parameters such as the average quality score of the sequence, number and length of homopolymers, number of mismatches in the primers or total length of the sequence are tested to determine whether the sequence represents a true DNA fragment or a sequencing artefact. Chimeric sequences represent a different type of error: during PCR amplification, synthesis from a given template might be interrupted and then restarted using a different template that shares a certain degree of homology with the first template, resulting in a final DNA fragment that is formed from (at least) two different templates. Different tools exist to detect and remove chimeric sequences64–66. However, as these sequences are usually equally distributed across all samples that are subjected to the same protocol, chimaeras will primarily affect estimates of species richness (total number of OTUs in a given sample), not estimates of relative species diversity between samples.
Bioinformatics data analysis tools
After DNA sequences have been acquired, they must be analysed and interpreted. The large amount of data produced in modern investigations requires sophisticated analysis tools; direct manipulation of the data, such as manually aligning DNA sequences, is no longer feasible. There are many approaches for analysing such data. Here we distinguish between the analysis of targeted amplicon and metagenomic microbiome data, where the latter includes a sampling of the entire complement of the DNA that is present in the microbiome rather than relying on inference of gene content from the organisms present (FIG. 1).
Analysis tools for targeted amplicon data
For targeted amplicon sequencing (for example, sequencing of 16S rDNA profiles), tools such as QIIME67, mothur66 and VAMPS were developed to allow researchers to compare and analyse microbial communities using large amounts of DNA sequence data. As noted above, high-throughput sequencing technologies introduce errors, or ‘noise’, into sequence data. For 454 pyrosequencing technology, there are approaches to reduce these errors or to ‘denoise’ the sequencing results by clustering the flowgrams produced by the sequencer into a smaller number of DNA sequences that were probably present in the original biological samples. This can be done with tools such as AmpliconNoise64 and Denoiser69, which can be applied individually or used within the context of QIIME, or they can be applied using mothur-based reimplementations.
One important decision to make when performing analysis is whether to use the original, published version of a tool or whether to use a tool integrator’s implementation. In QIIME, our laboratories use the original implementations; this makes installation more difficult but guarantees the use of ‘brand-name’ tools. In mothur, the algorithms are rewritten from scratch, which makes the package easier to install and, in many cases, makes it run faster, but it does not guarantee that the same results will be produced. Other tools may use either of these two strategies or a mixture of them.
Assigning amplicon sequences to operational taxonomic units
DNA sequences, whether or not they have been denoised, are then typically clustered into OTUs. This process is a proxy for assigning DNA sequences to microbial species and is an important step for the analysis of diversity using a variety of species-based or OTU-based ecological metrics. Because most microbial species cannot yet be cultured in the laboratory69, diversity estimates are generally based on DNA sequence data that are grouped into these OTUs. OTUs can be defined as containing as much diversity as a given taxonomic level (for example, species or genus) by changing the sequence similarity threshold. However, the sequence similarity thresholds used are imprecise measures of an imprecise concept of a microbial ‘species’, and the sequence identity of, for example, the V2 region of the 16S rDNA does not exactly reflect the sequence identity of the entire gene70. An alternative to grouping sequence data using OTU thresholds to cluster the sequences without any external information (also known as de novo OTU picking) is to analyse only those sequences that can be formally assigned to known taxa or to analyse only those sequences that are highly similar to known taxa. This alternative approach is known as reference-based OTU clustering (or picking), and it frequently yields similar patterns to de novo approaches. In reference-based OTU picking, new DNA sequences are compared to one of a variety of reference databases of typically long, high-quality DNA sequences, such as those that are maintained by greengenes15 or by SILVA16. Reference-based approaches have several key advantages over the de novo approaches that have just been described, although they lack the latter’s suitability for exploring uncharted territory in microbiome space. They can be especially useful for combining sequence data from different regions of the 16S rDNA gene by mapping disparate regions onto a database of full-length reference sequences or for combining sequence data that are generated from different sequencing technologies. In such cases, de novo OTU picking would not be appropriate, as identical microorganisms might wrongly be assigned to different OTUs based solely on differences in sequencing technology or in the DNA region chosen for amplification.
The reference-based approach to OTU generation is increasingly valuable as the extent of publicly available data expands because it allows new investigations to be interpreted in the context of existing studies. Picking OTUs against a reference database can also reduce the impact of chimeric sequences and noisy data. For example, one study resequenced the microbiota of two individuals using Illumina sequencing and compared community composition over time to previous results obtained on the same individuals by pyrosequencing11. These approaches are becoming increasingly important, as databases such as VAMPS, MG-RAST71 and the QIIME database aggregate sequences that are produced by different platforms and technologies, although such comparisons can be highly effective, even using existing techniques11.
As the amount of sequence data for microbial communities grows exponentially, it is becoming increasingly clear that detailed information about samples (metadata) is crucial for enabling comparison of studies in databases and for allowing meta-analyses of the sequence data. The Genomic Standards Consortium aims to make standard the collection and reporting of detailed and standardized ‘metadata’ about the sequences. Such data would include clinical information about the subjects and technical information about how the DNA extraction, sequencing and other steps were performed. Towards this goal, the Genomic Standards Consortium has recently introduced standards, such as Minimum Information about Any (x) Sequence (MIxS)72, that allow these important factors to be defined. These standards have already been adopted by large projects, such as the Human Microbiome Project73 and the Earth Microbiome Project74.
Inferring phylogenetic relationships
The phylogenetic relationships among DNA sequences can also be inferred, either by using an existing reference database with an associated phylogeny (such as greengenes or SILVA) or by inferring the phylogeny de novo using tools such as NAST75,76 (for sequence alignment) and FastTree77 (for phylogeny inference from aligned sequences). Phylogenies allow the use of phylogenetically aware analysis, such as UniFrac78 and Phylogenetic Diversity79, which have been extensively used. Again, such tools for phylogeny inference can be used in isolation or within the context of QIIME, mothur or other pipelines. Even after sequences have been assigned to OTUs and possibly related to one another using a phylogenetic tree, the scale of data is still extensive. Various approaches exist to interpret such data to reveal meaningful patterns in microbial diversity. Typical approaches include applying metrics of community-wide similarity and using analyses such as principal coordinates analysis (PCoA) to visualize the relationships between microbial communities. The generation of rarefaction curves to display within-community diversity (α-diversity) is also common, as are various approaches to assess informative taxa across communities and across categories of communities. VAMPS, QIIME and mothur all perform such analyses and frequently produce publication-ready figures11,80.
Analytical tools for metagenomic data
In addition to taxonomic analyses using marker genes, microbial community studies are increasingly using metagenomic sequencing to assess community membership in diverse environments8,10,81–83. One additional benefit of metagenomic sequencing is that it yields information on the encoded functional potential in the community DNA, and this functional profile can help to generate hypotheses on community dynamics and metabolic properties.
To determine the phylogenetic membership of microbial communities based on metagenomic sequences, several freely available and popular software packages compare the metagenomic reads to a variety of full genome sequences using BLAST or interpolated Markov models. The identity of the best match then determines the likely phylogenetic origin of the sequence84,85. Another alternative is to find and extract informative phylogenetic markers from the metagenomic reads, which can be processed with similar methods to targeted gene surveys86,87. However, the taxonomic assignments from arbitrary metagenomic fragments remain a big challenge as much of the novelty in metagenomes still corresponds to organisms that lack a representative sequenced genome, and complementing metagenomic analyses with 16S rDNA analyses for which much larger reference databases exist are often useful. One advantage of metagenomic approaches is their ability to discriminate strains of common species by gene content beyond the resolution that is possible with 16S rDNA sequences8, although this approach requires high coverage and thus cannot be applied to rarer members of the community.
Functional annotation of metagenomic sequences
Community functional potential is almost always analysed by comparing the metagenomic sequences to large databases of metabolic annotations88–90. Depending on the sequencing strategy, coverage and community complexity, the sequences can either be used directly as short fragments, or they can be assembled into larger contigs for gene calling before annotation (for a review of metagenomic assembly, see REF. 91). Some software packages exist to tie together various components, and although no single standard exists yet, the Genomic Standards Consortium92 is currently working towards a consistent way for describing and comparing approaches. Depending on the need for customization and computational resources, annotation pipelines can be downloaded and run on a local computer cluster (for example, SmashCommunity93), or a user can upload their sequence data to a Web server and use a pre-structured pipeline, such as MG-RAST71, CAMERA94 or IMG/M95. One major benefit of online resources such as MG-RAST is the ability to compare publicly available metagenomic data sets, allowing users to perform comparative metagenomic analysis of their samples against a huge variety of environmental and host-associated metagenomic projects.
As with targeted gene surveys, one of the greatest challenges for understanding metagenomic data sets is the identification of significantly different features between communities. Just like targeted gene surveys, the large number of DNA sequences and functional groups requires paying careful attention to false discovery rates and good knowledge of suitable statistical tests — simple parametric statistics are rarely appropriate. Additionally, it can be difficult to understand the biological importance of a list of gene functions that are differentially represented between two communities or between several groups of communities. An increasing number of tools and strategies are being developed that allow the identification of substantial differences in functional groups in metagenomes and, importantly, they can aggregate these gene-level differences into metabolic pathways that are differentially represented46,96,97.
It should be noted that many DNA sequences from both metagenomic and targeted gene studies originate from poorly characterized genes and microorganisms, and our understanding of many of the microbial guests that are harboured in the human body is still shallow. There are thus ongoing efforts to sequence the complete genomes of a wide variety of microorganisms, in part to provide a framework from which to interpret DNA sequences from microbiome studies. These additional reference genomes that are provided by large sequencing efforts, such as the Human Microbiome Project, are allowing more insight into the nature of microorganisms that are identified in targeted gene surveys and are aiding in the functional annotation and assembly of metagenomic sequences. The availability of a larger number of reference genomes will also have a dramatic influence in our understanding of disease-associated strains and pathogenicity, as well as in providing a clearer picture of microbial evolution. Finally, the accumulation of more reference genomes will result in a faster pace at which we can reconstruct new genomes from short reads by using co-assembly methods98.
Conclusions and prospectus
Characterizing the taxonomic and functional characteristics of the human microbiome, despite considerable variation in methodology, is providing a much richer picture of our ‘normal’ microbial symbionts and, for several body sites, the association between microbial communities and human disease.
One theme that is common to both taxonomic and functional analyses of the human microbiome is that, in order to find associations between genes, organisms and physiological or disease states, it is more informative to use larger numbers of samples than it is to sequence each sample at greater coverage. Depending on the subtlety of the patterns to be discovered, surprisingly few sequences per sample may be needed to reveal them99. However, associations that involve rare species and rare genes will require far deeper sampling than is currently typical100. For example, we know that many pathogens can be introduced with inocula consisting of only a handful of cells: the discovery of rare, persistent pathogens (either at the level of species or at the level of strains) or of rare genes, such as virulence factors, might require several additional orders of magnitude of sequencing effort and/or finer resolution of sampling sites. These deeper investigations might involve directly sampling gut mucosa rather than relying on stools as a proxy or sampling specific and anatomically defined regions of skin. Relevant to this issue is the possibility that rare ‘keystone’ genes or species that are themselves undetectable might produce widespread ripples in common members of the microbiota that are more readily detectable. For example, sampling a cubic kilometre of Yellowstone National Park would be unlikely to reveal much wolf DNA, but we know that wolves are crucial in structuring this ecosystem because their role is reflected in the distribution of common plants, animals and even fish101. In this context, it is especially important to distinguish correlations between the presence or abundance of genes and species with physiological states and causation of those states by these species. Causality has been established in a few cases: stool transplantations from one human to another can cure persistent Clostridium difficile infections102, and transfer of microbial communities together with associated physiological states has been seen between different mice103 or even between humans and mice104. However, manipulating the microbiome of humans involves overcoming substantial bureaucratic obstacles, which, in our view, are excessive, given the prevalence of unverified and largely unregulated products that aim to manipulate the microbiome and are marketed directly to non-expert consumers. Thus, we can expect much of this work to take place in animal models for the foreseeable future. Whether prospective time series studies will reveal which associations between genes or microorganisms and physiological states are causal and which are side effects remains an important unresolved question.
In this Review, we have focused on 16S rDNA and metagenomic studies, which tell us about community membership and functional capacity. However, studies of gene expression at the RNA and protein level, of metabolites and of specific groups of lipids, carbohydrates and other markers will be increasingly important, as will studies of complete genomes of individual species in their natural environments. An especially exciting prospect is the ability to assemble personalized culture collections that describe the variability within an individual person105. Such investigations will allow more detailed manipulation of these communities, such as leave-one-out analyses and other perturbations, allowing us to untangle the effects of specific genes and organisms within complex communities. Manipulations of culture collections should help to determine which level of ‘multi-omics’ analysis is most useful for identifying biomarkers of human disease and also for generally increasing our understanding of the associations between microorganisms and human health.
Acknowledgments
This Review covers work in the Knight laboratory that is supported by the US National Institutes of Health (NIH), the Bill and Melinda Gates Foundation, the Crohn’s and Colitis Foundation and the Howard Hughes Medical Institutes. D.G. was supported by a grant from the NIH (NIHU54HG004969), the Crohn’s and Colitis Foundation of America and the Juvenile Diabetes Research Foundation.
Abbreviations
- Microbiota
The collection of microbial organisms from a defined environment, such as a human gut.
- Microbiome
The collection of genes that are harboured by microbiota.
- Metagenomics
The study of the collective genome of microorganisms from an environment. Shotgun metagenomics refers to the approach of shearing DNA that have been extracted from the environment and sequencing the small fragments.
- Amplicon
An amplified fragment of DNA from a region of a marker gene (such as 16S rDNA) that is generated by PCR.
- Bead beating
A process used to lyse cells and to disrupt larger structures before DNA extraction.
- Paired-end sequencing
An approach used in some sequencing platforms in which a single DNA clone is subjected to sequencing reads that originate from each of a set of primers, such that the direction of each sequencing reaction is directed to the origin of the other.
- Functional profiling approaches
Studies in which the genomic DNA of the microbiome is assessed for functional potential.
- Jumping libraries
Libraries that use molecular biology techniques to join together the ends of a larger DNA fragment, allowing sequencing on platforms that can only sequence a shorter fragment length. For example, 10 kb fragments might be reduced to 200 bases from each end, giving a final fragment size of 400 bp that can undergo paired-end sequencing.
- Operational taxonomic units (OTUs)
Sequences are generally collapsed into OTUs based on sequence similarity thresholds for downstream analyses. The typical threshold is 97%, and this is taken as a proxy for species level divergence, although what constitutes a microbial species remains an open debate.
- Chimeric sequence
An artificial sequence that juxtaposes gene regions from two or more unrelated organisms. It is produced by recombination between two or more DNA molecules during PCR amplification.
- Homopolymers
Sequences that contain repetitions of identical bases.
- Metadata
Information associated with sequences, including environmental conditions and the time and location of the sample collection site.
- Principal coordinates analysis (PCoA)
A multivariate technique used in microbiome studies to visualize the relationships among communities. Each community is represented by a point in typically two- or three- dimensional space, and similar communities are located close to one another in the resulting PCoA plot.
- Rarefaction curves
Plots of community diversity versus depth of sequencing (or, generally, observation). They are used to assess the amount of diversity and the extent to which it has been sampled at a given depth of sequencing.
- Interpolated Markov models
A bioinformatics technique used here to classify DNA sequences using patterns of k-mer nucleotide strings that are present in a within a genome database.
- Leave-one-out analyses
Studies of a microbial community that lacks one of its constituent microbial taxa.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aas J, Gessert CE, Bakken JS. Recurrent Clostridium difficile colitis: case series involving 18 patients treated with donor stool administered via a nasogastric tube. Clin Infect Dis. 2003;36:580–585. doi: 10.1086/367657. [DOI] [PubMed] [Google Scholar]
- 4.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 5.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome–host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson J, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser MJ. Harnessing the power of the human microbiome. Proc Natl Acad Sci USA. 2010;107:6125–6126. doi: 10.1073/pnas.1002112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. This is a large-scale study aimed at characterizing the functionality encoded in the gut microbiome. This work defined a minimal set of functions that are present in all of the sampled individuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. This paper was the first to establish that the microbial communities harboured across the human body are personalized but vary substantially across body sites and over time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. This is the densest time-series analysis of variation in the human microbiota that has been carried out so far. This study also proved the usefulness of newer DNA sequencers to provide deeper insights into the microbiota by recapturing previous results in variability across body sites and time using a different sequencing technology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 13.Maron PA, Ranjard L, Mougel C, Lemanceau P. Metaproteomics: a new approach for studying functional microbial ecology. Microb Ecol. 2007;53:486–493. doi: 10.1007/s00248-006-9196-8. [DOI] [PubMed] [Google Scholar]
- 14.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. The authors of this paper suggest a link between a person’s microbiome and their ability to metabolize a common drug, paracetamol (acetaminophen) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellemain E, et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi H, Sakamoto M, Benno Y. Evaluation of three different forward primers by terminal restriction fragment length polymorphism analysis for determination of fecal Bifidobacterium spp. in healthy subjects. Microbiol Immunol. 2004;48:1–6. doi: 10.1111/j.1348-0421.2004.tb03481.x. [DOI] [PubMed] [Google Scholar]
- 20.Bergmann GT, et al. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 2008;36:e120. doi: 10.1093/nar/gkn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walters WA, et al. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics. 2011;27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchesi JR. Prokaryotic and eukaryotic diversity of the human gut. Adv Appl Microbiol. 2010;72:43–62. doi: 10.1016/S0065-2164(10)72002-5. [DOI] [PubMed] [Google Scholar]
- 24.Parfrey LW, Walters WA, Knight R. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front Microbiol. 2011;2:153. doi: 10.3389/fmicb.2011.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ott SJ, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 26.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vestheim H, Jarman SN. Blocking primers to enhance PCR amplification of rare sequences in mixed samples — a case study on prey DNA in Antarctic krill stomachs. Front Zool. 2008;5:12. doi: 10.1186/1742-9994-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes M, Rohwer F. In: Metagenomics of the Human Body. Nelson KE, editor. Springer; New York: 2011. pp. 63–77. [Google Scholar]
- 29.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 30.Breitbart M, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. This paper describes a two-year longitudinal study of the development of the gut microbiota in an infant. This work provides a detailed analysis of the relationship between life events and changes in microbiome composition and function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso JG, Knight R, Kelley ST. Host-associated and free-living phage communities differ profoundly in phylogenetic composition. PLoS ONE. 2011;6:e16900. doi: 10.1371/journal.pone.0016900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willner D, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McOrist AL, Jackson M, Bird AR. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J Microbiol Methods. 2002;50:131–139. doi: 10.1016/s0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 38.Wang RF, Beggs ML, Erickson BD, Cerniglia CE. DNA microarray analysis of predominant human intestinal bacteria in fecal samples. Mol Cell probes. 2004;18:223–234. doi: 10.1016/j.mcp.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu GD, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206–206. doi: 10.1186/1471-2180-10-206. This study shows that long-term dietary patterns are associated with particular enterotypes. Bacteroides spp. were associated with a Western-like diet that is rich in proteins and animal fats, whereas Prevotella spp. were linked with high-carbohydrate diets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol Lett. 2010;307:80–86. doi: 10.1111/j.1574-6968.2010.01965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou HW, et al. BIPES, a cost-effective high-throughput method for assessing microbial diversity. ISME J. 2011;5:741–749. doi: 10.1038/ismej.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hummelen R, et al. Deep sequencing of the vaginal microbiota of women with HIV. PLoS ONE. 2010;5:e12078. doi: 10.1371/journal.pone.0012078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazarevic V, et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009;79:266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gloor GB, et al. Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS ONE. 2010;5:e15406. doi: 10.1371/journal.pone.0015406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartram AK, Lynch MD, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl Environ Microbiol. 2011;77:3846–3852. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Claesson MJ, et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilbert JA, Dupont CL. Microbial metagenomics: beyond the genome. Ann Rev Mar Sci. 2011;3:347–371. doi: 10.1146/annurev-marine-120709-142811. [DOI] [PubMed] [Google Scholar]
- 56.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 57.Rodrigue S, et al. Unlocking short read sequencing for metagenomics. PLoS ONE. 2010;5:e11840. doi: 10.1371/journal.pone.0011840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldberg SMD, et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc Natl Acad Sci USA. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- 62.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. doi: 10.1371/journal.pone.0027310. in the press. [DOI] [PMC free article] [PubMed]
- 63.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC bioinformatics. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. This paper introduces QIIME, an open-source software tool that performs the complete analysis of microbial communities. Among other functions, QIIME implements quality filtering of the input raw reads, OTU picking, α- and β-diversity estimates and prediction of OTUs that are significantly associated with categories in the data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nature Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 70.Schloss PD. The effects of alignment quality, distance calculation method, sequence filtering, and region on the analysis of 16S rRNA gene-based studies. PLoS Comput Biol. 2010;6:e1000844. doi: 10.1371/journal.pcbi.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meyer F, et al. The metagenomics RAST server — a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yilmaz P, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nature Biotech. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilbert JA, et al. The Earth Microbiome Project: meeting report of the “1 EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6 2010. Stand Genomic Sci. 2010;3:249–253. doi: 10.4056/aigs.1443528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DeSantis TZ, Jr, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. This study introduces UniFrac, a phylogenetically aware measure of similarity, and one of the most widely used methods to establish the extent to which different microbial communities resemble each other. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evolutionary Bioinform Online. 2006;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- 80.Morowitz MJ, et al. Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci USA. 2011;108:1128–1133. doi: 10.1073/pnas.1010992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 82.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. In this study, faecal microbiomes were found to cluster into three distinct groups (‘enterotypes’) with minimal overlap. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brady A, Salzberg S. PhymmBL expanded: confidence scores, custom databases, parallelization and more. Nature Methods. 2011;8:367. doi: 10.1038/nmeth0511-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mitra S, et al. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinformatics. 2011;12:S21. doi: 10.1186/1471-2105-12-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharpton TJ, et al. PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data. PLoS Comput Biol. 2011;7:e1001061. doi: 10.1371/journal.pcbi.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.von Mering C, et al. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- 88.Muller J, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Glass E, et al. Meeting report from the Genomic Standards Consortium (GSC) Workshop 10. Stand Genom Sci. 2010;3:225–231. doi: 10.4056/sigs.1423520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arumugam M, Harrington ED, Foerstner KU, Raes J, Bork P. SmashCommunity: a metagenomic annotation and analysis tool. Bioinformatics. 2010;26:2977–2978. doi: 10.1093/bioinformatics/btq536. [DOI] [PubMed] [Google Scholar]
- 94.Sun S, et al. Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 2011;39:D546–D551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Markowitz VM, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kristiansson E, Hugenholtz P, Dalevi D. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737–2738. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- 97.Liu B, Pop M. MetaPath: identifying differentially abundant metabolic pathways in metagenomic datasets. BMC Proc. 2011;5:S9. doi: 10.1186/1753-6561-5-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen K, Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput Biol. 2005;1:106–112. doi: 10.1371/journal.pcbi.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuczynski J, et al. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nature Methods. 2010;7:813–819. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. ISME J. 2008;2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- 101.Mcpeek MA, Mcpeek MA. The consequences of changing the top predator in a food web: a comparative experimental approach. Ecol Monogr. 1998;68:1–23. [Google Scholar]
- 102.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 103.West TE, et al. Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes Immun. 2011;2011:1–9. doi: 10.1038/gene.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. This paper showed that a substantial proportion of an individual’s gut microbiota can be recaptured using anaerobic culturing conditions, both in vitro and in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006;44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zoetendal EG, et al. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brotman RM, Ravel J, Cone RA, Zenilman JM. Rapid fluctuation of the vaginal microbiota measured by Gram stain analysis. Sex Transm Infect. 2010;86:297–302. doi: 10.1136/sti.2009.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]