Summary
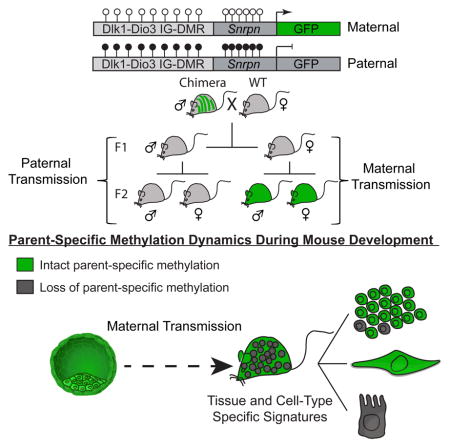
Parent-specific differentially methylated regions (DMRs) are established during gametogenesis and regulate parent-specific expression of imprinted genes. Monoallelic expression of imprinted genes is essential for development suggesting that imprints are faithfully maintained in embryo and adult. To test this hypothesis, we targeted a reporter for genomic methylation to the imprinted Dlk1-Dio3 intergenic DMR (IG-DMR) to assess the methylation of both parental alleles at single cell resolution. Biallelic gain or loss of IGDMR methylation occurred in a small fraction of mouse embryonic stem cells significantly affecting developmental potency. Mice carrying the reporter in either parental allele showed striking parent-specific changes in IG-DMR methylation causing substantial and consistent tissue- and cell type-dependent signatures in embryos and postnatal animals. Furthermore, dynamics in DNA methylation persisted during adult neurogenesis resulting in inter-individual diversity. This substantial cell-cell DNA methylation heterogeneity implies that dynamic DNA methylation variations in the adult may be of functional importance.
Graphical Abstract
Introduction
Parental imprinting is a heritable epigenetic mechanism resulting in parent-specific monoallelic expression of subset of genes (Ferguson-Smith, 2011; Reik and Walter, 2001) and such imprinting is essential during early mammalian development (McGrath and Solter, 1984; Surani and Barton, 1983). While methylation imprints established during gametogenesis are thought to be stable in development, complex tissue-specific expression of imprinted genes can occur in the developing embryo (Barton et al., 1991; Thomson and Solter, 1988), with possible functional consequences in the animal (Davies et al., 2005; Frost and Moore, 2010; Wilkinson et al., 2007). Due to their monoallelic nature, imprinted genes are specifically susceptible to alterations that may be caused by loss-of-function mutations or by epimutations in regulatory elements. Indeed, Loss-of-imprinting (LOI) correlates with mild to severe developmental abnormalities, organ malfunctions, behavior anomalies and cancer (Avior et al., 2016; Peters, 2014; Robertson, 2005; Yamazawa et al., 2010).
DNA methylation is central for the regulation of parental imprinting as gamete-specific differentially methylated regions (DMRs) act in cis to regulate the monoallelic parent-of-origin expression of multiple imprinted genes (Barlow and Bartolomei, 2014). Following fertilization, imprinted DMRs are protected from global de-methylation and de novo methylation in somatic cells with the exception of primordial germ cells, where all methylation imprints are removed and re-established in a sex-dependent manner during gametogenesis (Lee et al., 2014; Reik, 2007). Recent advances in sequencing technologies facilitated single-base resolution DNA methylation maps of multiple embryonic and adult tissues (Hon et al., 2013; Roadmap Epigenomics et al., 2015; Ziller et al., 2013), enabling insights into the stability of imprinted DMRs in adult tissues and the identification of novel imprinted DMRs in both humans (Court et al., 2014; Stelzer et al., 2013) and mice (Xie et al., 2012). It is believed that following fertilization, imprinted DMRs are mostly maintained by the activity of Dnmt1 (Li et al., 1993; Tucker et al., 1996) and that loss of parent-specific methylation is stochastic and may contribute to disease (Ferguson-Smith, 2011; Reik, 2007; Reik and Walter, 2001; Robertson, 2005). Nevertheless, because of the “snapshot” nature of sequencing data, present understanding of imprint maintenance during embryonic development and in adult tissues is limited and precludes the assessment of tissues and cell-type heterogeneity at single cell resolution.
The imprinted Dlk1-Dio3 locus on mouse chromosome 12 is characterized by the reciprocal expression of maternal non-coding transcripts and paternal protein coding genes regulated by both cis (Lin et al., 2003) and trans (Cockett et al., 1996; Seitz et al., 2003) acting mechanisms. The intergenic DMR (IG-DMR) serves as an imprinted control center regulating parent-specific expression of genes in this locus (da Rocha et al., 2008; Lin et al., 2003). Mice with uniparental disomy and genetic manipulations of the locus have substantiated that proper imprinting is essential for normal development, with LOI resulting in early embryonic lethality (Georgiades et al., 2000; Lin et al., 2007; Lin et al., 2003; Tevendale et al., 2006). Targeted deletions of individual genes in Dlk1-Dio3 locus lead to complex abnormalities in the embryo and postnatal animal and include cartilage, bone, muscle and placenta defects (Andersen et al., 2013; Sekita et al., 2008; Takahashi et al., 2009), obesity (Moon et al., 2002), metabolic and behavioral dysfunctions (Labialle et al., 2014; Qian et al., 2016; Sittig and Redei, 2014).
We have recently established a Reporter of Genomic Methylation (RGM) that relies on an imprinted gene promoter (Snrpn) driving a fluorescent protein (Stelzer and Jaenisch, 2015; Stelzer et al., 2015). Here, we utilized RGM to facilitate a comprehensive study of the dynamics of imprinted DMRs in embryos and adult mice. RGM was targeted in mouse embryonic stem (mES) cells to each allele of the Dlk1-Dio3 IG-DMR. Aberrant methylation at the IG-DMR strongly affected developmental potency in chimera assays. Furthermore, we identify sex-dependent differences in the degree and kinetics of paternal allele demethylation, with blastocyst-derive female mESCs displaying rapid demethylation during early passages. Mice carrying the reporter in either allele were used to assess the maintenance of imprints in embryos and adult mice. Surprisingly, methylation changes at the Dlk1-Dio3 DMR were found to be dynamic in most tissues of the embryo and the postnatal animal. In particularly, methylation imprints varied at the single cell level during adult neurogenesis resulting in inter-individual diversity and epigenetic variability.
Results
Allele-specific targeting of Dlk1-Dio3 IG-DMR
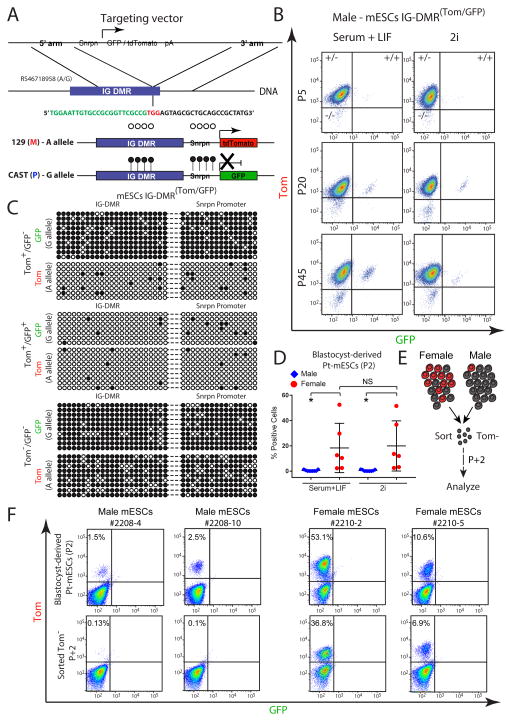
We utilized CRISPR/Cas9 mediated gene editing in F1 hybrid 129XCastaneous (CAST) male mES cells to target Snrpn-GFP or Snrpn-tdTomato (Tom) to each allele of the Dlk1-Dio3 IG-DMR (Figure 1A). The IG-DMR acquires paternal methylation during spermatogenesis while the maternal allele is hypomethylated in the oocyte (da Rocha et al., 2008). Consistent with this notion, cells targeted with Snrpn-Tom to the maternal allele and Snrpn-GFP to the paternal allele (IG-DMRTom/GFP) expressed the Tom reporter but not the GFP reporter (Figures 1B and S1A). Bisulfite sequencing of targeted cell lines demonstrated that while the maternally-targeted Tom allele was hypomethylated in the IG-DMR and downstream Snrpn promoter regions, the paternally-targeted GFP reporter allele exhibited high levels of DNA methylation which spread from the IG-DMR region into the Snrpn promoter (Figure 1C), resulting in its repression. During expansion of targeted cell lines, a small fraction of cells emerged that were either double positive or double negative for reporter expression (Figure 1B). These sub-populations slightly increased during consecutive passages suggesting that they do not confer significant growth advantage (Figures 1B and S1B). Bisulfite sequencing, of sorted double-positive and double negative cells indicated hypomethylation or hypermethylation, respectively, of both parental alleles as well as the Snrpn promoters (Figures 1C). Notably, during prolonged culturing of the sorted cell populations a new population of cells emerged that had switched the allelic reporter activity repressing the maternal Tom allele and activating the paternal GFP (Figure S1C). Thus, all these data demonstrated that reporter activity faithfully reflects parent-specific gain or loss of DNA methylation at the Dlk1-Dio3 IG-DMR and that insertion of RGM does not affect the methylation levels of adjacent sequences. Recent studies have shown that culturing mESCs with inhibitors of MEK and GSK3 (2i) results in global hypomethylation (Ficz et al., 2013; Habibi et al., 2013). When cultured in standard 2i culture conditions, we observed no significant increase of double-positive cells compared with culturing in serum and LIF (Figures 1B and S1B).
Figure 1. Allele-specific targeting of the Dlk1-Dio3 IG-DMR.
(A) Schematic representation of CRISPR/Cas-mediated allele-specific targeting of Snrpn-GFP or Snrpn-Tom, adjacent to the IG-DMR region; green sequence - endogenous IG-DMR region; black sequence - targeting CRISPR; red sequence - PAM recognition site.
(B) Flow cytometric analysis of GFP/Tom reporter ES cells at different passages, cultured in serum+LIF or 2i.
(C) Allele-specific bisulfite sequencing was performed on sorted mES IG-DMR Tom+/GFP−, IG-DMR Tom+/GFP+ and Tom−/GFP− cells. Each row represents a distinct PCR amplicon (marked with dashed line) that includes the endogenous IG-DMR (left) and the downstream integrated Snrpn promoter region (right); open circles – unmethylated CpGs; black circles – methylated CpGs.
(D) Dot plot showing the percentage of GFP/Tomato positive cells in passage 2 (P2) male and female mESCs cultured in Serum+LIF or 2i, as measured by flow cytometry. Black lines indicate mean ± SD for each group. Statistical differences between genotypes were calculated using one-way ANOVA; * P<0.05; NS – not significant; Pt - paternally transmitted.
(E) Schematic diagram for sorting and analyzing paternally transmitted (Pt) Tomato negative (Tom−) male or female mESCs presented in (F).
(F) Flow cytometric analysis of the proportion of Tom-positive cells in passage 2 (P2) male and female mESCs (upper panel) and in sorted Tom− cells following two consecutive passages (lower panel). Pt - paternally transmitted.
To investigate whether the in vitro loss of parent-specific methylation also occurs in newly-derived mESCs, we isolated the inner cell mass (ICMs) from blastocysts carrying the paternally transmitted (Pt) GFP or Tomato reporter (see Figure S1D and Experimental procedures). As documented for targeted male cell lines (Figures 1B and S1B), newly isolated male mESCs exhibited rare and stable population of cells with aberrant paternal reporter activity (Figures 1D). Nevertheless, and in strike contrast, female mESCs were significantly more likely to activate paternal reporter activity with some cell lines exhibiting more than 50% GFP or Tom positive cells (Figure 1D). To test whether the observed variation reflects intrinsic sex-specific differences, male and female mESCs harboring the Tomato reporter in the paternal allele of the IG-DMR were sorted for Tomato negative cells and analyzed in subsequent passages (Figure 1E). Only two passages following sorting, female but not male cells showed robust reactivation of the reporter (Figure 1F). Culturing female cell lines in 2i showed no significant increase of positive cells compared with culturing in serum and LIF (Figures 1D), suggesting that X chromosome number (2 X in female vs. 1 X in male cells) but not culture conditions, play a role in the rapid demethylation of the IG-DMR as was observed previously (Zvetkova et al., 2005). In summary, our data suggest that female mouse ESCs with two X chromosomes exhibit rapid demethylation of the paternal allele of the IG-DMR as revealed by reporter activity.
Allele specific methylation, gene expression and reporter activity
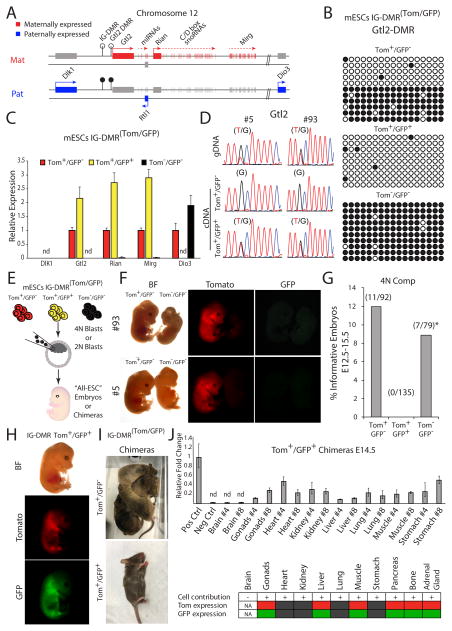
The Dlk1-Dio3 imprinted locus comprises multiple maternally expressed non-coding genes with unknown functions, including the lincRNA Gtl2 and large clusters of C/D box snoRNAs and miRNAs. Additionally, three protein coding genes being expressed exclusively from the paternal allele (Figure 2A). These include the atypical Notch ligand delta-like homologue 1 (Dlk1), a retrotransposon-like Rtl1 and the type 3 iodothyronine deiodinase (Dio3) (da Rocha et al., 2008). The IG-DMR serves as a cis acting regulatory center that establishes post-zygotic “secondary” DMRs such as in the promoter of Gtl2 (Figure 2A). We tested the methylation levels associated with the Gtl2 promoter DMR in the three IG-DMRTom/GFP cell populations. Figure 2B shows that the methylation state of the IG-DMR corresponded to that of the downstream Gtl2 promoter DMR, suggesting that the mechanism that mediates the establishment of Gtl2 DMR is functional in mES cells. Quantitative PCR on representative genes in the locus demonstrated that IG-DMR methylation strictly correlated with the expression patterns of maternal and paternal genes (Figure 2C). Thus, hypomethylation of the paternal allele (Tom+/GFP+) resulted in a near 2-fold increase in maternal gene expression, and loss-of expression of the paternal gene Dio3. Conversely, hypermethylation of the maternal alleles (Tom−/GFP−) resulted in complete repression of all maternal genes and a 2-fold increase in the expression of Dio3 as compared to cells with intact parent-of-origin methylation levels (Tom+/GFP−). Furthermore, utilizing a heterozygous SNP in the Gtl2 coding region demonstrated maternal monoallelic expression of Gtl2 in control IG-DMR Tom+/GFP− cells, while hypomethylated (Tom+/GFP+) cells exhibited biallelic Gtl2 expression (Figure 2D), consistent with 2-fold increase in expression (Figure 2C). We conclude that IG-DMR methylation reporter activity strictly correlates with parent-specific gene expression of multiple genes in the Dlk1-Dio3 region. Expression of other imprinted genes such as H19, PEG3, Snrpn was not altered in cells with aberrant IG-DMR methylation (Figure S1E).
Figure 2. Functional consequences of parent-specific loss of methylation in the ID-DMR region.
(A) Organization of imprinted genes in the mouse Dlk1-Dio3 locus; open lollipops – unmethylated region; black lollipops – methylated region.
(B) Bisulfite sequencing was performed on Gtl2 promoter DMR in distinct mESCs IG-DMR(Tom/GFP) sorted cell populations.
(C) Quantitative real-time PCR (qRT-PCR) of the mean relative fold change ± s.d of representative genes in the Dlk1-Dio3 region in three mES IG-DMR(Tom/GFP) sorted cells from 2 independently targeted cell lines. Data were normalized to Gapdh housekeeping control; nd – not detected.
(D) Sequencing of Gtl2 in two independent mESC IG-DMR(Tom/GFP) lines. Heterozygous SNP was identified in the genomic DNA (gDNA); monoallelic vs. biallelic expression was evaluated in the complimentary DNA (cDNA).
(E) Schematic representation of blastocyst injection strategy.
(F) Representative images of E13.5 4n complementation embryos, obtained from two independent mESC IG-DMR Tom−/GFP− and control mESC IG-DMR Tom+/GFP− lines.
(G) Summary of 4n embryo injections; * all embryos analyzed exhibited muscle and brain phenotypes.
(H) Representative images of mES IG-DMR Tom+/GFP+ cell contribution to E14.5 chimeric embryos and (I) postnatal mice.
(J) Upper panel - qRT-PCR detection of mES IG-DMR Tom+/GFP+ cell contribution to different organs in E14.5 chimeric embryos. Samples were normalized to ultra-conserved noncoding element in the mouse genome. Shown are mean relative fold change ± s.d of GFP detection in two embryos (#4, #8) compared with GFP positive (Pos Ctrl) and WT cells (Neg Ctrl); nd – not detected. Lower panel – summary of Tom and GFP expression in different organs of chimeric embryos.
Dlk1-Dio3 loss-of-imprinting affects developmental potency of ES cells
To assess whether Dlk1-Dio3 LOI would affect the developmental potential of ES cells we utilized tetraploid complementation (Tam and Rossant, 2003), the most stringent assay for developmental potency (Figure 2E). Embryos were analyzed at embryonic days 12.5 to 15.5 (E12.5–15.5). Figure 2F shows that while control IG-DMR Tom+/GFP− cells generated normal embryos with comparable frequencies to previous reports (Buganim et al., 2014), aberrantly hypermethylated IG-DMR Tom−/GFP− embryos exhibited growth defects at midgestation that included severe brain malformations and muscle defects whereas 4-n embryos from biallelically hypomethylated IG-DMR Tom+/GFP+ ES cells died prior to gastrulation (Figure 2G). Biallelically hypomethylated (Tom+/GFP+) cells, when injected into 2n host blastocysts, contributed to chimeric embryos and postnatal animals though with lower efficiency than control IG-DMR Tom+/GFP− ES cells (Figures 2H and 2I). Quantitative PCR detected the presence of donor cells in all tissues of chimeric embryos, except in brain (Figure 2J). Notably, some tissues maintained the expression of both fluorescent markers, while other tissues, with evident contribution of donor cells, (e.g. kidney, heart and lung) appeared double negative Tom−/GFP− indicating biallelic hypermethylation of the IG-DMR (Figures 2J and S2). Our results demonstrate that incorporation of both IG-DMR Tom−/GFP− and Tom+/GFP+ cells into chimeric embryos results in developmental defects, while IG-DMR Tom+/GFP+ cells display a more severe phenotype with lack of contribution to the brain in chimeric embryos.
Parent-specific imprints are maintained faithfully in some but not other tissues
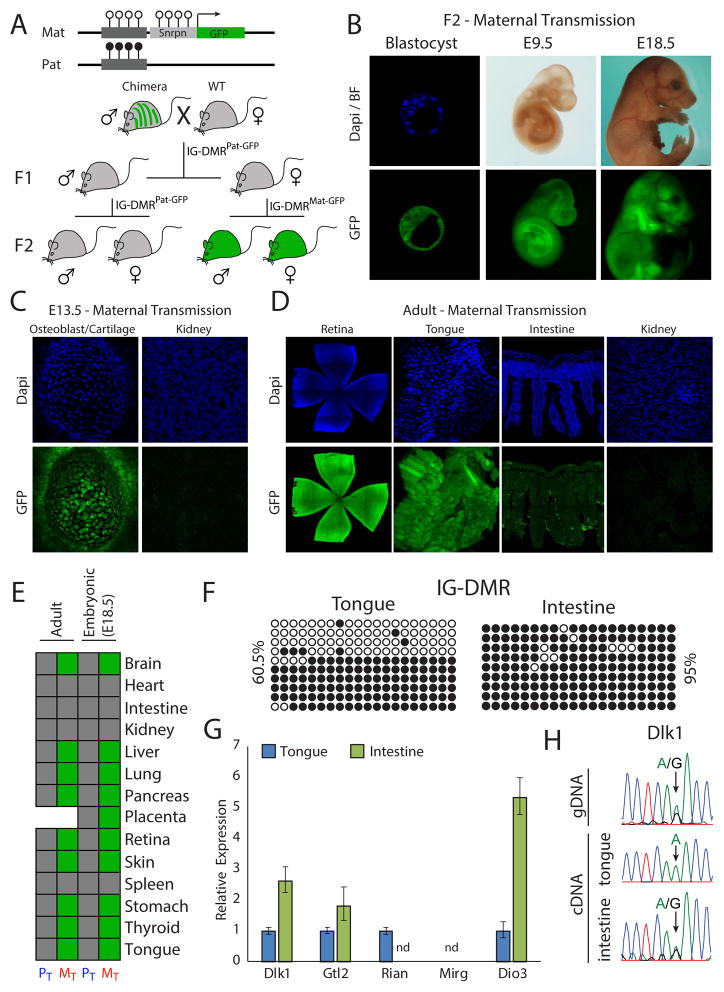
In order to study parent-specific methylation dynamics in vivo, cells with a maternal Snrpn-GFP reporter were injected into blastocysts to generate chimeras, which were bred to obtain transgenic males (Figure 3A). Mice carrying the reporter allele were born at the expected Mendelian ratio, implying that the reporter had no adverse effect. Since the IG-DMR is methylated during spermatogenesis, first generation (F1) males and females carrying the reporter allele were expected to be GFP negative. When maternally and paternally transmitted F2 embryos were analyzed (IG-DMRMat-GFP and IG-DMRPat-GFP, respectively), IG-DMRMat-GFP blastocysts were positive while IG-DMRPat-GFP blastocysts negative for GFP expression (Figures 3A, 3B, S3A and S3B), indicating proper parent-specific reporter expression. All IG-DMRMat-GFP embryos expressed the GFP reporter throughout development (Figures 3B, S3C and S3D) but close examination revealed differential GFP activity between some tissues (Figure 3C). Furthermore, tissues that repressed the GFP reporter in the developing embryo such as kidney heart and intestine persistently silenced the IG-DMRMat-GFP allele in adult animals (Figures 3D and 3E).
Figure 3. Generation of IG-DMRGFP reporter mice reveals tissue-specific reporter activity.
(A) Mating scheme for generation of parent-specific IG-DMRGFP reporter mice.
(B) Representative images of F2 IG-DMRMat-GFP embryos at different developmental stages. Dapi and anti-GFP staining; (C) embryonic and (D) adult tissues obtained from 5–7 week old mice.
(E) Summary of tissue-specific reporter activity in Maternal Transmitted (MT) and Paternal Transmitted (PT) E18.5 embryos (n=13) and 5–7 weeks old adult mice (n=6). Stitched pictures are shown for both retina and intestine.
(F) Bisulfite sequencing of the IG-DMR region in WT-tongue and intestine. Shown are percentages of methylated CpGs.
(G) qRT-PCR of the mean relative fold change ± s.d of representative genes in the Dlk1-Dio3 region in tongue and intestine from two WT-mice. Expression was normalized to Gapdh; nd – not detected.
(H) Sequencing analysis of heterozygous SNP identified in the Dlk1 coding region was performed on cDNAs obtained from WT-tongue and intestine tissues.
We tested whether reporter activity faithfully reflected the methylation patterns in Wild-Type (WT) untargeted mice. The IG-DMR region was hypermethylated in intestine consistent with maternal reporter silencing, whereas the region was hemi-methylated in tongue consistent with maternal reporter expression (Figures 3E and 3F). Gene expression analysis demonstrated complete downregulation of the maternally expressed gene Rian in intestine, consistent with hypermethylation of the IG-DMR region. Another maternal gene Gtl2 was found to be expressed in both intestine and tongue, corroborating independent regulation by its secondary promoter DMR (Figures 3G and 2A). Paternally expressed genes (Dlk1 and Dio3) exhibited elevated expression levels in intestine as compared with tongue (Figure 3G), suggesting tissues specific differences in regulation of gene expression. Consistent with methylation signatures, proper monoallelic expression in tongue and biallelic expression in intestine was identified using an informative SNP in the Dlk1 coding region (Figure 3H).
Cell type dependent imprinting
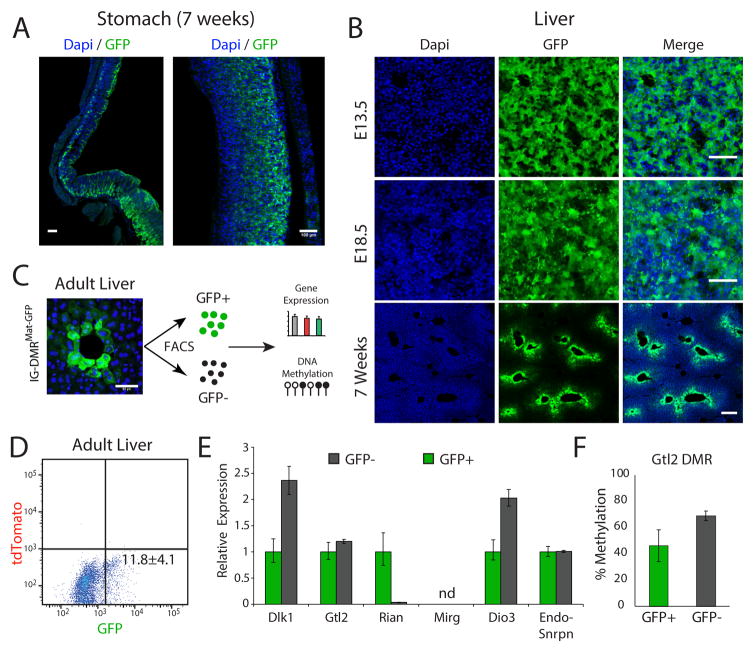
In addition to tissue-specific imprinting, adult tissues revealed cell-type dependent reporter expression in IG-DMRMat-GFP animals. Figure 4A shows selective expression of GFP in some but not other cells in the stomach, as well as regional heterogeneity. In the liver, reporter activity was gradually decreased with age and was restricted to epithelial cells surrounding the liver bile duct that retained proper GFP expression (Figure 4B). To further analyze the differences between the two cell populations, we used Fluorescence-activated Cell Sorting (FACS) to isolate GFP+ cells from adult mouse livers. Figures 4C–E shows a small fraction of GFP+ cells with a similar expression level of endogenous Snrpn, and Gtl2 in GFP+ cells compared with GFP− cells that displayed a twofold increase of paternally expressed genes Dlk1 and Dio3 and silencing of the maternally expressed gene Rian (Figure 4E). We performed bisulfite sequencing of the Gtl2-associated promoter DMR in the two cell populations and identified intermediate methylation levels (Figure 4F). Thus, our results support regulation of Gtl2 by its promoter DMR independent of the IG-DMR methylation, a finding consistent with its methylation state in ES cells (Figure 2B). While it was previously speculated that Rian and Mirg may be further processed from a large non-coding transcript originating from Gtl2 promoter (Royo and Cavaille, 2008), our data suggest that these transcripts are independently regulated.
Figure 4. Heterogeneous reporter activity in adult IG-DMRGFP/Mat tissues.
(A) Whole-mount stitching (left) and region specific (right) images of Dapi and anti-GFP staining in adult stomach sections; bar = 100μm.
(B) Representative images of Dapi and anti-GFP staining in liver sections of embryos and adults; bar=50μm.
(C) Single-cell suspension was established from 5 weeks old IG-DMRMat-GFP liver tissues, following cell sorting and DNA/RNA extraction; bar = 50μm.
(D) Flow cytometric of GFP positive cells, mean ± s.d of two independent livers.
(E) qRT-PCR analysis of representative genes in the Dlk1-Dio3 region in sorted IG-DMRMat-GFP liver cells. Shown is mean relative fold change ± s.d of two biological replicates; nd – not detected.
(F) Bisulfite sequencing of Gtl2 promoter DMR in sorted GFP positive and GFP negative IG-DMRMat-GFP liver cells. Shown are mean methylation levels ± s.d of two biological replicates. For each sample, more than 10 amplicons were sequenced to calculate the percentage of methylated CpGs.
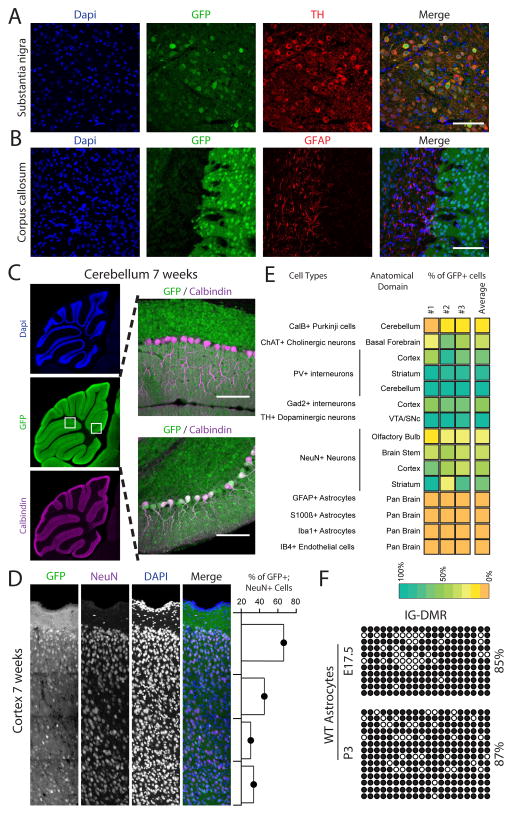
Imprinted genes in the Dlk1-Dio3 region are highly enriched in the brain. In addition, we show that loss of parent-specific methylation in the IG-DMR region results in marked brain phenotypes. At the macroscopic level the brain of 5–7 weeks old mice expressed the RGM reporter in a parent-of-origin specific pattern (Figure S4A). However, similar to the stomach and liver, close examination revealed consistent variations in GFP expression between different anatomical regions of the brain (Figure S4B) with overall variations being associated with cell-type specific reporter activity. Thus, while some cell types such as dopaminergic neurons robustly expressed GFP, other cell types such as astrocytes were GFP negative (Figures 5A, 5B and Figures S5–7). In addition to cell-type dependent GFP expression, we also noticed considerable heterogeneity within some cell-types. Figure 5c shows that while most Calbindin positive Purkinje cell were GFP negative, some Cerebellum lobes contained groups of adjacent GFP positive Purkinje cells (Figures S8A and S8B). Staining for the NeuN neuronal marker identified cellular variation in GFP expression associated with different cortical layers with the external granular and pyramidal layers containing high numbers of NeuN+GFP+ neurons, while the internal granular and pyramidal layers exhibited a high fraction of NeuN+GFP− cells (Figure 5D). Figure 5E summarizes the anatomical and cell type specific methylation differences. Given the high expression levels of multiple imprinted regulatory transcripts in the Dlk1-Dio3 region, these cell-type dependent differences in IG-DMR methylation may result in substantial gene expression differences between cell types and anatomical regions (See Figures S5–8). To validate the reporter activity in untargeted WT cells, we isolated pre- and post-natal astrocytes and performed bisulfite sequencing. Figure 5F shows that fetal and postnatal astrocytes, consistent with lack of IG-DMRMat-GFP reporter expression, exhibited hypermethylation of the IG-DMR region. The downstream Gtl2 promoter DMR identified hemimethylated levels, suggesting that Gtl2 maintains monoallelic regulation (Figure S9A).
Figure 5. Cell-type specific reporter activity in the adult brain.
(A–C) Representative images of brain sections from 7 weeks old mice stained with Dapi (blue), anti-GFP (green), anti-Tyrosine Hydroxylase (TH red), anti-Glial Fibrillary Acidic Protein (GFAP, red) and anti-Calbindin (purple); bar = 100μm (A) Overlap between GFP and TH is shown in the Substantia nigra region; (B) GFP and GFAP in the Corpus collosum are mutually exclusive. (C) Whole-mount stitching (left) and region specific (right) images demonstrate cellular mosaicism in the Cerebellum Purkinje cells: most cells are Calbindin+GFP− (right upper image), some lobes contain double positive Calbindin+GFP+ cells (right lower image); bar = 500μm (left images); 100μm (right images).
(D) Representative stitching images of 7 weeks old IG-DMRMat-GFP cortical layers stained with Dapi (blue), anti-GFP (green) and anti-NeuN (purple). Shown are percentages of GFP+NeuN+ neurons for each layer (right panel); bar = 250μm.
(E) Heat map summarizing the percentage of overlap between different cell type markers and GFP as measured in different brain anatomical regions. Shown are mean values of three independent IG-DMRMat-GFP brains.
(F) Bisulfite sequencing performed on the IG-DMR region in WT astrocytes isolated from E17.5 and P3 brains. Shown are percentages of methylated CpGs.
Maintenance of imprinting in the adult brain is variable at the single cell level
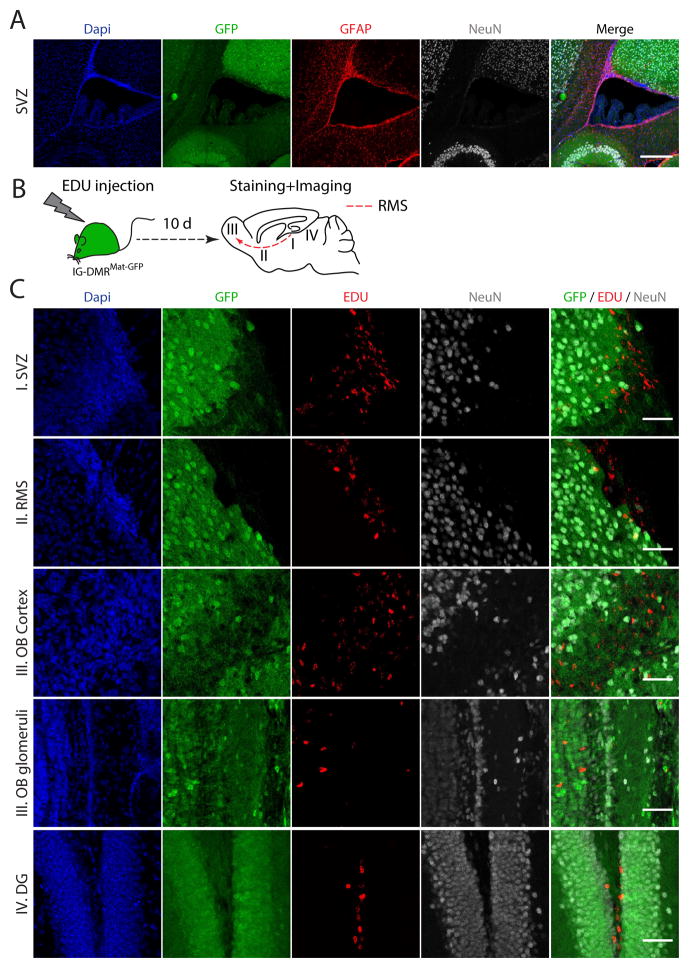
A recent report demonstrated biallelic expression of Dlk1 in neural stem cells (NSCs) and astrocytes in the post-natal neurogenic niche suggested to be mediated by hypermethylation of the IG-DMR region (Ferron et al., 2011). In agreement with these findings, GFAP+ cells residing in the Subventricular Zone (SVZ) ependymal wall were found to be GFP negative in adult IG-DMRMat-GFP brains (Figures 6A and S9B). This was in contrast to overall high reporter expression in the E13.5 SVG suggesting that the imprinting status of the IG-DMR changes in pre- vs. post-natal NSCs (Figure S9C). As NSCs migrate along the rostral migratory stream (RMS) to replenish the olfactory bulb (OB) neurons, we hypothesized that GFP negative NSCs may contribute to neuronal heterogeneity over time. Consistent with this notion, we identified NeuN+GFP− cells in the adult OB (Figures 5E and S9D). To study whether gain of maternal IG-DMR methylation in adult NSCs is irreversible, five week old mice were injected with EDU, a nucleoside analog of thymidine that allows marking of dividing cells and their post-mitotic daughters, and stained for EDU after 10 days (Figure 6B). Figure 6C shows that the vast majority of EDU+ labeled cells repressed GFP expression, consistent with biallelic hypermethylation of the IG-DMR region. GFP repression was also identified in EDU+ NSCs located at the SVZ, in EDU+ cells along the RMS and in EDU+ cells that migrated to the OB cortex and glomeruli. Furthermore, EDU+ cells originating from the Dentate Gyrus in the subgranular zone (SGZ) were GFP negative, suggesting that adult neurogenesis in the SGZ may contribute to neuronal heterogeneity in the hippocampus over-time (Fig. 6c). These results suggest that hypermethylation of the IG-DMR in NSCs is irreversible, potentially contributing to neuronal epigenetic variability over time.
Figure 6. Parent-specific DNA methylation in neural progenitors of the adult brain.
(A) Staining of the Sub Ventricular Zone (SVZ) in 7 weeks old IG-DMRMat-GFP brain with DAPI (blue), anti-GFP (green), anti-GFAP (red) and anti-NeuN (gray); bar=250μm. Shown are stitched images.
(B) Schematic representation of EDU labeling of 5 week old IG-DMRMat-GFP mice.
(C) Representative images of EDU positive cells in different anatomical regions of the brain. Shown are staining with Dapi (blue), anti-GFP (green), anti-EDU (red) and anti-NeuN (gray) in the SVZ, RMS, Olfactory Bulb (OB) cortex and glomeruli and in the Dentate Gyrus (DG), demonstrating that EDU+ and GFP+ cells are mutually exclusive; Bar = 50μm.
Discussion
In this study we utilized RGM (Stelzer et al., 2015) to report on parent-specific methylation changes of the Dlk1-Dio3 DMR. The RGM reporter was used to isolate sub-populations of ES cells that either had methylated the maternal allele or demethylated the paternal allele, which allowed assessing the consequence on developmental potency: when injected into 4n host embryos, the double negative cells generated only abnormal E13.5 embryos whereas injection of double positive ES cells lead to pre-gastrulation death indicating that loss of imprinting at this locus in mESCs results in impaired developmental potency. The generation of transgenic mice carrying the reporter in the maternal and paternal allele identified striking parent- and tissue-specific changes in IG-DMR methylation during development resulting in tissue- and cell type-dependent methylation signatures in the embryo and adult. Significantly, methylation changes were dynamic in tissues of the postnatal animal. This was particularly evident during adult neurogenesis resulting in inter-individual diversity and epigenetic variability at the single cell level.
Current understanding of the status of imprinted DMRs during development and in adult tissues is based on extensive molecular studies and high-resolution sequencing maps. Recent advancements in single cell sequencing (Smallwood et al., 2014) and allele specific RNA-FISH (Hansen and van Oudenaarden, 2013) technologies hold the promise for elucidating single cell parent-of-origin methylation and expression. Such methodology was recently used to uncover allele-specific expression heterogeneity of H19/Igf2 in single cells in mutant animals (Ginart et al., 2016). However, a serious limitation of current methods is that they provide only “snapshots” of bulk cell populations, thus precluding the evaluation of DNA methylation dynamics at single-cell resolution. Here we provide a systematic single-cell analysis of parent-specific methylation dynamics during mouse development. We show that, unlike the deleterious effects of loss-of parent-specific methylation in mESCs, during embryonic development the IG-DMR region is subjected to dynamic methylation changes in a tissue- and cell type-dependent manner. These methylation patterns persist in adult tissues consistent with the notion that gain of parent-specific methylation is irreversible. Although the full impact of parent-specific methylation dynamics during development and in postnatal animals remain to be identified, the consistent tissue and cellular patterns documented here in multiple animals, favors a rather regulated and non-stochastic process. In support of this notion, we show that methylation-mediated silencing of the IG-DMR reporter in tissues such as intestine or cell types such as astrocytes, does not simply silence the gene at this locus but rather regulates gene dosage as revealed by the biallelic expression of Dlk1 in the intestine.
Maternal deletion of the mouse IG-DMR region is comparable to biallelic hypermethylation of that region and was shown to result in prenatal lethality (Lin et al., 2007; Lin et al., 2003). Furthermore, induced pluripotent stem cells (iPSCS) hypermethylated in both IG-DMR parental alleles failed to generate “all-iPSCs mice” using 4n complementation (Stadtfeld et al., 2010) in agreement with prenatal-death of biallelically hypermethylated embryos, described here. The consequence of biallelic hypomethylation of the IG-DMR had not been assessed previously, as it had not been possible to generate such cells using classical genetics. Here we show that biallelically hypomethylated IG-DMR ES cells displayed reciprocal upregulation of maternal genes and repression of paternal genes in the Dlk1-Dio3 region. These cumulative gene expression perturbations resulted in pre-gastrulation death of 4n embryos, affecting an earlier developmental window than biallelic hypermethylation of the locus. In 2n chimeric embryos the biallelically hypomethylated cells contributed to many tissues with the notable exception of the brain. We also detected significant differences in the rate of acquiring and in the extent of aberrant paternal demethylation between male and female ES cells consistent with previous results that showed global demethylation in XX ES cells (Zvetkova et al., 2005).
Genes in the Dlk1-Dio3 locus are highly expressed in the brain, an organ that was previously associated with complex parent-of-origin effects (Davies et al., 2005; Ferron et al., 2015; Perez et al., 2015; Sittig and Redei, 2014; Wilkinson et al., 2007; Xie et al., 2012). A recent report suggested that gain or loss of DNA methylation in the IG-DMR region may be regulated in a dynamic manner in the adult neurogenic niche (Ferron et al., 2011). Consistent with this notion, we show striking cell-type dependent variation in IG-DMR methylation in the adult brain. Furthermore, our data suggest that loss of parent-specific methylation in adult NSCs, actively shapes the brain epigenome over time. Given the potential dosage-effects on dozens of regulatory genes in the Dlk1-Dio3 region, this epigenetic heterogeneity may account for substantial gene expression differences during ageing. Future studies combining allele-specific expression in single cells and transgenic animals, will allow to elucidate the full impact of parent specific methylation heterogeneity on gene dosage in vivo. Our results may provide a general framework for elucidating the contribution of dynamic changes in epigenetic state to gene dosage in normal developmental context, as well as in disease. The substantial cell-to-cell epigenetic heterogeneity illustrates the limitations of bulk approaches to study dynamic epigenetic variations.
Experimental Procedures
Reporter Cell lines
To generate IG-DMR reporter cell lines, targeting vectors and CRISPR/Cas9 were transfected into 129XCast F1 mouse embryonic stem cells (mESCs) using Xfect mESC transfection reagent (Clontech), according to the provider’s protocol. 48 hours following transfection, cells were FACS sorted for either GFP or tdTomato expression and plated on MEF feeder plates. Single colonies were analyzed for proper allelic integration by southern blot and PCR analysis. Clones carrying the Snrpn-tdTomato reporter targeted into the IG-DMR maternal 129-allele were re-transfected with Snrpn-GFP reporter vector to target the IG-DMR paternal Cast-allele, to establish double targeted cells (see complete list of primers in Table S1). To establish Blastocyst-derived mESCs, males carrying IG-DMR-Snrpn-GFP or IG-DMR-Snrpn-Tomato methylation reporter were crossed with BDF1 females following blastocyst isolation. ICM-derived mESCs were obtained according to previously established protocols (Markoulaki et al., 2008).
mESCs Cell Culture
Targeted mouse embryonic stem cells (mESCs) were cultured on irradiated mouse embryonic fibroblasts (MEFs) with standard ESCs medium: (500 ml) DMEM supplemented with 10% FBS (Hyclone), 10 ug recombinant leukemia inhibitory factor (LIF), 0.1 mM beta-mercaptoethanol (Sigma-Aldrich), penicillin/streptomycin1mM, L-glutamine and 1% nonessential amino acids (all from Invitrogen). For experiments in 2i culture conditions, mESCs were cultured on gelatin-coated plates with N2B27 + 2i + LIF medium containing: (500 ml), 240 ml DMEM/F12 (Invitrogen; 11320), 240 ml Neurobasal media (Invitrogen; 21103), 5 ml N2 supplement (Invitrogen; 17502048), 10 ml B27 supplement (Invitrogen; 17504044), 10 ug recombinant LIF, 0.1 mM beta-mercaptoethanol (Sigma-Aldrich), penicillin/streptomycin,1mM L-glutamine and 1% nonessential amino acids (all from Invitrogen), 50 ug/ml BSA (Sigma), PD0325901 (Stemgent, 1 uM), CHIR99021 (Stemgent, 3 uM).
Tetraploid and Diploid Embryo Injections
All blastocyst injections were performed with B6D2F2 (C57Bl/6xDBA) host embryos. To obtain tetraploid (4n) blastocysts, electrofusion was performed at approximately 44–47 h post hCG using a BEX LF-301 cell fusion device (Protech International Inc., Boerne, TX). Both 4n and 2n embryos were otherwise treated the same and cultured in Evolve® KSOMaa (Zenith Biotech, Guilford, CT) until they formed blastocysts (94–98 h after hCG injection) at which point they were placed in a drop of Evolve® w/HEPES KSOMaa (Zenith) medium under mineral oil for injection. A flat tip microinjection pipette with an internal diameter of 16 μm (Origio Inc, Charlottesville, VA) was used to introduce 10–12 cells into the blastocoel cavity. Within 1–2 h after injection, blastocysts were transferred to day 2.5 recipient CD1 Elite females (15–20 blastocysts per female).
Generation of reporter mice
Male chimeras carrying IG-DMR-Snrpn-GFP methylation reporter were crossed with BDF1 females. Male and female offspring carrying the paternally transmitted allele were bred to obtain offspring carrying a maternally or paternally transmitted allele in the F2 generation (See Figure 2a). F2 offspring harboring the reporter allele were analyzed at different ages. Mice were handled in accordance with institutional guidelines and approved by the Committee on Animal Care (CAC) and Department of Comparative Medicine (DCM) of Massachusetts Institute of Technology.
Tissue processing and immunohistochemistry
Neonatal and adult mice were perfused via a transcardial route with 4% paraformaldehyde (PFA)/PBS. E9.5–E18.5 embryos were fixed by overnight immersion in 4% PFA/PBS at 4°C. Fixed tissues and embryos were dissected and either imaged intact or sectioned with a vibratome (Leica VT1100) at 100–150 um or a cryostat (Leica) at 15–50 um thickness followed by immunohistochemical analysis. For vibratome sectioning, tissues were embedded with 3% agarose gel. For cryosectioning, tissues were equilibrated in 30% sucrose/PBS prior to embedding in Optimal Cutting Temperature (OCT) compound. Immunostaining procedures for tissue sections were previously described (Wu et al., 2014). Briefly, sections were permeablized with PBST (1 x PBS solution with 0.5% Triton X-100) for 1 hour at RT before blocking with 10% Normal Donkey Serum (NDS) in PBST. Sections were then incubated with appropriately diluted primary antibodies in PBST with 5% NDS for 12–24 hours at 4 °C, washed with PBST for 3 times at room temperature and then incubated with desired secondary antibodies in TBST with 5% NDS and DAPI to counter-stain the nuclei. Sections were washed 3 times with PBST before mounted onto slides with Fluoromount G (SouthernBiotech).
The following antibodies were used in this study: Chicken anti-GFP (1:1000, Aves Labs), Mouse anti-NeuN (1:1000, EMD Millipore), Mouse anti-GFAP (1:1000, Sigma Aldrich), Rabbit anti-GFAP (1:1000, Dako), Rabbit anti-Iba1 (1:1000, Wako), Rabbit anti-S100β (1:1000, Dako), Rabbit anti-Pax6 (1:1000, EMD Millipore), Rabbit anti-Calbindin (1:1000, Swant), Rabbit anti-PV (1:1000, Swant), Rabbit anti-GAD2 (1:1000, Sigma Aldrich), Rabbit anti-TH (1:1000, EMD Millipore), goat anti-ChAT (1:1000, EMD Millipore), 647 alexa-conjugated GS-IB4 (1:500, Thermo Fisher).
Microscopy and image analysis
Images were captured on a Zeiss LSM710 confocal microscope and processed with Zen software, ImageJ/Fiji, and Adobe Photoshop. For imaging based quantification, unless otherwise specified, 3 representative images from different mice were quantified manually and data were plotted with Graphpad.
Flow Cytometry
To assess the proportion of GFP and tdTomato in the established reporter cell lines, mESCs were treated with EDTA to obtain single-cells suspension, and assessed on the LSR II SORP, LSRFortessa SORP or FACSCanto II.
Bisulfite Conversion, PCR and Sequencing
Bisulfite conversion of DNA was established using the EpiTect Bisulfite Kit (Qiagen) following the manufacturer’s instructions. The resulting modified DNA was amplified by first round of nested PCR, following a second round using loci specific PCR primers (see complete list of primers in Table S1). The first round of nested PCR was done as follows: 94 °C for 4 min; 55 °C for 2 min; 72 °C for 2 min; Repeat steps 1–3 1X; 94 °C for 1 min; 55 °C for 2 min; 72 °C for 2 min; Repeat steps 5–7 35X; 72 °C for 5 min; Hold 12 °C. The second round of PCR was as follows: 95 °C for 4 min; 94 °C for 1 min; 55 °C for 2 min; 72 °C for 2 min; Repeat steps 2–4 35X; 72 °C for 5 min; Hold 12°C. The resulting amplified products were gel-purified, subcloned into a pCR2.1-TOPO-TA cloning vector (Life technologies), and sequenced.
Reverse transcription of RNA and quantitative real-time PCR
RNA was isolated using the RNeasy Mini Kit (Qiagen) including on-column DNase digest to remove genomic DNA. Reverse transcription was performed on 0.5–1μg of total RNA using random hexamer primers and SuperScript® III First-Strand Synthesis SuperMix (Life technologies) according to the manufacturer’s instructions. All PCR reactions were performed in a 96-well plate on a ABI 7900 HT Fast Real-Time PCR system (Applied Biosystems) using FAST SYBR Green Master Mix (Applied Biosystems). Relative quantification of gene expression was calculated using Gapdh primers or primers amplifying ultraconserved mouse genomic region. (see complete list of primers in Table S1).
Supplementary Material
Acknowledgments
We thank Patti Wisniewski and Colin Zollo for FACS analyses and cell sorting, Wendy Salmon for assistance with confocal imaging, and Maria Mihaylova for liver cells dissociation. The authors would like to thank Frank Soldner, Malkiel Cohen, Robert N. Plasschaert and Shawn Liu for critically reading the manuscript. This study was supported by NIH grant HD 045022. Y.S. is supported by a Human Frontier Postdoctoral Fellowship. H.W. is supported by a NARSAD Young Investigator Fellowship (Grant number 22950) and. RJ is an advisor to Stemgent and a cofounder of Fate Therapeutics and Fulcrum Therapeutics.
Footnotes
Author Contributions
Y.S. and R.J conceived the idea for this project. Y.S. and H.W. designed and conducted experiments and interpreted the data. Y.SG. assisted with bisulfite sequencing and SNP analysis. C.S.S. assisted with targeting of mESCs, and S.M. conducted 2N and 4N blastocyst injections. Y.S. and R.J. wrote the manuscript with input from all other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen DC, Laborda J, Baladron V, Kassem M, Sheikh SP, Jensen CH. Dual role of delta-like 1 homolog (DLK1) in skeletal muscle development and adult muscle regeneration. Development. 2013;140:3743–3753. doi: 10.1242/dev.095810. [DOI] [PubMed] [Google Scholar]
- Avior Y, Sagi I, Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat Rev Mol Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton SC, Ferguson-Smith AC, Fundele R, Surani MA. Influence of paternally imprinted genes on development. Development. 1991;113:679–687. doi: 10.1242/dev.113.2.679. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Markoulaki S, van Wietmarschen N, Hoke H, Wu T, Ganz K, Akhtar-Zaidi B, He Y, Abraham BJ, Porubsky D, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD, Nielsen DM, Georges M. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- Court F, Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, Sugahara N, Simon C, Moore H, Harness JV, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29:421–430. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- Ferron SR, Charalambous M, Radford E, McEwen K, Wildner H, Hind E, Morante-Redolat JM, Laborda J, Guillemot F, Bauer SR, et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2011;475:381–385. doi: 10.1038/nature10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron SR, Radford EJ, Domingo-Muelas A, Kleine I, Ramme A, Gray D, Sandovici I, Constancia M, Ward A, Menheniott TR, et al. Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat Commun. 2015;6:8265. doi: 10.1038/ncomms9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J, et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Watkins M, Surani MA, Ferguson-Smith AC. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development. 2000;127:4719–4728. doi: 10.1242/dev.127.21.4719. [DOI] [PubMed] [Google Scholar]
- Ginart P, Kalish JM, Jiang CL, Yu AC, Bartolomei MS, Raj A. Visualizing allele-specific expression in single cells reveals epigenetic mosaicism in an H19 loss-of-imprinting mutant. Genes Dev. 2016;30:567–578. doi: 10.1101/gad.275958.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Hansen CH, van Oudenaarden A. Allele-specific detection of single mRNA molecules in situ. Nat Methods. 2013;10:869–871. doi: 10.1038/nmeth.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labialle S, Marty V, Bortolin-Cavaille ML, Hoareau-Osman M, Pradere JP, Valet P, Martin PG, Cavaille J. The miR-379/miR-410 cluster at the imprinted Dlk1-Dio3 domain controls neonatal metabolic adaptation. EMBO J. 2014;33:2216–2230. doi: 10.15252/embj.201387038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Hore TA, Reik W. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell. 2014;14:710–719. doi: 10.1016/j.stem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Lin SP, Coan P, da Rocha ST, Seitz H, Cavaille J, Teng PW, Takada S, Ferguson-Smith AC. Differential regulation of imprinting in the murine embryo and placenta by the Dlk1-Dio3 imprinting control region. Development. 2007;134:417–426. doi: 10.1242/dev.02726. [DOI] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Markoulaki S, Meissner A, Jaenisch R. Somatic cell nuclear transfer and derivation of embryonic stem cells in the mouse. Methods. 2008;45:101–114. doi: 10.1016/j.ymeth.2008.04.002. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol. 2002;22:5585–5592. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JD, Rubinstein ND, Fernandez DE, Santoro SW, Needleman LA, Ho-Shing O, Choi JJ, Zirlinger M, Chen SK, Liu JS, et al. Quantitative and functional interrogation of parent-of-origin allelic expression biases in the brain. Elife. 2015;4:e07860. doi: 10.7554/eLife.07860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15:517–530. doi: 10.1038/nrg3766. [DOI] [PubMed] [Google Scholar]
- Qian P, He XC, Paulson A, Li Z, Tao F, Perry JM, Guo F, Zhao M, Zhi L, Venkatraman A, et al. The Dlk1-Gtl2 Locus Preserves LT-HSC Function by Inhibiting the PI3K-mTOR Pathway to Restrict Mitochondrial Metabolism. Cell Stem Cell. 2016;18:214–228. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100:149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, Hino T, Suzuki-Migishima R, Kohda T, Ogura A, et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 2008;40:243–248. doi: 10.1038/ng.2007.51. [DOI] [PubMed] [Google Scholar]
- Sittig LJ, Redei EE. Fine-tuning notes in the behavioral symphony: parent-of-origin allelic gene expression in the brain. Adv Genet. 2014;86:93–106. doi: 10.1016/B978-0-12-800222-3.00005-X. [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer Y, Jaenisch R. Monitoring Dynamics of DNA Methylation at Single-Cell Resolution during Development and Disease. Cold Spring Harb Symp Quant Biol. 2015 doi: 10.1101/sqb.2015.80.027334. [DOI] [PubMed] [Google Scholar]
- Stelzer Y, Ronen D, Bock C, Boyle P, Meissner A, Benvenisty N. Identification of novel imprinted differentially methylated regions by global analysis of human-parthenogenetic-induced pluripotent stem cells. Stem Cell Reports. 2013;1:79–89. doi: 10.1016/j.stemcr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer Y, Shivalila CS, Soldner F, Markoulaki S, Jaenisch R. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell. 2015;163:218–229. doi: 10.1016/j.cell.2015.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Barton SC. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science. 1983;222:1034–1036. doi: 10.1126/science.6648518. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Okamoto A, Kobayashi R, Shirai M, Obata Y, Ogawa H, Sotomaru Y, Kono T. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum Mol Genet. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]
- Tam PP, Rossant J. Mouse embryonic chimeras: tools for studying mammalian development. Development. 2003;130:6155–6163. doi: 10.1242/dev.00893. [DOI] [PubMed] [Google Scholar]
- Tevendale M, Watkins M, Rasberry C, Cattanach B, Ferguson-Smith AC. Analysis of mouse conceptuses with uniparental duplication/deficiency for distal chromosome 12: comparison with chromosome 12 uniparental disomy and implications for genomic imprinting. Cytogenet Genome Res. 2006;113:215–222. doi: 10.1159/000090835. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Solter D. The developmental fate of androgenetic, parthenogenetic, and gynogenetic cells in chimeric gastrulating mouse embryos. Genes Dev. 1988;2:1344–1351. doi: 10.1101/gad.2.10.1344. [DOI] [PubMed] [Google Scholar]
- Tucker KL, Beard C, Dausmann J, Jackson-Grusby L, Laird PW, Lei H, Li E, Jaenisch R. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- Wilkinson LS, Davies W, Isles AR. Genomic imprinting effects on brain development and function. Nat Rev Neurosci. 2007;8:832–843. doi: 10.1038/nrn2235. [DOI] [PubMed] [Google Scholar]
- Wu H, Luo J, Yu H, Rattner A, Mo A, Wang Y, Smallwood PM, Erlanger B, Wheelan SJ, Nathans J. Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron. 2014;81:103–119. doi: 10.1016/j.neuron.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazawa K, Ogata T, Ferguson-Smith AC. Uniparental disomy and human disease: an overview. Am J Med Genet C Semin Med Genet. 2010;154C:329–334. doi: 10.1002/ajmg.c.30270. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.